Abstract
Targeting tumor marker genes by RNA trans‐splicing is a promising means to induce tumor cell‐specific death. Using a screening system we designed RNA trans‐splicing molecules (RTM) specifically binding the pre‐mRNA of SLCO1B3, a marker gene in epidermolysis bullosa associated squamous cell carcinoma (EB‐SCC). Specific trans‐splicing, results in the fusion of the endogenous target mRNA of SLCO1B3 and the coding sequence of the suicide gene, provided by the RTM. SLCO1B3‐specific RTMs containing HSV‐tk were analyzed regarding their trans‐splicing potential in a heterologous context using a SLCO1B3 expressing minigene (SLCO1B3‐MG). Expression of the chimeric SLCO1B3‐tk was detected by semi‐quantitative RT‐PCR and Western blot analysis. Cell viability and apoptosis assays confirmed that the RTMs induced suicide gene‐mediated apoptosis in SLCO1B3‐MG expressing cells. The lead RTM also showed its potential to facilitate a trans‐splicing reaction into the endogenous SLCO1B3 pre‐mRNA in EB‐SCC cells resulting in tk‐mediated apoptosis. We assume that the pre‐selection of RTMs by our inducible cell‐death system accelerates the design of optimal RTMs capable to induce tumor specific cell death in skin cancer cells.
Keywords: Cancer gene therapy, RNA trans‐splicing, Epidermolysis bullosa, Squamous cell carcinoma, Herpes simplex virus ‐ thymidine kinase (HSV‐tk)
Highlights
The marker gene SLCO1B3 can be targeted by RNA trans‐splicing molecules (RTMs).
Accurate RNA trans‐splicing leads to the fusion of SLCO1B3 and HSV‐thymidine kinase.
Screening system accelerates the construction of RTMs with improved functionality.
The expression of the SLCO1B3‐HSV‐tk fusion protein induces apoptosis in target cells.
Abbreviations
- BD
binding domain
- BP
branch point
- DT-A
diphtheria toxin subunit A
- EB
epidermolysis bullosa
- EB-SCC
epidermolysis bullosa-associated squamous cell carcinoma
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GCV
ganciclovir
- GCVTP
ganciclovir triphosphate
- HSV
herpes simplex virus
- IRES
internal ribosomal entry site
- MG
minigene
- MMP9
matrix metalloproteinase 9
- MTT
3-(4,5-Dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide
- PPT
poly pyrimidine tract
- RDEB
recessive dystrophic epidermolysis bullosa
- RTM
RNA trans-splicing molecule
- RTMm1, RTMm2
RTM mutated 1, RTM mutated 2
- RTMmut
splicing deficient RTM
- RTMorg
RTM original
- SCC
squamous cell carcinoma
- SLCO1B3
solute carrier organic anion transporter family member 1B3
- SMaRT
spliceosome-mediated RNA trans-splicing
- SqRT-PCR
semi-quantitative real time PCR
- SS
splice site
- βhCG6
β-subunit of human gonadotropin gene 6
- STS
segmental trans-splicing
- tk
thymidine kinase
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
1. Introduction
Most gene therapy strategies for tumor elimination focus on producing toxin‐mediated death of the tumor cells. A transgene encoding a toxin is delivered into the tumor cells, resulting in death of the cells. Inefficient expression of the therapeutic gene due to suboptimal delivery methodology and low targeting specificity toward tumor cells remain the major obstacle to this technology (Altaner, 2008). An established suicide gene is the thymidine kinase (tk) gene from herpes simplex virus (HSV). This enzyme exhibits high substrate affinity for the nucleoside analog ganciclovir (GCV) (Elion et al., 1977). The nontoxic prodrug GCV is converted into the metabolite GCV triphosphate (GCVTP), which produces an intracellular toxic effect (Fillat et al., 2003): upon phosphorylation, GCV is incorporated into replicating DNA where it blocks DNA polymerization, resulting in cell death by apoptosis (Altaner, 2008; Hamel et al., 1996).
RNA trans‐splicing is a technology to combine exogenous genetic information into a target mRNA by exploiting the cell's endogenous spliceosome. The trans‐splicing reaction is facilitated by RNA trans‐splicing molecules (RTMs) consisting of a binding domain (BD) for gene targeting, a splicing domain for efficient trans‐splicing, and a coding domain comprising the sequence to be introduced. RTM binding to the target pre‐mRNA induces specific trans‐splicing between both molecules, resulting in a hybrid gene product consisting of the endogenous mRNA and the coding sequence provided by the RTM. This system, known as spliceosome‐mediated RNA trans‐splicing (SMaRT), has already been applied in vitro and in vivo to correct mutated genes in several genetic diseases, including epidermolysis bullosa (EB) (Wally et al., 2012; Koller et al., 2011; Murauer et al., 2011; Wally et al., 2010), cystic fibrosis (Liu et al., 2005; Song et al., 2009), hemophilia A (Chao et al., 2003), and spinal muscular atrophy (Coady and Lorson, 2010). In addition, SMaRT can be utilized to produce high levels of therapeutic proteins and antibodies in vivo by generating chimeric molecules (Wang et al., 2009).
Because RTMs provide cell‐specific expression by binding to a defined target gene, RNA trans‐splicing has also been considered for cancer gene therapy. For example, Puttaraju et al. (1999) demonstrated accurate trans‐splicing between the β‐subunit of human chorionic gonadotropin gene 6 and an RTM in vitro and in vivo (Puttaraju et al., 1999). More recently, we provided proof of principle for SMaRT in a suicide gene‐therapy approach in recessive dystrophic EB‐associated squamous cell carcinoma (RDEB‐SCC). In that study we described the construction of an RTM targeting the MMP‐9 gene, which is over‐expressed in cultured EB cancer cells, and delivery of the exotoxin streptolysin O. Trans‐splicing between the RTM and the target gene led to toxin‐mediated death preferentially of EB cancer cells compared to a non‐cancerous EB cell line (Gruber et al., 2011).
Since non‐specific trans‐ (Kikumori et al., 2001) and cis‐splicing events (Murauer et al., 2013; Wang et al., 2009) within the expression vector have already been observed further attention has to be turned on specificity aspects when using trans‐splicing in a suicide gene study in vivo. Therefore, for clinical application of suicide RTMs in EB cancer, the efficiency and specificity have to be improved so as to provide increased cell‐killing ability with as few side effects as possible. Therefore, we used RTMs containing the clinically evaluated HSV‐tk‐GCV system (Immonen et al., 2004; Nasu et al., 2007; Schwarzenberger et al., 2011; Xu et al., 2009). In a double‐transfection system, we analyzed RTMs targeting the solute carrier organic anion transporter family member 1B3 (SLCO1B3), a marker gene associated with various human cancers, including colorectal adenocarcinomas (Lee et al., 2008), breast (Muto et al., 2007) and lung cancer (Nagai et al., 2012) and, most importantly for our purposes, RDEB‐SCC (Cole et al., 2009). Here we describe a suicide‐RTM screening system which provides information on the ability of individual RTMs to induce tumor‐specific cell death. RTM sequence optimization improved the specificity of the trans‐splicing reaction, thereby identifying a potent RTM for subsequent in vivo applications in RDEB‐SCC.
2. Materials and methods
2.1. RTM screening constructs
2.1.1. SLCO1B3 target molecule
The target molecule harbors the 5′ coding region of AcGFP (nt1–nt336), a functional 5′ splice site (ag/gtaag) and nucleotides 4–1200 (first 3 nucleotides of intron 3 were removed to avoid the creation of a possible competitive 5′ splice site) of intron 3 of SLCO1B3. The intron 3 portion was amplified by PCR using Gotaq DNA polymerase (Promega), genomic DNA of a healthy donor, and an intron‐specific primer pair (fw: 5′‐gatcgatatcgaatgggttttatattttcaaactaaaataagttaatggaaaattttt‐3′, rv: 5′‐gagagcggccgcgatttgaatatacatttctcaaaagaagacatacaaatagc‐3′) incorporating restriction sites for cloning. The PCR product was ligated into the expression vector pcDNA 3.1D/V5‐His‐TOPO downstream of the 5′ AcGFP sequence using the restriction sites for EcoRV and NotI. Gel‐extractions of amplified PCR products were performed using a GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare). Plasmid preparations were performed using a Plasmid Mini Prep Kit (Sigma–Aldrich), according to the manufacturer's protocol. Sequence analysis of all plasmids and PCR products was carried out using a 3130 ABI Prism automated sequencer and ABI PRISM dye terminator cycle sequencing kit (Applied Biosystems).
2.1.2. RNA trans‐splicing molecule
The RTM backbone consists of a splicing domain carrying a short spacer region and 3′ splicing elements (branch point, polypyrimidine tract, 3′ acceptor splice site) for efficient splicing, and a coding domain incorporating the missing 3′ coding sequence of AcGFP (nt337–nt720) and the full‐length DsRed gene expressed under the translational control of an internal ribosomal entry site (IRES) (Dallinger et al., 2003; Gruber et al., 2011). A highly diverse RTM library specific for intron 3 of SLCO1B3 was created according to Bauer et al. (2013). The cloning procedure of the binding domain (BD) library includes PCR amplification of the intron 3 portion of SLCO1B3, fragmentation of the PCR products by sonication (∼10 min on ice) and finally, cloning of the resulting end‐repaired (DNATerminator® End Repair Kit, Lucigen Corporation) fragments into the RTM vector, upstream of the splicing domain using the restriction site for HpaI.
2.2. Constructs for the HSV‐tk‐based inducible cell death system
2.2.1. SLCO1B3 minigene
To simulate the endogenous trans‐splicing scenario as closely as possible, a SLCO1B3 minigene (SLCO1B3‐MG) was constructed which consists of the first coding exon of SLCO1B3 (exon 3: 84 nucleotides) and the first 1200 bases of intron 3 (due to the huge size of the entire intron; 39,205 nucleotides). The exon/intron 3 region of SLCO1B3 was PCR‐amplified from genomic DNA of a healthy donor using Gotaq DNA polymerase (Promega) and a specific primer pair (fw: 5′‐gatcaagcttatggaccaacatcaacatttgaataaaacagc‐3′, rv: 5′‐gagagcggccgcgatttgaatatacatttctcaaaagaagacatacaaatagc‐3′). The resulting PCR product was cloned into the pcDNA 3.1D/V5‐His‐TOPO vector (Invitrogen) using the restriction sites for HindIII and NotI.
2.2.2. RTMs
First, one of the most efficient BDs, evaluated in our fluorescence‐based screening system, was cloned into the pIRES2‐AcGFP1 vector (Clontech) together with the 3′ splicing elements using the restriction sites for EcoRI and PstI. The coding sequence (CDS) of the thymidine kinase gene from herpes simplex virus (HSV‐tk) without the start codon was amplified from the HAX1‐targeting vector, kindly provided by Dr. Peckl‐Schmid (Peckl‐Schmid et al., 2010). Prior to PCR amplification the PstI restriction site within the CDS of HSV‐tk was mutated using the QuikChange Lightning site‐directed mutagenesis kit (Stratagene), according to the manufacturer's protocol. We used a forward primer including a PstI restriction site (5′‐ctagctgcagcccacgctactgcgggttta‐3′), a reverse primer including a BamHI restriction site (5′‐gagagaggatcctcagttagcctcccccatctc‐3′) and Pfu turbo polymerase (Stratagene) for PCR amplification. The resulting PCR product was further subcloned into the RTM vector.
For RTM optimization, start codons and potential cryptic splice sites upstream of the splicing domain were modified with the QuikChange Lightning site‐directed mutagenesis kit (Stratagene) resulting in three different RTMs: RTMorg, RTMm1, RTMm2. The sequence of each construct is provided in Supplementary Figure 1.
2.2.3. Positive control
As a positive control we used a plasmid expressing a fusion protein of SLCO1B3 and HSV‐tk (SLCO1B3‐tk fusion) representing the accurate trans‐splicing product derived from RTM and SLCO1B3‐MG. To create this construct, exon 3 of SLCO1B3 was amplified from SLCO1B3‐MG using the Pfu turbo polymerase and the following primers (fw: 5′‐gagagaattcatggaccaacatcaacattt‐3′, rv: 5′‐ctagctgcagcttgaatccattgcagcgtc‐3′). The PCR product was cloned into the RTM‐vector using EcoRI and PstI restriction sites, thereby removing the binding and splicing domains of RTM. Finally, the remaining PstI restriction site was removed from the CDS by using a QuikChange Lightning site‐directed mutagenesis kit (Stratagene).
2.3. Constructs for retroviral delivery
2.3.1. RTM
The RTM sequence (including BD, spacer and tk coding region) was PCR amplified using a pIRES2‐AcGFP1 vector specific forward primer (5′‐gatcggatcccgctagcgctaccggactcagatctcg‐3′), a tk specific reverse primer (5′‐gatcgcggccgctcagttagcctcccccatctcc‐3′), the Pfu turbo polymerase (Stratagene) and RTMm2 as template for PCR amplification. The resulting PCR product was cloned into the retroviral vector pMXs‐IRES‐Blasticidin (Cell Biolabs) using the restriction sites for BamHI and NotI.
2.3.2. RTMmut
An RTM with an inactive splice site served as negative control. A point mutation at the 3′ splice site was inserted using the QuikChange Lightning site‐directed mutagenesis kit (Stratagene) to convert the sequence from cag/c to cgg/c.
2.3.3. Positive control
The SLCO1B3‐tk fusion was PCR amplified from the pIRES2‐AcGFP1 vector (screening procedure) carrying the SLCO1B3‐tk fusion using a SLCO1B3 exon 3 specific forward and a tk specific reverse primer (fw: 5′‐ctaggaattcatggaccaacatcaacatt‐3′, rv: 5′‐gatcgcggccgctcagttagcctcccccatctcc‐3′) and the Pfu turbo polymerase (Stratagene). The resulting PCR product was cloned into the pMXs‐IRES‐Blasticidin vector (Cell Biolabs) using the restriction sites for EcoRI and NotI.
2.3.4. FLAG constructs
A FLAG‐tag for detection of the trans‐splicing product on protein level was cloned into all constructs (RTMm2, RTMmut and positive control) by PCR amplification using the forward primers mentioned above and a tk specific reverse primer (5′‐gatcGCGGCCGCTCAtttatcatcatcatctttataatcGTTAGCCTCCCCCATCTCC‐3′) including the FLAG epitope and finally cloning of the resulting PCR products into the pMXs‐IRES‐Blasticidin (Cell Biolabs) vector as described above.
2.4. Cell culture and transfections
For all screening experiments the human embryonic kidney cell line HEK293AD (Stratagene) was used. HEK293AD cells were grown in DMEM supplemented with 10% FCS and 100U/ml penicillin/streptomycin (Biochrom) at 37 °C and 5% CO2 in a humidified incubator. The cells were passaged every 4 days by Trypsin‐EDTA (Biochrom) treatment following centrifugation at 250 g for 5 min. SCCRDEB2 cells (previously described by Watt et al. (2011)), were routinely grown in DMEM/Ham's F‐12 (2:1) (Hyclone) containing 10% serum and growth factors according to the protocol of Rheinwald and Green (1975). For better understanding, SCCRDEB2 cells are termed EB‐SCC cells throughout this paper. Plasmid transfection in HEK293AD was performed in a 60 mm tissue culture dish at a 60% cell density using the jetPEI reagent (Polyplus‐transfection SA) according to the manufacturer's protocol.
2.5. Retroviral RTM delivery into EB‐SCCs
For production of retroviral particles, the Phoenix™ Retrovirus Expression System (Orbigen) was used. As previously described, Phoenix cells were transfected at a confluence of approximately 60% with 10 μg of viral plasmid using a standard calciumphosphate transfection and cultured over night at 37 °C and 5% CO2 (Murauer et al., 2011). After 12–16 h the cells were washed once with PBS and further cultivated in fresh medium at 32 °C and 5% CO2. After 48 h cell culture supernatant containing infectious viral particles was harvested every 8 h. Medium containing viral particles was filtered through a 0.22 μm filter to remove cell debris and added to EB‐SCCs twice within 48 h. To increase transduction efficiency 5 μg/ml Polybrene were added to the virus and spin‐oculation was performed for 1.5 h at 600 g and 32 °C. The whole transduction procedure was performed at 32 °C and 5% CO2. 24 h after the last transduction step, cells were washed 4 times with PBS and cultivated under standard cell culture conditions. For the selection of transduced cells, cell culture medium was supplemented with 10 μg/ml Blasticidin.
2.6. RNA isolation and cDNA synthesis
RNA was isolated from treated HEK293AD cells two days post‐transfection using an RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Purified RNA (2 μg) was digested with DNase I for 30 min at room temperature (RT) and then used as a template for cDNA synthesis using the iScript™cDNA Synthesis Kit (Bio‐Rad). For cDNA synthesis, a mixture of oligo(dT) and random hexamer primers was provided.
2.7. Semi‐quantitative RT‐PCR (SqRT‐PCR)
SqRT‐PCR was performed to detect SLCO1B3‐tk fusion transcripts in EB‐SCC or HEK293AD cells co‐transfected with SLCO1B3‐MG and RTM. For PCR analysis, an SLCO1B3‐specific forward primer (5′‐ggaccaacatcaacatttgaataaaacagcagag‐3′), an HSV‐tk‐specific reverse primer (5′‐gtaagtcatcggctcgggta‐3′ or 5′‐agatgttcgcgattgtctcggaa‐3′), cDNA of treated cells and GoTaq®qPCR Master Mix (Promega) were included. For amplification of SLCO1B3 and SLCO1B3‐MG transcripts the following primer combinations were used i.) SLCO1B3: fw: 5′‐gggtgaatgcccaagagata‐3′, rv: 5′‐attgactggaaacccattgc‐3′, ii.) SLCO1B3‐MG: fw: 5′‐ggaccaacatcaacatttgaataaaacagcagag‐3′, rv: 5′‐gattaaaaactgatttaaatcaatataggg‐3′. The PCR was performed under the following conditions with a Bio‐Rad CFX96™ system: 95 °C for 2min, and 40 cycles of 20sec at 95 °C, 20sec at 64 °C, and 20sec at 72 °C. The experiment was carried out in duplicates and repeated two times. Correct PCR products were verified by direct sequencing.
2.8. Western blot analysis
For immunostaining of the SLCO1B3‐tk fusion protein, HEK293AD cells were resuspended in RIPA lysis buffer (Santa Cruz) and the extracted proteins were separated on a NuPAGE 4–12% BisTris gel (1.0 mm × 12 well, Invitrogen) under denaturating conditions for 2 h at 120 V. After equilibration of the SDS‐gel in standard blotting buffer for up to 30 min at RT, the proteins were electro‐blotted onto a nitrocellulose membrane (Amersham Hybon‐ECL, GE Healthcare) for 75 min at 0.25 A. Subsequently, the membrane was soaked in blocking buffer (5% milk powder in Tris buffered saline + Tween (TBS‐T)) for 1 h at RT and incubated overnight at 4 °C with first antibodies: goat anti‐HSV‐tk1 IgG (diluted 1:1000 in TBS‐T) (Santa Cruz Biotechnology) and rabbit anti α‐actinin IgG (diluted 1:1000–1:5000 in TBS‐T) (Santa Cruz Biotechnology). After three washings steps with TBS‐T, the membrane was incubated with the secondary antibodies, polyclonal rabbit anti‐goat HRP conjugate (diluted 1:200–1:1000 in TBS‐T, Dako) and HRP‐labeled Envision+ anti‐rabbit antibody (diluted 1:500–1:1000 in TBS‐T, Dako), respectively. The blot was washed three times and visualization of specific protein bands was performed using an Immun‐Star WesternC Kit (Bio‐Rad) and a ChemiDoc XRS Imager (Bio‐Rad).
2.9. Immunoprecipitation
For endogenous detection of the trans‐splicing protein, SLCO1B3‐tk, immunoprecipitation was performed. Briefly, transduced EB‐SCC cells (3 × 106) were resuspended in lysis buffer (25 mM Tris, 140 mM NaCl, 1 mM EDTA, 0.5% IPGAL) and incubated for 45 min at 4 °C rotating. 1 μg of anti‐FLAG antibody (Sigma–Aldrich) was added to the cleared lysate and incubated over night at 4 °C rotating. Then, the cell lysates were added to 30 μl protein G sepharose (GE Healthcare) and incubated for additional 2 h at 4 °C rotating. After three washing steps of sepharose G (once with lysis buffer and twice with PBS), the bound protein was denatured from the polymer and transferred to SDS‐Page, followed by Western blot analysis described in detail above. Additionally, FLAG fusion proteins were isolated from RTMm2 transduced RDEB‐SCC cells (2 × 106) by adding 40 μl of anti‐FLAG M2 Affinity Gel (Sigma–Aldrich) and a subsequent incubation over night at 4 °C rotating. After three washing steps the detection of SLCO1B3‐tk fusion proteins was performed as described in the Western blot analysis section.
2.10. Cell viability assay
Cell viability after RTM treatment was assessed by MTT (3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide) assay (Sigma). Forty‐eight hours after RTM transfection, HEK296AD cells were seeded into a 96‐well plate (10,000cells/well) and treated with 100 μM GCV (Cymevene®, Roche) the next day and incubated for 72 h. 20 μl of MTT (5 g/L in PBS) were added to each well (200 μl) and incubated for approximately 2 h at 37 °C. Then, all media was removed and the cells were lysed with 100 μl DMSO/glycine (6 vol DMSO + 1 vol 0.1 M glycine/NaOH, pH 10.2). After 10 min incubation on a plate shaker (500 rpm) the absorbance of the resultant formazan product was measured at 492 nm/620 nm with a plate photometer (Tecan). The percentage of cell viability was calculated by the equation: (OD GCV‐treated/OD GCV‐untreated) × 100/negative control (OD GCV‐treated/OD GCV‐untreated). HEK293AD cells transfected with SLCO1B3‐MG alone or EB‐SCC cells served as a negative control in this assay. Statistical analysis was performed with GraphPad Prism 5.03 software.
2.11. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
Detection of apoptosis was performed using an In Situ Cell Death Detection Kit, TMR red (Roche) according to the manufacturer's protocol. Briefly, cells were washed with 1× phosphate‐buffered saline (PBS), fixed with 2% paraformaldehyde for 60 min at RT and permeabilized with 0.1% BSA/0.2% Triton X‐100 in PBS for 5–10 min on ice. After 3 washing steps, the cells were incubated with TUNEL reaction solution for 1 h at 37 °C in the dark and finally analyzed by fluorescence microscopy (Axiophot, Carl Zeiss) and flow cytometry (FC500, Beckman Coulter).
3. Results
3.1. Screening for an efficient RTM binding domain
A binding domain (BD) screen accelerates the construction of a highly efficient RTM for endogenous applications. Exon 3 of SLCO1B3 gene constitutes the first coding exon, therefore intron 3 was examined for potent RTM binding sites. Moreover the huge size of intron 3 of 39,205 nt did not allow a screening procedure covering the whole intronic sequence, so we used the first 1200 nt of intron 3 for cloning into the target vector. Using our fluorescence‐based RTM screening procedure (Bauer et al., 2013; Murauer et al., 2013), we were able to select a promising BD specific for intron 3 of SLCO1B3 out of a pool of randomly created BDs with various binding localizations within the target intron (Supplementary Figure 2).
Binding of an RTM to intron 3 of the target molecule induces fusion of the split segments of AcGFP by 3′ RNA trans‐splicing, manifested in the expression of the full‐length reporter in co‐transfected cells (Figure 1A). In total, 22 individual 3′ RTMs, binding all over the first 1200 nt of intron 3 of SLCO1B3, were tested for their trans‐splicing efficiency by flow cytometric analysis after co‐transfection with the designed target molecule into HEK293AD cells. The intensity of the fluorescence signal and the amount of AcGFP‐expressing cells correlate with the trans‐splicing efficiency of the transfected RTM (The geometric means of GFP expression, as a measure of trans‐splicing efficiency of each RTM is given in Supplementary Table 1). The analysis revealed RTM_1 (binding position in intron 3: nt1012–nt1118) as one of the most efficient RTMs, and thus it was selected for the initial tests in the tk‐based inducible cell‐death system (Figure 1B+C). The short BD of RTM_1 probably reduces unwanted direct HSV‐tk expression from the RTM backbone as the amount of in frame ATGs and cryptic 5′ splice sites, leading to new potential start sites, are decreased significantly.
Figure 1.
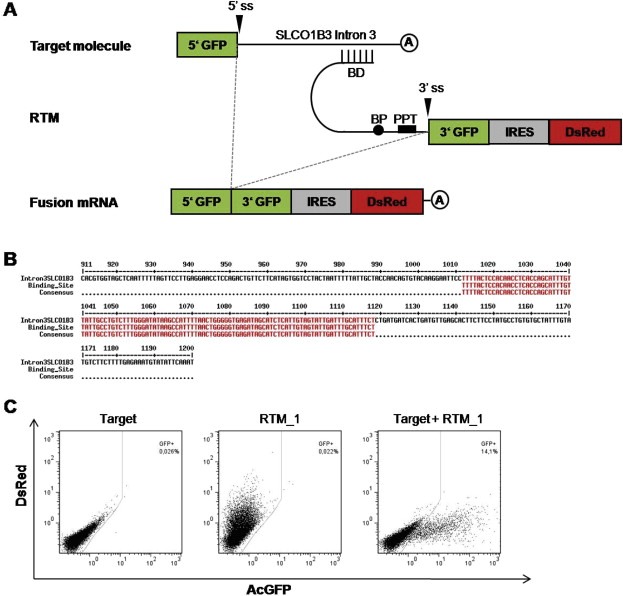
Evaluation of a specific binding domain (BD) for efficient trans‐splicing. (A) Schematic diagram of the screening procedure used for evaluation of BDs. (B) Binding position of RTM_1 within the target intron 3 of SLCO1B3 (nt1012‐nt1118). (C) Analysis of AcGFP expression by flow cytometry in HEK293AD cells transfected with the target molecule, RTM_1, or both the target molecule + RTM_1. Only cells co‐transfected with the target molecule and RTM_1 showed green fluorescence signal, resulting from fusion of both parts of the split AcGFP gene by RNA trans‐splicing. BD: Binding Domain, BP: Branch Point, PPT: Poly Pyrimidine Tract, 3′ ss: 3′ splice site.
3.2. Trans‐splicing between SLCO1B3‐MG and RTM is accurate
To further evaluate the specificity of RTMs in a suicide gene therapy approach we cloned the BD from RTM_1 into the suicide RTM plasmid containing the tk sequence from HSV. The sequence upstream of the splicing domain was mutated to reduce potential cryptic splice sites and start codons, resulting in 3 different RTMs, termed RTMorg, RTMm1 and RTMm2. Detailed information on sequence modifications of the RTMs is provided in Supplementary Figure 1.
To mimic the endogenous RDEB‐SCC marker expression in heterologous HEK293AD cells, we designed a minigene (MG) vector carrying the first coding exon (exon 3) of the tumor marker gene SLCO1B3 as well as the first 1200 bases of intron 3. Intron 3 was selected as RTM binding site due to the fact that the fusion of the first coding exon of the gene and HSV‐tk by accurate trans‐splicing minimizes the size of the resulting fusion product and probably maintains its functionality. HSV‐tk‐ and SLCO1B3‐tk‐expressing vectors served as positive controls (Figure 2A). Upon correct trans‐splicing of the RTM with the MG pre‐mRNA, a fusion mRNA of SLCO1B3 exon 3 and HSV‐tk is generated, encoding a fusion protein that is approximately 3 kDa larger than the original HSV‐tk (Figure 2B).
Figure 2.
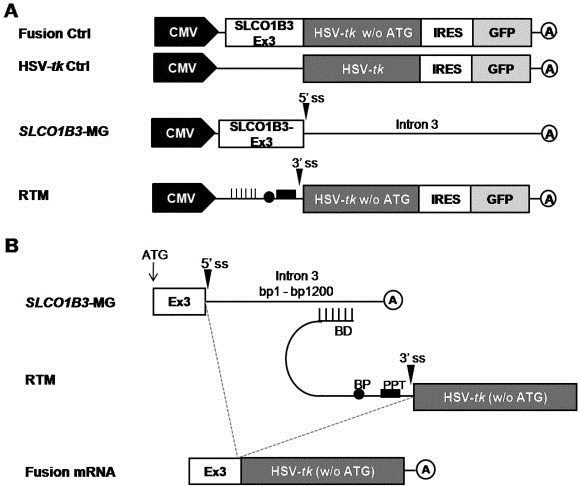
(A) Constructs used in the experimental procedure. (B) Schematic diagram of the double‐transfection system used for RTM evaluation. Correct trans‐splicing of the RTM to SLCO1B3‐MG results in a fusion mRNA consisting of exon 3 of SLCO1B3 and tk. BD: Binding Domain, BP: Branch Point, PPT: Poly Pyrimidine Tract, 3′ ss: 3′ splice site, A: polyadenylation signal.
3.3. The fusion protein of SLCO1B3‐tk exhibits suicide gene activity
Next we determined whether the fusion protein that results from the trans‐splicing reaction between SLCO1B3‐MG and the RTM is expressed correctly and still possesses its functionality as a suicide gene. To this end, we created the chimeric cDNA of SLCO1B3 exon 3 and HSV‐tk and confirmed its expression in whole‐cell lysates of HEK293AD cells by Western blot analysis (Figure 3B, lanes 9, 10). The SLCO1B3‐tk fusion protein was detected at the predicted molecular weight of 40 kDa, 3 kDa larger than the original HSV‐tk (37 kDa). Furthermore, the biologic activity of SLCO1B3‐tk fusion protein was assessed in an MTT cell viability assay (Figure 4A). HEK293AD cells were transfected with the plasmid HSV‐tk or the fusion construct and treated with 100 μM GCV. The SLCO1B3‐tk fusion protein was able to convert the prodrug GCV into the toxic metabolite GCV triphosphate (GCVTP) which induces cell death, producing a reduction of cell viability similar that caused by the original HSV‐tk. Therefore, these data confirmed the functionality of the fusion construct that is expected to result from the trans‐splicing procedure in our experimental setup.
Figure 3.
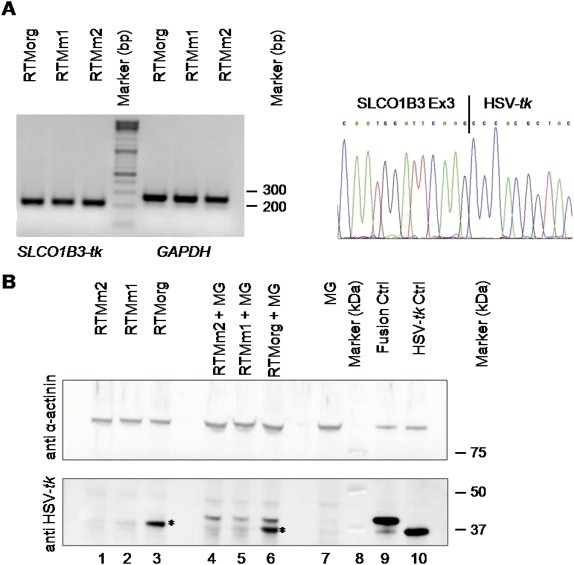
Detection of accurate trans‐splicing on the mRNA and protein level. (A) SqRT‐PCR analysis of HEK293AD cells double‐transfected with SLCO1B3‐MG and either the original RTM (RTMorg), or the mutated RTMs (RTMm1 and RTMm2). The correct PCR product of the fusion mRNA of SLCO1B3 and HSV‐tk (205 bp) is shown. GAPDH was used as housekeeping gene. All PCR products were verified by direct sequencing, demonstrating correct trans‐splicing between SLCO1B3‐MG and the RTMs on the mRNA level. A representative figure shows the sequence of the SLCO1B3‐HSV‐tk junction (right panel). (B) Sequence optimization leads to reduced background expression of RTMs as shown by Western blot analysis: Upper panel: α‐actinin staining (100 kDa) was used as a loading control. Lower panel: Western blot analysis demonstrated the expression of the SLCO1B3‐fusion protein (40 kDa) in all cell populations double‐transfected with the SLCO1B3‐MG (MG) and each RTM [lane 4, 5, 6]. Sequence optimized RTMm1 and RTMm2 showed greatly reduced background expression of HSV‐tk (37 kDa) in both single [lane 1, 2] and double [lane 4, 5] transfected HEK293 cells as compared to the RTMorg [lane 3, 6] (marked by asterisk). Cells transfected with the MG alone [lane 7], either the fusion [lane 9] or HSV‐tk [lane 10] construct served as negative and positive controls, respectively. Lane 8: Precision Plus Protein WesternC Standards (Biorad).
Figure 4.
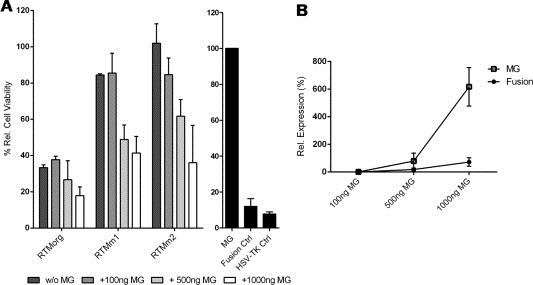
Biological activity of the fusion protein SLCO1B3 and HSV‐tk resulting from accurate trans‐splicing. (A) MTT assay was performed to measure reduction in cell viability. 100 ng of each RTM as well as different concentrations of SLCO1B3‐MG (100 ng, 500 ng, 1000 ng) were transfected into HEK293AD cells and the cells were treated with 100 μM GCV for 72 h. HEK293AD cells transfected with SLCO1B3‐MG alone, the SLCO1B3‐tk fusion or HSV‐tk served as negative and positive controls, respectively. The mean ± SD of two independent experiments is shown. Six replicates of each sample were measured in every experiment. The percentage of cell viability was calculated by the equation: (OD GCV‐treated/OD GCV‐untreated) × 100/negative control (OD GCV‐treated/OD GCV‐untreated). (B) SqRT‐PCR analysis of HEK293AD cells co‐transfected with RTMm2 (100 ng) and different amounts of SLCO1B3‐MG (100 ng, 500 ng, 1000 ng). Both, the transcripts of SLCO1B3‐MG and the fusion mRNA of SLCO1B3 and HSV‐tk were amplified. GAPDH was used as housekeeping gene. Data were calculated relative to the transcript level in HEK293AD cells transfected with 100 ng MG and 100 ng RTMm2.
3.4. Detection of SLCO1B3‐tk fusion at the mRNA and protein level
To investigate accurate trans‐splicing between SLCO1B3‐MG and each individual RTM (RTMorg, RTMm1, RTMm2) we first performed sqRT‐PCR. We detected the correct fusion mRNA in all three co‐transfected cell populations, represented by a 205 bp PCR product on an agarose gel (Figure 3A). In addition we performed Western blot analysis to confirm synthesis of the trans‐spliced protein product. As shown in Figure 3B, high background expression of HSV‐tk (37 kDa) was observed in cells transfected with RTMorg alone, whereas very low background expression was seen in cells transfected with the improved RTMs, RTMm1 and RTMm2. Furthermore, double transfection of cells with RTMorg plus the MG resulted in generation of both the trans‐spliced fusion protein (40 kDa) and the background HSV‐tk protein (37 kDa). In cells double‐transfected with RTMm1 or RTMm2 and the MG, sufficient expression of the trans‐spliced fusion protein was observed, while hardly any background expression of HSV‐tk was detected. These data indicate that improvement of RTMs by sequence optimization is an important step for reducing unwanted expression of HSV‐tk.
3.5. Suicide gene induction by RNA trans‐splicing
Trans‐splicing provides gene‐ and cell‐specificity, thereby reducing unwanted side effects that accompany other toxin‐mediated cell‐death approaches. In the present system, cell death is mainly induced in cells expressing the up‐regulated tumor marker gene SLCO1B3. Expression of the fusion constructs at the protein level (Figure 3B) correlates with the reduction of cell viability measured in MTT assays (Figure 4A). Single transfection of RTMorg induced apoptosis in HEK293AD cells, which can be explained by the presence of alternative start codons (ATGs) and splice sites upstream of and within the BD. These would allow alternative in‐frame tk proteins to be expressed from the RTM vector alone, leading to a toxic effect in GCV‐treated cells. However, by deleting the superfluous ATGs and potential cryptic splice sites within and upstream of the BD sequence of the RTM by site‐directed mutagenesis, we were able to reduce this unwanted expression significantly. RTMm1 and RTMm2 alone showed only a minimal ability to induce apoptosis using 100 ng plasmid per transfection. Furthermore, addition of different amounts of SLCO1B3‐MG (100 ng, 500 ng, 1000 ng) led to fusion of SLCO1B3 exon 3 with the tk sequence of the RTM by trans‐splicing, resulting in expression of a functional SLCO1B3‐tk fusion protein in transfected cells. Both RTMm1 and RTMm2 induced a clear dose‐dependent reduction in cell viability after MG addition (Figure 4A). Additionally, data achieved by sqRT‐PCR confirmed that the increased levels of SLCO1B3‐MG expression go along with an enhanced trans‐splicing efficiency of RTMm2, manifested in the expression of SLCO1B3‐tk fusion transcripts (Figure 4B). The TUNEL assay performed on transfected HEK293AD cells underlines the reliability and functionality of the inducible cell‐death system by trans‐splicing constructs (Figure 5A). Co‐transfection of SLCO1B3‐MG (1 μg) and RTMm2 (100 ng) into HEK293AD cells induced apoptosis in up to 63.8% of cells, as revealed by flow cytometry. This level of killing is comparable with the cell lethality rate reached by using the SLCO1B3‐tk fusion control vector. Light microscopy performed on HEK293AD cells co‐transfected with SLCO1B3‐MG and RTMm2 also showed reduced numbers of healthy cells in comparison to cells transfected with RTMm2 or the SLCO1B3‐MG alone (Figure 5B).
Figure 5.
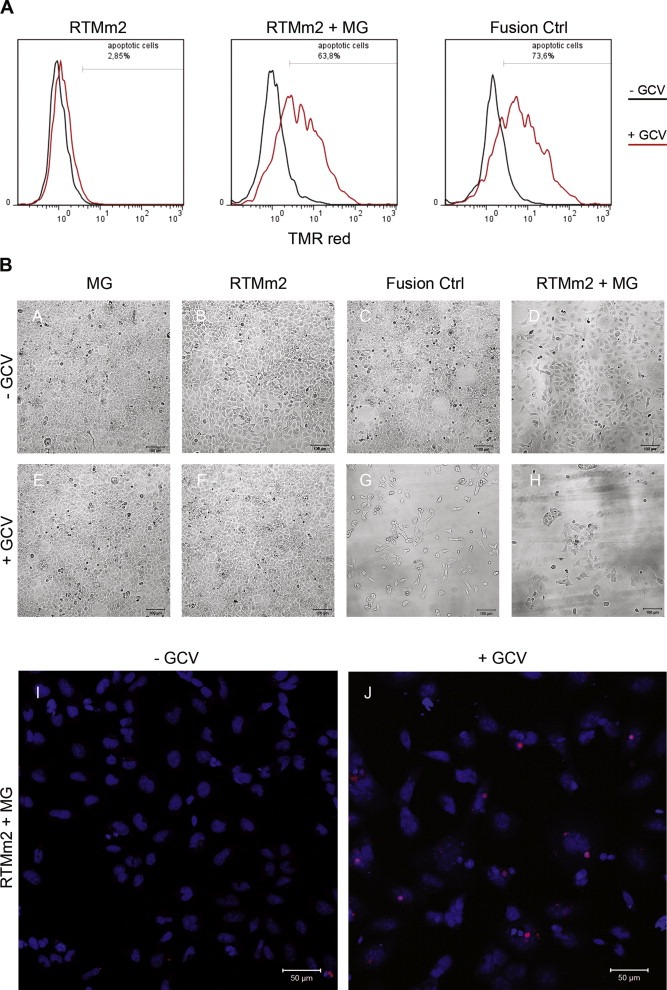
Detection of cell death after RNA trans‐splicing. (A) In situ detection of cell death by TUNEL assay of HEK293AD cells treated with RTMm2 and RTMm2+MG was assessed by flow cytometric analysis. The SLCO1B3‐tk fusion served as positive control. Overlay of TMR red positive cells is shown. The black line represents the GCV‐untreated and the red line the GCV‐treated cell populations. (B) The reduction in cell number of transfected and GCV‐treated (100 μM) HEK293 cells (E–H) in comparison to GCV‐untreated cells (A–D) was visualized by light microscopy analysis. A decrease in cell viability of SLCO1B3‐tk and RTMm2+SLCO1B3‐MG transfected cells 72 h after addition of GCV was detected. TMR red staining (TUNEL assay) and nuclear counterstaining with DAPI of RTMm2 and SLCO1B3‐MG co‐transfected HEK293 cells revealed a significant higher rate of apoptotic cells after GCV treatment (J) in comparison to the level seen in the GCV‐untreated cell population (I).
3.6. Pre‐evaluated RTMm2 induced trans‐splicing reaction with endogenous SLCO1B3 in EB‐SCC cells
In order to demonstrate the functionality of the pre‐evaluated RTMm2 also on the endogenous level, we tested its trans‐splicing ability in EB‐SCC cells, over‐expressing SLCO1B3 (South A.P.; Manuscript in preparation). To simplify the monitoring of the trans‐splicing efficiency manifested in the expression of the SLCO1B3‐tk fusion protein over a longer period of time, RTMm2 was cloned into a retroviral vector and transduced into EB‐SCC cells. A retroviral vector carrying the SLCO1B3‐tk fusion gene and a splicing deficient RTMmut (3′ splice site was removed from RTMm2 by site directed mutagenesis) were included as positive and negative controls in this experiment, respectively. RTMm2 transduction of the tumor cells resulted in a long lasting expression of the RTM facilitating the detection of the SLCO1B3‐tk fusion molecule on RNA and protein level. The SLCO1B3‐tk fusion was detected in RTMm2 transduced EB‐SCC cells by sqRT‐PCR, demonstrating the endogenous functionality of our designed RTM with a relative trans‐splicing efficiency of 4.9% in comparison to total amplified SLCO1B3 transcripts. RTMmut did not induce a trans‐splicing reaction (Figure 6A + B). We were able to confirm the translation of the SLCO1B3‐tk fusion by immunoprecipitation using two different purification methods, via protein G sepharose or FLAG affinity gel (Figure 6C). Finally, we analyzed the functionality of SLCO1B3‐tk fusion protein in transduced EB‐SCC cells upon GCV treatment (Figure 6D). MTT assays demonstrated a reduction of cell viability in fusion Ctrl (to 19.5%) and RTMm2 (to 45%) transduced cells in comparison to untreated EB‐SCC cells, attesting to the functionality of the fusion protein also on the endogenous level. RTMm2 showed a significant reduction in cell viability compared to RTMmut (p = 0.006), indicating induction of apoptosis in EB‐SCC cells via trans‐splicing specific suicide gene transfer.
Figure 6.
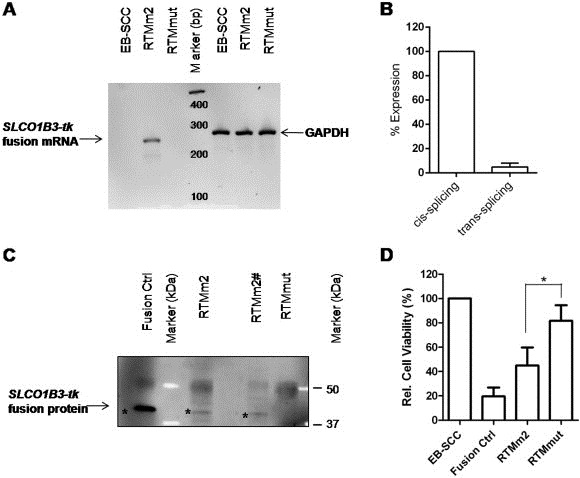
Trans‐splicing into endogenous SLCO1B3 pre‐mRNA. (A) SqRT‐PCR confirmed a correct trans‐splicing product (245 bp) only in RTMm2 transduced EB‐SCC cells. The housekeeping gene GAPDH served as control for quality and quantity. PCR products were verified by direct sequencing (data not shown). (B) mRNA level of endogenous trans‐splicing compared to cis‐splicing within the SLCO1B3 gene in RTMm2 transduced EB‐SCC cells. Specific PCR products were amplified by sqRT‐PCR and trans‐splicing efficiency (total SLCO1B3‐tk transcripts) was calculated relative to the cis‐splicing level (total SLCO1B3 transcripts = 100%). GAPDH was used as housekeeping gene (C) Detection of SLCO1B3‐tk fusion protein (marked by asterisk) by immunoprecipitation (using anti‐FLAG antibody) in combination with Western blot analysis (using anti‐HSV‐tk antibody) in cells containing transduced RTMm2 or the fusion ctrl. In contrast, no specific protein was isolated from RTMmut transduced cells. Fusion proteins were purified using protein G sepharose (Fusion Ctrl, RTMm2 and RTMmut) or FLAG affinity gel (RTMm2#) and visualized with anti‐HSV‐tk antibody by Western blot analysis. (D) Expression of SLCO1B3‐tk fusion protein resulted in reduced cell survival as detected by MTT assay. The mean of at least 4 different experiments ± SD is shown. *P < 0.05 Student's t test, unpaired, two‐tailed.
4. Discussion
In this work we present a screening system accelerating the design of “suicide” RTMs capable to induce apoptosis in tumor cells by accurate trans‐splicing into the tumor marker SLCO1B3.
Targeted elimination of tumor cells is challenging for many reasons. One significant hurdle is finding a means to discriminate between normal and malignant cells. We expedite spliceosome mediated RNA trans‐splicing (SMaRT) as a promising approach for cancer cell targeting. RNA trans‐splicing is a technology that exhibits its potential in different applications: such as therapeutic gene correction, production of therapeutic proteins, molecular imaging and suicide gene therapy approaches (as reviewed by Wally et al., 2012). A trans‐splicing‐mediated approach for tumor cell targeting was previously demonstrated by Nakayama et al. applying a “segmental trans‐splicing” (STS) strategy. In this work the 5′ and 3′ fragments of the Shigatoxin gene were separately introduced into tumors, combined in the target cells by accurate trans‐splicing leading to the subsequent expression of a Shigatoxin protein and a marked suppression of tumor size over time (Nakayama et al., 2005). However, the delivery of two vectors may hamper the chance of a therapeutic success. Therefore we utilize the increased endogenous expression of a tumor marker gene to maintain the trans‐splicing‐mediated expression of a fusion protein exhibiting cell killing potential predominantly in tumor cells. This should increase the tumor‐specific expression of the toxin as preferable those cells are targeted by the delivered RNA trans‐splicing molecule (RTM), in which the marker gene is expressed at high levels.
The feasibility of 3′ trans‐splicing technology in a suicide gene therapy approach was initially demonstrated by Puttaraju et al. (1999). They have designed an RTM capable to trans‐splice the coding sequence of diphtheria toxin (DT‐A) to the tumor marker gene β‐subunit of human chorionic gonadotropin gene 6 (βhCG6). After transfecting tumor bearing athymic mice (H1299 human lung cancer cells) with the RTM a correct splicing product consisting of βhCG6 and DT‐A was detected on the mRNA level. More recently we also provided proof of principle to this technology in cancer cells (SCC) from patients suffering from recessive dystrophic epidermolysis bullosa (RDEB) (Gruber et al., 2011). Utilizing a fluorescence based screening system we defined an RTM targeting the matrix metalloproteinase 9 (MMP‐9), which was shown to be an upregulated mRNA transcript in RDEB‐SCC in vitro. The RTM accomplished accurate trans‐splicing on the endogenous mRNA level in cell culture thereby generating a new chimeric fusion mRNA consisting of exon 1 of MMP‐9 and the suicide gene HSV‐tk encoded by the RTM. Furthermore, cell death was induced in MMP‐9 positive cancer cells by the RTM, delivering the coding sequence of exotoxin streptolysin O, indicating target‐specific RNA replacement.
However, since non‐specific trans‐ and cis‐splicing events induced by a designed RTM can reduce the pool of specific and functional RTMs significantly, our present work concentrates on the construction and optimization of RTMs in order to reduce these unwanted side effects which have to be eradicated prior to in vivo investigations.
First, we constructed an RTM targeting the gene SLCO1B3, coding for the organic anion transporter polypeptide OATP1B3 (Kalliokoski and Niemi, 2009) which was shown to be an upregulated mRNA population in EB‐SCC cells. OATP1B3 is mainly expressed at the membrane of human hepatocytes and act as influx transporters for its substrates from blood (Niemi, 2007). An increased expression of SLCO1B3 mRNA was previously observed in colorectal cancer, where it provides apoptotic resistance to colon cancer cells (Lee et al., 2008) and more recently, also in head and neck SCC tumor tissue (Zolk et al., 2013). Although, SLCO1B3 demonstrated to be a potent target for the evaluation and improvement of RTMs in our suicide gene therapy approach, some aspects have to be considered when used in a more translational perspective. For systemic application, the targeting and killing of SLCO1B3‐expressing hepatocytes by the RTM can be prevented by, e.g. the usage of a skin specific K14 promoter (Staggers et al., 1995; Hosseini et al., 2010). An alternative route for a possible application might also constitute the local injection of the RTM into the epidermal tumor tissue. However, the most promising RTM introduction has to be evaluated in future in vivo studies which can be viral‐ or non‐viral‐based. Additionally, these experiments will also answer the question of possible side effects in non‐cancerogenous cells/tissue with a lack of SLCO1B3 expression.
Due to the huge size of SLCO1B3 intron 3 (39,205 nt) we only screened the first 1,200 nt for potent RTM binding sites. To date, few rational parameters are known for the design of a BD, and future screening experiments may also identify even more potent BDs in the 3′ part of intron 3. Recently, Murauer et al. suggested that by blocking the endogenous 3′ intron/exon junction the splicing machinery may favor the use of 3′ ss provided by the RTM leading to increased trans‐splicing efficiency (Murauer et al., 2013). However, due to the huge size of our target intron we preferred to select the 5′ region of intron 3 near the first 5′ coding exon for RTM binding.
The two critical points concerning development of a suicide RTM are: (i) the occurrence of nonspecific trans‐splicing events between the RTM and pre‐mRNAs of other genes; and (ii) direct expression of the suicide gene from the RTM vector or functional suicide gene expression through cis‐splicing within the RTM backbone sequence. In this work we increased the trans‐splicing specificity by modifying the RTM's BD and deleting ATGs and cis‐splice sites upstream of the suicide gene (HSV‐tk) sequence within the RTM. The HSV‐tk system provides an easy monitoring of the trans‐splicing efficiency and specificity by the RTM as the toxicity of the tk gene is only activated after ganciclovir treatment of the cells. A significant reduction of background tk expression for both RTMm1 and RTMm2 was detected by Western blot analysis and confirmed by MTT assays. Additionally, GCV treatment of HEK293AD cells co‐transfected with RTM and SLCO1B3‐MG led to a significant reduction of cell viability in comparison to cells transfected with SLCO1B3‐MG or an RTM alone. Although the HSV‐tk system is known to show a “bystander effect” (Freeman et al., 1993) which may influence our experimental system, we clearly observed a reduction of cell viability after the SLCO1B3‐MG addition in a dose dependent manner. The cell viability was found to decrease as the amount of SLCO1B3‐MG‐expressing plasmid increased, owing to higher trans‐splicing rates in the nucleus. However, the current inducible cell‐death system has to be further optimized due to persistent, albeit significantly reduced background tk expression caused by the high amounts of RTM transfected in the heterologous system. In addition, our results support the construction of RTMs for subsequent tests in endogenous model systems since optimized RTMm2 demonstrated its functionality also in EB‐SCC cells. Here we used a retroviral vector system for delivery and stable expression of the RTM to ease the monitoring and detection of the SLCO1B3‐tk fusion protein resulting from accurate trans‐splicing in EB‐SCC cells. Although retro‐ or lentiviral vectors are previously described as transport systems for RTMs in gene correction studies, where a long term restoration of mutated genes is favored (Murauer et al., 2011; Wally et al., 2010; Avale et al., 2013), we assume that for a suicide gene therapy approach a viral integration system might not be necessary because cell killing by a strong toxin do not require long lasting expression of the RTM.
We detected a relative trans‐splicing efficiency (SLCO1B3‐tk fusion transcripts versus total SLCO1B3 transcripts) of about 5%. Still we observed a significant reduction of cell viability of RTM transduced tumor cells after ganciclovir treatment. These data indicate that in this cancer suicide gene approach the trans‐splicing efficiency plays a minor role, as compared to the effects of the toxin. In future experiments variations in RTM administration, RTM delivery systems or the selected toxin for apoptosis induction will provide further information concerning the efficiency, specificity and safety of the RNA trans‐splicing‐induced cell death system in comparison to conventional cancer therapy approaches already applied. Taken together, these results confirm the potential of SMaRT to induce the death of target cells expressing SLCO1B3 and we suggest SMaRT as a promising tool to target and eliminate specific tumor cells in a suicide gene therapy approach to treat patients suffering from EB‐SCC.
Funding
This work was supported by DEBRA Austria.
Supporting information
Supplementary data
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.08.005.
Gruber Christina, Koller Ulrich, Murauer Eva M., Hainzl Stefan, Hüttner Clemens, Kocher Thomas, South Andrew P., Hintner Helmut, Bauer Johann W., (2013), The design and optimization of RNA trans‐splicing molecules for skin cancer therapy, Molecular Oncology, 7, doi: 10.1016/j.molonc.2013.08.005.
Contributor Information
Christina Gruber, Email: c.gruber@salk.at.
Ulrich Koller, Email: u.koller@salk.at.
References
- Altaner, C. , 2008. Prodrug cancer gene therapy. Cancer Lett.. 270, 191–201. [DOI] [PubMed] [Google Scholar]
- Avale, M.E. , Rodríguez-Martín, T. , Gallo, J.M. , 2013. Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum. Mol. Genet.. 22, 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, J.W. , Murauer, E.M. , Wally, V. , Koller, U. , 2013. RNA trans-splicing for genodermatoses. Methods Mol. Biol.. 961, 441–455. [DOI] [PubMed] [Google Scholar]
- Chao, H. , Mansfield, S.G. , Bartel, R.C. , Hiriyanna, S. , Mitchell, L.G. , Garcia-Blanco, M.A. , Walsh, C.E. , 2003. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med.. 9, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Coady, T.H. , Lorson, C.L. , 2010. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J. Neurosci.. 30, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, C.L. , Pourreyron, C. , Foerster, J. , Purdie, K. , Salas-Alanis, J.C. , Murrell, D.F. , Bruckner-Tuderman, L. , McGrath, J.A. , Leigh, I.M. , South, A.P. , 2009. SLCO1B3 expression in recessive dystrophic epidermolysis bullosa associated squamous cell carcinoma is reduced by recombinant type VII collagen. J. Invest Dermatol.. 129, S55 [Google Scholar]
- Dallinger, G. , Puttaraju, M. , Mitchell, L.G. , Yancey, K.B. , Yee, C. , Klausegger, A. , Hintner, H. , Bauer, J.W. , 2003. Development of spliceosome-mediated RNA trans-splicing (SMaRT) for the correction of inherited skin diseases. Exp. Dermatol.. 12, 37–46. [DOI] [PubMed] [Google Scholar]
- Elion, G.B. , Furman, P.A. , Fyfe, J.A. , de Miranda, P. , Beauchamp, L. , Schaeffer, H.J. , 1977. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl) guanine. Proc. Natl. Acad. Sci. U S A. 74, 5716–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillat, C. , Carrio, M. , Cascante, A. , Sangro, B. , 2003. Suicide gene therapy mediated by the Herpes Simplex virus thymidine kinase gene/Ganciclovir system: fifteen years of application. Curr. Gene Ther.. 3, 13–26. [DOI] [PubMed] [Google Scholar]
- Freeman, S.M. , Abboud, C.N. , Whartenby, K.A. , Packman, C.H. , Koeplin, D.S. , Moolten, F.L. , Abraham, G.N. , 1993. The “bystander effect”: tumor regression when a fraction of the tumor mass is genetically modified. Cancer Res.. 53, 5274–5283. [PubMed] [Google Scholar]
- Gruber, C. , Gratz, I.K. , Murauer, E.M. , Mayr, E. , Koller, U. , Bruckner-Tuderman, L. , Meneguzzi, G. , Hintner, H. , Bauer, J.W. , 2011. Spliceosome-mediated RNA trans-splicing facilitates targeted delivery of suicide genes to cancer cells. Mol. Cancer Ther.. 10, 233–241. [DOI] [PubMed] [Google Scholar]
- Hamel, W. , Magnelli, L. , Chiarugi, V.P. , Israel, M.A. , 1996. Herpes simplex virus thymidine kinase/ganciclovir-mediated apoptotic death of bystander cells. Cancer Res.. 56, 2697–2702. [PubMed] [Google Scholar]
- Hosseini, S.J. , Zomorodipour, A. , Jalal, R. , Sabouni, F. , Ataei, F. , 2010. A study of the expression of functional human coagulation factor IX in keratinocytes using a nonviral vector regulated by K14 promoter. Appl. Biochem. Biotechnol.. 162, 1599–1611. [DOI] [PubMed] [Google Scholar]
- Immonen, A. , Vapalahti, M. , Tyynela, K. , Hurskainen, H. , Sandmair, A. , Vanninen, R. , Langford, G. , Murray, N. , Yla-Herttuala, S. , 2004. AdvHSV-tk gene therapy with intravenous ganciclovir improves survival in human malignant glioma: a randomised, controlled study. Mol. Ther.. 10, 967–972. [DOI] [PubMed] [Google Scholar]
- Kalliokoski, A. , Niemi, M. , 2009. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol.. 158, 693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikumori, T. , Cote, G.J. , Gagel, R.F. , 2001. Promiscuity of pre-mRNA spliceosome-mediated trans splicing: a problem for gene therapy?. Hum. Gene Ther.. 12, 1429–1441. [DOI] [PubMed] [Google Scholar]
- Koller, U. , Wally, V. , Mitchell, L.G. , Klausegger, A. , Murauer, E.M. , Mayr, E. , Gruber, C. , Hainzl, S. , Hintner, H. , Bauer, J.W. , 2011. A novel screening system improves genetic correction by internal exon replacement. Nucleic Acids Res.. 39, e108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. , Belkhiri, A. , Lockhart, A.C. , Merchant, N. , Glaeser, H. , Harris, E.I. , Washington, M.K. , Brunt, E.M. , Zaika, A. , Kim, R.B. , El Rifai, W. , 2008. Overexpression of OATP1B3 confers apoptotic resistance in colon cancer. Cancer Res.. 68, 10315–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Luo, M. , Zhang, L.N. , Yan, Z. , Zak, R. , Ding, W. , Mansfield, S.G. , Mitchell, L.G. , Engelhardt, J.F. , 2005. Spliceosome-mediated RNA trans-splicing with recombinant adeno-associated virus partially restores cystic fibrosis transmembrane conductance regulator function to polarized human cystic fibrosis airway epithelial cells. Hum. Gene Ther.. 16, 1116–1123. [DOI] [PubMed] [Google Scholar]
- Murauer, E.M. , Gache, Y. , Gratz, I.K. , Klausegger, A. , Muss, W. , Gruber, C. , Meneguzzi, G. , Hintner, H. , Bauer, J.W. , 2011. Functional correction of type VII collagen expression in dystrophic epidermolysis bullosa. J. Invest Dermatol.. 131, 74–83. [DOI] [PubMed] [Google Scholar]
- Murauer, E.M. , Koller, U. , Hainzl, S. , Wally, V. , Bauer, J.W. , 2013. A reporter-based screen to identify potent 3' trans-splicing molecules for endogenous RNA repair. Hum. Gene Ther. Methods. 24, 19–27. [DOI] [PubMed] [Google Scholar]
- Muto, M. , Onogawa, T. , Suzuki, T. , Ishida, T. , Rikiyama, T. , Katayose, Y. , Ohuchi, N. , Sasano, H. , Abe, T. , Unno, M. , 2007. Human liver-specific organic anion transporter-2 is a potent prognostic factor for human breast carcinoma. Cancer Sci.. 98, 1570–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, M. , Furihata, T. , Matsumoto, S. , Ishii, S. , Motohashi, S. , Yoshino, I. , Ugajin, M. , Miyajima, A. , Matsumoto, S. , Chiba, K. , 2012. Identification of a new organic anion transporting polypeptide 1B3 mRNA isoform primarily expressed in human cancerous tissues and cells. Biochem. Biophys. Res. Commun.. 418, 818–823. [DOI] [PubMed] [Google Scholar]
- Nakayama, K. , Pergolizzi, R.G. , Crystal, R.G. , 2005. Gene transfer-mediated pre-mRNA segmental trans-splicing as a strategy to deliver intracellular toxins for cancer therapy. Cancer Res.. 65, 254–263. [PubMed] [Google Scholar]
- Nasu, Y. , Saika, T. , Ebara, S. , Kusaka, N. , Kaku, H. , Abarzua, F. , Manabe, D. , Thompson, T.C. , Kumon, H. , 2007. Suicide gene therapy with adenoviral delivery of HSV-tK gene for patients with local recurrence of prostate cancer after hormonal therapy. Mol. Ther.. 15, 834–840. [DOI] [PubMed] [Google Scholar]
- Niemi, M. , 2007. Role of OATP transporters in the disposition of drugs. Pharmacogenomics. 8, 787–802. [DOI] [PubMed] [Google Scholar]
- Peckl-Schmid, D. , Wolkerstorfer, S. , Konigsberger, S. , Achatz-Straussberger, G. , Feichtner, S. , Schwaiger, E. , Zaborsky, N. , Huemer, M. , Gratz, I.K. , Schibli, R. , Lamers, M. , Crameri, R. , Moser, K. , Luger, E.O. , Achatz, G. , 2010. HAX1 deficiency: impact on lymphopoiesis and B-cell development. Eur. J. Immunol.. 40, 3161–3172. [DOI] [PubMed] [Google Scholar]
- Puttaraju, M. , Jamison, S.F. , Mansfield, S.G. , Garcia-Blanco, M.A. , Mitchell, L.G. , 1999. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol.. 17, 246–252. [DOI] [PubMed] [Google Scholar]
- Rheinwald, J.G. , Green, H. , 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 6, 331–343. [DOI] [PubMed] [Google Scholar]
- Schwarzenberger, P. , Byrne, P. , Gaumer, R. , Norton, J. , Harrison, L. , Marrogi, A. , Kolls, J.K. , 2011. Treatment of mesothelioma with gene-modified PA1STK cells and ganciclovir: a phase I study. Cancer Gene Ther.. 18, 906–912. [DOI] [PubMed] [Google Scholar]
- Song, Y. , Lou, H.H. , Boyer, J.L. , Limberis, M.P. , Vandenberghe, L.H. , Hackett, N.R. , Leopold, P.L. , Wilson, J.M. , Crystal, R.G. , 2009. Functional cystic fibrosis transmembrane conductance regulator expression in cystic fibrosis airway epithelial cells by AAV6.2-mediated segmental trans-splicing. Hum. Gene Ther.. 20, 267–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staggers, W.R. , Paterson, A.J. , Kudlow, J.E. , 1995. Sequence of the functional human keratin K14 promoter. Gene. 153, 297–298. [DOI] [PubMed] [Google Scholar]
- Wally, V. , Brunner, M. , Lettner, T. , Wagner, M. , Koller, U. , Trost, A. , Murauer, E.M. , Hainzl, S. , Hintner, H. , Bauer, J.W. , 2010. K14 mRNA reprogramming for dominant epidermolysis bullosa simplex. Hum. Mol. Genet.. 19, 4715–4725. [DOI] [PubMed] [Google Scholar]
- Wally, V. , Murauer, E.M. , Bauer, J.W. , 2012. Spliceosome-mediated trans-splicing: the therapeutic cut and paste. J. Invest Dermatol.. 132, 1959–1966. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Mansfield, S.G. , Cote, C.A. , Jiang, P.D. , Weng, K. , Amar, M.J. , Brewer, B.H. , Remaley, A.T. , McGarrity, G.J. , Garcia-Blanco, M.A. , Puttaraju, M. , 2009. Trans-splicing into highly abundant albumin transcripts for production of therapeutic proteins in vivo. Mol. Ther.. 17, 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, S.A. , Pourreyron, C. , Purdie, K. , Hogan, C. , Cole, C.L. , Foster, N. , Pratt, N. , Bourdon, J.C. , Appleyard, V. , Murray, K. , Thompson, A.M. , Mao, X. , Mein, C. , Bruckner-Tuderman, L. , Evans, A. , McGrath, J.A. , Proby, C.M. , Foerster, J. , Leigh, I.M. , South, A.P. , 2011. Integrative mRNA profiling comparing cultured primary cells with clinical samples reveals PLK1 and C20orf20 as therapeutic targets in cutaneous squamous cell carcinoma. Oncogene. 30, 4666–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Li, S. , Li, X.L. , Guo, Y. , Zou, B.Y. , Xu, R. , Liao, H. , Zhao, H.Y. , Zhang, Y. , Guan, Z.Z. , Zhang, L. , 2009. Phase I and biodistribution study of recombinant adenovirus vector-mediated herpes simplex virus thymidine kinase gene and ganciclovir administration in patients with head and neck cancer and other malignant tumors. Cancer Gene Ther.. 16, 723–730. [DOI] [PubMed] [Google Scholar]
- Zolk, O. , Schnepf, R. , Muschler, M. , Fromm, M.F. , Wendler, O. , Traxdorf, M. , Iro, M. , Zenk, H. , 2013. Transporter gene expression in human head and neck squamous cell carcinoma and associated epigenetic regulatory mechanisms. Am. J. Pathol.. 182, 234–243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


