Abstract
Breast cancer is a clinically heterogeneous disease, which necessitates a variety of treatments and leads to different outcomes. As an example, only some women will benefit from chemotherapy. Identifying patients who will respond to chemotherapy and thereby improve their long‐term survival has important implications to treatment protocols and outcomes, while identifying non responders may enable these patients to avail themselves of other investigational approaches or other potentially effective treatments. In this study, serum metabolite profiling was performed to identify potential biomarker candidates that can predict response to neoadjuvant chemotherapy for breast cancer. Metabolic profiles of serum from patients with complete (n = 8), partial (n = 14) and no response (n = 6) to chemotherapy were studied using a combination of nuclear magnetic resonance (NMR) spectroscopy, liquid chromatography–mass spectrometry (LC–MS) and statistical analysis methods. The concentrations of four metabolites, three (threonine, isoleucine, glutamine) from NMR and one (linolenic acid) from LC–MS were significantly different when comparing response to chemotherapy. A prediction model developed by combining NMR and MS derived metabolites correctly identified 80% of the patients whose tumors did not show complete response to chemotherapy. These results show promise for larger studies that could result in more personalized treatment protocols for breast cancer patients.
Keywords: Breast cancer, Neoadjuvant chemotherapy, Therapy response, Metabolomics, NMR, LC‐;MS
Highlights
-
►
Metabolomics differentiates response to neoadjuvant breast cancer chemotherapy.
-
►
Four serum metabolites are found to correlate with response to chemotherapy.
-
►
A 4‐metabolite model identifies 80% of the patients not showing complete response.
-
►
Additional studies on larger patient cohorts are needed to validate the findings.
Abbreviations
- NMR
nuclear magnetic resonance
- LC–MS
liquid chromatography–mass spectrometry
- pCR
pathologic complete response
- SD
stable disease
- PR
partial response
- PLS-DA
partial least squares discriminant analysis
- ROC
receiver operating characteristic
- CV
cross validation
1. Introduction
Breast cancer, although histologically similar, is clinically a very heterogeneous disease, which results in a range of treatment effectiveness and outcomes (Paik et al., 2004). Neoadjuvant chemotherapy can significantly benefit breast cancer patients; however, the varied response to such therapy means that a significant proportion of the patient population is subjected to ineffective treatment while at the same time being exposed to the therapy's toxicities (Wolmark et al., 2001). Pathologic complete response (pCR), which is defined as the disappearance of the invasive cancer cells in the breast after chemotherapy, is used to evaluate patient response and is strongly associated with improved long‐term survival rates (Bear et al., 2004; Fisher et al., 1998; Kuerer et al., 1999). Unfortunately, less than 30% of patients overall show complete response to neoadjuvant chemotherapy (Jones and Smith, 2006). An ability to predict response to chemotherapeutic agents should enable development of personalized treatment protocols, improving survival rates and reducing unnecessary exposure of patients to toxic drugs.
Research focused on finding useful molecular or clinical predictors of pCR to neoadjuvant chemotherapy in breast cancer is relatively sparse. Imaging studies, such as magnetic resonance imaging (MRI) (Padhani et al., 2006) and scintimammography (Marshall et al., 2005; Sciuto et al., 2002) were proposed to predict pathological responses to neoadjuvant chemotherapy, but they are somewhat limited by low sensitivity combined with high costs. High levels of MUC‐1 antigen (CA 15.3) in pre‐treatment serum and its fall after chemotherapy can predict responses as well (Al‐Azawi et al., 2006) but many patients do not exhibit elevation of this marker before treatment and hence it is not helpful for such patients (Kurebayashi et al., 2003). Approaches using genomics and immunohistochemistry have been explored to find serum and tissue biomarkers (Bear et al., 2003; Guarneri et al., 2006; Rouzier et al., 2005; van't Veer et al., 2002). It has been shown that gene signatures such as HER2 overexpression/amplification and lack of ER expression were associated with pCR and certain neoadjuvant chemotherapy regimens (Chen et al., 2011; Gianni et al., 2005; Thuerigen et al., 2006). Other molecular markers such as tumor RNA (Parissenti et al., 2010), glucose‐regulated protein (GRP78) (Lee et al., 2011) and hormone receptors (Van Poznak et al., 2002; Wang et al., 2009) have also been identified as potential predictors of pCR. However, suboptimal performance is a major issue that limits their wide applicability. Circulating tumor cells (CTC) have also been established as providing outcome predictions from particular therapies; however CTCs can be detected in less than 30% of early stage breast cancer patients, which limits their clinical applicability (Hayes and Smerage, 2008).
As an alternative approach for biomarker discovery, metabolomics (or metabolite profiling) enables identification of small‐molecule metabolites in biofluids and tissues that are sensitive to altered pathology (Lindon et al., 2004; Nicholson et al., 1999; Nicholson and Wilson, 2003). High‐throughput analytical techniques of nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) combined with multivariate statistical analyses provide information on a large number of metabolites, including those that have altered levels between healthy subjects and patients with various diseases including cancer (Lanza et al., 2010; Pan and Raftery, 2007; Zhang et al., 2012). So far, the metabolomics based approaches have been used in a large variety of applications, including early disease detection, drug response, toxicity and nutritional studies, and basic systems biology (Clayton et al., 2006; Gowda et al., 2008; Griffin and Kauppinen, 2007; Serkova and Niemann, 2006; Sreekumar et al., 2009; Zhang et al., 2008). Compared with other biomarker discovery approaches for breast cancer, metabolomics provides a strong link between genotype and phenotype, and may provide some insight into oncogenesis. Also, once established, tests based on metabolic profiles are relatively inexpensive, rapid and can be automated (Spratlin et al., 2009).
A growing number of metabolomics studies are contributing toward an improved understanding of breast cancer, and these advances have been reviewed (Claudino and Quattrone, 2007; Gowda et al., 2008; Oakman et al., 2011a). Many of the studies have focused on identifying altered metabolic levels in breast cancer cells or tissues, and relating these changes to their associated metabolic pathways (Cheng et al., 1998; Sitter et al., 2002; Whitehead and Kieber‐Emmons, 2005; Bathen et al., 2007; Sitter et al., 2006; Yang et al., 2007). A very recent study using metabolic profiling of numerous human cancer cell lines found a high correlation between breast cancer (and other cancer) proliferation and the glycine biosynthetic pathway (Jain et al., 2012). Previously, differences between normal and metastatic mammary epithelial cell lines, including up‐regulation of fatty acid synthesis and alterations in glycolysis, the TCA cycle and others, were detected using 13C stable isotopic label tracing by 2D NMR and GC–MS methods (Yang et al., 2007). Breast cancer tumors could be separated from non‐involved tissues based on intensities from spectra generated by high‐resolution magic angle spinning (HR‐MAS) NMR spectroscopy with a sensitivity of 83% and a specificity of 100%. Some metabolites such as choline and glycine were found to be significantly upregulated in tumors larger than 2 cm (Sitter et al., 2006). In another NMR study, a multivariate statistical model based on 67 urinary metabolites successfully identified all the breast cancer patients with high specificity (93%) (Slupsky et al., 2010). Breast cancer prognostic factors, such as estrogen and progesterone receptor status, could be predicted by HR‐MAS NMR based metabolomics on tissue samples (Giskeodegard et al., 2010). Metastatic breast cancer patients could be differentiated from early stage patients with 72% prediction accuracy using serum samples detected by NMR based metabolomics (Oakman et al., 2011b). For identifying breast cancer recurrence, a predictive model built on 11 biomarkers detected by combining NMR and two‐dimensional gas chromatography–mass spectrometry (GC × GC–MS), provided 86% sensitivity and 84% specificity (Asiago et al., 2010).
In this study, we use a metabolomics approach to predict the response to chemotherapy in the neoadjuvant setting. Serum samples from 28 patients obtained before preoperative chemotherapy have been studied using a combination of NMR, liquid chromatography–mass spectrometry (LC–MS) and multivariate statistics methods. Four metabolites that were identified from NMR and MS methods are well correlated with pCR. A statistical model built based on these metabolites predicts pCR with high sensitivity and specificity.
2. Experiments
2.1. Chemicals
For NMR experiments, deuterium oxide (D2O, 99.9% D) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). Sodium azide (NaN3) and the sodium salt of trimethylsilylpropionic acid‐d4 (TSP) were purchased from Sigma–Aldrich (Milwaukee, WI). For LC–MS experiments, two internal standards (tridecanoic acid and chlorophenylalanine) and linolenic acid were purchased from Sigma–Aldrich (analytical grade, St. Louis, MO). HPLC‐grade methanol and acetic acid were purchased from Fisher Scientific (Pittsburgh, PA).
2.2. Patients and serum samples
The patients were enrolled into the study between 2005 and 2008 (Table 1; see also Supplementary Table S1), and were treated according to current guidelines for neoadjuvant therapy. Those patients with locally advance breast cancer and eligible for neoadjuvant chemotherapy were enrolled in this study. All patients were recruited and treated at the Department of Gynecology and Obstetrics, University of Tuebingen. The criteria for patient selection included: 1) signed informed consent and willingness to comply with requirements; 2) at least 18 years old; 3) histopathologically verified lesions of locally advanced breast cancer; 4) indication for neoadjuvant chemotherapy; 5) consent to neoadjuvant chemotherapy; 6) negative result for a serum human chorionic gonadotropin pregnancy test before start of the study for women of childbearing potential; 7) ability to consent to study participation. Patient's study data were collected in case report forms (CRF). For each included patient a CRF was completed. Documents which identify the patient (e.g. the patient identification log and the signed informed consent) were maintained in confidence by the investigator. More detailed information about the sample collection can be found in the supplementary materials.
Table 1.
Summary of clinicopathological characteristics of the breast cancer patients studied.
| Patient characteristics | pCR | PR | SD | |
|---|---|---|---|---|
| Number of patients | 8 | 14 | 6 | |
| Average age (range) | 49.5 (37–60) | 48.7 (34–64) | 43.7 (37–59) | |
| Stage | T1 | 0 | 0 | 2 |
| T2 | 6 | 10 | 3 | |
| T3 | 2 | 1 | 1 | |
| T4 | 0 | 3 | 0 | |
| N0 | 6 | 1 | 2 | |
| N1 | 1 | 4 | 2 | |
| N+ | 1 | 9 | 0 | |
| NX | 0 | 0 | 2 | |
| M0 | 8 | 12 | 5 | |
| MX | 0 | 2 | 1 | |
| Menopause status | pre | 4 | 7 | 4 |
| post | 4 | 6 | 1 | |
| N/A | 0 | 1 | 1 | |
| ER status | pos | 3 | 12 | 4 |
| neg | 5 | 2 | 2 | |
| PR status | pos | 5 | 11 | 5 |
| neg | 3 | 3 | 1 | |
| Her2 status | pos | 5 | 5 | 3 |
| neg | 3 | 9 | 3 | |
| Grading | G1 | 0 | 0 | 1 |
| G2 | 6 | 10 | 5 | |
| G2‐3 | 1 | 0 | 0 | |
| G3 | 1 | 4 | 0 | |
pCR: pathologic complete response; PR: partial response; SD: stable disease.
The response to neoadjuvant chemotherapy in patients with locally advanced breast cancer was based on the correlation of magnetic resonance imaging (MRI), two dimensional (2D)/three dimensional (3D) ultrasound (US) and mammography (MG) with histopathology (study code BCD‐001, funded by the BMBF, Germany). Based on their response, patients were categorized as having pathologic complete response (pCR), partial response (PR) or stable disease (SD). Here, pCR is defined as the disappearance of all tumor deposits; PR indicates a reduction of tumor volume exceeding 50%; while tumor reduction less than 50% or increase of volume up to 25% is scored as stable disease (SD) (Neubauer et al., 2008). The response of the tumors to the neoadjuvant chemotherapy was evaluated pathologically by classifying the regressive changes using a semiquantitative scoring system from 0 to 4 (0 = no effect, 1 = resorption and tumor sclerosis, 2 = minimal residual invasive tumor [<0.5 cm], 3 = residual noninvasive tumor only, 4 = no tumor detectable) according to the tumor regression grading described by Sinn et al. (1994). A consultant pathologist (U. Vogel) blinded to clinical outcome reviewed all paired biopsy and surgical specimens. Samples were de‐identified in compliance with a protocol approved by the IRB committees at both Eberhard‐Karls‐University of Tuebingen and Purdue University.
Serum samples from all 28 breast cancer patients with baseline samples available before neoadjuvant chemotherapy were obtained. Clinical and histopathological data are listed in Table 1 and Supplementary Table S1. Of these patients, 8 belonged to pCR group, 6 to SD group and 14 to PR group (Table 1 and S1). Blood (2 × 8 mL) was drawn 45 min before the start of the chemotherapy. The collected blood was allowed to clot for 45 min at room temperature and centrifuged for 10 min at 3000 rpm at room temperature; the upper serum phase was then isolated, aliquoted and frozen at −80 °C until further use.
Patients were treated according to the design of GeparQuattro including 4 three weekly EC cycles consisting of Epirubicin (90 mg/m2), with simultaneous administration of Cyclophosphamide (600 mg/m2). Subsequently, patients received Docetaxel (100 mg/m2) in three weekly intervals. Patients with Her2/neu positive tumors were also treated with Trastuzumab (6 mg/kg, i.v.) every three weeks, starting with a loading dose of 8 mg/kg i.v. on day 1 of the first EC‐cycle.
2.3. NMR spectroscopy
Frozen serum samples were thawed and 530 μL was mixed with 5 μL 0.1% NaN3 solution. A 60 μL 0.5 mM TSP solution was utilized as an internal standard. 1H 1D NMR experiments were performed using the CPMG (Carr–Purcell–Meiboom–Gill) pulse sequence coupled with water presaturation on a Bruker Avance‐500 spectrometer equipped with a TXI gradient cryoprobe. 128 scans with 16 k time domain data points were collected using a spectral width of 6000 Hz. An exponential weighting function corresponding to 1.0 Hz line broadening was applied to the time domain data before Fourier transformation. The spectra were then phased, baseline corrected and referenced to alanine (δ = 1.48 ppm) using Bruker Topspin 3.0 software.
2.4. Liquid chromatography resolved mass spectrometry (LC–MS)
Frozen serum samples (100 μL) were thawed, and protein was precipitated by adding 200 μL methanol containing two internal standards, tridecanoic acid and chlorophenylalanine, used to monitor extraction efficiency. The solutions were centrifuged at 13,200 rpm for 30 min; the resulting supernatants were dried under vacuum and reconstituted in 25 μL methanol/water (1:1). A pooled sample, which was a mixture of small random volumes from all 28 samples, was extracted using the same procedure as above. This sample was used as a quality control (QC) and was analyzed after every 10 patient samples. LC–MS analysis was performed using an Agilent 6520 HPLC‐QTOF system (Agilent Technologies, Santa Clara, CA) consisting of an Agilent 1200 SL liquid chromatograph coupled with a time‐of‐flight (TOF) mass spectrometer. The reconstituted serum samples were gradient‐eluted at 600 μL/min using (A) 0.2% acetic acid in water and (B) 0.2% acetic acid in methanol (2% B to 98% B in 13 min, 98% B for 6 min). A 3 μL sample aliquot was injected onto a 2.1 × 50 mm Agilent Zorbax Extend‐C18 1.8 μm particle column with a 2.1 × 30 mm Agilent Zorbax SB‐C 8 3.5 μm particle guard column heated to 60 °C. Electrospray ionization (ESI) was used in positive mode. The MS interface capillary was maintained at 325 °C, with a sheath gas flow of 9 L/min. The spray voltage for positive ion injection was 4.0 kV. The instrument was scanned over a range of 50–1000 m/z. Agilent MassHunter Workstation LC–TOF and QTOF Acquisition software (B.02.01) was used for automatic peak detection and mass spectrum deconvolution.
3. Data analysis
3.1. Patient characteristics
The three groups of patients have similar age distributions. Although the pCR cohort has the highest average age (49.5) and the SD cohort has the lowest (43.7), both groups have almost the same age range (37–60 for pCR and 37–59 for SD). In terms of cancer stage and grade, most cancer patients in all three cohorts were in the T1–T2 stage and G2 grade, which represent 75% of pCR patients (n = 6), 71% of PR (n = 10) and 83% of SD patients (n = 5), respectively. All three cohorts contain both pre and post‐menopausal patients. In the pCR and PR groups, pre and post‐menopausal patients are evenly distributed (pre:post = 4:4 and 7:6 for pCR and PR, respectively), while SD cohorts have more pre‐menopausal patients (pre:post = 4:1 for SD). All three groups have diverse distributions of ER/PR/Her2 statuses. Most of the samples (n = 24) are from ER or PR positive women. And almost half of the samples (n = 13) are Her2 positive.
3.2. Data pre‐processing
From the 1H NMR spectra, 27 spectral regions with metabolites peaks were identified and integrated after local baseline correction, and normalized to the total sum of all the metabolites. In order to identify distinguishing metabolite biomarkers, the integral for each metabolite was statistically compared using the Student's t‐test between different groups of patients.
The LC–MS data were processed using Agilent's MassHunter Qualitative Analysis software (version B.03.01). A list of ion intensities for each peak detected was generated, matching m/z and retention time (RT) for each ion. Agilent MassHunter Workstation Mass Profiler Professional software (version B.02.00) was then used for compound peak alignment and removal of any peaks with missing values (ion intensity = 1) in more than one sample from any group; 115 metabolites passed this filter. Internal standard peaks were also removed. Finally, the Agilent Formula Database (Agilent 2010) was used for metabolite identification by matching the accurate mass spectrum to a database of compounds. The Student's t‐test was performed between pCR and SD samples, and 9 metabolites with low p‐values (<0.1) (Supplementary Table S2) were selected as biomarker candidates for further statistical analysis.
3.3. Statistical analysis
The statistical analysis for this metabolomics study was performed retrospectively after sample collection. We performed comparisons by computing p‐values between all 3 pairs of the responding groups (SD, PR, pCR) to find any difference or separation between these groups.
All 27 metabolite peak integrals from NMR and 9 LC–MS metabolites with p < 0.1 were imported into Matlab software (Mathworks) installed with the PLS toolbox (Eigenvector Research, Inc., version 4.0) for partial least squares (PLS) analysis to obtain clustering information and identify outliers. The same software was also used for partial least squares discriminant analysis (PLS‐DA) modeling. Leave‐one‐out cross validation (CV) was chosen, and the number of latent variables were selected according to the root mean square error of CV procedure. The R statistical package (version 2.8.0) was used to generate receiver operating characteristics (ROC) curves and box‐and‐whisker plots.
4. Results
In order to visualize the intrinsic grouping of samples and identify outliers, PLS analysis was performed using the 27 NMR derived metabolite regions, and the results of this analysis are shown in the Supplementary Figure S1. The results showed that the SD patient group has one outlier, which was omitted from further analysis; the new PLS score plot thus obtained is depicted in Figure 1(a). Both pCR and SD groups were separated along the first two latent variables (LV1 and LV2), while the PR group lies between pCR and SD. Similar analysis performed using the 9 metabolites with p < 0.1 obtained from LC–MS data was also performed, and the results are shown in Figure 1(b). As observed for the NMR data, metabolites derived from LC–MS also separated pCR from the SD group of patients. However, again the clustering for the PR group was not as well defined.
Figure 1.
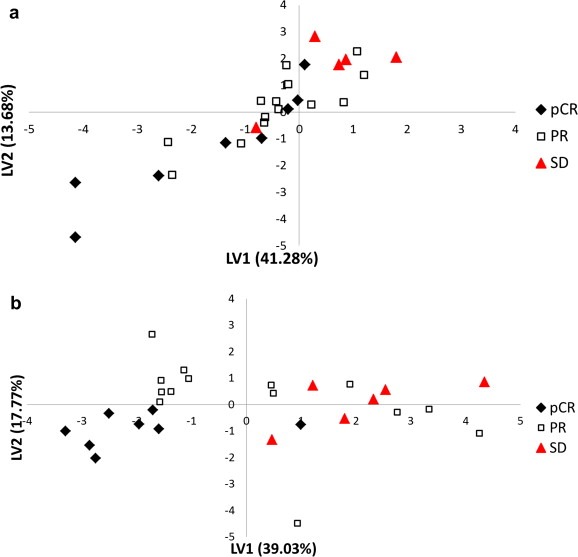
PLS score plot based on (a) 27 metabolites detected by NMR and (b) 9 metabolites detected by LC–MS for all the patients. In (a), one outlier was deleted. The LV label on the two axes stands for latent variable. pCR: pathologic complete response; PR: partial response; SD: stable disease.
Comparison of the NMR data between different groups of patients using the Student's t‐test showed four metabolites to be statistically significant (p < 0.05) (Table 2). One‐way ANOVA was not used here since the objective is to see the difference between each pair of groups. These p‐values indicate that levels of three metabolites, isoleucine, threonine and glutamine, were significantly different between pCR and SD groups and the levels of two metabolites, threonine and glutamine, were different between PR and SD. Only one metabolite, histidine, differed significantly between pCR and PR. The LC–MS data showed that the most statistically differentiating compounds found were long‐chain lipids or fatty acids. The most interesting of these, linolenic acid, was validated using a pure, commercially obtained compound. This metabolite separated pCR from SD samples perfectly, which makes combining linolenic acid with other LC–MS detected markers unnecessary for model building. Statistical analysis shows linolenic acid to be significantly different between pCR and SD groups (p < 0.01). Performance of these metabolites was evaluated individually using receiver operating characteristic (ROC) curves and box‐and‐whisker plots. Figure 2 shows box‐and‐whisker plots for the five metabolites (4 from NMR and 1 from LC–MS) and Supplementary Figure S2 shows ROC curves along with area under the ROC (AUROC) values. The concentration distribution for all the metabolites except histidine showed a consistent trend from pCR to PR to SD; while threonine, glutamine and linolenic acid increased, isoleucine decreased. Further, four metabolites, threonine, glutamine, isoleucine and linolenic acid each showed a minimum AUROC of 0.80 in classifying pCR and SD patients, while only linolenic acid distinguished pCR from both SD and PR patients with an AUROC of greater than 0.80 (Supplementary Figure S2). As mentioned above, linolenic acid distinctly separated pCR from SD with an AUROC = 1 (Table 3).
Table 2.
Summary of NMR metabolites having low p‐values.
| Chemical shift (δ) | Multiplicity | Assignment | p‐Value (pCR vs. SD) | p‐Value (pCR vs. PR) | p‐Value (PR vs. SD) |
|---|---|---|---|---|---|
| 4.24 | m | Threonine | 0.04 | 0.28 | 0.01 |
| 1.00 | s | Isoleucine | 0.04 | 0.10 | 0.20 |
| 2.09 | m | Glutamine | 0.01 | 0.30 | 0.02 |
| 7.07 | s | Histidine | 0.29 | 0.01 | 0.54 |
Figure 2.
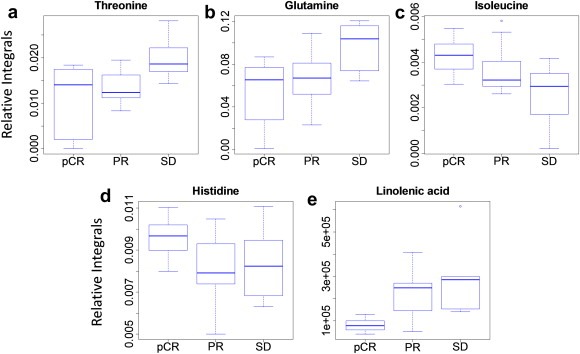
Individual metabolite box‐and‐whisker plots for the different groups of patients: (a) threonine; (b) glutamine; (c) isoleucine; (d) histidine; and (e) linolenic acid.
Table 3.
Summary of data for linolenic acid detected by LC–MS.
| Compound name | Formula | Mass calculated (Da) | Mass detected (Da) | Delta mass (ppm) | RT detected (min) | p‐Value(pCR vs. SD) | p‐Value(pCR vs. PR) | p‐Value(PR vs. SD) |
|---|---|---|---|---|---|---|---|---|
| Linolenic acid | C18H30O2 | 278.2246 | 278.2247 | −0.36 | 11.44 | 0.03 | 0.0002 | 0.32 |
Further analysis focused on evaluating the performance of the metabolites in combination. First, we combined the 3 NMR markers, threonine, glutamine and isoleucine that distinguished pCR and SD, performed PLS‐DA and built a statistical classification model. Leave‐one‐out cross validation was used to reduce overfitting and estimate model accuracy. Figure 3 shows prediction results using this model for different patient groups. The AUROC for distinguishing pCR and SD was 0.81. However, as with the individual metabolites, the performance for distinguishing pCR from the other two groups, PR and SD, was relatively poor (AUROC = 0.72).
Figure 3.
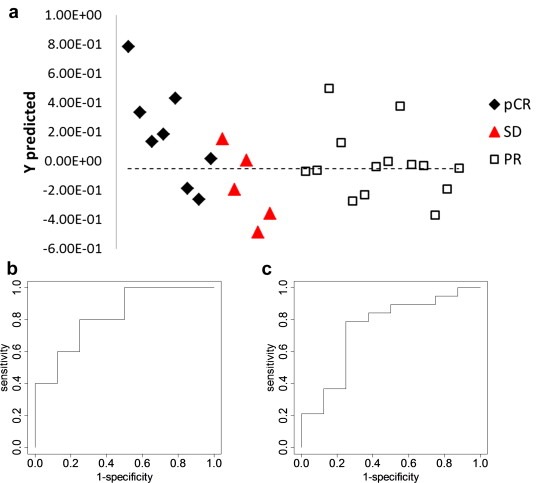
(a) Predictions results for the PLS‐DA model based on the 3 metabolite markers (threonine, isoleucine and glutamine) detected by NMR; (b) ROC curve for pCR vs. SD using the cross‐validated predicted class values (AUROC = 0.81); (c) ROC curve for pCR vs. the other groups using the cross‐validated predicted class values (AUROC = 0.72).
Analysis was also performed combining three NMR derived markers (threonine, glutamine and isoleucine) with LC–MS detected linolenic acid. The results from the statistical analysis are shown in Figure 4. The model provides 100% selectivity and 80% sensitivity for the prediction of pCR vs. SD with an AUROC of 0.95, which is significantly better compared to the classification offered by the 3 marker model (Figure 3). PR samples were not well classified by either model.
Figure 4.
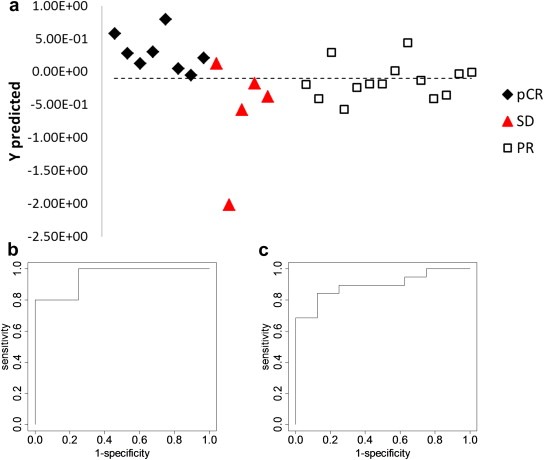
(a) Prediction results for the PLS‐DA model based on combining isoleucine, glutamine, and threonine detected by NMR and linolenic acid detected by LC–MS; (b) ROC curve for pCR vs. SD using the cross‐validated predicted class values (AUROC = 0.95); (c) ROC curve for pCR vs. the other two groups combined using the cross‐validated predicted class values (AUROC = 0.89).
5. Discussion
In this study, we present a new metabolomics approach for predicting the outcome of breast cancer neoadjuvant chemotherapy. By combining two different analytical platforms, NMR and LC–MS, three amino acids and one fatty acid are shown to be highly correlated with pCR. Although the clinical and histopathological parameters of the patients and tumors in each response group are quite heterogeneous, the serum samples still fall into distinct clusters. In order to investigate the impact of different subtypes of breast cancer on the performance of four markers we used for model building, p‐values from the Student's t‐test were obtained by comparing samples from different ER/PR/Her2 status for pCR, PR, SD. All of these sample data are summarized in Supplementary Table S3. No p‐value less than 0.05 can be found, indicating that none of the 4 markers is significantly altered by any of these parameters in the data set.
The resulting prediction model has high sensitivity and specificity. Considering the challenges involved in predicting the outcome of almost any cancer treatment, this promising method might open a new approach for selecting the use of certain cancer treatments or even provide better personalized treatment options. Interestingly, however, as seen from the comparison of the clinical data and clustering of the patient samples in the statistical analysis based on blood metabolites (Table 1, Supplementary Table S1 and Figure 4), the derived metabolite biomarkers provide a strong response to chemotherapy and do not indicate an association with any sub types such as ER, PR or Her2 status. These results suggest that altered metabolic pathways associated with the derived biomarkers are unconnected to ER, PR or Her2 status. However, studies on larger patient cohorts are required to substantiate these findings and identify metabolic link between the pathologically different subtypes, if any.
The performance of linolenic acid detected by LC–MS analysis was particularly striking, and showed an AUROC of 0.86 for the classification of pCR from PR and SD, and 1.0 for the classification of pCR from SD (Figure S2). In addition, the combination of MS and NMR detected metabolites provides a number of advantages including enabling identification of additional distinguishing metabolites that provide better insights into the cellular metabolism and provides potentially a more robust model that can perform effectively when tested with a larger cohort of patients. Four metabolites derived from the combination of NMR and MS distinguished the three breast cancer patient groups, pCR, PR and SD with good performance. The predictive model thus developed by combining the four metabolites correctly separated pCR from SD patients with 100% specificity and 80% sensitivity (Figure 4). Such a high degree of sample classification achieved through the serum derived metabolites is particularly noteworthy at a time when the prediction of the response to breast cancer neoadjuvant chemotherapy continues to be challenging. We found that PR samples show large variance in the values for most of the markers we identified. Some PR patients tend to be predicted as pCR; while others cluster with SD samples. This effect may be due to the heterogeneous results of chemotherapy for these patients who are classified between SD and pCR. Pathologic response rates are surrogates of patient outcome. It will be important in a larger cohort of patients to determine if the PRs who were predicted as pCR have a better clinical outcome than those predicted to be SD.
To the best of our knowledge, this is the first study to combine NMR and MS to identify altered metabolites in the serum for use as predictors of response to neoadjuvant chemotherapy. Altered levels of the four metabolites identified in this study, threonine, glutamine, isoleucine and linolenic acid that predict response to neoadjuvant chemotherapy represent changes in the metabolic activity of several pathways. Information about the association of specific metabolic pathways with response prediction remains to be clearly understood. However, numerous studies have reported altered levels of blood serum/plasma amino acids in breast cancer patients and those results match with the association of the amino acids found in the present study (Asiago et al., 2010; Lai et al., 2005; Zhang et al., 2011). For example, a study using blood plasma showed increased concentration of threonine in breast cancer (Kubota et al., 1992). In addition, investigations of breast cancer cells showed increased levels of threonine, under hypoxic conditions, and such an increase is attributed to impaired protein synthesis due to hypoxia (Weljie et al., 2011). Increased glutamine levels in blood plasma have been reported for breast cancer, in addition to esophageal and liver cancers (Gao et al., 2009; Proenza et al., 2003; Zhang et al., 2011). Glutamine is an essential nutrient for the growth of cancer cells, and the catabolism of glutamine is a major route for the proliferation of mammalian cells. Many types of cancer cells are shown to depend on high rates of glutamine uptake and metabolism (Coles and Johnstone, 1962; Eagle, 1955; Wise et al., 2008). An association of isoleucine that we find in the present study has not been reported previously for breast cancer. However, its altered levels in blood plasma are known in liver and colorectal cancers (Georgiannos et al., 1995; Watanabe et al., 1984). Identification of the fatty acid, linolenic acid, is also consistent with several earlier investigations, which showed significant upregulation of fatty acids in breast cancer (Baron et al., 2004; Boros et al., 2002; Yang et al., 2007).
Thus, although, three of the four distinguishing metabolites, threonine, glutamine and linolenic acid, have previously been shown to have links with breast cancer, the mechanism of their down‐regulation in patients that responded to chemotherapy (pCR group) compared to those that do not (SD group) is not well understood. The results, however, suggest that metabolites in the serum of breast cancer patients are indicators of tumor/host metabolism and that they can predict both sensitivity and resistance to chemotherapy a priori. This is consistent with previous reports that indicate some hypoxic cancer cells exhibit higher levels of threonine and that high levels of fatty acids found in malignant breast cancer cells are also associated with clinically aggressive tumor resistance behavior (Kuhajda, 2000; Weljie et al., 2011; Zeng et al., 2010). Moreover, it is now known that some cancer cells display addiction to glutamine. Glutamine is a major oxidizable substrate for tumor cells, which require glutamine for survival (Fuchs and Bode, 2006; Wise and Thompson, 2010; Yuneva et al., 2007). Hence, the higher concentration of glutamine observed for patients (SD group) that did not respond to chemotherapy appears to favor tumor survival and growth, and offer resistance to chemotherapy.
6. Conclusion
We present a prediction model for the outcome of breast cancer neoadjuvant chemotherapy based on metabolic profiling studies that combine NMR and LC–MS methods. A combination of four metabolites, three detected by NMR (threonine, glutamine and isoleucine), and one by MS (linolenic acid) distinguish groups of patients with no, partial or complete response. While this pilot study was focused on a small patient cohort, it clearly indicates that several blood‐based metabolite markers are sensitive to response, and that the approach is promising for predicting the response to chemotherapy. Studies using larger patient cohorts will be needed to substantiate the results and determine any association of the derived metabolite markers with pathologically different sub types. Validation of these results using a larger and independent sample cohort, and identification of additional metabolite markers, will provide better insights into the pathology at the molecular level that lead to different response outcomes for the three groups of patients. In addition, considering its strong performance as a biomarker in the present study, linolenic acid and possibly other fatty acids might be of particular interest for further validation studies. This approach, which clearly differentiates patients that respond to drugs from those that do not, may provide a useful tool for a non‐invasive prognosis of the treatment outcome.
Conflict of interest
Daniel Raftery has an executive role at Matrix‐Bio, Inc.
Supporting information
The following is the supplementary material related to this article:
Supplementary data
Acknowledgments
This work was supported by Purdue University and the Purdue Research Foundation. SC is a grantee of the Breast Cancer Research Foundation and her efforts are dedicated to the memory of Evelyn H. Lauder. The authors also thank Dr. Shanaiah Murthy at Matrix‐Bio, Inc, and the Purdue Interdepartmental NMR Facility (PINMRF) for their assistance with NMR experiments.
Supplementary material 1.
1.1.
Supplementary material related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2012.10.003.
Wei Siwei, Liu Lingyan, Zhang Jian, Bowers Jeremiah, Gowda G.A. Nagana, Seeger Harald, Fehm Tanja, Neubauer Hans J., Vogel Ulrich, Clare Susan E., Raftery Daniel, (2013), Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer, Molecular Oncology, 7, doi: 10.1016/j.molonc.2012.10.003.
References
- Al-Azawi, D. , Kelly, G. , Myers, E. , McDermott, E.W. , Hill, A.D.K. , Duffy, M.J. , Higgins, N.O. , 2006. CA 15-3 is predictive of response and disease recurrence following treatment in locally advanced breast cancer. BMC Cancer 6, 220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiago, V.M. , Alvarado, L.Z. , Shanaiah, N. , Gowda, G.A.N. , Owusu-Sarfo, K. , Ballas, R.A. , Raftery, D. , 2010. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 70, 8309–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, A. , Migita, T. , Tang, D. , Loda, M. , 2004. Fatty acid synthase: a metabolic oncogene in prostate cancer?. J. Cell. Biochem. 91, 47–53. [DOI] [PubMed] [Google Scholar]
- Bathen, T.F. , Jensen, L.R. , Sitter, B. , Fjosne, H.E. , Halgunset, J. , Axelson, D.E. , Gribbestad, I.S. , Lundgren, S. , 2007. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade and hormone status. Breast Cancer Res. Treat. 104, 181–189. [DOI] [PubMed] [Google Scholar]
- Bear, H.D. , Anderson, S. , Brown, A. , Smith, R. , Mamounas, E.P. , Fisher, B. , Margolese, R. , Theoret, H. , Soran, A. , Wickerham, D.L. , Wolmark, N. , 2003. The effect on tumor response of adding sequential preoperative docetaxel to preoperative doxorubicin and cyclophosphamide: preliminary results from national surgical adjuvant breast and bowel project protocol B-27. J. Clin. Oncol. 21, 4165–4174. [DOI] [PubMed] [Google Scholar]
- Bear, H.D. , Anderson, S. , Smith, R.E. , Robidoux, A. , Kahlenberg, M.S. , Margolese, R.G. , Dakhil, S.R. , Pajon, E.R. , Hoehn, J.L. , Mamounas, E.P. , Geyer, C.E. , Julian, T.B. , Wolmark, N. , 2004. A randomized trial comparing preoperative (preop) doxorubicin/cyclophosphamide (AC) to preop AC followed by preop docetaxel (T) and to preop AC followed by postoperative (postop) T in patients (pts) with operable carcinoma of the breast: results of NSABP B-27. Breast Cancer Res. Treat. 88, S16 [Google Scholar]
- Boros, L.G. , Cascante, M. , Lee, W.N.P. , 2002. Metabolic profiling of cell growth and death in cancer: applications in drug discovery. Drug Discov. Today 7, 364–372. [DOI] [PubMed] [Google Scholar]
- Chen, Y.Z. , Chen, C.M. , Yang, B.L. , Xu, Q.H. , Wu, F. , Liu, F. , Ye, X. , Meng, X. , Mougin, B. , Liu, G.Y. , Shen, Z.Z. , Shao, Z.M. , Wu, J.O. , 2011. Estrogen receptor-related genes as an important panel of predictors for breast cancer response to neoadjuvant chemotherapy. Cancer Lett. 302, 63–68. [DOI] [PubMed] [Google Scholar]
- Cheng, L.L. , Chang, I.W. , Smith, B.L. , Gonzalez, R.G. , 1998. Evaluating human breast ductal carcinomas with high-resolution magic-angle spinning proton magnetic resonance spectroscopy. J. Magn. Reson. 135, 194–202. [DOI] [PubMed] [Google Scholar]
- Claudino, W.M. , Quattrone, A. , Biganzolim, L. , Pestrin, M. , Bertini, I. , Di Leo, A. , 2007. Metabolomics: available results, current research projects in breast cancer, and future applications. J. Clin. Oncol. 25, 2840–2846. [DOI] [PubMed] [Google Scholar]
- Clayton, T.A. , Lindon, J.C. , Cloarec, O. , Antti, H. , Charuel, C. , Hanton, G. , Provost, J.P. , Le Net, J.L. , Baker, D. , Walley, R.J. , Everett, J.R. , Nicholson, J.K. , 2006. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 440, 1073–1077. [DOI] [PubMed] [Google Scholar]
- Coles, N.W. , Johnstone, R.M. , 1962. Glutamine metabolism in Ehrlich ascites-carcinoma cells. Biochem. J. 83, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle, H. , 1955. Nutrition needs of mammalian cells in tissue culture. Science 122, 501–504. [DOI] [PubMed] [Google Scholar]
- Fisher, B. , Bryant, J. , Wolmark, N. , Mamounas, E. , Brown, A. , Fisher, E.R. , Wickerham, D.L. , Begovic, M. , DeCillis, A. , Robidoux, A. , Margolese, R.G. , Cruz, A.B. , Hoehn, J.L. , Lees, A.W. , Dimitrov, N.V. , Bear, H.D. , 1998. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J. Clin. Oncol. 16, 2672–2685. [DOI] [PubMed] [Google Scholar]
- Fuchs, B.C. , Bode, B.P. , 2006. Stressing out over survival: glutamine as an apoptotic modulator. J. Surg. Res. 131, 26–40. [DOI] [PubMed] [Google Scholar]
- Gao, H.C. , Lu, Q. , Liu, X. , Cong, H. , Zhao, L.C. , Wang, H.M. , Lin, D.H. , 2009. Application of (1)H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 100, 782–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiannos, S.N. , Weston, P.M.T. , Goode, A.W. , 1995. Correlation between albuminuria and positively charged amino-acids in gastrointestinal cancer. Int. Surg. 80, 49–52. [PubMed] [Google Scholar]
- Gianni, L. , Zambetti, M. , Clark, K. , Baker, J. , Cronin, M. , Wu, J. , Mariani, G. , Rodriguez, J. , Carcangiu, M. , Watson, D. , Valagussa, P. , Rouzier, R. , Symmans, W.F. , Ross, J.S. , Hortobagyi, G.N. , Pusztai, L. , Shak, S. , 2005. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J. Clin. Oncol. 23, 7265–7277. [DOI] [PubMed] [Google Scholar]
- Giskeodegard, G.F. , Grinde, M.T. , Sitter, B. , Axelson, D.E. , Lundgren, S. , Fjosne, H.E. , Dahl, S. , Gribbestad, I.S. , Bathen, T.F. , 2010. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J. Proteome Res. 9, 972–979. [DOI] [PubMed] [Google Scholar]
- Gowda, G.A.N. , Zhang, S.C. , Gu, H.W. , Asiago, V. , Shanaiah, N. , Raftery, D. , 2008. Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 8, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, J.L. , Kauppinen, R.A. , 2007. Tumour metabolomics in animal models of human cancer. J. Proteome Res. 6, 498–505. [DOI] [PubMed] [Google Scholar]
- Guarneri, V. , Broglio, K. , Kau, S.W. , Cristofanilli, M. , Buzdar, A.U. , Valero, V. , Buchholz, T. , Meric, F. , Middleton, L. , Hortobagyi, G.N. , Gonzalez-Angulo, A.M. , 2006. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J. Clin. Oncol. 24, 1037–1044. [DOI] [PubMed] [Google Scholar]
- Hayes, D.F. , Smerage, J. , 2008. Is there a role for circulating tumor cells in the management of breast cancer?. Clin. Cancer Res. 14, 3646–3650. 18559576 [Google Scholar]
- Jain, M. , Nilsson, R. , Sharma, S. , Madhusudhan, N. , Kitami, T. , Souza, A.L. , Kafri, R. , Kirschner, M.W. , Clish, C.B. , Mootha, V.K. , 2012. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.L. , Smith, I.E. , 2006. Neoadjuvant treatment for early-stage breast cancer: opportunities to assess tumour response. Lancet Oncol. 7, 869–874. [DOI] [PubMed] [Google Scholar]
- Kubota, A. , Meguid, M.M. , Hitch, D.C. , 1992. Amino-acid profiles correlate diagnostically with organ site in 3 kinds of malignant-tumors. Cancer 69, 2343–2348. [DOI] [PubMed] [Google Scholar]
- Kuerer, H.M. , Newman, L.A. , Smith, T.L. , Ames, F.C. , Hunt, K.K. , Dhingra, K. , Theriault, R.L. , Singh, G. , Binkley, S.M. , Sneige, N. , Buchholz, T.A. , Ross, M.I. , McNeese, M.D. , Buzdar, A.U. , Hortobagyi, G.N. , Singletary, S.E. , 1999. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J. Clin. Oncol. 17, 460–469. [DOI] [PubMed] [Google Scholar]
- Kuhajda, F.P. , 2000. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16, 202–208. [DOI] [PubMed] [Google Scholar]
- Kurebayashi, J. , Yamamoto, Y. , Tanaka, K. , Kohno, N. , Kurosumi, M. , Moriya, T. , Nishimura, R. , Ogawa, Y. , Taguchi, T. , 2003. Significance of serum carcinoembryonic antigen and CA 15-3 in monitoring advanced breast cancer patients treated with systemic therapy: a large-scale retrospective study. Breast Cancer 10, 38–44. [DOI] [PubMed] [Google Scholar]
- Lai, H.S. , Lee, J.C. , Lee, P.H. , Wang, S.T. , Chen, W.J. , 2005. Plasma free amino acid profile in cancer patients. Semin. Cancer Biol. 15, 267–276. [DOI] [PubMed] [Google Scholar]
- Lanza, I.R. , Zhang, S.C. , Ward, L.E. , Karakelides, H. , Raftery, D. , Nair, K.S. , 2010. Quantitative metabolomics by (1)H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One 5, e10538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. , Nichols, P. , Groshen, S. , Spicer, D. , Lee, A.S. , 2011. GRP78 as potential predictor for breast cancer response to adjuvant taxane therapy. Int. J. Cancer 128, 726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon, J.C. , Holmes, E. , Nicholson, J.K. , 2004. Metabonomics and its role in drug development and disease diagnosis. Expert Rev. Mol. Diagn. 4, 189–199. [DOI] [PubMed] [Google Scholar]
- Marshall, C. , Eremin, J. , El-Sheemy, M. , Eremin, O. , Griffiths, P.A. , 2005. Monitoring the response of large (> 3 cm) and locally advanced (T3-4, N0-2) breast cancer to neoadjuvant chemotherapy using Tc-99m-Sestamibi uptake. Nucl. Med. Commun. 26, 9–15. [DOI] [PubMed] [Google Scholar]
- Neubauer, H. , Gall, C. , Vogel, U. , Hornung, R. , Wallwiener, D. , Solomayer, E. , Fehm, T. , 2008. Changes in tumour biological markers during primary systemic chemotherapy (PST). Anticancer Res. 28, 1797–1804. [PubMed] [Google Scholar]
- Nicholson, J.K. , Lindon, J.C. , Holmes, E. , 1999. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189. [DOI] [PubMed] [Google Scholar]
- Nicholson, J.K. , Wilson, I.D. , 2003. Understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2, 668–676. [DOI] [PubMed] [Google Scholar]
- Oakman, C. , Tenori, L. , Biganzoli, L. , Santarpia, L. , Cappadona, S. , Luchinat, C. , Di Leo, A. , 2011. Uncovering the metabolic fingerprint of breast cancer. Int. J. Biochem. Cell. Biol. 43, 1010–1020. [DOI] [PubMed] [Google Scholar]
- Oakman, C. , Tenori, L. , Claudino, W.M. , Cappadona, S. , Nepi, S. , Battaglia, A. , Bernini, P. , Zafarana, E. , Saccenti, E. , Fornier, M. , Morris, P.G. , Biganzoli, L. , Luchinat, C. , Bertini, I. , Di Leo, A. , 2011. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann. Oncol. 22, 1295–1301. [DOI] [PubMed] [Google Scholar]
- Padhani, A.R. , Hayes, C. , Assersohn, L. , Powles, T. , Makris, A. , Suckling, J. , Leach, M.O. , Husband, J.E. , 2006. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: initial clinical results. Radiology 239, 361–374. [DOI] [PubMed] [Google Scholar]
- Paik, S. , Shak, S. , Tang, G. , Kim, C. , Baker, J. , Cronin, M. , Baehner, F.L. , Walker, M.G. , Watson, D. , Park, T. , Hiller, W. , Fisher, E.R. , Wickerham, D.L. , Bryant, J. , Wolmark, N. , 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826. [DOI] [PubMed] [Google Scholar]
- Pan, Z.Z. , Raftery, D. , 2007. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 387, 525–527. [DOI] [PubMed] [Google Scholar]
- Parissenti, A.M. , Chapman, J.A.W. , Kahn, H.J. , Guo, B.Q. , Han, L. , O'Brien, P. , Clemons, M.P. , Jong, R. , Dent, R. , Fitzgerald, B. , Pritchard, K.I. , Shepherd, L.E. , Trudeau, M.E. , 2010. Association of low tumor RNA integrity with response to chemotherapy in breast cancer patients. Breast Cancer Res. Treat. 119, 347–356. [DOI] [PubMed] [Google Scholar]
- Proenza, A.M. , Oliver, J. , Palou, A. , Roca, P. , 2003. Breast and lung cancer are associated with a decrease in blood cell amino acid content. J. Nutr. Biochem. 14, 133–138. [DOI] [PubMed] [Google Scholar]
- Rouzier, R. , Perou, C.M. , Symmans, W.F. , Ibrahim, N. , Cristofanilli, M. , Anderson, K. , Hess, K.R. , Stec, J. , Ayers, M. , Wagner, P. , Morandi, P. , Fan, C. , Rabiul, I. , Ross, J.S. , Hortobagyi, G.N. , Pusztai, L. , 2005. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 11, 5678–5685. [DOI] [PubMed] [Google Scholar]
- Sciuto, R. , Pasqualoni, R. , Bergomi, S. , Petrilli, G. , Vici, P. , Belli, F. , Botti, C. , Mottolese, M. , Maini, C.L. , 2002. Prognostic value of Tc-99m-sestamibi washout in predicting response of locally advanced breast cancer to neoadjuvant chemotherapy. J. Nucl. Med. 43, 745–751. [PubMed] [Google Scholar]
- Serkova, N.J. , Niemann, C.U. , 2006. Pattern recognition and biomarker validation using quantitative H-1-NMR-based metabolomics. Expert Rev. Mol. Diagn. 6, 717–731. [DOI] [PubMed] [Google Scholar]
- Sinn, H.P. , Schmid, H. , Junkermann, H. , Huober, J. , Leppien, G. , Kaufmann, M. , Bastert, G. , Otto, H.F. , 1994. [Histologic regression of breast cancer after primary (neoadjuvant) chemotherapy]. Geburtshilfe Frauenheilkd 54, 552–558. [DOI] [PubMed] [Google Scholar]
- Sitter, B. , Sonnewald, U. , Spraul, M. , Fjosne, H.E. , Gribbestad, I.S. , 2002. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 15, 327–337. [DOI] [PubMed] [Google Scholar]
- Sitter, B. , Lundgren, S. , Bathen, T.F. , Halgunset, J. , Fjosne, H.E. , Gribbestad, I.S. , 2006. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 19, 30–40. [DOI] [PubMed] [Google Scholar]
- Slupsky, C.M. , Steed, H. , Wells, T.H. , Dabbs, K. , Schepansky, A. , Capstick, V. , Faught, W. , Sawyer, M.B. , 2010. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 16, 5835–5841. [DOI] [PubMed] [Google Scholar]
- Spratlin, J.L. , Serkova, N.J. , Eckhardt, S.G. , 2009. Clinical applications of metabolomics in oncology: a review. Clin. Cancer Res. 15, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar, A. , Poisson, L.M. , Rajendiran, T.M. , Khan, A.P. , Cao, Q. , Yu, J.D. , Laxman, B. , Mehra, R. , Lonigro, R.J. , Li, Y. , Nyati, M.K. , Ahsan, A. , Kalyana-Sundaram, S. , Han, B. , Cao, X.H. , Byun, J. , Omenn, G.S. , Ghosh, D. , Pennathur, S. , Alexander, D.C. , Berger, A. , Shuster, J.R. , Wei, J.T. , Varambally, S. , Beecher, C. , Chinnaiyan, A.M. , 2009. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Thuerigen, O. , Schneeweiss, A. , Toedt, G. , Warnat, P. , Hahn, M. , Kramer, H. , Brors, B. , Rudlowski, C. , Benner, A. , Schuetz, F. , Tews, B. , Eils, R. , Sinn, H.P. , Sohn, C. , Lichter, P. , 2006. Gene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancer. J. Clin. Oncol. 24, 1839–1845. [DOI] [PubMed] [Google Scholar]
- van't Veer, L.J. , Dai, H.Y. , van de Vijver, M.J. , He, Y.D.D. , Hart, A.A.M. , Mao, M. , Peterse, H.L. , van der Kooy, K. , Marton, M.J. , Witteveen, A.T. , Schreiber, G.J. , Kerkhoven, R.M. , Roberts, C. , Linsley, P.S. , Bernards, R. , Friend, S.H. , 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536. [DOI] [PubMed] [Google Scholar]
- Van Poznak, C. , Tan, L. , Panageas, K.S. , Arroyo, C.D. , Hudis, C. , Norton, L. , Seidman, A.D. , 2002. Assessment of molecular markers of clinical sensitivity to single-agent taxane therapy for metastatic breast cancer. J. Clin. Oncol. 20, 2319–2326. [DOI] [PubMed] [Google Scholar]
- Wang, L.B. , Jiang, Z.N. , Sui, M.H. , Shen, J.G. , Xu, C.Y. , Fan, W.M. , 2009. The potential biomarkers in predicting pathologic response of breast cancer to three different chemotherapy regimens: a case control study. BMC Cancer 9, 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, A. , Higashi, T. , Sakata, T. , Nagashima, H. , 1984. Serum amino-acid levels in patients with hepatocellular-carcinoma. Cancer 54, 1875–1882. [DOI] [PubMed] [Google Scholar]
- Weljie, A.M. , Bondareva, A. , Zang, P. , Jirik, F.R. , 2011. H-1 NMR metabolomics identification of markers of hypoxia-induced metabolic shifts in a breast cancer model system. J. Biomol. NMR 49, 185–193. [DOI] [PubMed] [Google Scholar]
- Wise, D.R. , DeBerardinis, R.J. , Mancuso, A. , Sayed, N. , Zhang, X.Y. , Pfeiffer, H.K. , Nissim, I. , Daikhin, E. , Yudkoff, M. , McMahon, S.B. , Thompson, C.B. , 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A 105, 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, D.R. , Thompson, C.B. , 2010. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, T.L. , Kieber-Emmons, T. , 2005. Applying in vitro NMR spectroscopy and 1H NMR metabonomics to breast cancer characterization and detection. Prog. NMR Spectroc. 47, 165–174. [Google Scholar]
- Wolmark, N. , Wang, J. , Mamounas, E. , Bryant, J. , Fisher, B. , 2001. Preoperative chemotherapy in patients with operable breast cancer: nine-year results from National Surgical Adjuvant Breast and Bowel Project B-18. J. Natl. Cancer Inst. Monogr. 96–102. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Richardson, A.D. , Smith, J.W. , Osterman, A. , 2007. Comparative metabolomics of breast cancer. Pac. Symp. Biocomput. 181–192. [PubMed] [Google Scholar]
- Yuneva, M. , Zamboni, N. , Oefner, P. , Sachidanandam, R. , Lazebnik, Y. , 2007. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell. Biol. 178, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Biernacka, K.M. , Holly, J.M.P. , Jarrett, C. , Morrison, A.A. , Morgan, A. , Winters, Z.E. , Foulstone, E.J. , Shield, J.P. , Perks, C.M. , 2010. Hyperglycaemia confers resistance to chemotherapy on breast cancer cells: the role of fatty acid synthase. Endocr. Relat. Canc. 17, 539–551. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Bowers, J. , Liu, L. , Wei, S. , Gowda, G.A. , Hammoud, Z. , Raftery, D. , 2012. Esophageal cancer metabolite biomarkers detected by LC-MS and NMR methods. PLoS One 7, e30181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Liu, L. , Wei, S. , Gowda, G.A.N. , Hammoud, Z. , Kesler, K.A. , Raftery, D. , 2011. Metabolomics study of esophageal adenocarcinoma. J. Thorac. Cardiovasc. Surg. 141, 469–475. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Gowda, G.A.N. , Asiago, V. , Shanaiah, N. , Barbas, C. , Raftery, D. , 2008. Correlative and quantitative H-1 NMR-based metabolomics reveals specific metabolic pathway disturbances in diabetic rats. Anal. Biochem. 383, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary material related to this article:
Supplementary data


