Abstract
Ductal carcinoma in situ (DCIS) is an intraductal neoplastic proliferation of epithelial cells that is separated from the breast stroma by an intact layer of basement membrane and myoepithelial cells. DCIS is a non‐obligate precursor of invasive breast cancer, and up to 40% of these lesions progress to invasive disease if untreated. Currently, it is not possible to predict accurately which DCIS would be more likely to progress to invasive breast cancer as neither the significant drivers of the invasive transition have been identified, nor has the clinical utility of tests predicting the likelihood of progression been demonstrated. Although molecular studies have shown that qualitatively, synchronous DCIS and invasive breast cancers are remarkably similar, there is burgeoning evidence to demonstrate that intra‐tumor genetic heterogeneity is observed in a subset of DCIS, and that the process of progression to invasive disease may constitute an ‘evolutionary bottleneck’, resulting in the selection of subsets of tumor cells with specific genetic and/or epigenetic aberrations. Here we review the clinical challenge posed by DCIS, the contribution of the microenvironment and genetic aberrations to the progression from in situ to invasive breast cancer, the emerging evidence of the impact of intra‐tumor genetic heterogeneity on this process, and strategies to combat this heterogeneity.
Keywords: Breast cancer, Intraductal, Intra-tumor genetic heterogeneity, Darwinian evolution, Genomics
Highlights
Ductal carcinoma in situ (DCIS) is a non‐obligate precursor of breast cancer.
Validated predictors of DCIS‐invasive breast cancer (IBC) progression are yet to be developed.
At least some DCIS are composed of mosaics of cancer cells with distinct genetic aberrations.
DCIS and synchronous IBC may harbor distinct genetic aberrations.
Progression from DCIS to IBC may follow a Darwinian evolutionary model.
1. Introduction
Ductal carcinoma in situ (DCIS) is defined as a premalignant proliferation of neoplastic epithelial cells contained within the lumen of mammary ducts. DCIS are lined by a layer of semi‐continuous myoepithelial cells and surrounded by an intact basement membrane (Lopez‐Garcia et al., 2010). For several decades it has been accepted that DCIS constitutes a non‐obligate precursor of invasive ductal carcinoma. In 1973, Wellings and Jensen proposed that non‐malignant ductal lesions are precursors of invasive breast cancer (IBC) based on the evidence of gradual histological continuity observed in normal and abnormal breast tissues (Wellings and Jensen, 1973). This theory is supported by a plethora of succeeding work aiming to characterize DCIS and IBC at a molecular level, which have revealed their genetic similarity and likely common origin (Buerger et al., 1999, 1999, 2010, 1998).
Clinical observational studies have further corroborated the hypothesis that DCIS is a precursor of IBC. Perhaps the most persuasive evidence to suggest that DCIS and IBC are progressive stages of an evolutionary continuum is that they affect the same anatomical site. Longitudinal studies of patients with DCIS managed with biopsy only revealed that 20–50% of this population later developed IBC in the same quadrant of the same breast as the original DCIS (Page et al., 1995, 1982, 2005). DCIS is found adjacent to invasive disease in the vast majority of IBCs at the time of diagnosis (Evans et al., 1997; Fisher et al., 1975), where it was thought to be the precursor lesion, however the coexistence of DCIS with IBC varies according to the subtype of breast cancer (Abdel‐Fatah et al., 2007). DCIS can be classified into similar molecular subtypes as IBC based primarily on the expression patterns of ER, PR, HER2, EGFR and cytokeratin 5/6 (Bryan et al., 2006; Clark et al., 2011; Livasy et al., 2007; Muggerud et al., 2010), and associated in situ and invasive components often, but not always (see below), exhibit a similar immunophenotype (Steinman et al., 2007; Tamimi et al., 2008). Also, nuclear grade is generally concordant between in situ and invasive components of invasive carcinomas, which have comparable nuclear morphology (Giardina et al., 2003) and DNA ploidy (Ottesen, 2003).
DCIS has become a formidable clinical challenge due to its increasing incidence. In fact, 54,944 diagnoses of DCIS are expected in 2013 according to the American Cancer Society, up from 45,900 cases in 2010, and now DCIS accounts for approximately 20% of all breast cancers (American Cancer Society, 2013). This rapid increase in the incidence of DCIS parallels the introduction of mammography screening, as the majority of DCIS lesions are detected upon stereostatic biopsy of mammographic microcalcifications (Virnig et al., 2010). Although there have been numerous efforts to develop clinical or molecular tests (Solin et al., 2013) to predict which patients are likely to develop invasive disease following a diagnosis of DCIS, there is currently no test with demonstrated clinical utility to identify this population. Hence, the vast majority of patients are still subjected to surgical treatment followed by radiation and/or prophylactic systemic therapies (e.g. tamoxifen). Further, despite extensive efforts to unravel the biological underpinnings of the phenomenon of progression from in situ to invasive disease and to develop biomarkers predictive of the likelihood of progression to IBC, several biological aspects of the transition from in situ to invasive disease have yet to be elucidated and there are still no histopathological or molecular markers to predict accurately the progression from DCIS to IBC.
Here we discuss the biological processes that are likely to play a role in the progression from in situ to invasive disease, the challenges posed by intra‐tumor genetic heterogeneity and potential strategies to develop biomarkers to define the subsets of DCIS that are likely to progress to IBC.
2. The role of the microenvironment in progression from DCIS to IBC
The theories of progression from DCIS to IBC fall broadly into two categories: those, which consider invasiveness as an acquired behavior that stems from genetic aberrations occurring in the neoplastic cells, and those, which suggest it is independent of additional genetic changes within the lesion. The most popular ‘non‐genetic’ theory is that the microenvironment or tumor stroma actively drives tumor progression. Several studies have examined the pro‐invasive influence of the extracellular matrix (ECM) and non‐tumoral cells on DCIS. For example, Lyons and colleagues (Lyons et al., 2011) suggested that mammary gland involution post‐pregnancy is a driving force of tumor progression as a result of re‐modeling of the ECM. Using a murine model of postpartum involution, the authors observed that MCF10ADCIS.com cells subjected to the involution microenvironment formed large tumors with increased intra‐tumor collagen deposition and expression of cyclooxygenase‐2 (COX‐2). In this model, both fibrillar collagen and COX‐2 were required for the acquisition of an invasive phenotype, which could be at least in part blocked by non‐steroidal anti‐inflammatory drugs (Lyons et al., 2011). Hu and colleagues also found that stromal fibroblasts increased invasion in a xenograft model of DCIS in part by increasing expression of COX‐2 in tumor epithelial cells (Hu et al., 2009). Other studies have linked elevated stromal cell expression of Lysyl oxidases (LOXs), a family of ECM modifying enzymes, to invasion and metastasis (Barker et al., 2011; Levental et al., 2009).
Gene expression profiling has revealed that substantial changes occur during progression from DCIS to IBC in various cell types of the tumor microenvironment, including fibroblasts, myoepithelial cells and leukocytes (Allinen et al., 2004; Ma et al., 2009; Vargas et al., 2012). The underlying causes of these differences in gene expression are still unclear. As expected, no clonal genetic aberrations were detected in the myoepithelial cells and fibroblasts surrounding DCIS or IBC (Allinen et al., 2004; Qiu et al., 2008). Consequently, it has been suggested that epigenetic alterations in the stroma may be involved in the progression from in situ to IBC (Hu et al., 2005).
Histologically, the major difference between DCIS and IBC is that the former retain an outer layer of myoepithelial cells and an intact basement membrane. A commonly touted theory of progression is that the normal myoepithelium acts as a ‘gatekeeper’, exerting tumor suppressive effects on the in situ lesion, and that it is the loss of this suppressive environment that unleashes the progression to invasive disease (Barsky and Karlin, 2005; Gudjonsson et al., 2005; Place et al., 2011; Polyak and Hu, 2005). As well as forming a physical barrier to invasion, myoepithelial cells also secrete various ECM components and protease inhibitors, such as Maspin, proposed to inhibit the invasive capacity of DCIS in a paracrine manner (Barsky and Karlin, 2005, 2006, 1997). Hu and colleagues provided support for this theory upon finding that co‐transplantation of normal myoepithelial cells prevented occurrence of invasive disease in a xenograft model of human DCIS, whereas fibroblasts enhanced tumor growth and invasion (Hu et al., 2008). Such mechanisms of progression may partially explain the lack of differences detected between DCIS and IBC at the genomic level.
Epigenetic alterations, namely inheritable changes in gene expression that do not involve changes in DNA sequences, within neoplastic cells could also account for the current lack of significant differences detected between DCIS and IBC at DNA sequence level. DNA methylation commonly increases from normal breast epithelium to DCIS, however the majority of studies have found similar levels of promoter hypermethylation in DCIS and IBC (Moelans et al., 2011b; Park et al., 2011; Verschuur‐Maes et al., 2012). This suggests that alterations in DNA methylation are early events in breast carcinogenesis rather than key factors in the transition to invasive disease. Consistent with this observation, an increase in the number of methylated genes has been documented in columnar cell lesions, the earliest morphologically identifiable precursor of breast cancer, as compared to normal breast epithelium, however no differences between the number of methylated genes between columnar cell lesions, DCIS and IBC were observed (Verschuur‐Maes et al., 2012). Given the recent data from the Encyclopedia of DNA Elements (ENCODE)(Encode Project Consortium, 2012) suggesting that methylation is likely to be a consequence rather than a cause of gene silencing (Sproul et al., 2011), alternative epigenetic events may be even more important in the progression from in situ to IBC than methylation. In fact, changes in chromatin states could play a role in progression as global changes in histone modifications that mark heterochromatin and euchromatin have been implicated in epithelial‐to‐mesenchymal transition (EMT)(McDonald et al., 2011), which is reported to be associated with progression to IBC from DCIS (Knudsen et al., 2012). Therefore, studies investigating the role of epigenetic alterations in the progression from in situ to IBC are warranted.
3. Genetic alterations in the progression to invasive disease
Numerous lines of evidence demonstrate that DCIS is a non‐obligate precursor of IBC and that DCIS harbors genetic aberrations similar to those found in synchronous and metachronous IBC developing in the same quadrant. Importantly, however, robust transcriptomic or genomic signatures to distinguish DCIS from IBC have proven elusive. Several gene expression profiling analyses of premalignant, preinvasive and IBCs to determine whether gene expression patterns could be used for diagnostic or prognostic purposes have been performed. These studies have shown that at the transcriptomic level, preinvasive lesions and invasive breast cancer of the same histological grade display remarkably similar gene expression patterns and that it is not possible to identify gene signatures that discern between the pathological stages of DCIS and IBC robustly (Ma et al., 2003; Vincent‐Salomon et al., 2008). Overall, analyses of chromosomal aberrations by array‐based comparative genomic hybridization (aCGH) have been no more fruitful in clearly discriminating DCIS from IBC (Gao et al., 2009; Liao et al., 2012; Yao et al., 2006). DCIS and invasive components from the same patients are frequently found to be closely related not only on the basis of their gene expression but also gene copy number aberrations (Johnson et al., 2012; Lee et al., 2012; Liao et al., 2012; Moelans et al., 2011a; Porter et al., 2003). Although these findings lend further support to the notion that DCIS is a precursor of IBC, they have also been interpreted as evidence to suggest that progression from in situ to invasive disease is not necessarily driven by specific genetic aberrations in DCIS cells.
Although DCIS and IBC appear genetically similar, some qualitative differences have been found between matched DCIS and IBC (Table 1). Studies on the prevalence of HER2 overexpression and HER2 gene amplification provide examples where clear genomic differences are observed between adjacent invasive tumors and in situ lesions or within in situ lesions (Figure 1). In a small percentage of HER2‐positive tumors amplification of the HER2 locus was found to be present in the DCIS but not the associated IBC (Burkhardt et al., 2010; Latta et al., 2002; Park et al., 2006), and some cases of DCIS showed overt heterogeneity of HER2 overexpression within the DCIS component (Figure 1). These data indicate that either HER2 amplification may have been lost during progression to invasive disease or that the invasive component arose from a clone in the DCIS that did not harbor HER2 amplification in the first place. Recent studies have found evidence for convergent phenotypic evolution in tumor progression and metastasis (Gerlinger et al., 2012), and it is plausible that progression from DCIS to IBC also constitutes a convergent phenotype (i.e. it may be caused by numerous genetic and/or epigenetic mechanisms, Figure 2A). In fact, the hypothesis that progression from in situ to IBC constitutes a convergent phenotype provides a potential explanation for the negative results in the genomic and transcriptomic comparisons between DCIS and IBC.
Table Table 1.
Common gene amplifications found to be distinct between DCIS and synchronous adjacent IBC.
| Gene | Study | Number of paired DCIS and IBC cases | % cases restricted to DCIS | % cases restricted to IBC | % cases present in both components | % of cases absent in both components |
|---|---|---|---|---|---|---|
| MYC | Jang et al. (2012) | 203 | 1 | 3 | 8.9 | 87.1 |
| Robanus‐Maandag et al. (2003) | 12 | 0 | 1 | NA | NA | |
| 14 | 0 | 5 | 4 | 5 | ||
| Burkhardt et al. (2010) | 135 (71 analyzable) | 5.6 | 0 | 5.6 | 88.7 | |
| HER2 | Jang et al. (2012) | 203 | 1 | 0 | 22.8 | 76.2 |
| Burkhardt et al. (2010) | 135 (74 analyzable) | 5.4 | 0 | 16.2 | 78.4 | |
| Park et al. (2006) | 270 | 1.5 | 0 | 28.5 | 70 | |
| FGFR1 | Jang et al. (2012) | 203 | 0 | 3.1 | 10.3 | 86.6 |
| CCND1 | Jang et al. (2012) | 203 | 1.5 | 0.5 | 15.9 | 82.1 |
| Burkhardt et al. (2010) | 135 (73 analyzable) | 0 | 2.7 | 11.0 | 86.3 |
Latta et al. (Latta et al., 2002) reported complete concordance in HER2 amplification between the in situ and invasive components in all but one case analyzed by fluorescence in situ hybridization (FISH); in this case HER2 amplification was found to be present in the in situ but absent in the invasive component (total number of paired DCIS and IBC cases analyzed by FISH not available). DCIS, ductal carcinoma in situ; IBC, invasive breast cancer; NA, not available.
Figure 1.
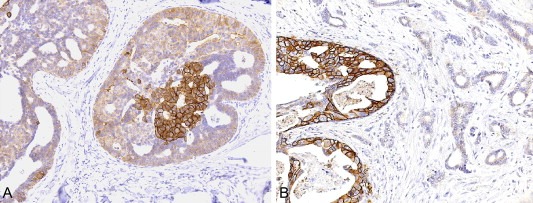
Heterogeneous expression of HER2 in breast cancer. (A) Heterogeneous expression of HER2 in neoplastic cells of a ductal carcinoma in situ; note that only a subpopulation of the cancer cells in one duct display strong, membranous expression of HER2. (B) Representative micrograph illustrating a ductal carcinoma in situ composed of cells displaying HER2 membranous expression associated with a HER2‐negative invasive ductal carcinoma.
Figure 2.
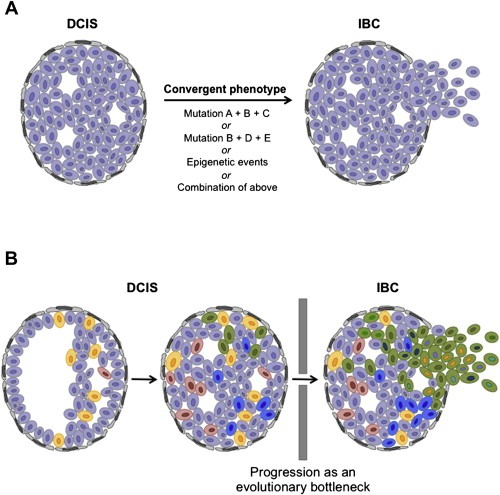
Hypothetical models of progression from in situ to invasive breast cancer. (A) Progression from DCIS to IBC as a convergent phenotype, where several combinations of somatic genetic and/or epigenetic aberrations result in the acquisition of the biological properties required for cancer cells to progress from in situ to invasive disease (i.e. the genetic/epigenetic aberrations selected for are distinct between patients but all result in the progression to invasive disease). (B) Progression from DCIS to IBC as an evolutionary bottleneck. As DCIS develops, cells accumulate somatic mutations and copy number aberrations (depicted by color) to generate a heterogeneous lesion with distinct subclones harboring private mutations in addition to the founder genetic aberrations present in all neoplastic cells. Only subclones harboring a specific repertoire of genetic aberrations are selected and pass through the evolutionary bottleneck of progression to IBC. DCIS: ductal carcinoma in situ; IBC: invasive breast cancer.
The majority of studies comparing DCIS and IBC have not focused solely on synchronously occurring ipsilateral samples (Liao et al., 2012; Robanus‐Maandag et al., 2003). Such studies have suggested that there might be copy number aberrations associated with progression from DCIS to IBC, including MYC (Robanus‐Maandag et al., 2003), FGFR1 (Jang et al., 2012), and CCND1 (Burkhardt et al., 2010) gene amplification, which were shown to be more frequently observed in IBC than in DCIS (Table 1). It should be noted, however, that these genetic aberrations do not appear to be either necessary or sufficient for the acquisition of an invasive phenotype (Burkhardt et al., 2010; Jang et al., 2012; Liao et al., 2012; Park et al., 2006; Robanus‐Maandag et al., 2003). In fact, in some cases of paired DCIS and IBCs, when amplifications of the MYC and CCND1 loci have been found, they were reported to be restricted to the DCIS component, to the invasive component or to be concurrently present in both (Burkhardt et al., 2010; Jang et al., 2012; Robanus‐Maandag et al., 2003) (Table 1). The approaches employed so far, looking for common transcriptomic and/or genomic differences between matched synchronous DCIS and IBC, would fail to identify significant differences if progression to invasive disease is a convergent phenotype without a given mechanism to account for the majority of cases. Consistent with this hypothesis, recent studies based on pairwise comparisons between DCIS and IBC have revealed the existence of significant genetic differences, however these differences are distinct from patient to patient (Hernandez et al., 2012; Heselmeyer‐Haddad et al., 2012). One could posit that drivers of progression may be masked when DCIS and IBC are compared as groups and not matched entities if distinct drivers of progression are specific to individual cases or a small number of patients (Hernandez et al., 2012). It is also plausible that these distinct genetic and/or epigenetic aberrations may target similar or complementary pathways that would result in similar biological outputs (i.e. progression from in situ to invasive disease)(Yap et al., 2012). In addition, limitations in the types of the genetic aberrations detected by the techniques used (e.g. somatic point mutations and small insertions and deletions, somatic rearrangements and copy number aberrations) may have resulted in the apparent lack of genetic differences between DCIS and synchronous adjacent IDC.
4. Genetic heterogeneity and clonal evolution in DCIS and IBC
Recent in‐depth genetic analyses of synchronous ipsilateral DCIS and IBC have attempted to determine both intra‐tumor genetic heterogeneity in both components and genetic differences acquired during progression to IBC (Hernandez et al., 2012; Heselmeyer‐Haddad et al., 2012). Remarkably, both studies found not only extensive genetic heterogeneity, in the form of the percentage of neoplastic cells harboring amplification of specific loci in DCIS, but also presented some evidence supporting the idea of clonal selection during the progression toward invasive disease, suggesting that transition from DCIS to IBC may constitute an evolutionary bottleneck (Turner and Reis‐Filho, 2012; Yap et al., 2012) (Figure 2B).
Our group performed aCGH, fluorescence in situ hybridization (FISH) and Sequenom analysis on 13 synchronous adjacent DCIS and IBC sets and found that matched DCIS and IBC had strikingly similar genomic profiles (Hernandez et al., 2012), in agreement with the results of previous studies. Importantly, however, not only intra‐lesion genetic heterogeneity but also differences in the prevalence of specific amplifications and mutations between the DCIS and invasive samples were detected, consistent with a model where DCIS is composed of a mosaic of tumor cells that in addition to the founder genetic aberrations harbor private mutations, and that clonal selection does take place in the progression from in situ to invasive disease. For example, three of 13 microdissected cases harbored PIK3CA mutations which were restricted to the DCIS component in two cases, and in a third case the frequency of the PIK3CA mutant allele decreased from 49% to 25% in the IBC component (Hernandez et al., 2012).
Heselmeyer‐Haddad and colleagues used FISH to probe eight genomic loci frequently lost or gained in breast cancer in single cells microdissected from matched DCIS and IBC samples (Heselmeyer‐Haddad et al., 2012). Through the analysis of patterns of gains and losses in single cells it was also concluded that DCIS exhibit a high level of intra‐tumor genetic heterogeneity and that matched DCIS and IBC have similar but not identical patterns of genomic imbalances. Differences between matched pairs of DCIS and IBC were detected; in four cases a switch from DCIS to IBC in the modal clone was detected and associated with a gain of MYC in more than 50% of the cells they analyzed. This suggests that specific genetic aberrations are selected for during progression to IBC, but that these changes may vary from patient to patient, consistent with the hypothesis that progression from in situ to IBC may constitute a convergent phenotype.
These studies highlight the importance of i) comparing synchronous ipsilateral samples of DCIS and IBC, as the genetic differences observed between pairs of synchronous DCIS and IBC would probably not have been detected in unmatched samples, and ii) using methods that provide quantitative information on genotype and copy number status, as most differences would not have been detected if single cells had not been analyzed or if semi‐quantitative methods had not been employed. Taken together, these observations provide a tantalizing glimpse into the progression of DCIS to IBC, and support the theory that this process is a result of selection of subpopulations of neoplastic cells. Although previous work support the idea that intra‐tumor heterogeneity exists in DCIS (Aubele et al., 2000, 1999, 1996, 2008), these recent studies provide strong circumstantial evidence to suggest that progression from DCIS to IBC may occur, at least in some samples, by Darwinian selection.
5. Design of future studies of the progression of DCIS to IBC
To obtain conclusive answers as to whether progression from DCIS to IBC is a result of intra‐lesional genetic heterogeneity and clonal selection, it will be necessary to enter the rapidly expanding field of massively parallel sequencing. Over the past few years the cost and speed of DNA sequencing has plummeted facilitating an increase in the depth of sequencing (Mwenifumbo and Marra, 2013). The unprecedented level of genetic information provided by massively parallel sequencing has allowed for a progressively more detailed picture of inter‐ and intra‐tumor genetic heterogeneity and the process of tumor evolution. On an intra‐individual scale studies have found the existence of genetic differences between primary breast cancers and metastases that support the hypothesis of clonal evolution (Ding et al., 2010; Shah et al., 2009). Shared genomic aberrations exist between primary breast tumors and their metastatic deposits suggesting common ancestry, however the prevalence of specific mutations in the cells within each lesion have been shown to differ. This indicates that in the tumors studied, a subpopulation of cells from the primary tumor gave rise to the metastasis. In addition, aberrations have been found that were specific to the metastasis potentially indicating continued evolution (Ding et al., 2010; Shah et al., 2009). On an intra‐tumor level multi‐region sequencing of tumors has confirmed the existence of intra‐tumor genetic heterogeneity (Gerlinger et al., 2012, 2011, 2010), which is a pre‐requisite of clonal evolution.
Increased depth of sequencing provides the ability to detect point mutations and copy number alterations present in minute fractions of a tumor population (the exact fraction depends on numerous factors, including the number of sequencing reads, cellularity, allelic frequency, tumor ploidy, and patterns of gene copy number aberrations). This information when integrated by computational algorithms can be used to infer the prevalence of subclonal populations within a tumor, or between tumor sites to address cancer genome evolution (Nik‐Zainal et al., 2012; Shah et al., 2012). These methodologies have been used to elucidate the subclonal populations present in breast cancers and the likely order of occurrence of mutations in their evolution, dramatically increasing our knowledge of the mutational landscape of these cancers (Nik‐Zainal et al., 2012; Shah et al., 2012). Bioinformatic tools to analyze massively parallel sequencing data are continuously improving to allow more detailed analysis from sequencing from bulk tumor populations, with more sensitive methods to identify somatic point mutations (Cibulskis et al., 2013), and to determine tumor purity and cell ploidy (Carter et al., 2012). These methods were recently employed to elucidate the frequency of subclonal driver mutations present in chronic lymphocytic leukemia, and their effect on clinical outcome from whole exome sequencing data (Landau et al., 2013). These analyses have not only revealed that the presence of subclonal driver mutations is associated with poorer outcome, but also that chemotherapy acted as an evolutionary bottleneck, leading to selection and ulterior expansion of specific driving subclones (Landau et al., 2013). Based on the assumption that DCIS is the precursor lesion of IBC, one could exact a similar approach with matched DCIS and IBC, treating them as longitudinal time points. High‐depth whole genome or whole exome sequencing complemented by independent analyses of copy number and allelic frequencies would comprehensively address the level of genetic heterogeneity of DCIS and IBC and whether progression to invasive disease truly constitutes an evolutionary bottleneck.
Such analysis on bulk tumor material could be enhanced by complementary evaluations at the ultimate resolution of genetic heterogeneity, the single cell. To date, one can potentially determine the presence of genomic aberrations in subclonal populations of a tumor by analyzing copy number variations at a single cell level (Navin et al., 2011). These methods, combining subclonal analysis by copy number with the lineage analysis based on mutation frequency from deep sequencing of bulk tissue, have the potential to conclusively answer whether progression from in situ to IBC is results from the selection of a subpopulation of DCIS cells harboring a specific genotype (Figure 3).
Figure 3.
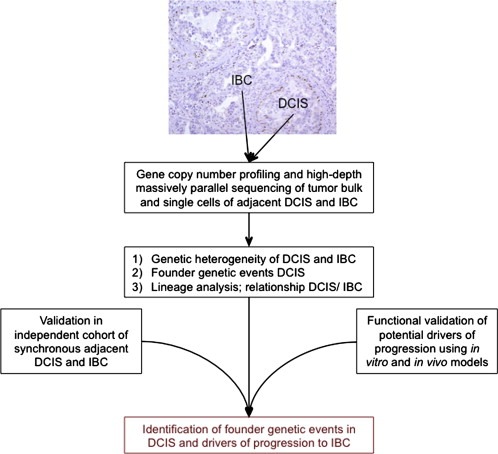
Schematic of a potential approach for the identification of founder genetic events in DCIS and genetic aberrations that drive progression from DCIS to IBC. DCIS: ductal carcinoma in situ; IBC: invasive breast cancer.
Following on from DNA sequencing, functional annotation of these findings could then be used to determine whether progression to IBC constitutes a convergent phenotype and whether specific signaling pathways or networks are involved. Additionally, in vitro and in vivo models of DCIS would be necessary to confirm the roles of potential drivers of progression, and to determine whether drivers may provide novel therapeutic targets or prognostic markers. Although such in vitro and in vivo models that accurately represent the biology of DCIS are scarce, a handful of cell lines and xenograft models have been used with some success to recapitulate a DCIS‐like phenotype. For example, the ER‐negative/HER2‐negative MCF10DCIS.com cell line (Miller et al., 2000) recapitulates DCIS‐like structures when grown in three‐dimensional cell culture models. A model for ER‐negative/HER2‐negative DCIS also exists in the HMT‐3522 series (Rizki et al., 2008). Models for ER‐positive in situ disease are more limited, however representatives of ER‐positive/HER2‐positive DCIS and HER2‐positive DCIS can be found in the 21T progression series and SUM225 cell line, respectively (Band et al., 1990; Behbod et al., 2009; Forozan et al., 1999; Souter et al., 2010). These cell line models of DCIS may be used in in vitro assays probing three‐dimensional mammary acinar structure, proliferation, apoptosis and invasion (Debnath et al., 2003; Imbalzano et al., 2009; Lee et al., 2012; Rizki et al., 2008) to annotate functionally potential drivers of progression from DCIS to IBC. In addition, DCIS cell lines have been used in xenograft studies to model progression from DCIS to IBC in vivo (Hu et al., 2009; Souter et al., 2010; Lyons et al., 2011), and recently Medina and colleagues (Behbod et al., 2009) developed a ‘mammary intraductal’ xenograft model, which involves injection of neoplastic cells directly into the mammary ducts via a cleaved nipple, rather than subcutaneous injection to more accurately recapitulate the histology of DCIS. This model, however, still faces the limitations posed by the use of xenografts, including the lack of a competent mouse immune system in the models analyzed and the fact that the tumor–microenvironment interactions between human DCIS cells and mouse myoepithelial and stromal cells may be distinct from those of human DCIS cells, myoepithelial cells and stromal cells, which may be overcome in part by the use of humanized stroma (Kuperwasser et al., 2004). Importantly, however, studies aiming to develop additional in vitro and in vivo models of DCIS and of the progression from in situ to invasive disease, which closely recapitulate the cardinal features of these lesions, are warranted.
6. Conclusion
The discovery of clinically useful biomarkers to differentiate women with DCIS at high or low risk of developing IBC has so far been confounded by the apparent lack of genomic and transcriptomic differences between the two. Recent studies based on medium‐throughput sequencing and fluorescence in situ hybridization hint at genomic differences and clonal evolution in progression to IBC. In fact, there is evidence to suggest that cancers may follow a Darwinian evolution and that several biological phenomena (e.g. resistance to specific targeted therapies, the process of metastatic dissemination, and progression from in situ to invasive disease) may constitute evolutionary bottlenecks. Consequently, the employment of novel technologies, including high depth massively parallel sequencing and single cell analyses, is an important next step to discern the contribution of genomic alterations and Darwinian evolution to the transition from DCIS to IBC.
Conflict of interest
The authors have no conflicts of interest to declare.
Cowell Catherine F., Weigelt Britta, Sakr Rita A., Ng Charlotte K.Y., Hicks James, King Tari A., Reis-Filho Jorge S., (2013), Progression from ductal carcinoma in situ to invasive breast cancer: Revisited, Molecular Oncology, 7, doi: 10.1016/j.molonc.2013.07.005.
Contributor Information
Britta Weigelt, Email: weigeltb@mskcc.org.
Jorge S. Reis-Filho, Email: reisfilj@mskcc.org
References
- Abdel-Fatah, T.M. , Powe, D.G. , Hodi, Z. , Lee, A.H. , Reis-Filho, J.S. , Ellis, I.O. , 2007. High frequency of coexistence of columnar cell lesions, lobular neoplasia, and low grade ductal carcinoma in situ with invasive tubular carcinoma and invasive lobular carcinoma. The American Journal of Surgical Pathology. 31, 417–426. [DOI] [PubMed] [Google Scholar]
- Allinen, M. , Beroukhim, R. , Cai, L. , Brennan, C. , Lahti-Domenici, J. , Huang, H. , Porter, D. , Hu, M. , Chin, L. , Richardson, A. , Schnitt, S. , Sellers, W.R. , Polyak, K. , 2004. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 6, 17–32. [DOI] [PubMed] [Google Scholar]
- American Cancer Society, 2013. Cancer Facts and Figures 2013 American Cancer Society; Atlanta, GA: [Google Scholar]
- Aubele, M. , Cummings, M. , Walsch, A. , Zitzelsberger, H. , Nahrig, J. , Hofler, H. , Werner, M. , 2000. Heterogeneous chromosomal aberrations in intraductal breast lesions adjacent to invasive carcinoma. Analytical Cellular Pathology. 20, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubele, M. , Mattis, A. , Zitzelsberger, H. , Walch, A. , Kremer, M. , Hutzler, P. , Hofler, H. , Werner, M. , 1999. Intratumoral heterogeneity in breast carcinoma revealed by laser-microdissection and comparative genomic hybridization. Cancer Genetics and Cytogenetics. 110, 94–102. [DOI] [PubMed] [Google Scholar]
- Band, V. , Zajchowski, D. , Swisshelm, K. , Trask, D. , Kulesa, V. , Cohen, C. , Connolly, J. , Sager, R. , 1990. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Research. 50, 7351–7357. [PubMed] [Google Scholar]
- Barker, H.E. , Chang, J. , Cox, T.R. , Lang, G. , Bird, D. , Nicolau, M. , Evans, H.R. , Gartland, A. , Erler, J.T. , 2011. LOXL2-mediated matrix remodeling in metastasis and mammary gland involution. Cancer Research. 71, 1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsky, S.H. , Karlin, N.J. , 2005. Myoepithelial cells: autocrine and paracrine suppressors of breast cancer progression. Journal of Mammary Gland Biology and Neoplasia. 10, 249–260. [DOI] [PubMed] [Google Scholar]
- Barsky, S.H. , Karlin, N.J. , 2006. Mechanisms of disease: breast tumor pathogenesis and the role of the myoepithelial cell. Nature Clinical Practice Oncology. 3, 138–151. [DOI] [PubMed] [Google Scholar]
- Behbod, F. , Kittrell, F.S. , LaMarca, H. , Edwards, D. , Kerbawy, S. , Heestand, J.C. , Young, E. , Mukhopadhyay, P. , Yeh, H.W. , Allred, D.C. , Hu, M. , Polyak, K. , Rosen, J.M. , Medina, D. , 2009. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Research: BCR. 11, R66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, B.B. , Schnitt, S.J. , Collins, L.C. , 2006. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Modern Pathology. 19, 617–621. [DOI] [PubMed] [Google Scholar]
- Buerger, H. , Otterbach, F. , Simon, R. , Poremba, C. , Diallo, R. , Decker, T. , Riethdorf, L. , Brinkschmidt, C. , Dockhorn-Dworniczak, B. , Boecker, W. , 1999. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. The Journal of Pathology. 187, 396–402. [DOI] [PubMed] [Google Scholar]
- Buerger, H. , Otterbach, F. , Simon, R. , Schafer, K.L. , Poremba, C. , Diallo, R. , Brinkschmidt, C. , Dockhorn-Dworniczak, B. , Boecker, W. , 1999. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. The Journal of Pathology. 189, 521–526. [DOI] [PubMed] [Google Scholar]
- Burkhardt, L. , Grob, T.J. , Hermann, I. , Burandt, E. , Choschzick, M. , Janicke, F. , Muller, V. , Bokemeyer, C. , Simon, R. , Sauter, G. , Wilczak, W. , Lebeau, A. , 2010. Gene amplification in ductal carcinoma in situ of the breast. Breast Cancer Research and Treatment. 123, 757–765. [DOI] [PubMed] [Google Scholar]
- Carter, S.L. , Cibulskis, K. , Helman, E. , McKenna, A. , Shen, H. , Zack, T. , Laird, P.W. , Onofrio, R.C. , Winckler, W. , Weir, B.A. , Beroukhim, R. , Pellman, D. , Levine, D.A. , Lander, E.S. , Meyerson, M. , Getz, G. , 2012. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotechnology. 30, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis, K. , Lawrence, M.S. , Carter, S.L. , Sivachenko, A. , Jaffe, D. , Sougnez, C. , Gabriel, S. , Meyerson, M. , Lander, E.S. , Getz, G. , 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology. 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E. , Warwick, J. , Carpenter, R. , Bowen, R.L. , Duffy, S.W. , Jones, J.L. , 2011. Molecular subtyping of DCIS: heterogeneity of breast cancer reflected in pre-invasive disease. British Journal of Cancer. 104, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath, J. , Muthuswamy, S.K. , Brugge, J.S. , 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30, 256–268. [DOI] [PubMed] [Google Scholar]
- Ding, L. , Ellis, M.J. , Li, S. , Larson, D.E. , Chen, K. , Wallis, J.W. , Harris, C.C. , McLellan, M.D. , Fulton, R.S. , Fulton, L.L. , Abbott, R.M. , Hoog, J. , Dooling, D.J. , Koboldt, D.C. , Schmidt, H. , Kalicki, J. , Zhang, Q. , Chen, L. , Lin, L. , Wendl, M.C. , McMichael, J.F. , Magrini, V.J. , Cook, L. , McGrath, S.D. , Vickery, T.L. , Appelbaum, E. , Deschryver, K. , Davies, S. , Guintoli, T. , Lin, L. , Crowder, R. , Tao, Y. , Snider, J.E. , Smith, S.M. , Dukes, A.F. , Sanderson, G.E. , Pohl, C.S. , Delehaunty, K.D. , Fronick, C.C. , Pape, K.A. , Reed, J.S. , Robinson, J.S. , Hodges, J.S. , Schierding, W. , Dees, N.D. , Shen, D. , Locke, D.P. , Wiechert, M.E. , Eldred, J.M. , Peck, J.B. , Oberkfell, B.J. , Lolofie, J.T. , Du, F. , Hawkins, A.E. , O'Laughlin, M.D. , Bernard, K.E. , Cunningham, M. , Elliott, G. , Mason, M.D. , Thompson, D.M. , Ivanovich, J.L. , Goodfellow, P.J. , Perou, C.M. , Weinstock, G.M. , Aft, R. , Watson, M. , Ley, T.J. , Wilson, R.K. , Mardis, E.R. , 2010. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 464, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, A.J. , Pinder, S.E. , Snead, D.R. , Wilson, A.R. , Ellis, I.O. , Elston, C.W. , 1997. The detection of ductal carcinoma in situ at mammographic screening enables the diagnosis of small, grade 3 invasive tumours. British Journal of Cancer. 75, 542–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, E.R. , Gregorio, R.M. , Fisher, B. , Redmond, C. , Vellios, F. , Sommers, S.C. , 1975. The pathology of invasive breast cancer. A syllabus derived from findings of the National Surgical Adjuvant Breast Project (protocol no. 4). Cancer. 36, 1–85. [DOI] [PubMed] [Google Scholar]
- Forozan, F. , Veldman, R. , Ammerman, C.A. , Parsa, N.Z. , Kallioniemi, A. , Kallioniemi, O.P. , Ethier, S.P. , 1999. Molecular cytogenetic analysis of 11 new breast cancer cell lines. British Journal of Cancer. 81, 1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. , Szumel, R. , Marsh, C. , Zhou, W. , Gabrielson, E. , 1996. Genetic progression, histological grade, and allelic loss in ductal carcinoma in situ of the breast. Cancer Research. 56, 5260–5265. [PubMed] [Google Scholar]
- Gao, Y. , Niu, Y. , Wang, X. , Wei, L. , Lu, S. , 2009. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. Journal of Molecular Medicine. 87, 145–152. [DOI] [PubMed] [Google Scholar]
- Gerlinger, M. , Rowan, A.J. , Horswell, S. , Larkin, J. , Endesfelder, D. , Gronroos, E. , Martinez, P. , Matthews, N. , Stewart, A. , Tarpey, P. , Varela, I. , Phillimore, B. , Begum, S. , McDonald, N.Q. , Butler, A. , Jones, D. , Raine, K. , Latimer, C. , Santos, C.R. , Nohadani, M. , Eklund, A.C. , Spencer-Dene, B. , Clark, G. , Pickering, L. , Stamp, G. , Gore, M. , Szallasi, Z. , Downward, J. , Futreal, P.A. , Swanton, C. , 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England Journal of Medicine. 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina, C. , Serio, G. , Lepore, G. , Lettini, T. , Dalena, A.M. , Pennella, A. , D'Eredita, G. , Valente, T. , Ricco, R. , 2003. Pure ductal carcinoma in situ and in situ component of ductal invasive carcinoma of the breast. A preliminary morphometric study. Journal of Experimental & Clinical Cancer Research: CR. 22, 279–288. [PubMed] [Google Scholar]
- Gudjonsson, T. , Adriance, M.C. , Sternlicht, M.D. , Petersen, O.W. , Bissell, M.J. , 2005. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. Journal of Mammary Gland Biology and Neoplasia. 10, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, L. , Wilkerson, P.M. , Lambros, M.B. , Campion-Flora, A. , Rodrigues, D.N. , Gauthier, A. , Cabral, C. , Pawar, V. , Mackay, A. , A'Hern, R. , Marchio, C. , Palacios, J. , Natrajan, R. , Weigelt, B. , Reis-Filho, J.S. , 2012. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. The Journal of Pathology. 227, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heselmeyer-Haddad, K. , Berroa Garcia, L.Y. , Bradley, A. , Ortiz-Melendez, C. , Lee, W.J. , Christensen, R. , Prindiville, S.A. , Calzone, K.A. , Soballe, P.W. , Hu, Y. , Chowdhury, S.A. , Schwartz, R. , Schaffer, A.A. , Ried, T. , 2012. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. The American Journal of Pathology. 181, 1807–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M. , Peluffo, G. , Chen, H. , Gelman, R. , Schnitt, S. , Polyak, K. , 2009. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proceedings of the National Academy of Sciences of the United States of America. 106, 3372–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M. , Yao, J. , Cai, L. , Bachman, K.E. , van den Brule, F. , Velculescu, V. , Polyak, K. , 2005. Distinct epigenetic changes in the stromal cells of breast cancers. Nature Genetics. 37, 899–905. [DOI] [PubMed] [Google Scholar]
- Hu, M. , Yao, J. , Carroll, D.K. , Weremowicz, S. , Chen, H. , Carrasco, D. , Richardson, A. , Violette, S. , Nikolskaya, T. , Nikolsky, Y. , Bauerlein, E.L. , Hahn, W.C. , Gelman, R.S. , Allred, C. , Bissell, M.J. , Schnitt, S. , Polyak, K. , 2008. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 13, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano, K.M. , Tatarkova, I. , Imbalzano, A.N. , Nickerson, J.A. , 2009. Increasingly transformed MCF-10A cells have a progressively tumor-like phenotype in three-dimensional basement membrane culture. Cancer Cell International. 9, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, M.H. , Kim, E.J. , Choi, Y. , Lee, H.E. , Kim, Y.J. , Kim, J.H. , Kang, E. , Kim, S.W. , Kim, I.A. , Park, S.Y. , 2012. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Research: BCR. 14, R115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.E. , Gorringe, K.L. , Thompson, E.R. , Opeskin, K. , Boyle, S.E. , Wang, Y. , Hill, P. , Mann, G.B. , Campbell, I.G. , 2012. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Research and Treatment. 133, 889–898. [DOI] [PubMed] [Google Scholar]
- Knudsen, E.S. , Ertel, A. , Davicioni, E. , Kline, J. , Schwartz, G.F. , Witkiewicz, A.K. , 2012. Progression of ductal carcinoma in situ to invasive breast cancer is associated with gene expression programs of EMT and myoepithelia. Breast Cancer Research and Treatment. 133, 1009–1024. [DOI] [PubMed] [Google Scholar]
- Kuperwasser, C. , Chavarria, T. , Wu, M. , Magrane, G. , Gray, J.W. , Carey, L. , Richardson, A. , Weinberg, R.A. , 2004. Reconstruction of functionally normal and malignant human breast tissues in mice. Proceedings of the National Academy of Sciences of the United States of America. 101, 4966–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau, D.A. , Carter, S.L. , Stojanov, P. , McKenna, A. , Stevenson, K. , Lawrence, M.S. , Sougnez, C. , Stewart, C. , Sivachenko, A. , Wang, L. , Wan, Y. , Zhang, W. , Shukla, S.A. , Vartanov, A. , Fernandes, S.M. , Saksena, G. , Cibulskis, K. , Tesar, B. , Gabriel, S. , Hacohen, N. , Meyerson, M. , Lander, E.S. , Neuberg, D. , Brown, J.R. , Getz, G. , Wu, C.J. , 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 152, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta, E.K. , Tjan, S. , Parkes, R.K. , O'Malley, F.P. , 2002. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Modern Pathology. 15, 1318–1325. [DOI] [PubMed] [Google Scholar]
- Lee, S. , Stewart, S. , Nagtegaal, I. , Luo, J. , Wu, Y. , Colditz, G. , Medina, D. , Allred, D.C. , 2012. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Research. 72, 4574–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental, K.R. , Yu, H. , Kass, L. , Lakins, J.N. , Egeblad, M. , Erler, J.T. , Fong, S.F. , Csiszar, K. , Giaccia, A. , Weninger, W. , Yamauchi, M. , Gasser, D.L. , Weaver, V.M. , 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139, 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, S. , Desouki, M.M. , Gaile, D.P. , Shepherd, L. , Nowak, N.J. , Conroy, J. , Barry, W.T. , Geradts, J. , 2012. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes, Chromosomes & Cancer. 51, 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livasy, C.A. , Perou, C.M. , Karaca, G. , Cowan, D.W. , Maia, D. , Jackson, S. , Tse, C.K. , Nyante, S. , Millikan, R.C. , 2007. Identification of a basal-like subtype of breast ductal carcinoma in situ. Human Pathology. 38, 197–204. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia, M.A. , Geyer, F.C. , Lacroix-Triki, M. , Marchio, C. , Reis-Filho, J.S. , 2010. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 57, 171–192. [DOI] [PubMed] [Google Scholar]
- Lyons, T.R. , O'Brien, J. , Borges, V.F. , Conklin, M.W. , Keely, P.J. , Eliceiri, K.W. , Marusyk, A. , Tan, A.C. , Schedin, P. , 2011. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nature Medicine. 17, 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.J. , Dahiya, S. , Richardson, E. , Erlander, M. , Sgroi, D.C. , 2009. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Research: BCR. 11, R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.J. , Salunga, R. , Tuggle, J.T. , Gaudet, J. , Enright, E. , McQuary, P. , Payette, T. , Pistone, M. , Stecker, K. , Zhang, B.M. , Zhou, Y.X. , Varnholt, H. , Smith, B. , Gadd, M. , Chatfield, E. , Kessler, J. , Baer, T.M. , Erlander, M.G. , Sgroi, D.C. , 2003. Gene expression profiles of human breast cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 100, 5974–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, O.G. , Wu, H. , Timp, W. , Doi, A. , Feinberg, A.P. , 2011. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature Structural & Molecular Biology. 18, 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, F.R. , Santner, S.J. , Tait, L. , Dawson, P.J. , 2000. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. Journal of the National Cancer Institute. 92, 1185–1186. [DOI] [PubMed] [Google Scholar]
- Moelans, C.B. , de Wegers, R.A. , Monsuurs, H.N. , Maess, A.H. , van Diest, P.J. , 2011. Molecular differences between ductal carcinoma in situ and adjacent invasive breast carcinoma: a multiplex ligation-dependent probe amplification study. Cellular Oncology (Dordrecht). 34, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelans, C.B. , Verschuur-Maes, A.H. , van Diest, P.J. , 2011. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. The Journal of Pathology. 225, 222–231. [DOI] [PubMed] [Google Scholar]
- Muggerud, A.A. , Hallett, M. , Johnsen, H. , Kleivi, K. , Zhou, W. , Tahmasebpoor, S. , Amini, R.M. , Botling, J. , Borresen-Dale, A.L. , Sorlie, T. , Warnberg, F. , 2010. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Molecular Oncology. 4, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwenifumbo, J.C. , Marra, M.A. , 2013. Cancer genome-sequencing study design. Nature Reviews Genetics. 14, 321–332. [DOI] [PubMed] [Google Scholar]
- Navin, N. , Kendall, J. , Troge, J. , Andrews, P. , Rodgers, L. , McIndoo, J. , Cook, K. , Stepansky, A. , Levy, D. , Esposito, D. , Muthuswamy, L. , Krasnitz, A. , McCombie, W.R. , Hicks, J. , Wigler, M. , 2011. Tumour evolution inferred by single-cell sequencing. Nature. 472, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin, N. , Krasnitz, A. , Rodgers, L. , Cook, K. , Meth, J. , Kendall, J. , Riggs, M. , Eberling, Y. , Troge, J. , Grubor, V. , Levy, D. , Lundin, P. , Maner, S. , Zetterberg, A. , Hicks, J. , Wigler, M. , 2010. Inferring tumor progression from genomic heterogeneity. Genome Research. 20, 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S. , Van Loo, P. , Wedge, D.C. , Alexandrov, L.B. , Greenman, C.D. , Lau, K.W. , Raine, K. , Jones, D. , Marshall, J. , Ramakrishna, M. , Shlien, A. , Cooke, S.L. , Hinton, J. , Menzies, A. , Stebbings, L.A. , Leroy, C. , Jia, M. , Rance, R. , Mudie, L.J. , Gamble, S.J. , Stephens, P.J. , McLaren, S. , Tarpey, P.S. , Papaemmanuil, E. , Davies, H.R. , Varela, I. , McBride, D.J. , Bignell, G.R. , Leung, K. , Butler, A.P. , Teague, J.W. , Martin, S. , Jonsson, G. , Mariani, O. , Boyault, S. , Miron, P. , Fatima, A. , Langerod, A. , Aparicio, S.A. , Tutt, A. , Sieuwerts, A.M. , Borg, A. , Thomas, G. , Salomon, A.V. , Richardson, A.L. , Borresen-Dale, A.L. , Futreal, P.A. , Stratton, M.R. , Campbell, P.J. , Breast Cancer Working Group of the International Cancer Genome, C2012. The life history of 21 breast cancers. Cell. 149, 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, P. , Pekkel, V. , Fuqua, S.A. , Osborne, C.K. , Clark, G.M. , Allred, D.C. , 1998. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. Journal of the National Cancer Institute. 90, 697–703. [DOI] [PubMed] [Google Scholar]
- Ottesen, G.L. , 2003. Carcinoma in situ of the female breast. A clinico-pathological, immunohistological, and DNA ploidy study. APMIS Supplementum. 1–67. [PubMed] [Google Scholar]
- Page, D.L. , Dupont, W.D. , Rogers, L.W. , Jensen, R.A. , Schuyler, P.A. , 1995. Continued local recurrence of carcinoma 15–25 years after a diagnosis of low grade ductal carcinoma in situ of the breast treated only by biopsy. Cancer. 76, 1197–1200. [DOI] [PubMed] [Google Scholar]
- Page, D.L. , Dupont, W.D. , Rogers, L.W. , Landenberger, M. , 1982. Intraductal carcinoma of the breast: follow-up after biopsy only. Cancer. 49, 751–758. [DOI] [PubMed] [Google Scholar]
- Park, K. , Han, S. , Kim, H.J. , Kim, J. , Shin, E. , 2006. HER2 status in pure ductal carcinoma in situ and in the intraductal and invasive components of invasive ductal carcinoma determined by fluorescence in situ hybridization and immunohistochemistry. Histopathology. 48, 702–707. [DOI] [PubMed] [Google Scholar]
- Park, S.Y. , Kwon, H.J. , Lee, H.E. , Ryu, H.S. , Kim, S.W. , Kim, J.H. , Kim, I.A. , Jung, N. , Cho, N.Y. , Kang, G.H. , 2011. Promoter CpG island hypermethylation during breast cancer progression. Virchows Archiv. 458, 73–84. [DOI] [PubMed] [Google Scholar]
- Place, A.E. , Jin Huh, S. , Polyak, K. , 2011. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Research: BCR. 13, 227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak, K. , Hu, M. , 2005. Do myoepithelial cells hold the key for breast tumor progression?. Journal of Mammary Gland Biology and Neoplasia. 10, 231–247. [DOI] [PubMed] [Google Scholar]
- Porter, D. , Lahti-Domenici, J. , Keshaviah, A. , Bae, Y.K. , Argani, P. , Marks, J. , Richardson, A. , Cooper, A. , Strausberg, R. , Riggins, G.J. , Schnitt, S. , Gabrielson, E. , Gelman, R. , Polyak, K. , 2003. Molecular markers in ductal carcinoma in situ of the breast. Molecular Cancer Research: MCR. 1, 362–375. [PubMed] [Google Scholar]
- Qiu, W. , Hu, M. , Sridhar, A. , Opeskin, K. , Fox, S. , Shipitsin, M. , Trivett, M. , Thompson, E.R. , Ramakrishna, M. , Gorringe, K.L. , Polyak, K. , Haviv, I. , Campbell, I.G. , 2008. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nature Genetics. 40, 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki, A. , Weaver, V.M. , Lee, S.Y. , Rozenberg, G.I. , Chin, K. , Myers, C.A. , Bascom, J.L. , Mott, J.D. , Semeiks, J.R. , Grate, L.R. , Mian, I.S. , Borowsky, A.D. , Jensen, R.A. , Idowu, M.O. , Chen, F. , Chen, D.J. , Petersen, O.W. , Gray, J.W. , Bissell, M.J. , 2008. A human breast cell model of preinvasive to invasive transition. Cancer Research. 68, 1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robanus-Maandag, E.C. , Bosch, C.A. , Kristel, P.M. , Hart, A.A. , Faneyte, I.F. , Nederlof, P.M. , Peterse, J.L. , van de Vijver, M.J. , 2003. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. The Journal of Pathology. 201, 75–82. [DOI] [PubMed] [Google Scholar]
- Sanders, M.E. , Schuyler, P.A. , Dupont, W.D. , Page, D.L. , 2005. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer. 103, 2481–2484. [DOI] [PubMed] [Google Scholar]
- Shah, S.P. , Morin, R.D. , Khattra, J. , Prentice, L. , Pugh, T. , Burleigh, A. , Delaney, A. , Gelmon, K. , Guliany, R. , Senz, J. , Steidl, C. , Holt, R.A. , Jones, S. , Sun, M. , Leung, G. , Moore, R. , Severson, T. , Taylor, G.A. , Teschendorff, A.E. , Tse, K. , Turashvili, G. , Varhol, R. , Warren, R.L. , Watson, P. , Zhao, Y. , Caldas, C. , Huntsman, D. , Hirst, M. , Marra, M.A. , Aparicio, S. , 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 461, 809–813. [DOI] [PubMed] [Google Scholar]
- Shah, S.P. , Roth, A. , Goya, R. , Oloumi, A. , Ha, G. , Zhao, Y. , Turashvili, G. , Ding, J. , Tse, K. , Haffari, G. , Bashashati, A. , Prentice, L.M. , Khattra, J. , Burleigh, A. , Yap, D. , Bernard, V. , McPherson, A. , Shumansky, K. , Crisan, A. , Giuliany, R. , Heravi-Moussavi, A. , Rosner, J. , Lai, D. , Birol, I. , Varhol, R. , Tam, A. , Dhalla, N. , Zeng, T. , Ma, K. , Chan, S.K. , Griffith, M. , Moradian, A. , Cheng, S.W. , Morin, G.B. , Watson, P. , Gelmon, K. , Chia, S. , Chin, S.F. , Curtis, C. , Rueda, O.M. , Pharoah, P.D. , Damaraju, S. , Mackey, J. , Hoon, K. , Harkins, T. , Tadigotla, V. , Sigaroudinia, M. , Gascard, P. , Tlsty, T. , Costello, J.F. , Meyer, I.M. , Eaves, C.J. , Wasserman, W.W. , Jones, S. , Huntsman, D. , Hirst, M. , Caldas, C. , Marra, M.A. , Aparicio, S. , 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 486, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solin, L.J. , Gray, R. , Baehner, F.L. , Butler, S.M. , Hughes, L.L. , Yoshizawa, C. , Cherbavaz, D.B. , Shak, S. , Page, D.L. , Sledge, G.W. , Davidson, N.E. , Ingle, J.N. , Perez, E.A. , Wood, W.C. , Sparano, J.A. , Badve, S. , 2013. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. Journal of the National Cancer Institute. 105, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter, L.H. , Andrews, J.D. , Zhang, G. , Cook, A.C. , Postenka, C.O. , Al-Katib, W. , Leong, H.S. , Rodenhiser, D.I. , Chambers, A.F. , Tuck, A.B. , 2010. Human 21T breast epithelial cell lines mimic breast cancer progression in vivo and in vitro and show stage-specific gene expression patterns. Laboratory Investigation. 90, 1247–1258. [DOI] [PubMed] [Google Scholar]
- Sproul, D. , Nestor, C. , Culley, J. , Dickson, J.H. , Dixon, J.M. , Harrison, D.J. , Meehan, R.R. , Sims, A.H. , Ramsahoye, B.H. , 2011. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 108, 4364–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman, S. , Wang, J. , Bourne, P. , Yang, Q. , Tang, P. , 2007. Expression of cytokeratin markers, ER-alpha, PR, HER-2/neu, and EGFR in pure ductal carcinoma in situ (DCIS) and DCIS with co-existing invasive ductal carcinoma (IDC) of the breast. Annals of Clinical and Laboratory Science. 37, 127–134. [PubMed] [Google Scholar]
- Sternlicht, M.D. , Kedeshian, P. , Shao, Z.M. , Safarians, S. , Barsky, S.H. , 1997. The human myoepithelial cell is a natural tumor suppressor. Clinical Cancer Research. 3, 1949–1958. [PubMed] [Google Scholar]
- Tamimi, R.M. , Baer, H.J. , Marotti, J. , Galan, M. , Galaburda, L. , Fu, Y. , Deitz, A.C. , Connolly, J.L. , Schnitt, S.J. , Colditz, G.A. , Collins, L.C. , 2008. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Research: BCR. 10, R67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, N.C. , Reis-Filho, J.S. , 2012. Genetic heterogeneity and cancer drug resistance. The Lancet Oncology. 13, e178–185. [DOI] [PubMed] [Google Scholar]
- Vargas, A.C. , McCart Reed, A.E. , Waddell, N. , Lane, A. , Reid, L.E. , Smart, C.E. , Cocciardi, S. , da Silva, L. , Song, S. , Chenevix-Trench, G. , Simpson, P.T. , Lakhani, S.R. , 2012. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Research and Treatment. 135, 153–165. [DOI] [PubMed] [Google Scholar]
- Verschuur-Maes, A.H. , de Bruin, P.C. , van Diest, P.J. , 2012. Epigenetic progression of columnar cell lesions of the breast to invasive breast cancer. Breast Cancer Research and Treatment. 136, 705–715. [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon, A. , Lucchesi, C. , Gruel, N. , Raynal, V. , Pierron, G. , Goudefroye, R. , Reyal, F. , Radvanyi, F. , Salmon, R. , Thiery, J.P. , Sastre-Garau, X. , Sigal-Zafrani, B. , Fourquet, A. , Delattre, O. , breast cancer study group of the Institut, C2008. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clinical Cancer Research. 14, 1956–1965. [DOI] [PubMed] [Google Scholar]
- Virnig, B.A. , Tuttle, T.M. , Shamliyan, T. , Kane, R.L. , 2010. Ductal carcinoma in situ of the breast: a systematic review of incidence, treatment, and outcomes. Journal of the National Cancer Institute. 102, 170–178. [DOI] [PubMed] [Google Scholar]
- Wellings, S.R. , Jensen, H.M. , 1973. On the origin and progression of ductal carcinoma in the human breast. Journal of the National Cancer Institute. 50, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Yao, J. , Weremowicz, S. , Feng, B. , Gentleman, R.C. , Marks, J.R. , Gelman, R. , Brennan, C. , Polyak, K. , 2006. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Research. 66, 4065–4078. [DOI] [PubMed] [Google Scholar]
- Yap, T.A. , Gerlinger, M. , Futreal, P.A. , Pusztai, L. , Swanton, C. , 2012. Intratumor heterogeneity: seeing the wood for the trees. Science Translational Medicine. 4, 127ps110 [DOI] [PubMed] [Google Scholar]


