Abstract
The frequently altered phosphatidylinositol‐3‐kinase (PI3K)/Akt signaling pathway is involved in the regulation of cellular processes required for breast carcinogenesis. The aim of the project was to develop a method to identify hotspot mutations in the PIK3CA gene in circulating tumor cells (CTCs) of metastatic breast cancer (metBC) patients.
From 44 enrolled CTC‐positive metBC patients a total number of 57 peripheral blood samples were analysed by CellSearch®. Genomic DNA of enriched CTCs was isolated, amplified and analyzed for PIK3CA mutations in exons 9 and 20 which lead to E542K, E545K or H1047R amino acid changes and result in increased PI3K activity. The mutations were detected by using SNaPshot‐methodology comprising PCR amplification and single nucleotide primer extension.
SNaPshot analysis was established using genomic DNA from different breast cancer cell lines and then successfully transferred to investigate blood samples and single cells. Overall, twelve hotspot mutations in either exon 9/E545K (6/12, 50%) or exon 20/H1047R (6/12, 50%) could be determined within 9 out of 57 (15.8%) blood samples from 7 out of 44 (15.9%) patients; CTC counts ranged from 1 to 9748. PIK3CA variants E542K, E545G and E545A were not detected.
Analysing the PIK3CA genotype of CTCs has clinical relevance with respect to drug resistance, e.g. against HER2‐targeted therapy. The herein described approach including SNaPshot technology provides a simple method to characterize hotspot mutations within CTCs enriched from peripheral blood and can be easily adopted for analysing further therapeutically relevant SNPs.
Keywords: Circulating tumor cells, Metastatic breast cancer, PIK3CA mutations, SNaPshot assay
Highlights
The SNaPshot assay is a robust tool allowing SNP detection within PIK3CA on CTCs.
PIK3CA mutations in exon 9 and 20 were determined in CTCs of breast cancer patients.
9 out of 57 blood samples from 7 out of 44 patients harbored PIK3CA mutant alleles.
The SNaPshot assay can be adopted for further mutational analysis on CTCs.
Abbreviations
- APC
allophycocyanin
- CK-PE
cytokeratin-phycoerythrin
- CS
CellSearch®
- CTC(s)
circulating tumor cell(s)
- DAPI
4,6-diamidino-2-phenylindole
- DTCs
disseminated tumor cells
- ER
estrogen receptor
- FITC
fluorescein-isothiocyanate
- HER2
human epidermal growth factor receptor 2
- PI3K
phosphatidylinositol-3-kinase
- PTEN
phosphatase and tensin homolog
- PR
progesterone receptor
- SNP
single nucleotide polymorphism
- WGA
Whole Genome Amplification
1. Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death in females worldwide, accounting for 23% (1.38 million) of the total new cancer cases and 14% (458,400) of the total cancer deaths in 2008 (Jemal et al., 2010, 2011).
Up to now, breast cancer treatment decisions are based upon the histology of the primary tumor and its expression status of molecular markers such as estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). Although significant improvements in breast cancer treatment have been achieved within the last decades, the need for novel therapeutic approaches aiming at specific tumorigenic cells and their key oncogenic pathways has become obvious.
In several clinical studies, the detection and enumeration of CTCs from breast cancer patients has been established showing a correlation with decreased progression‐free and overall survival in operable (Ignatiadis et al., 2008; Xenidis et al., 2006) and advanced breast cancer (Cristofanilli et al., 2005). Molecular characterization of CTCs is essential to confirm their malignant origin and to identify diagnostically and therapeutically relevant targets which help to stratify cancer patients for individual therapies (Scher et al., 2009).
Breast cancer cells in primary tumors, CTCs as well as disseminated tumor cells (DTCs) vary phenotypically and functionally in established prognostic factors such as HER2, ER, PR and in gene expression profiles (Bozionellou et al., 2004; Meng et al., 2004; Müller and Pantel, 2009; Strati et al., 2011; Powell et al., 2012). These findings advocate for the usage of CTCs as ‘liquid biopsy’ to refine therapeutic regimens.
In breast cancer, the PI3K/PTEN/AKT pathway which is involved in the regulation of central cellular processes is often dysregulated, due to alterations in several pathway components including amplification of the HER2 gene locus, loss of PTEN (phosphatase and tensin homolog) protein function, and mutations of the PIK3CA gene (Bachman et al., 2004; Dunlap et al., 2010; Paradiso et al., 2007). PIK3CA gene variations result in different activation levels of PI3K (Araújo et al., 2010).
Class 1A PI3‐kinases are heterodimeric lipid kinases composed of a catalytic subunit (p110a), encoded by the PIK3CA gene and a regulatory subunit (p85a). Specific mutations within PIK3CA that cluster in hotspots located in exon 9 (E542/545K; catalytic domain) and exon 20 (H1047R; kinase domain) have been demonstrated to activate PI3K/AKT signaling and provide a strong selective growth advantage to the cell. This suggestion is accompanied by the fact that all three hotspot mutations confer in vitro and in vivo tumorigenicity (Bader et al., 2006; Ikenoue et al., 2005; Isakoff et al., 2005; Kang et al., 2005; Samuels et al., 2005; Zhao et al., 2005). These observations and the fact that specific inhibitors for PIK3CA are being developed emphasize the importance of analyzing the PIK3CA mutational status in CTCs, and relating this status to potential drug resistance, especially in those CTCs surviving HER2‐targeting therapy.
In order to detect PIK3CA mutations, PCR‐based screening methods and direct sequencing of PCR products have been applied (Board et al., 2008; Simi et al., 2008; Qiu et al., 2008). Here for the first time, we describe the application of a modified approach based on the SNaPshot‐technology published by Hurst et al. in order to simultaneously analyse the PIK3CA mutations E542K, E545 G/K and H1047R in CTCs of metBC patients and in single breast cancer cells. This assay allows a combined implementation of multiplex PCR amplification and multiplex primer extension for the targeted detection of several mutations in one approach (Hurst et al., 2009). We have used this technology because it enables a sensitive analysis of SNPs from single tumor cells in a multiplex approach.
2. Material and methods
2.1. Cell lines and cell culture
MCF‐7, SKBR‐3 and T47D breast cancer cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). SKBR‐3 cells were reported to contain a PIK3CA wild‐type gene, while MCF‐7 and T47D cells harbor heterozygous hotspot mutations in the PIK3CA gene: MCF‐7 is mutated in exon 9 (E545K), T47D in exon 20 (H1047R) (Hollestelle et al., 2007; Jensen et al., 2011; Kataoka et al., 2010; Weigelt et al., 2011). All cell lines were cultured in RPMI 1640 containing 10% fetal calf serum and 1% Penicillin‐Streptomycin (all Gibco, Karlsruhe, Germany). Culture medium for MCF‐7 was supplemented with 25 mM HEPES (Gibco); for T47D cells 10 mM HEPES, 1 mM sodium pyruvate (Gibco) and 0.45% D‐(+)Glucose solution (Sigma–Aldrich, Munich, Germany) was added. All cells were grown at 37 °C in a humidified atmosphere with 5% CO2. For spiking experiments, dilution series for MCF‐7, SKBR‐3 and T47D cells were performed and about 100 cells in PBS were spiked into 7.5 ml blood of healthy volunteers collected into CellSearch® (CS) CellSave preservative tubes (Veridex LLC, Raritan, NJ, USA).
2.2. Patients
Patients were recruited within a translational spin‐off project of the German Detect III trial which is investigating blood samples from primarily HER2‐negative metastatic breast cancer patients who harbor HER2‐positive CTCs (for more information see www.detect‐studien.de). In total, 57 blood samples (7.5 ml) from 44 CTC‐positive advanced metBC patients were collected into CellSave tubes and processed by CellSearch® (Veridex LLC, Raritan, NJ, USA). The clinical patient data are shown in Table 1, more detailed information is provided in the Supplemental data Table 1. Written informed consent was obtained from all participating patients and the study was approved by the local ethical committee (525/2011AMG1).
Table 1.
Clinical data of patients (primary tumor (PT) and CTCs).
| Characteristics | Total | In % | |
|---|---|---|---|
| Patients | 44 | 100.0 | |
| Age | |||
| Mean | 58.4 | ||
| Median | 59.0 | ||
| Range | 26–78 | ||
| Tumor size | |||
| T1 | 13 | 29.5 | |
| T2 | 15 | 34.1 | |
| T3 | 7 | 15.9 | |
| T4 | 8 | 18.2 | |
| Tx | 1 | 2.3 | |
| Nodal status | |||
| Negative | 11 | 25.0 | |
| Positive | 29 | 65.9 | |
| Nx | 4 | 9.1 | |
| M status of primary tumor at the time of diagnosis | |||
| M0 | 29 | 65.9 | |
| M1 | 7 | 15.9 | |
| Mx | 8 | 18.2 | |
| Histology | |||
| Ductal | 33 | 75.0 | |
| Lobular | 7 | 15.9 | |
| Others | 4 | 9.1 | |
| Grading | |||
| GI | 0 | 0 | |
| GII | 18 | 40.9 | |
| GIII | 21 | 47.7 | |
| Not determined | 5 | 11.4 | |
| ER status | |||
| Positive | 30 | 68.2 | |
| Negative | 12 | 27.3 | |
| Not determined | 2 | 4.5 | |
| PR status | |||
| Positive | 28 | 63.6 | |
| Negative | 14 | 31.8 | |
| Not determined | 2 | 4.5 | |
| HER2 status | |||
| Negative | 36 | 81.8 | |
| Not determined | 8 | 18.2 | |
| Patients with HER2+ CTC | 15 | 34.1 | |
| Range of CTC | 1–9748 | ||
2.3. Blood sample processing, detection of CTCs and assessment of HER2 expression
Isolation and enumeration of tumor cells from spiked blood samples obtained from healthy volunteers as well as from blood of metBC patients were processed by using the FDA‐approved CS. This system allows for the automated enrichment and immunostaining of CTCs in patients with metastatic breast, colon and prostate cancer. A total of 7.5 ml of whole blood was collected in a CellSave Preservative Tube and the CellSearch® Circulating Tumor Cell Kit (Veridex LLC, Raritan, NJ, USA) was applied for CTC enrichment and enumeration. Herein, immunomagnetic enrichment is achieved by using an anti‐EpCAM ferrofluid. Enriched cells are then fluorescently stained with the nucleic acid dye 4,6‐diamidino‐2‐phenylindole (DAPI) and labeled with monoclonal antibodies specific for epithelial cells (anti‐cytokeratins 8, 18, 19) and leukocytes (anti‐CD45). CTCs are subsequently identified by cytokeratin positivity/negativity for the leukocyte common antigen CD45 and nuclear DAPI staining to ensure the integrity of the nucleus. All CTC assessments were performed by trained and Veridex certified technicians. Blood samples were designated as positive when at least one CTC was present. HER2 expression of CTCs was characterized within the CS by the addition of a fluorescein‐labeled anti‐HER2 antibody (CS tumor phenotyping reagent HER2; Veridex), as described previously (Riethdorf et al., 2010). HER2‐specific immunostaining of CTCs was categorized into negative (0), weak (1+), moderate (2+) and strong (3+) (Riethdorf et al., 2010), whereas only HER2‐strong expressing CTCs were counted as HER2‐positive. Further blood sample processing after CTC/HER2 assessment can be found depicted in Figure 1.
Figure 1.
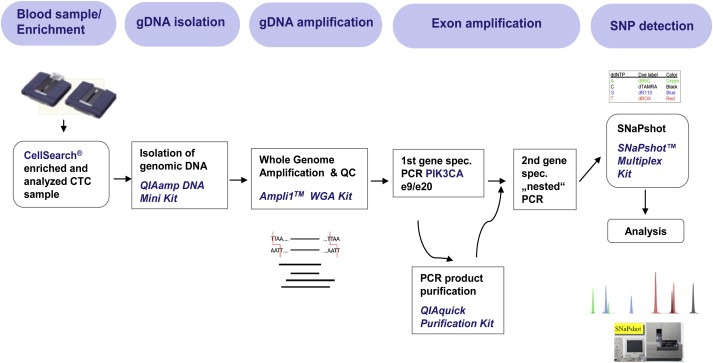
Workflow for the molecular characterization of circulating tumor cells by dint of CTC enrichment, Whole Genome Amplification and SNaPshot technology for PIK3CA.
2.4. Extraction of genomic DNA and Whole Genome Amplification (WGA)
DNA extraction from CS enriched tumor cells was obtained by using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Therefore, the entire contents of CTC‐positive CS cartridges were transferred into tubes, digested with Proteinase K and complete cell lysis was verified by visual control under a fluorescence microscope. Column‐bound DNA was eluted with 30 μl of nuclease‐free water and concentrated further to a final volume of 10 μl. 3 μl were employed for WGA using the Ampli1™ WGA Kit from Silicon Biosystems according to manufacturer's protocol. The WGA procedure is based on a ligation‐mediated PCR following a site‐specific DNA digestion. For quality control, 2 μl of Ampli1™ products were analysed utilising the Ampli1™ QC Kit (Silicon Biosystems, Bologna, Italy).
2.5. PCR
After WGA, the first gene‐specific PCR for PIK3CA exons 9 and 20 was performed in a volume of 25 μl containing 1× PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 5% DMSO, 1U Taq DNA polymerase (Life Technologies GmbH, Darmstadt, Germany) and 5 μl of whole genome amplified DNA as template. Primers were designed by Beckman Coulter to produce amplicons covering hotspot codons E542, E545 (exon 9) and H1047 (exon 20) (0.2 μM each; Table 2). Thermal cycler conditions were: 95 °C for 5min, 45 cycles of 95 °C for 45sec, 58 °C for 45sec, 72 °C for 45sec and finally 10min at 72 °C. PCR products were checked for quality and quantity by running 5 μl in a 3% agarose‐TBA gel. Remaining PCR products were purified using a PCR Purification Kit (Qiagen). Purified product (2.5 μl) was then further processed for a second exon 9 and exon 20 specific PCR (“nested PCR”) using nested PCR primers (Table 2) with the same PCR conditions as described above, only the number of PCR cycles was increased to 60.
Table 2.
Primer sequences for first gene‐specific, nested PCR amplification and for SNaPshot analysisa for exon 9 and exon 20 of PIK3CA.
| PIK3CA exon | Primer name | Sequence (5'–>3') | Product size (bp) |
|---|---|---|---|
| 9 | e9_131_left | GCTAGAGACAATGAATTAAGGGAAAA | 131 |
| e9_131_right | CTCCATTTTAGCACTTACCTGTGAC | ||
| 20 | e20_167_left | GCCAGAACTACAATCTTTTGATGAC | 167 |
| e20_167_right | CATGCTGTTTAATTGTGTGGAAG | ||
| 9 | e9_nested_left | GACAAAGAACAGCTCAAAGCAA | 110 |
| e9_nested_right | ATTTTAGCACTTACCTGTGAC | ||
| 20 | e20_nested_left | TTGATGACATTGCATACATTCG | 134 |
| e20_nested_right | GTGGAAGATCCAATCCATTT | ||
| PIK3CA exon | Mutation | Sequence (5'–>3')a | Size (bp) |
| 9 | E542K | T(19)TACACGAGATCCTCTCTCT | 38 |
| E545G | T(29)TCCTCTCTCTGAAATCACTG | 49 | |
| E545K | T(34)ATCCTCTCTCTGAAATCACT | 54 | |
| 20 | H1047R | T(46)TGAAACAAATGAATGATGCAC | 67 |
Primer sequences were published before (Hurst et al., 2009).
2.6. SNaPshot assay
The probes used in the SNaPshot reaction were adopted from Hurst et al. (2009) (Table 2). Probes anneal one nucleotide position 5′ of the potential mutated nucleotide on the template DNA and are extended by one base only due to the use of dideoxynucleotides (ddNTPs). Each of the four ddNTPs (ddATP, ddCTP, ddGTP, ddTTP) is labeled with a different fluorophore enabling them to be distinguished from each other. SNaPshot analysis was performed using the SNaPshot Multiplex Kit (Applied Biosystems, Carlsbad, US). Reactions were conducted in a total volume of 10 μl containing 4 μl PCR product (2 μl for each exon), 5 μl SNaPshot Multiplex Master Mix, and 1 μl of pooled SNaPshot probes (0.8 μM E542K, 2.3 μM E545G, 1.5 μM E545K, 1.5 μM H1047R). Primer extension was carried out in a thermal cycler (MJ Research, Watertown, US) using the following program: 25 cycles of 96 °C, 10sec (denaturation); 50 °C, 5sec (probe‐annealing) and 60 °C, 30sec (primer extension), then cooling down to 4 °C. In order to eliminate unincorporated ddNTPs, labeled extension products were treated with 0.5 μl Calf Intestinal Phosphatase (1 U/μl, NEB, Ipswich, MA) and incubated for 1 h at 37 °C, followed by a denaturation step at 75 °C for 15min. Subsequently, 10 μl of HiDi™ formamide and 0.6 μl of an internal size standard (Genescan‐120LIZ, both Applied Biosystems) were mixed with 2 μl extension product and transferred to an ABgene Thermo‐Fast 96 PCR Detection Plate (Thermo Scientific, Karlsruhe, Germany) for separation using an ABI PRISM 3100 Genetic Analyzer with a 36 cm length capillary and POP‐7™ polymer. SNP‐analysis was performed using GeneMapper 3.7 Software.
2.7. Deposition and preparation of single breast cancer cells
In order to amplify DNA in a low volume reaction format using single cells as DNA source AmpliGrid technology (Beckman Coulter, Munich, Germany) was applied. Single MCF‐7 cells were deposited in 1× PBS onto reaction sites of an AmpliGrid slide by dint of cell sorting via a BD FACSAria™ flow cytometer. After air‐drying the cells, a visual quality control was performed by inspecting Hoechst 33342 positive cells using fluorescence microscopy (Axioplan 2, Carl Zeiss GmbH, Göttingen, Germany). For DNA extraction from single cells the Beckman Coulter Cell Extraction Kit was used. The master mix for cell lysis comprises the following components: 1 μl lysis enzyme, 6 μl 10× reaction buffer and 53 μl nuclease‐free H2O. A volume of 0.75 μl was pipetted to the reaction side and the droplet was covered with 5 μl sealing solution before the loaded AmpliGrid was placed on the AmpliSpeed slide cycler (Beckman Coulter) to execute the cell extraction program (75 °C, 5min; 95 °C, 2min; RT). After cell lysis, gene‐specific PCR reactions with the primers listed in Table 2 were carried out on‐slide in a total volume of 1 μl. One half of the amplified products was used for quality control with the Agilent DNA 1000 kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany), the other half was employed for SNaPshot analysis.
3. Results
3.1. CTC assessment and determination of HER2 expression
Since the blood samples were used to set up the technique we – in contrast to the clinical cut‐off of ≥5 CTCs for metBC – considered samples CTC‐positive if ≥1 CTC was detected. In this study, 44 CTC‐positive metBC patients were enrolled and a total number of 57 blood samples was analysed. CTC counts ranged from 1 to 9748. CTCs were further examined for HER2 expression, with 15 out of 44 patients (34.1%) harboring at least one HER2‐positive (3+) CTC. In total, 23 of the 57 analysed samples (40.3%) contained HER2‐positive CTCs. For two patients (4.5%) HER2‐status was not assessed. In general, CTCs within patients differed strongly in HER2 expression. Figure 2 exemplifies different HER2 expression levels (negative (0), weak (1+), moderate (2+), strong (3+)) of CTCs enriched from patient 26.
Figure 2.
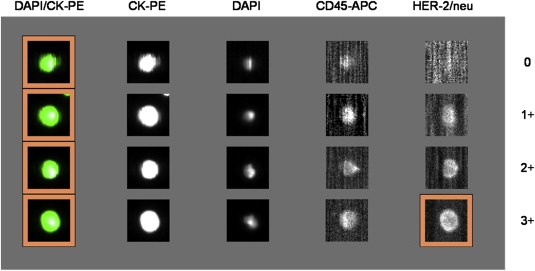
HER2 immunoscoring of CTCs from patient 26. CTCs show a cytokeratin signal (CK), are DAPI‐positive and CD45‐negative. HER2 expression of CTCs was determined using the FITC‐labeled anti‐HER2 antibody in the CellSearch® system. Intensities of HER2‐labeling was classified into negative (0), weak (1+), moderate (2+), and strong (3+); CK‐PE, cytokeratin‐phycoerythrin; DAPI, 4,6‐diamidino‐2‐phenylindole; APC, allophycocyanin; FITC, fluorescein‐isothiocyanate.
3.2. SNaPshot analysis using genomic DNA from breast cancer cell lines
Initially, the SNaPshot methodology was established using genomic DNA of breast cancer cell lines harboring different PIK3CA hotspot mutations of interest or carrying the wild‐type gene. MCF‐7 cells were chosen due to a mutation in exon 9 which results in an amino acid change at position E545 (E545K). T47D cells carry a point mutation in exon 20 leading to an amino acid change at position 1047 (H1047R). Finally, SKBR‐3 cells were reported to exhibit no PIK3CA mutations at these positions (Hollestelle et al., 2007; Jensen et al., 2011; Kataoka et al., 2010; Weigelt et al., 2011). In the beginning, primer sets for exon amplification described in Hurst et al. (2009) were used but gave non‐satisfying results within our setting. Therefore, we – in cooperation with Beckman Coulter – designed new PIK3CA primers to optimize DNA fragment amplification for exons 9 and 20. These primers were successfully implemented for single and duplex PCR amplification.
To detect SNPs the resulting amplicons for exons 9 and 20 were then used for PIK3CA SNaPshot analysis. Results are depicted in Figure 3 wherein the different colored peaks – generated by the Genemapper software – indicate which of the specific fluorescently labeled dideoxynucleotides was added during the primer extension reaction. SNPs can thus be identified by the peak color and size relative to the internal Genescan‐10LIZ size standard. The molecular weights of the SNaPshot extension products are influenced by the different fluorophores, thus mutant alleles are readily identified as they exhibit mobility shifts relative to the wild‐type alleles. In panel 1 of Figure 3 the electropherogram obtained for SNaPshot analysis of wild‐type PIK3CA in SKBR‐3 cells is depicted. The middle and bottom panels represent the peak patterns for heterozygous mutations in exon 9/E545K (MCF‐7) and exon 20/H1047R in T47D cells, respectively. To verify that SNaPshot is detecting the relevant SNPs, PCR products were sequenced (Figure 4).
Figure 3.
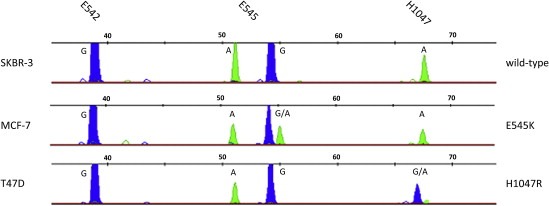
SNaPshot result for detection of hotspot mutations in the PIK3CA gene for genomic DNA of breast cancer cell lines. SNP detection in SKBR‐3 (wild‐type), MCF‐7 (E545K) and T47D (H1047R) performed on pooled singleplex PCRs for exon 9 and 20. Bases are represented by the following colorcode: A = green; G = blue.
Figure 4.
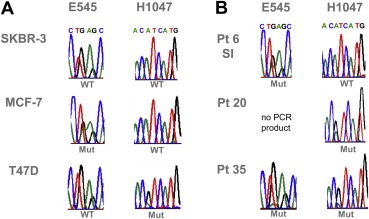
Sequences for PIK3CA exon 9 (E545) and exon 20 (H1047) derived from nested PCR products for spiked cell lines and patient (pt) samples. A. Sequencing results for breast cancer cell lines SKBR‐3 (WT), MCF‐7 (E545K) and T47D (H1047R). PCR products for MCF‐7 showed a heterozygous mutation at E545 where in codon GAG the G is replaced by an A resulting in AAG. Products for spiked T47D cells carried an H1047 mutation leading to CGT instead of CAT. B. Sequencing results for pts 6, 20 and 35. Sequence pattern of sample 1 (SI) from pt 6 shows a mutation in E545, the one from pt 20 harbored a mutation at position H1047, whereas amplified products of pt 35 showed both (E545K and H1047R, respectively).
3.3. SNaPshot analysis for spiked blood samples
According to the workflow depicted in Figure 1 the SNaPshot assay was then performed to detect PIK3CA mutations in breast cancer cell lines which were spiked into 7.5 ml of CellSave preservative blood from healthy donors. Electropherograms pictured in Figure 5 for about 100 MCF‐7 and T47D cells show the aforementioned mutant alleles for E545K (MCF‐7) and H1047R (T47D). Spiked SKBR‐3 cells exhibit the peak pattern for wild‐type PIK3CA at the examined positions.
Figure 5.
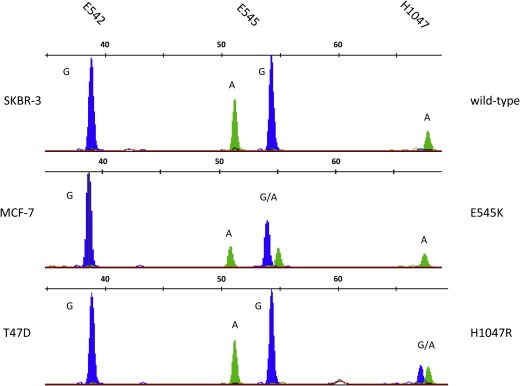
SNaPshot electropherograms for detection of hotspot mutations in the PIK3CA gene for spiked SKBR‐3, MCF‐7 and T47D breast cancer cells. SNP detection in about 100 SKBR‐3 (wild type), MCF‐7 (E545K) and T47D (H1047R) cells performed on pooled singleplex PCRs for exon 9 and 20. Bases are represented by the following colorcode: A = green; G = blue.
3.4. SNaPshot analysis of CTCs in blood from metBC patients
Based upon the established methodology on spiked blood samples, our SNaPshot protocol was applied to CTCs in 57 blood samples obtained from 44 metBC patients (Figure 6).
Figure 6.
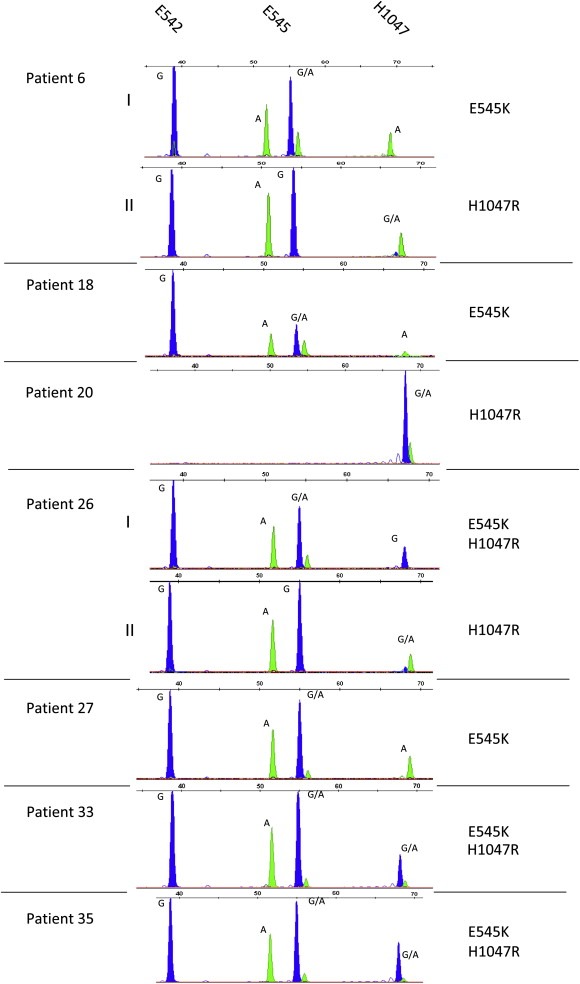
SNP analysis of hotspot mutations in the PIK3CA gene in circulating tumor cells from 7 metBC patients. SNaPshot electropherograms were obtained after CTC enrichment with the Veridex CellSearch® system, Whole Genome Amplification and gene‐specific PCRs for exon 9 and exon 20 of the PIK3CA gene. For patient (pt) 6 two blood samples were available exhibiting different mutations: sample I showed an E545K mutation while in sample II mutation H1047R was detected. For pt 26 PIK3CA mutations E545K and H1047R were observed separately in 2 samples. Pts 18 and 27 revealed mutant alleles at position E545, the sample of pt 20 was merely mutated in exon 20 (H1047R). In CTC samples for pts 33 and 35 mutations E545K and H1047R were observed. Incorporated bases are represented by the following colors: A = green; C = black; G = blue.
For patient 6 as well as for patient 26 two independent CTC‐positive blood samples were available. In both cases two different PIK3CA mutations, E545K and H1047R, were detected. Sample I of patient 6 (7 CTCs) exhibited an E545K mutation, and in sample II (7 CTCs) an H1047R mutation was detected. In sample I from patient 26 E545K and H1047R mutations were found simultaneously, with the H1047R mutation being detected in both ‐ the homozygous (sample I, 345 CTCs) and heterozygous (sample II, 487 CTCs) states. For patients 18 (121 CTCs) and 27 (a, not determined; see text below, Table 3), one blood sample each was assessed revealing a mutant PIK3CA allele at position E545 (E545K). E545K and H1047R mutations were observed simultaneously within CTC populations from single blood samples of patients 33 and 35 (7 and 2 CTCs). Finally, for patient 20 (4950 CTCs) a heterozygous mutation in H1047R was detected, whereas the mutational analysis for exon 20 could not be conducted due to failure of nested PCR for exon 9.
Table 3.
PIK3CA mutations detected in 7 metastatic breast cancer patients, CTC counts and HER2 status of CTCs within the patients.
| Patient | CTC count | Her2 status CTC patient | PIK3CA mutation |
|---|---|---|---|
| 6b | 7; 7 | Positive | H1047R; E545K |
| 18 | 121 | Positive | E545K |
| 20 | 4950 | Positive | H1047R |
| 26* | 345; 487 | Positive | H1047R; E545K/H1047R |
| 27 | a | Positive | E545K |
| 33 | 7 | Negative | E545K, H1047R |
| 35 | 2 | Positive | E545K, H1047R |
Not determined, because of ferrofluid in CS cartridge; but ≥ 1 CTC determined by visual microscopic inspection.
2 different blood samples were analyzed.
Overall, twelve hotspot mutations in either exon 9/E545K (6/12, 50%) or exon 20/H1047R (6/12, 50%) of PIK3CA were detected in 9 out of 57 (15.8%) blood samples from 7 out of 44 (15.9%) patients (Table 3). Point mutations resulting in E542K, E545G or E545A were not detected within our study. For patient 27 the exact number of CTCs could not be determined because of ferrofluid in the CS cartridge. However, CTCs were present according to standard microscopic inspection. Mutations could be detected in blood samples with as low as two CTCs.
3.5. SNaPshot analysis of single breast cancer cells deposited onto AmpliGrid slides
SNaPshot analysis was also performed after the deposition of single MCF‐7 cells onto AmpliGrids via cell sorting. Results gained for the mutational single cell analysis are illustrated in Figure 7.
Figure 7.
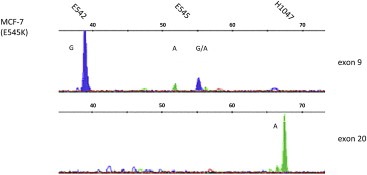
SNaPshot detection on single MCF‐7 breast cancer cells. Top: SNaPshot analysis performed on PCR product for PIK3CA exon 9 showing a heterozygous mutation at position E545K; bottom: result for SNP detection in exon 20 of single MCF‐7 cells; A = green, G = blue.
As expected the heterozygous mutation at position E545K in the PIK3CA exon 9 was detected, although peak height was lower than obtained in the aforementioned experiments.
4. Discussion
Up to now, the detection of relevant therapeutic targets in clinical practice is restricted to the primary tumor and success or failure of anti‐cancer therapies is only evaluated retrospectively by the absence or presence of metastases after surgery and therapy. Thus, there is an urgent need for biomarkers for real‐time personalized monitoring of the efficacy of systemic adjuvant therapy.
Molecular analysis of CTCs, which might reflect certain subpopulations of the primary tumor as well as cells forming metastases, could overcome current limitations. Therefore, we think that SNaPshot analysis of heterogeneous primary tumor samples is not suitable for prediction and that monitoring CTCs as “liquid biopsy” (Pantel et al., 2009) in contrast to analysis of the primary tumor can be utilized to determine the efficacy of chemotherapies and to gain deeper insights into the selection of resistant tumor cells under biological therapies. So far, the CS is the only FDA‐approved diagnostic test to automate the detection and enumeration of CTCs, for monitoring disease progression and therapy efficacy in metastatic prostate, colorectal and breast cancer (Cristofanilli et al., 2005). However, besides enumeration, molecular characterization of CTCs is mandatory to not only confirm their malignant origin. Discovering an association between gene expression profiles or expression of gene variants and clinical outcome, and identification of diagnostically and therapeutically relevant targets in CTCs may help stratify cancer patients for individual therapies (Sieuwerts et al., 2009). According to already published data (Fehm et al., 2010), we identified HER2‐positive CTCs in the blood samples of patients with HER2‐negative primary tumors (Table 1).
Furthermore, by dint of CTC analysis our knowledge of basic molecular pathways of invasion, migration and immune surveillance can be expanded and might contribute to the identification of metastatic stem cells with important implications for the development of improved therapies in the near future (Korkaya and Wicha, 2009). Assessing the presence of target antigens on CTCs could be considered as a real‐time biopsy allowing the evaluation of changes in tumour phenotype during the clinical course of the disease. A combination of highly sensitive multi‐parametric molecular methods and imaging has been evaluated very recently for the molecular characterization of CTCs (Punnoose et al., 2010). However, this has been hindered by the very limited sample amount available.
In this manuscript we describe the development of an approach to simultaneously determine several SNPs in the PIK3CA gene in order to characterize CTCs of metBC patients. This assay combines multiplex PCR amplifications of exons 9 and 20 of the PIK3CA gene combined with a multiplex primer extension assay allowing targeted detection of several mutations in one reaction. This so‐called SNaPshot technology has been published by Hurst et al. (2009). However, they applied it to characterize primary cancer tissues. We have modified the original approach to analyse – to our knowledge for the first time – rare CTCs out of breast cancer patients' blood extending it for single breast cancer cells. Regarding tumor heterogeneity and the fact that the correlation of molecular characteristics with outcome helps to identify predictive and prognostic parameters in breast cancer, the method comprising CTC enrichment, WGA and SNaPshot assay represents a novel research approach without being limited by the amount of cells.
Sensitivity tests showed that the SNaPshot technology could detect mutant DNA when it represents 5–10% of the total DNA achieving 100% concordance with results from high‐resolution melting analysis and sequencing (Hurst et al., 2009). In our experiments, the concordance rate between SNaPshot and sequencing was 100% as well (Figure 4). In literature, frequency distributions of exon 9 and exon 20 hotspot mutations among PIK3CA mutant breast cancer are summarized as 20% for E545K and 55% for H1047R, while E542K and E545G appear less frequent (11% and <1%) (Table 4; adapted from COSMIC, www.mycancergenome.com). For establishing the SNaPshot technology three cell lines were included displaying the wild type PIK3CA gene and harboring mutations leading to E545K (MCF‐7) and H1047R (T47D), respectively. An additional cell line comprising the E542K was not available in our laboratory, however, several publications described the existence of this mutation in a breast cancer cell line (BT483: Hollestelle et al., 2007; Jensen et al., 2011; Weigelt et al., 2011) and the feasibility of our SNaPshot assay at this position could be proven (wild type pattern ‐G instead of an A‐ is displayed).
Table 4.
Frequency of exon 9 and exon 20 hot spot mutations among PIK3CA mutant breast cancer (modified after www.mycancergenome.com).
| Gene | Exon | Location | Amino Acid position | Amino Acid change | Nucleotide change | Mutation frequency |
|---|---|---|---|---|---|---|
| PIK3CA | 9 | Helical domain | E542 | p.E542K | c.1624G > A | 11% |
| E545 | p.E545K | c.1633G > A | 20% | |||
| p.E545G | c.1634A > G | <1% | ||||
| 20 | Kinase domain | H1047 | p.H1047R | c.3140A > G | 55% |
Although the overall sample number we investigated was very low, we were able to detect mutations in 15.8% (9/57) of the CTC‐positive samples, with E545K and H1047R mutations occurring at the same frequency (each 50%, 6/12). Furthermore, occurrence of these somatic mutations seems to be dependent on the breast cancer subtype. For instance, 30% of hormone receptor–positive tumors exhibit PIK3CA mutations, while the incidence in triple‐negative breast cancer seems to be less frequent (3.3%, Stemke‐Hale et al., 2008). Within our collective comprising 7 patients having CTCs with mutations in the PIK3CA gene locus, one patient harbored a triple‐negative tumor, whereas the other 6 patients had hormone receptor‐positive primary tumors (Supplemental data Table 1).
Albeit the impact of these mutations concerning patient outcome is not completely clear yet, it has been reported that PI3K hyperactivity contributes to a lower response to trastuzumab and lapatinib treatment in patients with HER2‐positive tumors (Berns et al., 2007; Eichhorn et al., 2008; Serra et al., 2008; Kataoka et al., 2010) as well as to resistance to anti‐estrogen therapies. Furthermore, mutations regarding exon 20 of PIK3CA have been associated with poor prognosis (Lai et al., 2008). Jensen and co‐workers reported that PIK3CA gene mutations may be discordant between primary breast cancer and corresponding metastases (Jensen et al., 2011).
CTC population like its primary tumor is heterogenic in its cellular composition in terms of their phenotype and genotype. We observed this heterogeneity in blood samples obtained from different patients as well as in CTCs isolated from the same blood sample (Figure 2). Although we have not yet separated single CTCs in our experiments, we also observed heterogeneity in PIK3CA alleles supporting the published data and the need for single cell analysis.
In their recent publication Hurst et al. (2009) state that the identification of mutations using this approach is more straightforward compared to sequencing. In their hands, sequence analysis failed in 17% of the samples. One reason might be as the authors argue that especially in situations of low quality DNA, e.g. from FFPE‐tissue samples, the SNaPshot method has the advantage that the fluorescent signals are distributed over fewer peaks. This might also be beneficial regarding CTC analysis, since the CTCs identified using CS have been run through several staining, washing and fixation procedures.
For analysis of all the mutations by sequencing one would have to perform independent PCRs for the 2 exons, followed by 4 bidirectional sequencing reactions. In the SNaPshot assay two multiplex PCRs for exons 9 and 20 are followed by one multiplex detection assay and one capillary electrophoresis run.
One notable limitation of this particular SNaPshot technology might be that the application is limited to a small number of known SNPs. In order to investigate PIK3CA mutations which concentrate within only few hot spots this is feasible, since up to 10 SNPs can be screened at once. As aforementioned the sensitivity of the SNaPshot methodology was reported to be approximately 5% (Hurst et al., 2009); an even higher detection rate (up to 0.01%) might be achieved by the recently published BEAMing technology (Beads, Emulsification, Amplification, and Magnetics; Higgins et al., 2012) which is also based on PCR amplification of hot spot regions. However, neither the SNaPshot nor the BEAMing technique allows for the discovery of unknown mutations.
5. Conclusion
Taken together, the herein described assay provides a simple and inexpensive tool to determine variants of key signaling proteins in single cells that could readily be extended to analyze SNPs in other therapeutically relevant genes such as PTEN, ER or EGFR. The SNaPshot assay can be performed in high throughput, is robust and objective making it suitable for use in such a diagnostic setting. Regarding PIK3CA analysis, it may be used to further characterize e.g. HER2‐positive CTCs which are resistant against anti‐HER2 targeted therapy using trastuzumab. As specific inhibitors for PIK3CA become available, rapid screening of patient samples for mutations will be essential and the assay may be used to select breast cancer patients benefiting from such a treatment.
Funding
This project was partly funded by Novartis Pharma GmbH, Nuernberg, Germany.
Supporting information
Supplementary data
Acknowledgments
We thank Cornelia Grimmel (Department of Dermatology, Eberhard‐Karls‐University of Tuebingen) for excellent assistance in flow cytometry and Ann‐Kathrin Hauser and Claudia Schulte (both Department of Neurodegenerative Diseases, Tuebingen) for their support in SNaPshot analysis. Further, we would like to acknowledge Carolyn D. Hurst (Leeds Institute of Molecular Medicine, St James's University Hospital, Leeds, UK) for critical reading of the manuscript.
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.07.007.
Schneck Helen, Blassl Christina, Meier-Stiegen Franziska, Neves Rui Pedro, Janni Wolfgang, Fehm Tanja, Neubauer Hans, (2013), Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients, Molecular Oncology, 7, doi: 10.1016/j.molonc.2013.07.007.
Contributor Information
Helen Schneck, Email: Helschn84@gmail.com.
Christina Blassl, Email: Christina.Blassl@gmail.com.
Franziska Meier-Stiegen, Email: Franziska.Meier-Stiegen@med.uni-duesseldorf.de.
Rui Pedro Neves, Email: ruipedroneves@gmail.com.
Wolfgang Janni, Email: Wolfgang.Janni@uniklinik-ulm.de.
Tanja Fehm, Email: Tanja.Fehm@med.uni-duesseldorf.de.
Hans Neubauer, Email: hans.neubauer@med.uni-duesseldorf.de.
References
- Araújo, W.M. , Vidal, F.C.B. , Souza, W. , Freitas Junior, J. , Souza, W. , Morgado-Diaz, J. , 2010. PI3K/Akt and GSK-3β prevents in a differential fashion the malignant phenotype of colorectal cancer cells. J. Cancer Res. Clin. Oncol.. 136, 1773–1782. [DOI] [PubMed] [Google Scholar]
- Bachman, K.E. , Argani, P. , Samuels, Y. , Silliman, N. , Ptak, J. , Szabo, S. , 2004. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol. Ther.. 3, 772–775. [DOI] [PubMed] [Google Scholar]
- Bader, A.G. , Kang, S. , Vogt, P.K. , 2006. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl. Acad. Sci. U S A. 3, 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns, K. , Horlings, H.M. , Hennessy, B.T. , Madiredjo, M. , Hijmans, E.M. , Beelen, K. , 2007. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell.. 12, 395–402. [DOI] [PubMed] [Google Scholar]
- Board, R.E. , Thelwell, N.J. , Ravetto, P.F. , Little, S. , Ranson, M. , Dive, C. , 2008. Multiplexed assays for detection of mutations in PIK3CA. Clin. Chem.. 54, 757–760. [DOI] [PubMed] [Google Scholar]
- Bozionellou, V. , Mavroudis, D. , Perraki, M. , Papadopoulos, S. , Apostolaki, S. , Stathopoulos, E. , 2004. Trastuzumab administration can effectively target chemotherapy-resistant Cytokeratin-19 messenger RNA-positive tumor cells in the peripheral blood and bone marrow of patients with breast cancer. Clin. Cancer Res.. 10, 8185–8194. [DOI] [PubMed] [Google Scholar]
- Cristofanilli, M. , Hayes, D.F. , Budd, G.T. , Ellis, M.J. , Stopeck, A. , Reuben, J.M. , 2005. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol.. 23, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. , Le, C. , Shukla, A. , Patterson, J. , Presnell, A. , Heinrich, M. , 2010. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res. Treat. 120, 409–418. [DOI] [PubMed] [Google Scholar]
- Eichhorn, P.J.A. , Gili, M. , Scaltriti, M. , Serra, V. , Guzman, M. , Nijkamp, W. , 2008. Phosphatidylinositol 3-Kinase Hyperactivation results in lapatinib resistance that is reversed by the mTOR/Phosphatidylinositol 3-Kinase inhibitor NVP-BEZ235. Cancer Res.. 68, 9221–9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehm, T. , Müller, V. , Aktas, B. , Janni, W. , Schneeweiss, A. , Stickeler, E. , 2010. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res. Treat.. 124, 403–412. [DOI] [PubMed] [Google Scholar]
- Higgins, M.J. , Jelovac, D. , Barnathan, E. , Blair, B. , Slater, S. , Powers, P. , 2012. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res.. 18, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollestelle, A. , Elstrodt, F. , Nagel, J.H.A. , Kallemeijn, W.W. , Schutte, M.l. , 2007. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol. Cancer Res.. 5, 195–201. [DOI] [PubMed] [Google Scholar]
- Hurst, C.D. , Zuiverloon, T.C. , Hafner, C. , Zwarthoff, E.C. , Knowles, M.A. , 2009. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res. Notes. 2, 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis, M. , Kallergi, G. , Ntoulia, M. , Perraki, M. , Apostolaki, S. , Kafousi, M. , 2008. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, Mammaglobin a, and HER2 in early breast cancer. Clin. Cancer Res.. 14, 2593–2600. [DOI] [PubMed] [Google Scholar]
- Ikenoue, T. , Kanai, F. , Hikiba, Y. , Obata, T. , Tanaka, Y. , Imamura, J. , 2005. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res.. 65, 4562–4567. [DOI] [PubMed] [Google Scholar]
- Isakoff, S.J. , Engelman, J.A. , Irie, H.Y. , Luo, J. , Brachmann, S.M. , Pearline, R.V. , 2005. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res.. 65, 10992–11000. [DOI] [PubMed] [Google Scholar]
- Jemal, A. , Siegel, R. , Xu, J. , Ward, E. , 2010. Cancer statistics 2010. CA Cancer J. Clin.. 60, 277–300. [DOI] [PubMed] [Google Scholar]
- Jemal, A. , Bray, F. , Center, M.M. , Ferlay, J. , Ward, E. , Forman, D. , 2011. Global cancer statistics. CA Cancer J. Clin.. 61, 69–90. [DOI] [PubMed] [Google Scholar]
- Jensen, J. , Laenkholm, A. , Knoop, A. , Ewertz, M. , Bandaru, R. , Liu, W. , 2011. PIK3CA mutations may Be discordant between primary and corresponding metastatic disease in breast cancer. Clin. Cancer Res.. 17, 667–677. [DOI] [PubMed] [Google Scholar]
- Kang, S. , Bader, A.G. , Vogt, P.K. , 2005. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. U S A. 102, 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, Y. , Mukohara, T. , Shimada, H. , Saijo, N. , Hirai, M. , Minami, H. , 2010. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann. Oncol.. 21, 255–262. [DOI] [PubMed] [Google Scholar]
- Korkaya, H. , Wicha, M.S. , 2009. HER-2, Notch, and breast cancer stem cells: targeting an Axis of Evil. Clin. Cancer Res.. 15, 1845–1847. [DOI] [PubMed] [Google Scholar]
- Lai, Y. , Mau, B. , Cheng, W. , Chen, H. , Chiu, H. , Tzen, C. , 2008. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann. Surg. Oncol.. 15, 1064–1069. [DOI] [PubMed] [Google Scholar]
- Meng, S. , Tripathy, D. , Shete, S. , Ashfaq, R. , Haley, B. , Perkins, S. , 2004. HER-2 gene amplification can be acquired as breast cancer progresses. Proc. Natl. Acad. Sci. U S A. 101, 9393–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, V. , Pantel, K. , 2009. HER2 as marker for the detection of circulating tumor cells. Breast Cancer Res. Treat. 117, 535–537. [DOI] [PubMed] [Google Scholar]
- Pantel, K. , Alix-Panabieres, C. , Riethdorf, S. , 2009. Cancer micrometastases. Nat. Rev. Clin. Oncol.. 6, 339–351. [DOI] [PubMed] [Google Scholar]
- Paradiso, A. , Mangia, A. , Azzariti, A. , Tommasi, S. , 2007. Phosphatidylinositol 3-Kinase in breast cancer: where from here?. Clin. Cancer Res.. 13, 5988–5990. [DOI] [PubMed] [Google Scholar]
- Powell, A.A. , Talasaz, A.H. , Zhang, H. , Coram, M.A. , Reddy, A. , Deng, G. , May 7, 2012. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. [Epub ahead of print] PLoS One. 10.1371/journal.pone.0033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnoose, E.A. , Atwal, S.K. , Spoerke, J.M. , Savage, H. , Pandita, A. , Yeh, R. , 2010. Molecular biomarker analyses using circulating tumor cells. [Epub ahead of print] PLoS One. 10.1371/journal.pone.0012517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, W. , Tong, G.X. , Manolidis, S. , Close, L.G. , Assaad, A.M. , Su, G.H. , 2008. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. Int. J. Cancer. 122, 1189–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf, S. , Müller, V. , Zhang, L. , Rau, T. , Loibl, S. , Komor, M. , 2010. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the Neoadjuvant GeparQuattro trial. Clin. Cancer Res.. 16, 2634–2645. [DOI] [PubMed] [Google Scholar]
- Samuels, Y. , Diaz, L.A. , Schmidt-Kittler, O. , Cummins, J.M. , DeLong, L. , Cheong, I. , 2005. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell.. 7, 561–573. [DOI] [PubMed] [Google Scholar]
- Scher, H.I. , Jia, X. , Bono, J.S. , de, Fleisher, M. , Pienta, K.J. , Raghavan, D. , Heller, G. , 2009. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol.. 10, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra, V. , Markman, B. , Scaltriti, M. , Eichhorn, P.J.A. , Valero, V. , Guzman, M. , 2008. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res.. 68, 8022–8030. [DOI] [PubMed] [Google Scholar]
- Sieuwerts, A. , Kraan, J. , Bolt-de Vries, J. , Spoel, P. , Mostert, B. , Martens, J.M. , 2009. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res. Treat. 118, 455–468. [DOI] [PubMed] [Google Scholar]
- Simi, L. , Pratesi, N. , Vignoli, M. , Sestini, R. , Cianchi, F. , Valanzano, R. , 2008. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am. J. Clin. Pathol.. 130, 247–253. [DOI] [PubMed] [Google Scholar]
- Stemke-Hale, K. , Gonzalez-Angulo, A.M. , Lluch, A. , Neve, R.M. , Kuo, W. , Davies, M. , 2008. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res.. 68, 6084–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati, A. , Markou, A. , Parisi, C. , Politaki, E. , Mavroudis, D. , Georgoulias, V. , Lianidou, E. , 2011. Gene expression profile of circulating tumor cells in breast cancer by RT-qPCR. BMC Cancer. 11, 422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt, B. , Warne, P.H. , Downward, J. , 2011. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 30, 3222–3233. [DOI] [PubMed] [Google Scholar]
- Xenidis, N. , Perraki, M. , Kafousi, M. , Apostolaki, S. , Bolonaki, I. , Stathopoulou, A. , 2006. Predictive and prognostic value of peripheral blood Cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in Node-negative breast cancer patients. J. Clin. Oncol.. 24, 3756–3762. [DOI] [PubMed] [Google Scholar]
- Zhao, J.J. , Liu, Z. , Wang, L. , Shin, E. , Loda, M.F. , Roberts, T.M. , 2005. The oncogenic properties of mutant p110α and p110β phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc. Natl. Acad. Sci. U S A. 102, 18443–18448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


