Abstract
The NY‐ESO1 gene is a cancer/testis antigen considered to be suitable target for the immunotherapy of human malignancies. Despite the identification of the epigenetical silencing of the NY‐ESO1 gene in a large variety of tumors, the molecular mechanism involved in this phenomenon is not fully elucidated. In two non epithelial cancers (glioma and mesothelioma), we found that the epigenetic regulation of the NY‐ESO1 gene requires the sequential recruitment of the HDAC1‐mSin3a‐NCOR, Dnmt3b‐HDAC1‐Egr1 and Dnmt1‐PCNA‐UHRF1‐G9a complexes. Thus, our data illustrate the orchestration of a sequential epigenetic mechanism including the histone deacetylation and methylation, and the DNA methylation processes.
Keywords: Dnmt, Epigenetic, HDAC, NY-ESO1, Glioma, Mesothelioma
Highlights
-
►
NY‐ESO1 is a cancer/testis antigen.
-
►
NY‐ESO1 expression is considered to be suitable targets for the immunotherapy.
-
►
NY‐ESO1 is epigenetically silenced in several cancers (glioma and mesothelioma).
1. Introduction
In mammalian, aberrant epigenetic silencing of genes by promoter DNA (hyper)methylation, histone methylation and/or histone deacetylation plays an important role in the pathogenesis of cancer (Vaissière et al., 2008). Indeed, epigenetic silencing affects genes involved in a wide panel of cell function such as cell cycle control, apoptosis, cell signaling, tumor cell invasion and metastasis, angiogenesis and immune recognition. In this last case, the discovery of epigenetic silencing of tumor‐associated antigens and their potential re‐expression by using epigenetic drugs provides new developments to design chemo‐immunotherapeutic strategies for the treatment of patients with solid malignancies of different histology (Maio et al., 2003; Sigalotti et al., 2005). Among the cancer‐associated antigens, the epigenetic regulation of the several Cancer/Testis or Cancer/Germ‐line family of antigens was investigated in the last decades (Karpf, 2006). Several studies mention the fact that the epigenetic regulation of the NY‐ESO1 gene promoter in HCT116 cells implicates the cooperative action of two DNA methyltransferases, the Dnmt1 and Dnmt3b, and that the NY‐ESO1mRNA can be synergistically induced by the co‐treatment of glioma cells by a histone deacetylase inhibitor (HDACi, valproic acid) and a DNA methyltransferases inhibitor (Dnmti, 5aza‐2‐deoxycytidine) (James et al., 2006; Oi et al., 2009). However, the molecular mechanisms and actors involved in the epigenetic regulation of the NY‐ESO1 gene promoter are not fully elucidated.
2. Material and methods
2.1. Analyses of DNA methylation
DNA was extracted by using the QiaAmp DNA mini Kit (Qiagen, France). Methylated DNA ImmunoPrecipitation experiments were realized by using MeDIP kit (Diagenode, France).
2.2. Chromatin immunoprecipitation (ChIP) and re‐ChIP experiments
Briefly, chromatin was purified from cells after crosslinking with 1% formaldehyde for 10 min at room temperature. ChIP and re‐ChIP assays were performed by using the ChIP‐IT™ chromatin immunoprecipitation Kits and the re‐ChIP‐IT™ Kit (Active Motif, France), with indicated antibodies and primers. Positive controls to show the ChIP validity realized with the Dnmt3a, Sirt1, Sirt2, SetDB1, HDAC2, HDAC6, HDAC7, myc, Sp1, MBD1, MBD2, MDB, MeCP2, EZH2, KDM6A and p300 antibodies were illustrated in Figure S1.
2.3. Western blot and immunoprecipitation
In brief, proteins were size fractionated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose or PVDF membrane. Saturation and blotting were realized by using SNAP i.d Protein Detection System (Millipore, France). The detection of proteins was performed using ECL (GE‐Amersham Biosciences, France) and/or SuperSignal west femto Maximum Sensitivity (Pierce, France) chemiluminescence reagents. Bands were quantified using Quantity One quantification software (BioRad, France).
Immunoprecipitations were realized by using the Catch and Release® v2.0 Reversible Immunoprecipitation System (Millipore, France) with 4 μg of considered antibodies.
2.4. Quantitative PCR (qPCR)
RNA was isolated from cells using trizol. qPCR reactions were done in triplicate by using the Multiplex Quantitative PCR system (Mx4000), qPCR Core Reagent kit (Stratagene) according to manufacturer's instructions and by using primers specific of NY‐ESO1 (Oi et al., 2009).
2.5. Coupled ChIP experiment with NaturePURE™ affinity purification of protein complexes
The pcDNA3.2‐CapTEV/CT‐V5‐DEST‐HDAC1 vector was obtained by using HDAC1 Ultimate™ ORF clone and the Gatway® Cloning system (Life Technologies, France). After cellular transfection, crosslinking was performed by using formaldehyde (37% stock) to a final concentration of 1% (10 min at room temperature). Next, crosslinking was quenched by using glycine solution (to a final concentration of 125 nM). After 3 whases in ice‐cold PBS solution, cells were scrapped. ChIP‐It Shearing kit was next used to prepare chromatin (Active Motif, France). Chromatin was sonicated by using Bioruptor Sonicator (Diagenode, France). After equilibration of the Streptavidin Agarose column (according to the manufacturer's instruction, Life Technologies, France), 40 mg of sonicated chromatin was added on column (3 h at 4 °C). After washes, AcTEV™ Protease cleavage was performed overnight at 4 °C with gentle agitation on rotary shaker. Elution was performed by gravity flow into microcentrifuge tube according to the manufacturer's instructions (Life Technologies, France). Next, eluate was polled previous to be separate by using gel filtration chromatography (Bio‐Sil SEC Column, BioRad, France), to separate protein/DNA complexes based on their relative size. “DNA and protein samples” were realized from each fraction. “DNA samples” were realized according to the step D of the ChIP‐IT kit (Active Motif, France) previous to be used for qPCR analyses. “Protein samples” were obtained by using TCA precipitation (Add 1 volume of TCA solution to 4 volumes of sample, incubation 10 min at 4 °C, centrifugation 14,000 rpm, 5 min; remove supernatant, 3 pellet washes with 200 μL of cold acetone, Dry pellet by placing tube in 95 °C heat block for 5–10 min to drive off acetone), previous to be use for western blot analyses.
2.6. Activation of HLA‐A*0201‐restricted, NY‐ESO‐1‐specific CD8+ T‐cells
1·105 tumor cells were co‐cultured with 5·104 NY‐ESO‐1‐specific CD8+ T‐cells in complete medium containing 10 μg mL−1 brefeldin A (Sigma–Aldrich, France) for 5 h at 37 °C and washed. Cells were stained with allophycocyanin‐conjugated mouse anti‐human CD8 and phycoerythrin (PE)‐conjugated mouse anti‐human IFN‐γ monoclonal antibodies (BD Biosciences, Le Pont de Claix, France) using methods already described (Sigalotti et al., 2002). CD8 and IFN‐γ expression were analyzed by flow cytometry.
2.7. Statistical analyses
All the results shown in this article result and/or are representative of three independent experiments. In each of the biological repeats, each RNA sample was analyzed at least in triplicates; each DNA and protein sample was analyzed at least in duplicates. T test (GraphPad software) was realized to compare two values.
2.8. Supplemental data
The supplemental data include lists of antibodies and primers used in all experiments and six supplemental figures.
3. Results
3.1. Dnmt1/3b, HDAC1 and G9a: the epigenetic actors
To describe the molecular actors and mechanisms governing the epigenetic regulation of the NY‐ESO1 gene, we initially decided to use two types of cancer cells in which the promoter region of the NY‐ESO1 gene was already described as being methylated or unexpressed: the U251 cells i.e. a cell line representing the glioma and the Meso96 (M96) cells i.e. a cell line representing the mesothelioma (Oi et al., 2009; Sahin et al., 2000). We also used Mesenchymal Stem Cells (MSC) as positive control since these cells expressed NY‐ESO1 according to Cronwright et al. (2005). In addition, our choice was also supported by the fact that a significant number of glioma and mesothelioma cells were deficient in NY‐ESO1mRNA on contrary to MSC (Figure 1A).
Figure 1.
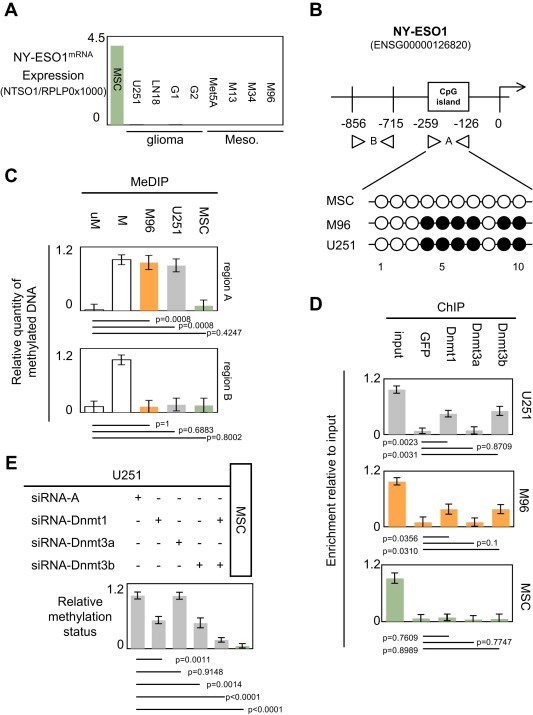
Dnmt1 and Dnmt3b participate to the epigenetic regulation of the NY‐ESO1 gene. (A) Expression level of NY‐ESO1mRNA in a panel of glioma and mesothelioma cells. Mesenchymal stem cell (MSC) was used as positive control i.e. cells expressing NY‐ESO1mRNA (Cronwright et al., 2005). U251 and LN18 are glioma cell lines, G1 and G2 are primary‐cultured tumor cells obtained from GBM, Met5A is mesothelial cell line, and M13, M34 and M96 are primary‐cultured tumor cells obtained from mesothelioma. (B) Schematic representation of the NY‐ESO1 gene. Methylated DNA immunoprecipitation experiments (MeDIP) were designed to estimate the methylation level of the −856/−715 and −259/−126 regions of the NY‐ESO1 gene i.e. one region including into CpG island and other not included into CpG island. Presence of CpG island was determined by using MethPrimer program. Methylation level of CG dinucleotides was determined by bisulfite sequencing (open circle: unmethylated, black circle: methylated). (C) Methylation status of the −856/−715 (bottom) and −259/−126 (top) regions of the NY‐ESO1 gene. uM: Unmethylated DNA: CpGenome™ Universal unmethylated DNA set, M: Methylated DNA : CpGenome™ Universal methylated DNA set, (Chemicon‐Millipore, France). (D) Identification by chromatin immunoprecipitation (ChIP) of the Dnmt recruited on the −259/−126 regions of the NY‐ESO1 gene. (E) Impact of the siRNA‐induced down‐regulation of Dnmt1, Dnmt3a and/or Dnmt3b on the methylation status of the −259/−126 region of the NY‐ESO1 gene. siRNA‐A is an aspecific siRNA used in control according to the manufacturer's instruction (Tebu‐Bio, Santa Cruz, France). The use of these siRNA was already monitored and described in previous report (Hervouet et al., 2010a).
The NY‐ESO1 gene harboring a CpG island located between the −259/−126 nucleotides (according to the MethPrimer program), we initially analyzed the methylation status of this region and the one of the −856/−715 region as control (Figure 1B). The choice of the −259/−126 is also supported by the fact that this regions includes Sp1 boxes and Sp1 transcriptionally regulates the NY‐ESO1mRNA expression (Kang et al., 2007).
Quantitative PCR realized from Methylated DNA Immunoprecipitation (MeDIP) experiments revealed that the −259/−126 region of NY‐ESO1 gene, but not the −856/−715 region, was methylated in the U251 and M96 cells and unmethylated in MSC cells (Figure 1C). In addition, bisulfite sequencing realized to investigate the methylation of CG dinucleotides localized on the −259/−126 region confirmed this point and revealed that the methylation concerns the CG referred as being 4–5–6–7–9 and 10 (Figure 1B).
To identify the Dnmt implicated into the methylation of the −259/−126 region of the NY‐ESO1 gene, we performed chromatin immunoprecipitation (ChIP) experiences in which antibodies were directed against the Dnmt1, Dnmt3a and the Dnmt3b. PCR products from the immunoprecipitated chromatin by the Dnmt1 and the Dnmt3b antibodies in ChIP experiments indicated that the Dnmt1 and Dnmt3b are recruited on the −259/−126 region of the NY‐ESO1 gene when this gene is methylated in the U251 and M96 cells (Figure 1D). Dnmt1 and Dnmt3b down‐regulations (by using siRNA) confirmed the fact that these two enzymes play a crucial role on the methylation of the −259/−126 region of the NY‐ESO1 gene (Figure 1E and S2) (Hervouet et al., 2010a). In addition, bisulfite sequencing indicated that the methylation of the CG referred as being 4–5–6–7 is affected by the siRNA‐Dnmt1 treatment while siRNA‐Dnmt3B treatment affected the CG dinucleotides referred as being 9 and 10 (Figure S3). Thus, our data support the idea that Dnmt1 and DNmt3b cooperate to methylate the −259/−126 region of the NY‐ESO1 gene.
Post‐translational modifications of histone also governing the epigenetic regulation of genes expression, we continued our study by analyzing the degree of global acetylation of the histone H3 and the level of histone H3 lysine56 acetylation of the −259/−126 region of the NY‐ESO1 gene. After immunoprecipitation of chromatin by an antibody directed against acetylated histone H3 (H3Ac), PCR amplification indicated that the histone H3 associated with the −259/−126 region of the NY‐ESO1 gene are very weakly acetylated in the U251 and M96 cells (Figure 2A). Similar experiments realized with an antibody directed against the K56‐acetylated histone H3 (H3K56Ac) indicated that the very weak acetylation of histone H3 concerned, in part, the lysine 56 (Figure 2A). Thus, this last result suggests that the −259/−126 region of the NY‐ESO1 gene is deacetylated by one or several HDACs or Sirtuins (Sirt). Hypothetically, a low level of acetylation of the NY‐ESO1−259/−126 gene could be explicated by an important recruitment of histone deacetylase on this gene. To investigate this hypothesis, we performed ChIP experiments using antibodies directed against several HDAC: HDAC1, HDAC2, HDAC6, HDAC7, Sirt1 and Sirt2 i.e directed a panel of histone deacetylase proteins. As illustratedin Figure 2B, we observed that only HDAC1 is recruited on the −259/−126 region of the NY‐ESO1 gene in the U251 and M96 cells. To confirm the role of HDAC1 into the deacetylation of histone H3 associated with the −259/−126 region of the NY‐ESO1 gene, we have analyzed the levels of H3K56Ac and H3Ac in U251 cells treated with siRNA directed against HDAC1. As illustrated in the Figures 2C and S4, we observed that the siRNA‐induced down‐expression of HDAC1 promoted the increase of the H3K56Ac and H3Ac levels. Thus, this data confirm that HDAC1 plays a crucial role into the deacetylation of histone H3 associated with the −259/−126 region of the NY‐ESO1 gene.
Figure 2.
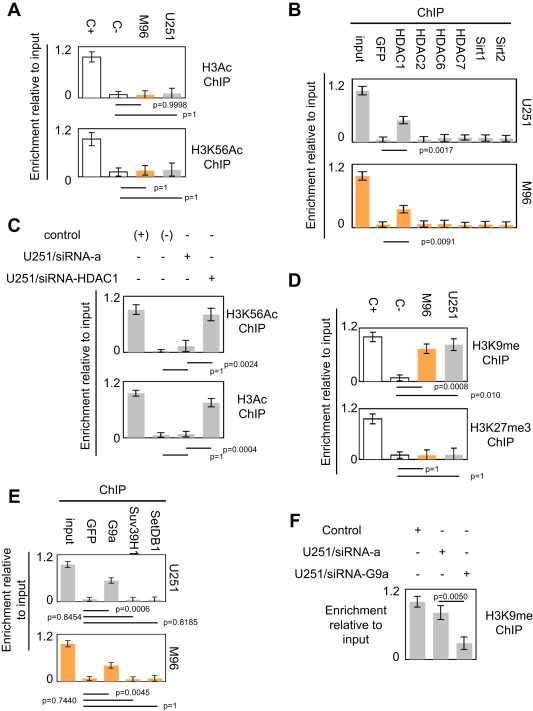
HDAC1 and G9a participate to the epigenetic regulation of the NY‐ESO1 gene. (A) Analyses of the degrees of acetylation of histone H3 included into −259/−126 regions of the NY‐ESO1 gene by acetylated chromatin immunoprecipitation method (AcChIP). Ct: positive control (Hela acid extract, Active Motif, France). (B) Identification by chromatin immunoprecipitation (ChIP) of the HDAC recruited on the −259/−126 regions of the NY‐ESO1 gene. Only the relative enrichment obtained with the antibody directed against HDAC1 is significantly superior to the enrichment obtained with the GFP antibody (p < 0.05, t test). (C) Impact of the siRNA‐induced down‐regulation of HDAC1 on the acetylation status of histone H3 associated with the −259/−126 region of the NY‐ESO1 gene. siRNA‐A is an aspecific siRNA used in control according to the manufacturer's instructions (Tebu‐Bio, Santa Cruz, France). (D) The degree of methylation of the lysine 9 of the histone H3 (K9H3) included into −259/−126 regions of the NY‐ESO1 gene was determined by K9H3 chromatin immunoprecipitation method (K9H3ChIP). Ct: positive control (Hela acid extract, Active Motif, France). (E) Identification by chromatin immunoprecipitation (ChIP) of the histone methylases recruited on the −259/−126 regions of the NY‐ESO1 gene. (C) Impact of the siRNA‐induced down‐regulation of G9a on the methylation status of histone H3 associated with the −259/−126 region of the NY‐ESO1 gene. siRNA‐A is an aspecific siRNA used in control according to the manufacturer's instruction (Tebu‐Bio, Santa Cruz, France).
We then complemented the epigenetic characterization of the −259/−126 region of the NY‐ESO1 gene by analyzing the degrees of the lysine 9 histone H3 methylation (H3K9me) and lysine 27 histone H3 trimethylation (H3K27me3). After immunoprecipitation of chromatin, PCR amplification indicated that histone H3 associated with the −259/−126 region of the NY‐ESO1 gene were methylated on lysine9 and unmethylated on lysine 27 in the U251 and M96 cells (Figure 2D). ChIP experiments realized with antibodies directed against G9a, SUV39H1 and SetDB1, three histone methyltransferases, revealed that G9a is co‐recruited on the −259/−126 region of the NY‐ESO1 gene, but not SUV39H1 and SetDB1 (Figure 2E). To confirm the role of G9a into the methylation of histone H3 associated with the −259/−126 region of the NY‐ESO1 gene, we have analyzed the level of H3K9me in U251 cells treated with siRNA directed against G9a. As illustrated in the Figures 2F and S5, we observed that the siRNA‐induced down‐expression of G9a promoted the decrease of H3K9me. Thus, this data confirm that G9a plays a crucial role into the methylation of histone H3 associated with the −259/−126 region of the NY‐ESO1 gene.
In conclusion, all these results suggest that HDAC1, G9a, Dnmt1 and Dnmt3b work, in concert, to promote the epigenetic regulation of the NY‐ESO1 gene.
3.2. Dnmt1/G9a, Dnmt3b/HDAC1, and HDAC1 work in multiprotein complexes
The HDAC1, G9a, Dnmt1 and Dnmt3b proteins being crucial actors of the epigenetic regulation of the NY‐ESO1 gene, we analyzed the impact of their siRNA‐induced down‐regulation on the level of NY‐ESO1mRNA expression. Strikingly, qPCR analyses revealed that the individual down‐regulation of HDAC1, G9a, Dnmt1 or Dnmt3b were not sufficient to promote the NY‐ESO1mRNA expression (Figure 3A). Based on these results, we then hypothesized that the HDAC1, G9a, Dnmt1 and Dnmt3b proteins cooperate to epigenetically regulate the expression of the NY‐ESO1 gene. Thus, we performed sequential ChIP experiments (re‐ChIP) to determine whether some of these proteins were simultaneously co‐recruited on the −259/−126 region of the NY‐ESO1 gene.
Figure 3.
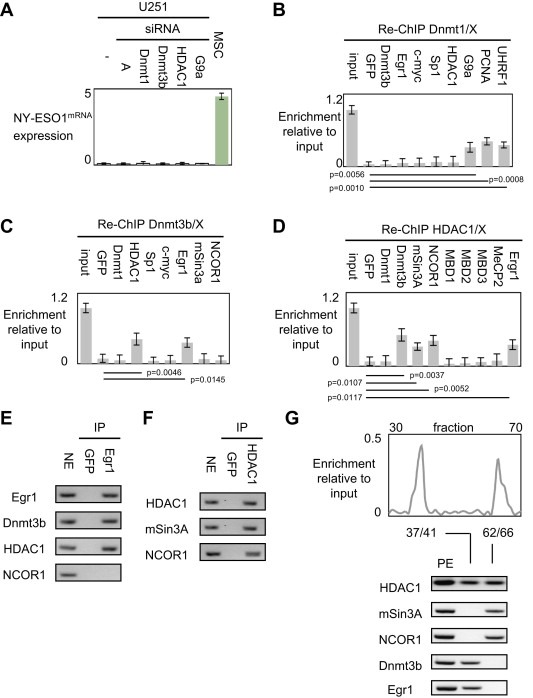
Identification of the actors of the epigenetic regulation of the NY‐ESO1 gene. (A) Impact of siRNA targeting Dnmt1, Dnmt3b, HDAC1 and G9a on the expression level of NY‐ESO1mRNA. None siRNA treatment induced the NY‐ESO1mRNA expression (all p > 0.05, t test). Mesenchymal stem cell (MSC) was used as positive control i.e. cells expressing NY‐ESO1mRNA. siRNA‐A is an aspecific siRNA used in control according to the manufacturer's instruction. (B–C–D). Sequential chromatin immunoprecipitation (reChIP) were performed by using indicated antibodies. GFP antibody was used as negative control for non‐specific ChIP. Only the relative enrichment obtained with the antibody directed against HDAC1 and Egr1 is significantly superior to the enrichment obtained with the GFP antibody (p < 0.05). (E–F–G). Immunoprecipitation (IP) monitoring the proteins interacting and co‐recruited with Dnmt1, Egr1 and HDAC1 respectively. NE: nuclear extract (positive controls for western blot analysis), GFP antibody was used to perform non‐specific IP (negative controls). Only the relative enrichment obtained with the antibody directed against Dnmt3b, mSin3A, NCOR1 and Egr1 is significantly superior to the enrichment obtained with the GFP antibody (p < 0.05, t test).
After a first ChIP realized with an antibody directed against Dnmt1, re‐ChIP experiments indicated that Dnmt1 is co‐recruited on the −259/−126 region of the NY‐ESO1 gene with PCNA, UHRF1 and G9a (i.e. with proteins composing the major complex promoting DNA methylation inheritance) but not with Dnmt3b and HDAC1 (Figure 3B), and not with transcription factors interacting with Dnmt1 potentially and having boxes of fixation into the −259/−126 region of the NY‐ESO1 gene (such as Sp1, Egr1 and/or c‐myc – Figure S6) (Bostick et al., 2007; Hervouet et al., 2010b; Sharif et al., 2007). Thus, these data indicated that Dnmt1, G9a, PCNA, UHRF1 were co‐recruited on the NY‐ESO1 gene. By taking into consideration the description of the Dnmt1‐PCNA‐UHRF1‐G9a complex, our data support the idea that the NY‐ESO1 gene is epigenetically regulated by the Dnmt1‐PCNA‐UHRF1‐G9a complex (Bostick et al., 2007; Sharif et al., 2007).
After a first ChIP realized with an antibody directed against Dnmt3b, re‐ChIP experiments showed that Dnmt3b is co‐recruited on the −259/−126 region of the NY‐ESO1 gene with HDAC1, and Egr1 but not with Dnmt1, c‐myc, Sp1, mSin3a or NCOR1 (Figure 3C). After a first ChIP realized with an antibody directed against HDAC1, re‐ChIP experiments revealed that HDAC1 is co‐recruited with Dnmt3b, mSin3a, NCOR1 and Egr1 but not with Dnmt1 or with proteins of the MBD family (Figure 3D).
In parallel with the reChIP experiments, we realized co‐immunoprecipitation (IP) analyses. Thus, our data indicated that G9a was co‐immunoprecipitated with Dnmt1, PCNA and UHRF1 (such as previously described), that Egr1 was co‐immunoprecipitated with Dnmt3b and HDAC1 but not with NCOR1, and that HDAC1 was co‐immunoprecipitated with Dnmt3b, NCOR1 and mSin3a (Figures 3E and F). Thus, these results suggest that the Dnmt3b‐HDAC1‐Egr1 and HDAC1‐mSin3A‐NCOR1 complexes participate to the epigenetic regulation of the NY‐ESO1 gene.
To confirm this idea, we finally coupled ChIP experiment with NaturePURE ™ affinity purification of protein complexes. Although EMSA is also a basic technical to analyze the proteins binding to known DNA oligonucleotide probes, we here used ChIP experiment with NaturePURE™ affinity purification in order to analyze the in vivo DNA/protein interaction (EMSA analyzes protein–DNA interactions in vitro) and to be independent of the use of antibody (In classical ChIP, ChIP grade antibody are required, and in EMSA, it's necessary to perform supershift assay with antibody to be certain of protein identity in a complex. On contrary, by coupling ChIP experiment with NaturePURE™ affinity purification of protein complexes, we do not need to use antibodies). For this application, we constructed the pcDNA3.2‐CapTEV/CT‐V5‐DEST‐HDAC1 vector according to the manufacturer's instructions (Invitrogen, France). After transfection of the U251 cells, cells are treated with formaldehyde to induce covalent protein:DNA crosslinks, lysed and sonicated to shear the chromatin into smaller fragments for purification by the NaturePURE™ affinity kit (Invitrogen, France). Using the NaturePURE™ affinity kit, the HDAC1‐including crosslinked complexes can be directly captured from the lysate through covalent binding. Stringent washing removes nonspecific proteins and DNA. After elution, crosslinking was reverted, DNA and proteins were purified. Thus, PCR analysis of the eluated fractions revealed the presence of the −259/−126 region of the NY‐ESO1 gene in fractions 37/41 and 62/66 (Figure 3G). After pooling these fractions, Western blot analyses showed the presence of HDAC1, Dnmt3b and Egr1 in fraction 37/41 and of HDAC1, NCOR1 and mSin3A in fraction 62/66 (Figure 3G).
Thus, all our experiments indicated that the multiprotein complexes, formed in part, by Dnmt1‐PCNA‐UHRF‐G9a, Dnmt3b‐HDAC1‐Egr1 and HDAC1‐mSin3A‐NCOR1 are the three most valuable players of the epigenetic regulation of the NY‐ESO1 gene.
3.3. Dynamic epigenetic regulation of the NY‐ESO1 gene
To determine the orchestration of the epigenetic regulation of the NY‐ESO1 gene by the Dnmt1‐PCNA‐UHRF1‐G9a, Dnmt3b‐HDAC1‐Egr1 and HDAC1‐mSin3A‐NCOR1 complexes, we performed a kinetic of ChIP experiments on G0/G1 synchronized cells (72 h of serum deprivation) initially treated with 2.5 μM α‐amanitin for 2 h (Figure 4A). ChIP experiments were realized with antibodies directed against 1) HDAC1 to monitor the dynamic recruitment of the Dnmt3b‐HDAC1‐Egr1 and HDAC1‐mSin3A‐NCOR1 complexes on the −259/−126 region of the NY‐ESO1 gene; 2) Dnmt3b to monitor the dynamic recruitment of the Dnmt3b‐HDAC1‐Egr1 complex on the −259/−126 region of the NY‐ESO1 gene; and 3) Dnmt1 to monitor the dynamic recruitment of the Dnmt1‐PCNA‐UHRF1‐G9a complex on the −259/−126 region of the NY‐ESO1 gene. Thus, we noted that the epigenetic regulation of the −259/−126 region of the NY‐ESO1 gene firstly included the recruitment of the HDAC1‐mSin3A‐NCOR1 complex, next the recruitment of the Dnmt3b‐HDAC1‐Egr1 complex, and finally the recruitment of the Dnmt1‐PCNA‐UHRF1‐G9a complex (Figure 4B). Thus, our data demonstrate that the epigenetic regulation of the −259/−126 region of the NY‐ESO1 gene is a dynamic process implicating 3 phases defined by the sequential recruitment of three different actors: HDAC1‐mSin3A‐NCOR1, Dnmt3b‐HDAC1‐Egr1, and Dnmt1‐PCNA‐UHRF1‐G9a and the promotion of the deacetylation of histone H3, the DNA methylation and the histone H3‐Lysine9 methylation.
Figure 4.
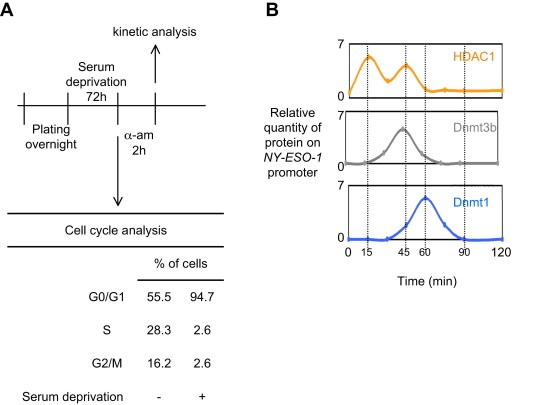
Orchestration of the epigenetic regulation of the NY‐ESO1 gene. (A) The kinetic of ChIP experiments was realized at indicated time after cell (U251) synchronization (72 h of serum deprivation) and α‐amanitin treatment. The amount of immunoprecipitated promoter were quantified by semi‐quantitative PCR and normalized to input according to the previous description (Hervouet et al., 2010b). Cell cycle phases were determined by using the NucleoCounter NC‐3000™ Kit (Chemometec, France). (B) Representation of the sequential epigenetic regulation of the NY‐ESO1 gene and of the effect of epigenetic drugs.
3.4. Impact of epigenetic drugs
Supported by the fact that the individual down‐regulation of the Dnmt1, Dnmt3b, HDAC1 or G9a proteins via siRNA strategies do not promote the NY‐ESO1mRNA expression, we decided to analyzed the effect of treatments combining or not “no‐specific” epigenetic drugs such as the 5‐aza‐2‐deoxycytidine (5aza) and/or valproic acid (VPA) drugs on the epigenetic marks regulating the NY‐ESO1mRNA expression. In addition of the study of the degree of DNA methylation, H3K56 acetylation, and H3K9 methylation, we also analyzed the degree of Sp1 recruitment on NY‐ESO1 gene since this gene includes Sp1 boxes and Sp1 transcriptionally regulates the NY‐ESO1mRNA expression (Kang et al., 2007).
Thus, we firstly observed that the VPA treatment of U251 cells was inefficient to promote the NY‐ESO1mRNA expression, despite the fact that this treatment induced a significant modification of the acetylation degree of Histone H3 (Figure 5A). Indeed, under this condition, NY‐ESO1 gene was still methylated, the degree of H3K9 methylation remains unchanged, and Sp1 was not recruited on NY‐ESO1 gene.
Figure 5.
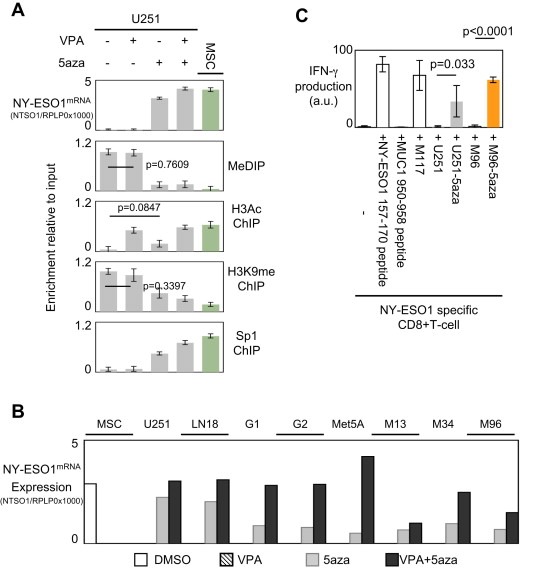
Impact of epigenetic drugs on the epigenetic regulation of the NY‐ESO1 gene and on the expression of this gene. (A) Impact of the valproic acid (VPA, 0.5 mM for 7 consecutive days) and/or 5aza‐deoxycytidine (5aza, 1 μM for 7 consecutive days) treatments on the NY‐ESO1mRNA expression and on the degree of DNA methylation, H3 acetylation and of K9H3me, of the −259/−126 region of the NY‐ESO1 gene. (B) Impact of the VPA, 5aza and VPA/5aza treatments on the NY‐ESO1mRNA expression in a panel of glioma and mesothelioma cells. (C) Recognition of 5‐aza‐2′‐deoxycytidine (5aza) treated, human leukocyte antigen (HLA)‐A*0201+ tumor cell lines by a HLA‐A*0201/NY‐ESO‐1(157–165)‐specific CD8+ T‐cell clone. IFN‐γ production by the NY‐ESO‐1‐specific CD8+ T‐cell clone in response to NY‐ESO‐1 peptide 157–170 or with an irrelevant peptide (MUC‐1 peptide 950–958), the autologous melanoma cell line (M117), a HLA‐A*0201+ glioblastoma cell line (U251) and a HLA‐A*0201+ MPM cell line (M96), treated or not with 0.5 mM 5‐aza.
Secondly, we noted that the 5aza treatment promoted the NY‐ESO1mRNA expression. Our data associated this situation with the decrease of the DNA methylation region of NY‐ESO1 gene, of the H3K9 methylation degree, a weak but significant increase of the H3 acetylation and with the recruitment of Sp1 on NY‐ESO1 gene (Figure 5A). More interestingly, we observed that the VPA+5aza treatment increased more the NY‐ESO1mRNA expression than the 5aza treatment. This point could be explaining by the fact that VPA+5aza treatment is more efficient than the 5aza treatment to promote the H3 acetylation. Thus, under this treatment, the recruitment of Sp1 was increased in comparison with the quantity of Sp1 binding NY‐ESO1 gene when U251 cells were treated with 5aza only (Figure 5A).
To check the efficiency of the VPA/5aza co‐treatment to promote the NY‐ESO1mRNA expression, we treated a panel of glioma and mesothelioma cells and primary cells. As expected, we observed that VPA/5aza was the most efficient treatment promoting the NY‐ESO1mRNA expression into the considered cells (Figure 5B).
Taken together, these data indicate that VPA/5aza is the most efficient treatment promoting the NY‐ESO1mRNA expression, but that the 5aza treatment is sufficient to promote NY‐ESO1mRNA expression.
Finally, we determined whether the expression of NY‐ESO1mRNA in U251 and M96 cells is associated with the expression of the NY‐ESO1 antigen. For this purpose, we tested the presence of the NY‐ESO1 antigen at the surface of the U251 and M96 cells by analyzing the activation state of a NY‐ESO‐1 specific CD8+ T‐cell clone in presence of U251 and M96 cells treated or not by 5aza, such as already described (Leclercq et al., 2011). Thus, in addition to evaluating the NY‐ESO1 expression at protein level, our study also evaluated the efficient presentation of the NY‐ESO1 protein at the surface of the cancer cells (U251 and M96). The activation state of a NY‐ESO1 specific CD8+ T‐cell clone in presence of U251 and M96 cells treated or not by 5aza is analyzed by measuring the IFN‐γ production by the CD8+T‐cell clone. IFN‐γ production by the clone was observed by intracytoplasmic staining. Clone specificity was demonstrated by incubating T‐cell clone with NY‐ESO1 peptide 157–170 or with an irrelevant peptide (MUC‐1 950–958). No IFN‐γ production was observed with T‐cell clone alone or incubating with irrelevant peptide (Figure 5C). Incubation of T‐cell clone with NY‐ESO1 peptide induced production of IFN‐γ by T‐cell clone. No T‐cell clone response was observed with untreated tumor cells. IFN‐γ production by the clone was observed only in response to 5‐aza treated cells. The level of activation of the clone in response to 5‐aza treated tumor cell lines and its level of recognition of the control autologous M117 melanoma cell line were then compared. All 5aza treated tumor cell lines were recognized, by the clone, similarly than the level of recognition of M117. These results confirmed that 5aza treatment is sufficient to induce the expression of enough NY‐ESO1 in cells to allow processing and presentation of this antigen on their surface.
4. Discussion
During the last decade, epigenetic mechanisms such as DNA methylation, histone acetylation and methylation processes, have been described as playing a crucial role into the gene expression. In the present report, we illustrate this dogma by describing the sequential epigenetic regulation of the NY‐ESO1 gene in which several proteins playing epigenetic function cooperate to govern the NY‐ESO1 gene expression (Figure 6). Indeed, our data clearly demonstrate that histone deacetylase HDAC1, two DNA methyltransferases Dnmt3b and Dnmt1, and histone methyltransferase G9a were implicated into the epigenetic regulation of the NY‐ESO1 gene. In addition to described that these proteins cooperate to epigenetically regulate the NY‐ESO1 gene via the formation of multiprotein complexes including an HDAC and a Dnmt (HDAC1/Dnmt3b) or a Dnmt and a histone methyltransferase (Dnmt1/G9a), our data described that this cooperation occurs into a sequential process. To date, there are only some other similar and recently published observations dissecting the existence of sequential or cyclical epigenetic mechanism. Thus, as in our case, the pS2 gene promoter is also regulated by dynamic epigenetic mechanisms (Métivier et al., 2008). However, in this case, the epigenetic regulation of the pS2 gene promoter only concerns its status of DNA methylation; while in our case, the epigenetic regulation of the NY‐ESO1 gene promoter is an example of the cooperation existing between the different machineries of DNA methylation (via the implication of the major complex promoting the DNA methylation inheritance i.e. the Dnmt1‐PCNA‐UHRF1‐G9a complex and a complex of de novo DNA methylation, the Dnmt3b‐HDAC1‐Egr1 complex), of histone deacetylation (through the implication of the Dnmt3b‐HDAC1‐Egr1 and HDAC1‐mSin3A‐NCOR1 complexes), and of histone methylation (via the G9a implication into the Dnmt1‐PCNA‐UHRF1‐G9a complex). More particularly and consistently with our previous results, the description of the molecular mechanisms of epigenetic regulation of the NY‐ESO1 gene promoter underlines the fact that DNA methylation could result from the cooperation between a targeted and an untargeted mechanism of DNA methylation represented by the Dnmt3b‐HDAC1‐Egr1 and the Dnmt1‐PCNA‐UHRF1‐G9a complexes, respectively (Hervouet et al., 2009, 2010b). Besides, the role of anchorage of Egr1 for the recruitment of Dnmt3b on the NY‐ESO1 gene was here confirmed by the fact that the Egr1 down‐regulation (by using siRNA approach) abrogated the recruitment of Dnmt3b on the NY‐ESO1 gene (Figure S7).
Figure 6.
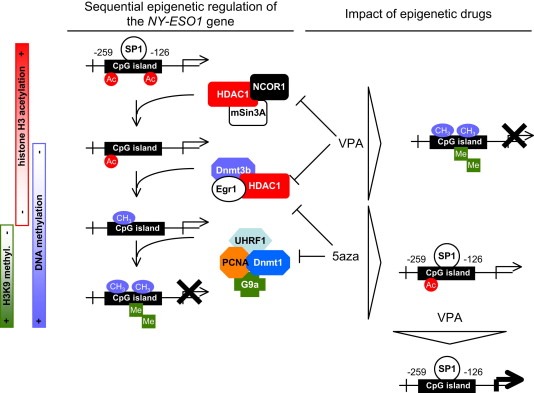
Schematic representation of the epigenetic regulation of the NY‐ESO1 gene.
Concerning the absence of detection of H3K27me on the NY‐ESO1 −259/−126 gene, our data confirm this point since neither EZH2 nor KDM6a (ie. a couple of histone methyltransferase and demethylase governing the H3K27me) (according to Greer and Shi, 2012) were recruited on the NY‐ESO1 −259/−126 gene (Greer and Shi, 2012) (Figure S8). Concerning the detection of H3K9me on the NY‐ESO1 −259/−126 gene, experiments of kinetic of ChIP indicated that KDM4A (but not LSD1, another histone demethylase) was recruited on the NY‐ESO1 −259/−126 gene (Figure S9). More interestingly, we noted that this recruitment is short and precedes the G9a recruitment. Thus, this last point explains why NY‐ESO1 −259/−126 appears as being H3K9me positive in our cells. The recruitment of HDAC1 on the NY‐ESO1 −259/−126 gene to promote the weak acetylation of this gene asks the question of the actor of acetylation of histones associated with this gene and the question of the time during which histones associated with the NY‐ESO1 −259/−126 gene is acetylated. To reply at these questions, we have analyzed the kinetic of H3Ac of histones associated with the NY‐ESO1 −259/−126 gene and the kinetic of recruitment of GCN5 and p300, two histone acetylases, on the NY‐ESO1 −259/−126 gene. As illustrated by the Figure S10, we noted that the H3Ac briefly increased at t = 10 min and t = 35 min i.e. when GCN5 (but not p300) is recruited on the gene. It also appears that these 2 periods of maximal recruitment of GCN5 above the period of HDAC1 recruitment. So this explains why NY‐ESO1 appears as very weakly acetylated. More generally, these data indicated that G9a/KDM4A and GCN5/HDAC1 are two couples of histone methyltransferase/demethylase and of histone acetylase/deacetylase playing a crucial role into the level of H3K9me and of H3Ac. In addition, the fact that HDAC1 play a role in the H3K56 could open a new way of regulation of H3K56Ac level since this epigenetic mark is usually regulated by HDAC1 in response to DNA damage or during the embryonic development (Dovey et al., 2010; Miller et al., 2010). This direct consideration or and the indirect role of HDAC1 and/or of the global level of H3 acetylation in the acetylation level of H3K56 (interplay between different epigenetic marks and proteins) are two ongoing subjects in the lab.
In terms of therapeutic implications, the identification of the molecular machinery of the epigenetic regulation of the NY‐ESO1 gene opens news promising and potent targeted treatment including epigenetics‐based therapy. Indeed, several reports mentioned the fact that the description of DNA methylation mechanisms governing the silencing of tumor suppressor genes and/or cancer/testis antigen could be permit to design successful therapeutic protocol anti‐glioma by using DNA hypomethylating agents or Dnmt inhibitors (Mund et al., 2005; Natsume et al., 2010). Thus, our data could serve as base for the development of new anti‐glioma and anti‐mesothelioma therapeutic protocols using epigenetics‐based therapy. Besides, we have recently published that 5‐aza‐2′‐deoxycytidine/valproate combination induces CTL response against mesothelioma (Leclercq et al., 2011). In addition, pre‐treatment of cells with epigenetic drugs into a vaccine therapy seems to be an interesting track since immunotherapeutic approaches of which vaccine therapy were developed in glioma and that we recently published that vaccination with epigenetically treated mesothelioma cells induces immunization and blocks tumor growth. (Guillot et al., 2011; Okada et al., 2009).
Finally, the restoration of the NY‐ESO1 gene expression (and the immune response associated with this expression) in glioma by epigenetic drugs can suggest that this treatment can induce the expression, in glioma, of other genes than NY‐ESO1 expressed in MSC. Thus, re‐expressing these genes can potentially promote the acquisition of mesenchymal stem‐like characters by glioma treated by epigenetic drugs. In other terms, our data ask the question to know if the 5aza/VPA treatment can promote the acquisition of mesenchymal stem‐like characters in GBM. Replying to this question appears as fundamental in a clinical point of view since, according to Verhaak et al.'s classification system, mesenchymal GBM were more likely to respond to radiotherapy than to first‐line chemotherapy (whereas classical GBMs were more likely to respond to first‐line chemotherapy than to radiotherapy). Thus, glioma presenting an unmethylated MGMT gene i.e. a glioma devoid of the presence of the most predictive marker of response to chemotherapy using temozolomide could be sensitive to other therapeutic approaches after its treatment by VPA/5aza treatment (Hegi et al., 2005). In a point of view of fundamental biology, replying to this question would return to identify molecular actor governing the acquisition of mesenchymal characters by tumor cells such as glioma. These points are two ongoing subjects in our lab.
5. Conclusion
To summarize, our data describe the actors of the epigenetic regulation of NY‐ESO1 gene, open promising antitumor of VPA/5aza treatment on mesothelioma and glioma (such as the one already described on rhabdomyosarcoma and medulloblastoma), and ask the question of the epigenetic role on the acquisition of mesenchymal characters by glioma cells (Ecke et al., 2009).
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Acknowledgments
This work was supported by grants from the Ligue contre le cancer Grand‐Ouest (et plus particulièrement par les comités départementaux de Loire‐Atlantique, de Vendée, d'Ile et Vilaine, Morbihan et Sarthe) and from the Association pour la Recherche et les Soins sur le mésothéliome ARSMESO44.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2012.11.004.
Cartron Pierre-François, Blanquart Christophe, Hervouet Eric, Gregoire Marc, Vallette François M., (2013), HDAC1‐mSin3a‐NCOR1, Dnmt3b‐HDAC1‐Egr1 and Dnmt1‐PCNA‐UHRF1‐G9a regulate the NY‐ESO1 gene expression, Molecular Oncology, 7, doi: 10.1016/j.molonc.2012.11.004.
References
- Bostick, M. , Kim, J. , Estève, P. , Clark, A. , Pradhan, S. , Jacobsen, S. , 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 27, 2187–2197. [DOI] [PubMed] [Google Scholar]
- Cronwright, G. , Le Blanc, K. , Götherström, C. , Darcy, P. , Ehnman, M. , Brodin, B. , 2005. Cancer/testis antigen expression in human mesenchymal stem cells: down-regulation of SSX impairs cell migration and matrix metalloproteinase 2 expression. Cancer Res. 65, 2207–2215. [DOI] [PubMed] [Google Scholar]
- Dovey, O. , Foster, C. , Cowley, S. , 2010. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecke, I. , Petry, F. , Rosenberger, A. , Tauber, S. , Mönkemeyer, S. , Hess, I. , Dullin, C. , Kimmina, S. , Pirngruber, J. , Johnsen, S. , Uhmann, A. , Nitzki, F. , Wojnowski, L. , Schulz-Schaeffer, W. , Witt, O. , Hahn, H. , 2009. Antitumor effects of a combined 5-aza-2'deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res. 69, 887–895. [DOI] [PubMed] [Google Scholar]
- Greer, E. , Shi, Y. , 2012. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot, F. , Boutin, B. , Blanquart, C. , Fonteneau, J. , Robard, M. , Grégoire, M. , Pouliquen, D. , 2011. Vaccination with epigenetically treated mesothelioma cells induces immunisation and blocks tumour growth. Vaccine 29, 5534–5543. [DOI] [PubMed] [Google Scholar]
- Hegi, M. , Diserens, A. , Gorlia, T. , Hamou, M. , de Tribolet, N. , Weller, M. , Kros, J. , Hainfellner, J. , Mason, W. , Mariani, L. , Bromberg, J. , Hau, P. , Mirimanoff, R. , Cairncross, J. , Janzer, R. , Stupp, R. , 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 991–1003. [DOI] [PubMed] [Google Scholar]
- Hervouet, E. , Vallette, F. , Cartron, P. , 2010. Impact of the DNA methyltransferases expression on the methylation status of apoptosis-associated genes in glioblastoma multiforme. Cell Death Dis. 1, 10.1038/cddis.2009.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet, E. , Vallette, F.M. , Cartron, P.F. , 2009. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics 4, [DOI] [PubMed] [Google Scholar]
- Hervouet, E. , Vallette, F.M. , Cartron, P.F. , 2010. Dnmt1/transcription factor interactions: an alternative mechanism of DNA methylation inheritance. Genes & Cancer 1, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S. , Link, P. , Karpf, A. , 2006. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene 25, 6975–6985. [DOI] [PubMed] [Google Scholar]
- Kang, Y. , Hong, J. , Chen, G. , Nguyen, D. , Schrump, D. , 2007. Dynamic transcriptional regulatory complexes including BORIS, CTCF and Sp1 modulate NY-ESO-1 expression in lung cancer cells. Oncogene 26, 4394–4403. [DOI] [PubMed] [Google Scholar]
- Karpf, A. , 2006. A potential role for epigenetic modulatory drugs in the enhancement of cancer/germ-line antigen vaccine efficacy. Epigenetics 1, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq, S. , Gueugnon, F. , Boutin, B. , Guillot, F. , Blanquart, C. , Rogel, A. , Padieu, M. , Pouliquen, D. , Fonteneau, J. , Grégoire, M. , 2011. 5-aza-2'-deoxycytidine/valproate combination induces CTL response against mesothelioma. Eur. Respir. J. 5, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Maio, M. , Coral, S. , Fratta, E. , Altomonte, M. , Sigalotti, L. , 2003. Epigenetic targets for immune intervention in human malignancies. Oncogene 22, 6484–6488. [DOI] [PubMed] [Google Scholar]
- Métivier, R. , Gallais, R. , Tiffoche, C. , Le Péron, C. , Jurkowska, R. , Carmouche, R. , Ibberson, D. , Barath, P. , Demay, F. , Reid, G. , Benes, V. , Jeltsch, A. , Gannon, F. , Salbert, G. , 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50. [DOI] [PubMed] [Google Scholar]
- Miller, K. , Tjeertes, J. , Coates, J. , Legube, G. , Polo, S. , Britton, S. , Jackson, S. , 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA non homologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund, C. , Brueckner, B. , Lyko, F. , 2005. Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: basic concepts and clinical applications. Epigenetics 1, 7–13. [DOI] [PubMed] [Google Scholar]
- Natsume, A. , Kondo, Y. , Ito, M. , Motomura, K. , Wakabayashi, T. , Yoshida, J. , 2010. Epigenetic aberrations and therapeutic implications in gliomas. Cancer Sci. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oi, S. , Natsume, A. , Ito, M. , Kondo, Y. , Shimato, S. , Maeda, Y. , Saito, K. , Wakabayashi, T. , 2009. Synergistic induction of NY-ESO-1 antigen expression by a novel histone deacetylase inhibitor, valproic acid, with 5-aza-2'-deoxycytidine in glioma cells. J. Neurooncol. 92, 15–22. [DOI] [PubMed] [Google Scholar]
- Okada, H. , Kohanbash, G. , Zhu, X. , Kastenhuber, E. , Hoji, A. , Ueda, R. , Fujita, M. , 2009. Immunotherapeutic approaches for glioma. Crit. Rev. Immunol. 29, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin, U. , Koslowski, M. , Türeci, O. , Eberle, T. , Zwick, C. , Romeike, B. , Moringlane, J. , Schwechheimer, K. , Feiden, W. , Pfreundschuh, M. , 2000. Expression of cancer testis genes in human brain tumors. Clin. Cancer Res. 6, 3916–3922. [PubMed] [Google Scholar]
- Sharif, J. , Muto, M. , Takebayashi, S. , Suetake, I. , Iwamatsu, A. , Endo, T. , Shinga, J. , Mizutani-Koseki, Y. , Toyoda, T. , Okamura, K. , Tajima, S. , Mitsuya, K. , Okano, M. , Koseki, H. , 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. [DOI] [PubMed] [Google Scholar]
- Sigalotti, L. , Coral, S. , Altomonte, M. , Natali, L. , Gaudino, G. , Cacciotti, P. , Libener, R. , Colizzi, F. , Vianale, G. , Martini, F. , Tognon, M. , Jungbluth, A. , Cebon, J. , Maraskovsky, E. , Mutti, L. , Maio, M. , 2002. Cancer testis antigens expression in mesothelioma: role of DNA methylation and bio immunotherapeutic implications. Br. J. Cancer 86, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalotti, L. , Coral, S. , Fratta, E. , Lamaj, E. , Danielli, R. , Di Giacomo, A. , Altomonte, M. , Maio, M. , 2005. Epigenetic modulation of solid tumors as a novel approach for cancer immunotherapy. Semin. Oncol. 32, 473–478. [DOI] [PubMed] [Google Scholar]
- Vaissière, T. , Sawan, C. , Herceg, Z. , 2008. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 659, 40–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data


