Abstract
miR‐21 expression in cancer tissue has been reported to be associated with the clinical outcome and activity of gemcitabine in pancreatic cancer. However, resection is possible in only a minority of patients due to the advanced stages often present at the time of diagnosis, and safely obtaining sufficient quantities of pancreatic tumor tissue for molecular analysis is difficult at the unresectable stages. In this study, we investigated whether the serum level of miR‐21 could be used as a predictor of chemosensitivity. We tested the levels of serum miR‐21 in a cohort of 177 cases of advanced pancreatic cancer who received gemcitabine‐based palliative chemotherapy. We found that a high level of miR‐21 in the serum was significantly correlated with a shortened time‐to‐progression (TTP) and a lower overall survival (OS). The serum miR‐21 level was an independent prognostic factor for both the TTP and the OS (HR 1.920; 95% CI, 1.274–2.903, p = 0.002 for TTP and HR 1.705; 95% CI, 1.147–2.535, p = 0.008 for OS). The results from a functional study showed that gemcitabine exposure down‐regulated miR‐21 expression and up‐regulated FasL expression. The increased FasL expression following gemcitabine treatment induced cancer cell apoptosis, whereas the ectopic expression of miR‐21 partially protected the cancer cells from gemcitabine‐induced apoptosis. Additionally, we confirmed that FasL was a direct target of miR‐21. Therefore, the serum level of miR‐21 may serve as a predictor of chemosensitivity in advanced pancreatic cancer. Additionally, we identified a new mechanism of chemoresistance mediated by the effects of miR‐21 on the FasL/Fas pathway.
Keywords: Serum miR-21, Advanced pancreatic cancer, Chemosensitivity, FasL
Highlights
-
►
Serum miR‐21 level predicts chemosensitivity in advanced pancreatic cancer.
-
►
High level of serum miR‐21 correlates with shortened time‐to‐progression.
-
►
miR‐21 confers chemoresistance of pancreatic cancer cells to gemcitabine.
-
►
FasL is a direct target of miR‐21.
Abbreviations
- FasL
Fas ligand
- PDAC
pancreatic duct adenocarcinoma
- TTP
time to progression
- OS
overall survival
- microRNA
miRNA
- qPCR
quantitative real-time PCR
- siRNA
small-interfering RNA
- 3′UTR
3′ untranslated region
1. Introduction
Pancreatic cancer is the fourth leading cause of cancer‐related death and has an overall 5‐year survival rate of less than 5% (Siegel et al., 2012). Surgical resection represents the only curative treatment option, but more than 80% of patients are diagnosed at late, inoperable stages (Li et al., 2004). Despite advances in clinical management, the high mortality rate of pancreatic cancer, which is almost equal to its incidence, remains largely unchanged (Siegel et al., 2012). Therefore, a great need exists for novel targets for the diagnosis and therapy of pancreatic cancer.
Gemcitabine has been approved as a first line of treatment for locally advanced and metastatic pancreatic cancer (Burris et al., 1997). However, most pancreatic cancer patients treated with gemcitabine do not have an objective response to treatment, and only a minority of patients obtain a stabilization of the disease or partial response to treatment (Burris et al., 1997). The mechanisms regulating pancreatic cancer cell growth and resistance to chemotherapy are poorly understood. In recent years, considerable interest has been placed in understanding the role of microRNAs in cancer. MicroRNAs (miRNAs) are a class of small, non‐coding, ∼22‐nt RNA molecules with functions in the post‐transcriptional regulation of gene expression. These molecules have been shown to bind to partially complementary sequences in mRNAs and down‐regulate protein expression by targeting the mRNAs for degradation and/or inhibiting mRNA translation (Kim et al., 2009). Growing evidence suggests that aberrant miRNA expression contributes to the development and progression of pancreatic cancer (Bloomston et al., 2007; Mardin and Mees, 2009; Szafranska et al., 2007). In addition, at present, the most promising applications of miRNAs may be for the prognosis prediction and response modification for anti‐tumor therapies in cancer (Hummel et al., 2010). For example, miR‐93 (Fu et al., 2012), miR‐203 (Li et al., 2011) and miR‐200 (Lee et al., 2011) have been shown to be strongly associated with the response to cisplatin chemoresistance. The miRNA expression signature is correlated with the time to progression after chemotherapy (Kim et al., 2011). In addition, several miRNAs have been shown to influence the sensitivity of cancer cells to chemotherapy. Inhibiting endogenous miR‐140 expression with a locked nucleic acid‐modified anti‐miRNA partially sensitized chemotherapy‐resistant colon cancer cells to 5‐FU treatment (Song et al., 2009), whereas the restoration of miR‐128 expression sensitized breast cancer cells to the proapoptotic and DNA‐damaging effects of doxorubicin (Zhu et al., 2011).
The expression of miR‐21 has been shown to be involved in the progression of several types of cancers (Yang et al., 2011; Kwak et al., 2011), including pancreatic cancer (Bloomston et al., 2007; Szafranska et al., 2007; Nagao et al., 2011), and the up‐regulation of miR‐21 expression has been shown to correlate with a lower cancer survival rate (Jamieson et al., 2011; Dillhoff et al., 2008). Recent studies also demonstrated that miR‐21 modulates the chemosensitivity of cancer cells primarily by targeting PTEN or PDCD4 (Ali et al., 2010; Moriyama et al., 2009; Giovannetti et al., 2010; Tomimaru et al., 2010). In addition, cancer tissue miR‐21 has been shown to be useful as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer (Hwang et al., 2010).
Because no clinical means of predicting whether patients will benefit from gemcitabine treatment in advanced/inoperable pancreatic cancer exist, in this study, we investigate whether the serum level of miR‐21 can be used as a predictor of chemosensitivity in advanced pancreatic cancer. In addition, we explore the effects of miR‐21 expression on gemcitabine sensitivity in pancreatic cancer cells and identify the target gene involved in miR‐21‐mediated chemoresistance.
2. Materials and methods
2.1. Patient characteristics and clinical features
This study included two cohorts of patients. In cohort 1, human pancreatic cancer samples were collected from 65 patients after surgical resection at the Huashan Hospital, Fudan University, Shanghai, China, from January 2003 to December 2005. All of the patients underwent a macroscopic curative resection. The resected primary tumors and lymph nodes were histologically examined with hematoxylin and eosin staining using the tumor‐node‐metastasis classification system. Histologically, all of the tumors were classified as invasive ductal adenocarcinomas. None of the patients had received neoadjuvant radiochemotherapy. Freshly frozen and/or formalin‐fixed paraffin‐embedded (FFPE) samples were obtained for the miR‐21 expression analysis. The characteristics of the patients in cohort 1 are shown in Supplementary Table 1. In cohort 2, a total of 177 patients with pathologically confirmed locally advanced or metastatic pancreatic adenocarcinoma were retrospectively recruited between January 2009 and December 2010 from the Fudan University Shanghai Cancer Center, Shanghai, China. The criteria for locally advanced disease included tumor invasion to the celiac trunk or superior mesenteric artery, or both, which corresponded to clinical stage III according to the International Union Against Cancer (6th edition). Standard radiological studies included contrast‐enhanced abdominal CT, magnetic resonance imaging (MRI) and/or MR‐cholangiopancreaticography (MRCP). All of the patients received gemcitabine‐based palliative chemotherapy. The characteristics of the patients in cohort 2 are shown in Supplementary Table 2. Each participant donated 5 ml of venous blood before initiating chemotherapy. The blood was collected in tubes with an EDTA anticoagulant and was separated into the serum and cellular fractions within 4 h after the sample collection. The serum was then stored immediately at −80 °C until analysis. This study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center, Shanghai, China, and written informed consent was obtained from each participant, in accordance with the institutional guidelines of our hospital.
2.2. Cell lines and mice
The human pancreatic cancer cell lines PANC‐1 and BxPC3 were obtained from the American Type Culture Collection and grown in complete growth medium according to the recommendations of the supplier. The two cultured cell lines were maintained in a humidified 5% CO2 atmosphere at 37 °C. Both the cell lines were regularly authenticated by examining their morphology and testing for the absence of mycoplasma contamination (MycoAlert, Lonza, Rockland, ME, USA). Four to 6‐week‐old female BALB/c‐nu/nu nude mice were obtained from the Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China) and housed in laminar flow cabinets under specific pathogen‐free conditions with food and water ad libitum. All of the mouse experiments were conducted in accordance with the guidelines of the NIH for the care and use of laboratory animals. The study protocol was also approved by the Committee on the Use of Live Animals in Teaching and Research, Fudan University, Shanghai, China.
2.3. RNA isolation
For the freshly frozen tissue samples or cell lines, total RNA was extracted using TRIzol® Reagent (Invitrogen, San Diego, CA), and the small‐sized RNAs were isolated using a mirVana miRNA Isolation Kit (Ambion, Inc.), both according to the manufacturers' instructions. For the FFPE samples, the total RNA was isolated using a RecoverAll Total Nucleic Acid Isolation Kit (Ambion) according to the manufacturer's instructions. For the serum microRNA isolation, 5 ml of whole blood was obtained from each patient. The plasma was extracted from the serum by centrifuging the whole blood at 3000 rpm for 10 min followed by a 15‐min high‐speed centrifugation at 12,000 × g to completely remove the cellular debris. The supernatant from the serum was then frozen in separate aliquots at −80 °C. Total RNA was extracted from 500 μl of serum using a mirVana PARIS Kit (Ambion, Inc.) according to the manufacturer's instructions for enrichment procedure for small RNAs. To allow for normalization of sample‐to‐sample variation in RNA isolation, synthetic Caenorhabditis elegans miRNA cel‐miR‐54 (purchased as a custom RNA oligonucleotide from Qiagen) was added (50 pmol/l in a 5‐μl total volume) to each denatured sample. The use of cel‐miR‐54 has previously been shown not to affect the detection of human miRNAs (Arroyo et al., 2011).
2.4. qRT‐PCR assays
Real‐time PCR measurements with specific TaqMan primers for miR‐21 were performed to determine the mean C T value for each sample. Briefly, 2 μl aliquot of enriched small RNAs from plasma samples were reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, San Diego, CA) according to manufacturer's protocol. Then 2 μl of the cDNA solution was used as template for the PCR stage. PCR reaction was performed using 10 μl TaqMan Universal Master Mix (2×) (Applied Biosystems, USA), 1 μl gene‐specific primers/probe, and nuclease‐free H2O in a final volume of 20 μl. No‐template controls for both RT step and PCR step were included to ensure target specific amplification. Quantitative PCR was run on a 7500 HT quantitative PCR machine (Applied Biosystems, USA). The TaqMan primers for miR‐21 and U6 are shown in Supplementary Table 4. The C T values of the different samples were compared using the ΔΔC T method as described previously (Giovannetti et al., 2010). U6 RNA served as an internal reference for the tissues and cell lines, and the synthetic cel‐miR‐54 was used as a control for the serum miR‐21 expression. Semi‐quantitative real‐time RT‐PCR using SYBR Green I was performed to compare the relative expression levels of specific mRNAs as described previously (Lekanne Deprez et al., 2002).
2.5. Oligonucleotide transfection
For these experiments, miRNA duplexes corresponding to the miR‐21 sequence were designed as previously described (Lim et al., 2005). The control RNA duplexes (termed NC for “negative control”) for the miRNA mimics and the small‐interfering RNAs (siRNAs) were not homologous to any known human gene sequence. The siRNAs were designed to target the human FasL transcript (GenBank Access No. NM_000639). For the in vivo tumorigenicity assay, all of the pyrimidine nucleotides in the NC or miR‐21 duplex were replaced by the corresponding 2′‐O‐methyl analogs to improve RNA stability as previously described (Su et al., 2009). All of the RNA oligoribonucleotides were purchased from Genepharma (Shanghai, China). The sequences were as follows: miR‐21 mimics, 5′‐UAGCUUAUCAGACUGAUGUUGA‐3′ and 5′‐AACAUCAGUCUGAUAAGCUAUU‐3′; NC, 5′‐UUCUCCGAACGUGUCACGUTT‐3′ and 5′‐ACGUGACACGUUCGGAGAATT‐3′; FasL siRNA, 5′‐GGTGGCCTTGTGATCAATGAAACT‐3′ (sense); anti‐miR‐21 (an inhibitor of miR‐21), 5′‐UCAACAUCAGUCUGAUAAGCUA‐3′; anti‐miR‐C (used as a negative control for anti‐miR‐21 in the antagonism experiment), 5′‐GUGGAUAUUGUUGCCAUCA‐3′. The oligonucleotide transfection was performed with the Lipofectamine 2000 reagent (Invitrogen). Fifty nanomoles per liter of the RNA duplex were used for each transfection.
2.6. Vector constructs
To generate the FasL expression vector, the open reading frame of the human FasL gene was amplified and cloned into the pcDNA3.0 vector. Luciferase constructs were generated by ligating oligonucleotides containing the wild‐type or mutant putative target site of the FasL 3′UTR into the Psi‐CHECK2 vector (Promega) downstream of a luciferase gene. HEK293 or PANC‐1 cells were cotransfected with 80 ng of the luciferase reporter plasmid, 40 ng of the pRL‐TK‐Renilla‐luciferase plasmid (Promega, Madison, WI), and the indicated RNAs at a final concentration of 20 nM. Twenty‐four hours after the transfection, the firefly and Renilla luciferase activities were measured using the Dual‐Luciferase Reporter Assay (Promega). Transfections were done in duplicate and repeated in three independent experiments.
2.7. Cell viability assays
To assess the chemosensitivity to gemcitabine treatment, the oligonucleotide‐transfected cells were plated onto a 96‐well plate (5000 cells/well) and incubated overnight under normal culture conditions. Then the cells were incubated with various concentrations of gemcitabine (2.5, 5, 10, or 20 nM for the PANC‐1 cells and 5, 10, 20, or 40 nM for the BxPC3 cells). After 72 h of treatment, the cells were subjected to the CCK‐8 assay. The number of viable cells was determined with a Cell Counting Kit (Dojindo Molecular Technologies, Inc., Gaithersburg, MD), which counted the number of living cells using WST‐8. The absorbance was determined at 490 nm.
2.8. Apoptosis assay
The level of apoptotic cell death was determined by Annexin V and/or PI staining using an Annexin V‐FITC Kit (BD Pharmingen, San Diego, CA). The cells were analyzed by flow cytometry to determine the percentage of cells undergoing apoptosis and necrosis among the cells treated with gemcitabine. The apoptosis assay was conducted using the protocol supplied by the manufacturer (BioVision Inc.). Briefly, the cells were trypsinized gently and resuspended in 500 μl of 1× binding buffer. Then, the cells were treated with 5 μl of Annexin V‐FITC and 5 μl of PI. After incubation for 10 min in the dark, each sample was analyzed immediately using a FACSCalibur flow cytometer (Becton Dickinson). Approximately 15,000 cells were detected for each sample. Data analysis was performed using the Cell Quest software, and the data were expressed as the percentage of apoptotic cells.
2.9. Western blot analysis
Western blotting was performed according to our previous report (Wang et al., 2009). Briefly, the cellular proteins were extracted from the cultured cells and quantitated using a bicinchoninic acid (BCA) assay kit (Pierce, Rockford, IL, USA) with BSA as a standard. Equal amounts of protein from the various cells were separated on a 10% SDS–PAGE gel and transferred to a nitrocellulose membrane (Bio‐Rad). The membrane was blocked with 5% non‐fat milk and subsequently incubated with primary antibodies. The target proteins were detected using an enhanced chemiluminescence (ECL) kit (Amersham Pharmacia Biotech, Uppsala, Sweden) and anti‐FasL (Epitomics) and anti‐GAPDH (ProteinTech Group, Inc., Chicago, IL) antibodies.
2.10. Tumorigenicity assays in nude mice and gemcitabine administration
PANC‐1 cells transfected with NC or miR‐21 (2 × 106 cells in 200 μl) were injected s.c. into the right axillas of individual BALB/c‐nu/nu nude mice. When the tumor sizes reached 5 mm in diameter, the mice with the NC‐ or miR‐21‐transfected cells were randomly divided into two groups. One group received gemcitabine treatment (120 mg/kg, i.p., d1, 5, 9, and 13), and the other group received an equal volume of normal saline as a control. Tumor growth was then examined daily for the following 4 weeks. The length and width of the tumors (in millimeters) were measured weekly with calipers. The tumor volume was calculated by the formula (a × b 2) × 0.5, where a and b were the long and short dimensions, respectively. The mice were sacrificed 4 weeks later, and the tumors were subsequently removed and weighed. Each group contained at least six mice.
2.11. Statistical analyses
ANOVA and Student's t‐test were used to analyze the results, which were expressed as the mean ± SD. The chi‐squared test was used to analyze the associations between miR‐21 expression and the clinical–pathological parameters. For cohort 1, the overall survival (OS) was defined as the interval between the date of surgery and death. For cohort 2, the overall survival (OS) was defined as the interval between the date of a definitive diagnosis and death. The TTP was defined as the time from the start of chemotherapy to the first report of progression. For the patients who had not experienced a progression at the time of death or last follow‐up, the TTP was censored at the date of death or the last follow‐up (Meng et al., 2012). The Kaplan–Meier method was used to compare the TTP and OS between patients in different groups, and the log‐rank test was used to estimate the differences in survival. Univariate and multivariate analyses based on the Cox proportional hazards regression model were performed using the SPSS 15.0 software (SPSS, Inc.). A p value < 0.05 was considered to be statistically significant.
3. Results
3.1. miR‐21 expression is up‐regulated in pancreatic cancer tissues and associated with a poor prognosis
Recently, miR‐21 expression was found to be up‐regulated in pancreatic cancer tissue (Giovannetti et al., 2010). Therefore, we first examined the miR‐21 expression level in 10 paired pancreatic cancer samples by TaqMan real‐time RT‐PCR. The results showed that miR‐21 expression was significantly higher in the cancerous tissues compared with the corresponding non‐cancerous tissues (p < 0.001; Figure 1A). To substantiate the significance of the miR‐21 expression pattern in pancreatic cancer progression, we detected the expression of miR‐21 in 65 resectable PDAC patients. The median miR‐21 expression level of the 65 cases was chosen as the cut‐off point for separating the miR‐21 low‐expression tumors (n = 33) from miR‐21 high‐expression tumors (n = 32). A Kaplan–Meier analysis revealed that the median survival of the patients with a high miR‐21 expression level was 12.0 months compared with a median survival of 32.0 months in the patients with a low miR‐21 expression level (log rank = 8.590, p = 0.003; Figure 1B). Collectively, these data suggest that the up‐regulation of miR‐21 may contribute to the development of pancreatic cancer, and a high expression level of miR‐21 is associated with a poor prognosis.
Figure 1.
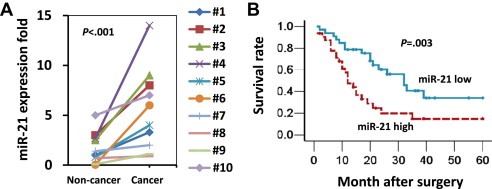
miR‐21 expression is up‐regulated in cancer tissue and correlates with shorter survival in patients with PDAC. (A) A comparison of miR‐21 expression levels in 10 paired pancreatic cancer and adjacent non‐cancer tissues by TaqMan real‐time PCR. The U6 RNA served as internal control. Statistical significance was calculated using Student's t‐test. (B) A Kaplan–Meier survival analysis of the PDAC patients grouped according to the expression level of miR‐21 in the cancer tissue. Mature miR‐21 expression was analyzed by TaqMan real‐time qPCR, and the median miR‐21 expression level of the 65 cases was chosen as the cut‐off point for separating the miR‐21 low‐expression tumors (n = 33) from miR‐21 high‐expression tumors (n = 32). The p‐value was determined using the log‐rank test.
3.2. A high level of serum miR‐21 is associated with a poor response to gemcitabine treatment in patients with advanced pancreatic cancer
Testing the miRNA levels in pancreatic cancer tissues is not a viable option for patients with an advanced disease because of the inherent difficulties in obtaining pancreatic tumor tissue. Therefore, we hypothesized that the level of miR‐21 in the serum could be used as a predictor of the chemosensitivity of advanced cases of pancreatic cancer toward palliative chemotherapy. To test this hypothesis, we examined the correlation between the serum level of miR‐21 and the cellular response to gemcitabine‐based chemotherapy. The levels of miR‐21 in a large cohort of 177 cases of advanced‐stage pancreatic cancer who received gemcitabine‐based palliative chemotherapy were examined by TaqMan real‐time PCR. The characteristics of the 177 cases are shown in Supplementary Table 2. The median expression level of miR‐21 of all of the samples was chosen as the cut‐off point for separating the tumors from patients with a low level of serum miR‐21 from the tumors from patients with a high level of serum miR‐21. A Kaplan–Meier analysis revealed that the time to progression (TTP) after gemcitabine treatment in the patients with a low miR‐21 expression level was 161 days, whereas the TTP in patients with a high miR‐21 expression level was 80 days (log rank = 13.620, p < 0.001; Figure 2A). Similarly, a low level of serum miR‐21 was significantly correlated with a longer overall survival (OS) in patients who received gemcitabine‐based palliative chemotherapy (348 days for a low miR‐21 level and 186 days for a high miR‐21 level; log rank = 9.981, p = 0.002, Figure 2B). To examine whether the serum miR‐21 level was associated with specific stages of the disease, the patients were classified according to the TNM staging. Of the 177 patients with advanced pancreatic cancer, the serum miR‐21 levels were significantly higher in the stage IV patients compared with the stage III patients (p = 0.008; Figure 2C). Because tumor stage affects prognosis (Figure 2D) and to exclude confounding effects, we then performed a Cox proportional hazards regression analysis. A multivariate analysis confirmed that the serum miR‐21 level was an independent prognostic factor for both the TTP and OS (HR 1.920; 95% CI, 1.274–2.903, p = 0.002 for TTP; and HR 1.705; 95% CI, 1.147–2.535, p = 0.008 for OS; Table 1). In both stage III and stage IV patients, a high level of miR‐21 in the serum was associated with a shortened TTP and OS (Figure 2E–F and Supplementary Table 3). Taken together, these results suggest that the serum level of miR‐21 can be used to predict the response of pancreatic cancer to gemcitabine treatment in patients with advanced stages of the disease.
Figure 2.
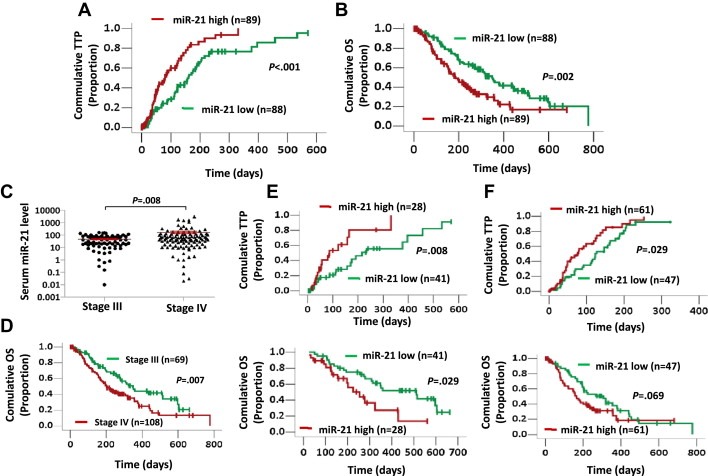
A high level of serum miR‐21 is associated with a poor response to gemcitabine treatment in patients with advanced pancreatic cancer. (A, B) Kaplan–Meier curves for the time to progression (TTP) (A) and overall survival (OS) (B) in a cohort of 177 advanced pancreatic cancer patients grouped by the serum level of miR‐21. The serum miR‐21 level was analyzed by TaqMan real‐time qPCR, and the median miR‐21 expression level was chosen as the cut‐off point for separating the miR‐21 low‐level cases (n = 88) from the miR‐21 high‐level cases (n = 89). (C) A comparison of the serum miR‐21 levels in stage III and stage IV tumors. The statistical significance of the differences between the two groups was calculated using Student's t‐test. (D) Kaplan–Meier curves for the pancreatic cancer patients with stage III (n = 69) or stage IV (n = 108) disease. (E, F) Kaplan–Meier curves for the TTP (top) and OS (bottom) of 69 stage III (E) and 108 stage IV (F) advanced pancreatic cancer patients grouped by the serum level of miR‐21. The p‐value was determined by the log‐rank test.
Table 1.
Univariate and multivariate Cox regression analyses of the serum miR‐21 level for the TTP and OS of patients with advanced pancreatic cancer (N = 177).
| Variables | TTP | OS | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | p‐value | Hazard ratio (95% CI) | p‐value | |
| Univariate analysis | ||||
| Age (<65 years vs. ≥65 years) | 0.950 (0.607–1.487) | 0.821 | 1.189 (0.804–1.758) | 0.385 |
| Gender (male vs. female) | 0.883 (0.590–1.320) | 0.543 | 0.810 (0.547–1.199) | 0.293 |
| Location (head vs. body/tail) | 1.319 (0.881–1.975) | 0.178 | 1.282 (0.872–1.883) | 0.206 |
| Stage (III vs. IV) | 2.097 (1.343–3.275) | 0.001 | 1.724 (1.152–2.578) | 0.008 |
| CA19‐9 (<500 vs. ≥500 IU/ml) | 1.696 (1.137–2.530) | 0.010 | 2.242 (1.514–3.321) | <0.001 |
| Serum miR‐21 level (low vs. high) | 2.124 (1.410–3.201) | <0.001 | 1.860 (1.259–2.750) | 0.002 |
| Multivariate analysis | ||||
| Stage (III vs. IV) | 0.008 (1.177–2.904) | 0.008 | 1.414 (0.935–2.140) | 0.101 |
| CA19‐9 (<500 vs. ≥500 IU/ml) | 1.526 (1.019–2.287) | 0.040 | 2.109 (1.420–3.133) | <0.001 |
| Serum miR‐21 level (low vs. high) | 1.920 (1.274–2.903) | 0.002 | 1.705 (1.147–2.535) | 0.008 |
Abbreviations: TTP, time to progression; OS, overall survival; CI, confidence interval.
p‐values in bold are statistically significant based on multivariate Cox regression analyses.
3.3. The up‐regulation of miR‐21 is associated with a decreased chemosensitivity to gemcitabine treatment due to an enhanced resistance to gemcitabine‐induced apoptosis
To determine the effects of miR‐21 overexpression on the sensitivity of cancer cells to gemcitabine treatment, pancreatic cancer cells were transfected with miR‐21 mimics to increase miR‐21 expression (Supplementary Figure 1). The transfected cells were then treated with increasing concentrations of gemcitabine, and cell growth was measured by the CCK‐8 assay 72 h later. As shown in Figure 3A, the growth of the cells transfected with the control mimics (NC) was manifestly inhibited by gemcitabine treatment while the inhibitory effect of gemcitabine was significantly reduced in the miR‐21‐transfected PANC‐1 and BxPC3 cells. To confirm the role of miR‐21 in chemoresistance, we examined whether the inhibition of miR‐21 expression correlated with an enhanced sensitivity to gemcitabine treatment. PANC‐1 and BxPC3 cells were transfected with an anti‐miR‐21 inhibitor to decrease the miR‐21 levels (Supplementary Figure 1) and then challenged with increasing concentrations of gemcitabine for 72 h. As shown in Figure 3B, the down‐regulation of miR‐21 expression rendered the pancreatic cancer cells more susceptible to the cytotoxic effects of gemcitabine treatment than the cells treated with a control inhibitor. With the use of Annexin V/PI double staining and flow cytometry, we also found that cells transfected with miR‐21 mimics exhibited lower levels of apoptosis upon gemcitabine treatment compared with the cells transfected with control mimics (Figure 3C). Additionally, an increase in the level of gemcitabine‐induced cellular apoptosis was observed in a loss‐of‐function study that used an anti‐miR‐21 inhibitor to significantly decrease endogenous miR‐21 expression (Figure 3C). We also performed an in vivo study to evaluate the effects of miR‐21 on the resistance to gemcitabine treatment. NC‐ and miR‐21‐transfected PANC‐1 cells were injected s.c. into the right axillas of nude mice, and the tumor‐bearing mice were treated with gemcitabine. We found that compared with the NC‐transfected cells, enhanced expression of miR‐21 significantly decreased gemcitabine‐induced tumor growth inhibition (Figure 3D). Taken together, these data indicate that increased expression of miR‐21 renders pancreatic cancer cells more resistant to gemcitabine treatment.
Figure 3.
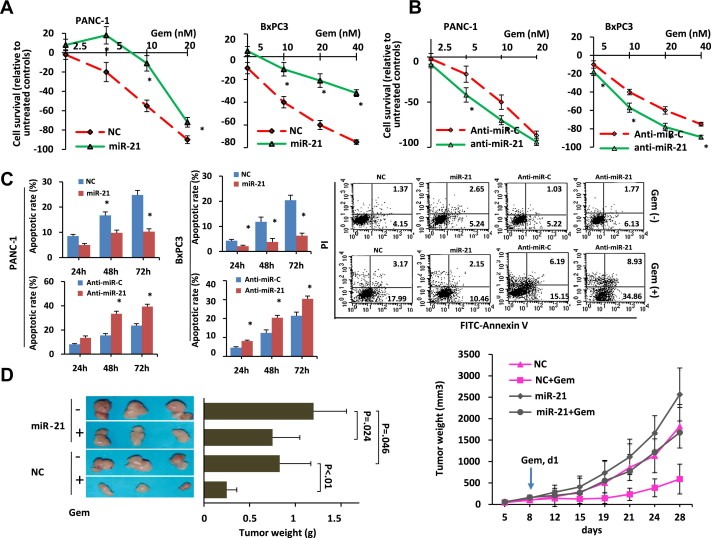
The up‐regulation of miR‐21 is associated with a decreased chemosensitivity to gemcitabine treatment due to an enhanced resistance to gemcitabine‐induced apoptosis. (A, B) Cells transfected with a miR‐21 mimic (A) or an anti‐miR‐21 inhibitor (B) were seeded in 96‐well plates (5000 cells/well), incubated for 24 h in culture medium, and then incubated for 72 h in the presence or absence of increasing gemcitabine concentrations. Cell viability was determined using the CCK‐8 assay. The results are presented as the means ± SD from three independent experiments. Statistical significance was calculated using Student's t‐test. NC represents the negative control for the miR‐21 mimics; anti‐miR‐C represents the negative control for the anti‐miR‐21 inhibitor. * = p < 0.05. (C) Cells transfected with a miR‐21 mimic or an anti‐miR‐21 inhibitor were treated with 10 nM gemcitabine for 24, 48, or 72 h. The cells were then harvested, subjected to Annexin/PI double staining, and analyzed by flow cytometry. The results are expressed as the percentage of apoptotic cells (means ± SD) from three independent experiments. Statistical significance was calculated using Student's t‐test. Representative FCM results are shown on the right. (D) The effect of miR‐21 expression on the chemosensitivity of a nude mouse xenograft model. NC‐ and miR‐21‐transfected PANC‐1 cells were injected s.c. into the right axilla of each mouse. The mice with NC or miR‐21‐transfected cells were randomly divided into two groups of six mice each. One group received gemcitabine treatment (10 mg/kg, i.p., d1, 5, 9, and 13), and the other group received an equal volume of normal saline as a control. The mice were sacrificed when the tumors reached 1.5 cm in diameter. The tumors were then removed and weighed. The tumor growth curves are shown on the right. The tumor weights are shown on the left, and the differences between the groups were compared using an ANOVA test. *p < 0.05.
3.4. FasL is a direct target of miR‐21 in pancreatic cancer
Next, we attempted to identify the relevant target mRNA of miR‐21. Using both TargetScan (http://www.targetscan.org) and Sanger (http://microrna.sanger.ac.uk) database searches, we found that the 3′UTRs of FasL mRNAs contain conserved miR‐21 binding sites (Figure 4A). In addition, previous studies have indicated that miR‐21 is involved in the apoptotic response by targeting FasL in specific cell types, including neurons and cardiocytes (Wang and Li, 2010; Sayed et al., 2010). To demonstrate whether miR‐21 directly regulates FasL in pancreatic cancer, we employed a dual‐luciferase reporter system. We cloned the 3′UTR of FasL into a reporter plasmid downstream of the luciferase gene to generate wild‐type and mutant FasL 3′UTR luciferase construct. The wild‐type and mutant 3′UTRs are shown in Figure 4B. Using this system, we found that the expression of miR‐21 significantly suppressed firefly luciferase reporter activity of the wild‐type FasL 3′UTR luciferase construct but not the mutant FasL 3′UTR luciferase construct. These results were observed in both HEK293 cells and the pancreatic cancer PANC‐1 cell line (Figure 4C), which indicates that miR‐21 suppresses FasL expression through the miRNA‐binding sequences within the FasL 3′UTR. Then we evaluated the FasL mRNA and protein levels in cells over‐expressing or under‐expressing miR‐21. We found that the over‐expression of miR‐21 reduced the endogenous expression of FasL both at the mRNA and protein level, whereas the downregulation of miR‐21 expression with an anti‐miR‐21 inhibitor increased the expression of FasL (Figure 4D). We also evaluated the correlation between miR‐21 expression and FasL during gemcitabine treatment and found an inverse correlation between the expression of miR‐21 and FasL. The expression of miR‐21 decreased following gemcitabine treatment; however, the expression of FasL mRNA significantly increased following gemcitabine treatment (Figure 4E). Therefore, the results from our study indicate that FasL is a direct target of miR‐21 in pancreatic cancer.
Figure 4.
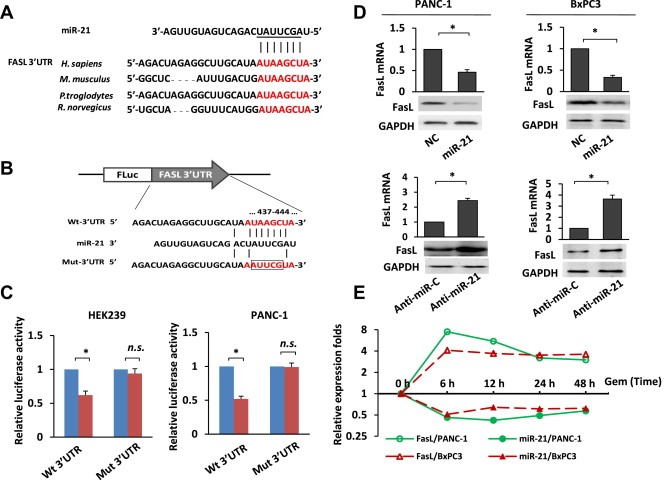
FasL is a direct target of miR‐21. (A) Alignment of the predicted miRNA binding sites in the 3′UTR of the FasL mRNA. (B) A human FasL 3′UTR fragment containing the wild‐type or mutant miR‐21‐binding sequence was cloned downstream of the luciferase reporter gene. (C) HEK293 and PANC‐1 cells were cotransfected with the negative control (NC) or miR‐21 mimics and the luciferase reporter construct containing the wild‐type or mutant FasL 3′UTR. For each experiment, the data were normalized to the luciferase activity detected in the cells transfected with the control vector, and the luciferase activity of the control vector was set to 1. n.s. = not significant. Statistical significance was calculated using Student's t‐test. (D) The effects of miR‐21 overexpression or suppression on endogenous FasL mRNA and protein expression as measured by real‐time PCR and western blotting respectively. GAPDH served as the internal control. Statistical significance was calculated using Student's t‐test. (E) The expression levels of the FasL mRNA and miR‐21 in PANC‐1 and BxPC3 cells at different time points following 10 nM gemcitabine treatment. *p < 0.05.
3.5. Decreased Fas/FasL signaling mediates miR‐21‐induced chemoresistance in pancreatic cancer
To confirm that miR‐21 confers chemoresistance by directly targeting FasL in pancreatic cancer, we tested the ability of Fas/FasL to trigger apoptosis in pancreatic cancer cells. Soluble FasL (sFasL), a recombinant form of the extracellular domain of the human FasL protein, was used in these experiments. We found that the addition of sFasL to pancreatic cancer cells triggered apoptosis in a dose‐dependent manner (Figure 5A). To determine whether the Fas death receptor pathway contributes to gemcitabine‐induced apoptosis, we used Fas‐Fc, a recombinant form of the extracellular domain of the human Fas protein, to inhibit cell apoptosis. We found that Fas‐Fc inhibited the cellular apoptosis induced by gemcitabine treatment (Figure 5B). Next, we studied the role of FasL in the miR‐21‐mediated inhibition of apoptosis. We found that the silencing of FasL by siRNA treatment in PANC‐1 cells significantly decreased FasL expression and gemcitabine‐induced apoptosis in the pancreatic cancer cells (Figure 5C), which was similar to the effects of miR‐21 over‐expression. In contrast, the ectopic expression of FasL significantly abrogated the miR‐21‐induced chemoresistance (Figure 5D), which indicates that FasL is a functional target of miR‐21. Taken together, these results suggest that miR‐21 expression suppresses FasL expression and protects pancreatic cancer cells from gemcitabine‐induced apoptosis (Figure 5E).
Figure 5.
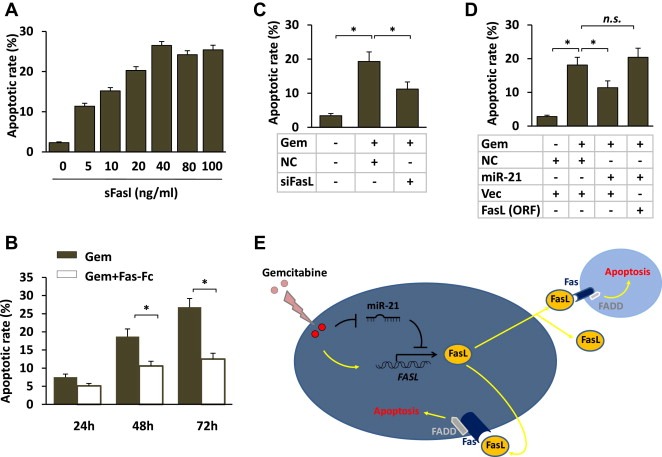
Deceased Fas/FasL signaling mediates the miR‐21‐induced chemoresistance in pancreatic cancer cells. (A) PANC‐1 cells were treated with increasing concentrations of soluble FasL (sFasL) as indicated. After 48 h, the percentage of apoptotic cells was evaluated by Annexin/PI double staining and analyzed by flow cytometry. The results are presented as the means ± SD from three independent experiments. (B) PANC‐1 cells were preincubated for 30 min with 5 μg/ml Fas‐Fc (an inhibitor of Fas activation by FasL) before being exposed to gemcitabine for 24, 48 or 72 h. The cells were then harvested, and the apoptotic cells were detected after 48 h using flow cytometry. The results are presented as the means ± SD from three independent experiments. (C) PANC‐1 cells were transfected with the negative control (NC) or siRNA targeting FasL and subsequently subjected to 10 nM gemcitabine treatment. After 72 h, the cells undergoing apoptosis were detected using flow cytometry. The results are presented as the means ± SD from three independent experiments. (D) PANC‐1 cells were transfected with the negative control (NC), miR‐21 mimic or FasL (open reading frame without the 3′UTR) expression vectors as indicated and subsequently subjected to 10 nM gemcitabine treatment. After 72 h, the cells undergoing apoptosis were detected using flow cytometry. The results are presented as the means ± SD from three independent experiments. ANOVA tests were used to determine the statistical significance of the differences among the groups. *p < 0.05. (E) A schematic of the role of the FasL/Fas pathway in miR‐21‐mediated chemoresistance.
4. Discussion
The development of resistance to chemotherapy is one of the major challenges in the treatment of patients with pancreatic cancer. In this study, we focused on the microRNA miR‐21, which is known to be involved in the progression of several types of cancers (Yang et al., 2011; Kwak et al., 2011). We report 2 significant, novel findings. First, in advanced pancreatic cancer, a high level of miR‐21 in the serum was closely associated with a poor response to palliative chemotherapy. Second, the effects of miR‐21 on chemosensitivity were mediated through the FasL/Fas pathway.
The results from our study demonstrate that a high level of serum miR‐21 was closely associated with a poor response to palliative chemotherapy. Many studies have shown that microRNAs can serve as predictors or modifiers of the chemotherapeutic response in different tumor types (Hummel et al., 2010). In fact, miR‐21 has been reported to be involved in the chemosensitivity of several types of cancers, including pancreatic cancer (Ali et al., 2010; Moriyama et al., 2009; Giovannetti et al., 2010; Tomimaru et al., 2010). For example, a high level of miR‐21 expression in cancer tissue is associated with a reduced disease‐free survival (DFS) or OS in patients who underwent a radical resection and adjuvant gemcitabine treatment (Hwang et al., 2010). However, most of these reports focused on studying the miRNA expression in cancerous tissues. Additionally, for pancreatic cancer, most studies have focused on resectable pancreatic cancer; however, resection is possible only in a minority of patients due to the advanced stages often present at the time of diagnosis (Li et al., 2004), and safely obtaining sufficient quantities of pancreatic tumor tissue for molecular analysis is difficult at unresectable stages. Therefore, non‐invasive methods for detection are warranted. We hypothesized that the serum level of miR‐21 could serve as a surrogate for the expression level of miR‐21 in the tumor tissue and be used as a predictor of the chemosensitivity of advanced stages of pancreatic cancer treated with palliative chemotherapy.
CA19‐9 is currently one of the most useful blood tests in differentiating benign from malignant pancreatic disorders, although it has limited value because several benign diseases and multiple types of adenocarcinoma, especially advanced gastrointestinal cancers, may give rise to elevated CA19‐9 levels (Steinberg, 1990; Duffy et al., 2010). However, one recent study has shown that the combination of miR‐16, miR‐196a and CA19‐9 was more effective then either miR‐16 + miR‐196a or CA19‐9 alone for pancreatic cancer diagnosis, especially in early tumor screening (Liu et al., 2012a). CA19‐9 has also been used for determining prognosis in pancreatic cancer (Boeck et al., 2006). Our previous study has confirmed that advanced pancreatic cancer patients with elevated levels of CA 19‐9 had a worse prognosis than those with low levels (Ouyang et al., 2011). Our present results agree with numerous earlier studies that CA19‐9 level is a very good indicator of OS (Supplementary Table 3). Multivariate Cox regression analyses indicated that serum miR‐21 and CA19‐9 levels were independent prognostic factors for both the TTP and the OS (Table 1). Importantly, CA19‐9 and miR‐21 have distinct prognostic values: CA19‐9 is an excellent prognostic indicator of OS; miR‐21 also is a prognostic indicator of OS, but miR‐21 is also a prognostic indicator of responsiveness to gemcitabine treatment.
Recent studies have shown that miRNAs are stably detectable in plasma/serum, and a strong correlation exists between tumor‐derived circulating miRNAs and miRNA directly within tumor cells (Mitchell et al., 2008; Chen et al., 2008). Circulating miRNAs may have potential as diagnostic, prognostic and predictive biomarkers, and these molecules may be useful as potential future therapeutic targets. The presence of miRNAs in the serum was first described in 2008 in patients with diffuse large B cell lymphoma (Lawrie et al., 2008). Serum miRNA expression profiles have subsequently been shown to have signatures that are related to tumor classification, diagnosis and state of disease progression. For example, miR‐92 has been shown to be significantly elevated in the plasma of patients with colorectal cancer (CRC) and serves as a non‐invasive molecular marker for CRC screening (Ng et al., 2009); A 4‐miRNA serum‐based signature may provide a simple profiling method of predicting the survival of non‐small‐cell lung cancer patients (Hu et al., 2010); and a plasma‐based microRNA panel has been shown to have considerable clinical value for the diagnosis of early‐stage hepatocellular carcinoma (Zhou et al., 2011). Increased serum miR‐21 has also been detected in pancreatic cancer (Liu et al., 2012b; LaConti et al., 2011; Kong et al., 2011). In this study, we report that the serum level of miR‐21 can be used to predict the chemosensitivity of advanced pancreatic cancer to gemcitabine treatment. Additionally, our study provides a new, non‐invasive, serum‐based molecular biomarker for the prediction of the response to gemcitabine treatment in patients with an advanced disease.
This study demonstrated that the effect of miR‐21 on the development of chemoresistance is mediated through the FasL/Fas pathway. FasL, a member of the TNF protein family, has been shown to induce apoptosis in cells that express the Fas receptor (Sun and Fink, 2007). The FasL/Fas death pathway is a key regulator of apoptosis that is induced by a variety of stimuli, including anti‐cancer agents (Chhipa and Bhat, 2007; Ginestier et al., 2010). Fas is a transmembrane cell surface receptor that triggers cell death (apoptosis) upon binding to FasL, which is an anchored cell membrane protein exposed to the extracellular space. The activation of these death receptors is triggered by cellular stress or death signals, and the activation of this pathway leads to the recruitment of the Fas‐associated death domain (FADD) and death domain (DD), which in turn induce the activation of caspase signaling and leads to apoptosis (Song et al., 2000). In our study, we also found that the addition of soluble FasL (sFasL, a recombinant form of the extracellular domain of the human FasL protein) to pancreatic cancer cells triggered apoptosis in a dose‐dependent manner, whereas the addition of Fas‐Fc (a recombinant form of the extracellular domain of the human Fas protein) partially blocked the cellular apoptosis induced by gemcitabine treatment. These results suggest that gemcitabine‐induced apoptosis in cancer cells is partially dependent on the Fas/FasL pathway. Because the 3′UTR region of the FasL mRNA contains conserved miR‐21 binding sites, we investigated the ability of miR‐21 to directly regulate FasL expression through these 3′UTR sites and found that mutation of these sites abrogated miR‐21‐mediated downregulation of FasL expression. Additionally, because gemcitabine treatment significantly up‐regulated FasL expression and down‐regulated miR‐21 expression, we hypothesized that FasL mediates the miR‐21‐induced chemoresistance in pancreatic cancer. Based on the observations that the silencing of FasL by siRNA treatment reduced the level of gemcitabine‐induced apoptosis in pancreatic cancer cells and that ectopic expression of FasL significantly abrogated the miR‐21‐induced chemoresistance, we concluded that FasL is a functional target of miR‐21.Therefore, the results from our study identify a new mechanism of miR‐21‐mediated chemoresistance in pancreatic cancer (Supplementary Figure 3).
Cumulatively, we demonstrated that the serum miR‐21 level can be used as a predictor of the chemosensitivity of pancreatic cancer to gemcitabine treatment. Because this approach is noninvasive and does not require a biopsy, it is especially useful for advanced pancreatic cancer. In addition, our results demonstrate the existence of a new mechanism of chemoresistance through the effects of miR‐21 on the FasL/Fas pathway.
Conflict of interest
The authors disclose no potential conflicts of interest.
Grant support
This study was supported by National Science Foundation of China (81001061); Shanghai Nature Science Fund, Shanghai, China (09ZR1406800); Doctoral Programs Foundation of Ministry of Education of China (20090071120076); Shanghai Science and Technology Committee Rising‐Star Program (11QA1401300); and Medical Talents Training Program of Health Bureau of Shanghai (XYQ2011008), and Fudan University 985 Research Grant.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgments
The authors would like to thank Dr. Li Zhang from Huadong Hospital, Fudan University, Shanghai for her precious advice. We also thank Prof. Zude Xu from Huashan Hospital, Fudan University, Shanghai for providing pancreatic cancer samples.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2012.10.011.
Wang Peng, Zhuang Liping, Zhang Juan, Fan Jie, Luo Jianmin, Chen Hao, Wang Kun, Liu Luming, Chen Zhen, Meng Zhiqiang, (2013), The serum miR‐21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR‐21 expression confers chemoresistance by targeting FasL, Molecular Oncology, 7, doi: 10.1016/j.molonc.2012.10.011.
Contributor Information
Zhen Chen, Email: cz.chenzhen@gmail.com.
Zhiqiang Meng, Email: mengzhq@sh163.net.
References
- Ali, S. , Ahmad, A. , Banerjee, S. , Padhye, S. , Dominiak, K. , Schaffert, J.M. , Wang, Z. , Philip, P.A. , Sarkar, F.H. , 2010. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 70, 3606–3617. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Arroyo, J.D. , Chevillet, J.R. , Kroh, E.M. , Ruf, I.K. , Pritchard, C.C. , Gibson, D.F. , Mitchell, P.S. , Bennett, C.F. , Pogosova-Agadjanyan, E.L. , Stirewalt, D.L. , Tait, J.F. , Tewari, M. , 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U S A 108, 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomston, M. , Frankel, W.L. , Petrocca, F. , Volinia, S. , Alder, H. , Hagan, J.P. , Liu, C.G. , Bhatt, D. , Taccioli, C. , Croce, C.M. , 2007. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297, 1901–1908. [DOI] [PubMed] [Google Scholar]
- Boeck, S. , Stieber, P. , Holdenrieder, S. , Wilkowski, R. , Heinemann, V. , 2006. Prognostic and therapeutic significance of carbohydrate antigen 19-9 as tumor marker in patients with pancreatic cancer. Oncology 70, 255–264. [DOI] [PubMed] [Google Scholar]
- Burris, H.A. , Moore, M.J. , Andersen, J. , Green, M.R. , Rothenberg, M.L. , Modiano, M.R. , Cripps, M.C. , Portenoy, R.K. , Storniolo, A.M. , Tarassoff, P. , Nelson, R. , Dorr, F.A. , Stephens, C.D. , Von Hoff, D.D. , 1997. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 15, 2403–2413. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Ba, Y. , Ma, L. , Cai, X. , Yin, Y. , Wang, K. , Guo, J. , Zhang, Y. , Chen, J. , Guo, X. , Li, Q. , Li, X. , Wang, W. , Wang, J. , Jiang, X. , Xiang, Y. , Xu, C. , Zheng, P. , Zhang, J. , Li, R. , Zhang, H. , Shang, X. , Gong, T. , Ning, G. , Zen, K. , Zhang, C.Y. , 2008. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006. [DOI] [PubMed] [Google Scholar]
- Chhipa, R.R. , Bhat, M.K. , 2007. Bystander killing of breast cancer MCF-7 cells by MDA-MB-231 cells exposed to 5-fluorouracil is mediated via Fas. J. Cell. Biochem. 101, 68–79. [DOI] [PubMed] [Google Scholar]
- Dillhoff, M. , Liu, J. , Frankel, W. , Croce, C. , Bloomston, M. , 2008. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J. Gastrointest. Surg. 12, 2171–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, M.J. , Sturgeon, C. , Lamerz, R. , Haglund, C. , Holubec, V.L. , Klapdor, R. , Nicolini, A. , Topolcan, O. , Heinemann, V. , 2010. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann. Oncol. 21, 441–447. [DOI] [PubMed] [Google Scholar]
- Fu, X. , Tian, J. , Zhang, L. , Chen, Y. , Hao, Q. , 2012. Involvement of microRNA-93, a new regulator of PTEN/Akt signaling pathway, in regulation of chemotherapeutic drug cisplatin chemosensitivity in ovarian cancer cells. FEBS Lett. 586, 1279–1286. [DOI] [PubMed] [Google Scholar]
- Ginestier, C. , Liu, S. , Diebel, M.E. , Korkaya, H. , Luo, M. , Brown, M. , Wicinski, J. , Cabaud, O. , Charafe-Jauffret, E. , Birnbaum, D. , Guan, J.L. , Dontu, G. , Wicha, M.S. , 2010. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J. Clin. Invest. 120, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti, E. , Funel, N. , Peters, G.J. , Del Chiaro, M. , Erozenci, L.A. , Vasile, E. , Leon, L.G. , Pollina, L.E. , Groen, A. , Falcone, A. , Danesi, R. , Campani, D. , Verheul, H.M. , Boggi, U. , 2010. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 70, 4528–4538. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Chen, X. , Zhao, Y. , Tian, T. , Jin, G. , Shu, Y. , Chen, Y. , Xu, L. , Zen, K. , Zhang, C. , Shen, H. , 2010. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28, 1721–1726. [DOI] [PubMed] [Google Scholar]
- Hummel, R. , Hussey, D.J. , Haier, J. , 2010. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur. J. Cancer 46, 298–311. [DOI] [PubMed] [Google Scholar]
- Hwang, J.H. , Voortman, J. , Giovannetti, E. , Steinberg, S.M. , Leon, L.G. , Kim, Y.T. , Funel, N. , Park, J.K. , Kim, M.A. , Kang, G.H. , Kim, S.W. , Del Chiaro, M. , Peters, G.J. , Giaccone, G. , 2010. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One 5, e10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson, N.B. , Morran, D.C. , Morton, J.P. , Ali, A. , Dickson, E.J. , Carter, C.R. , Sansom, O.J. , Evans, T.R. , McKay, C.J. , Oien, K.A. , 2011. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin. Cancer Res. 18, 534–545. [DOI] [PubMed] [Google Scholar]
- Kim, V.N. , Han, J. , Siomi, M.C. , 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. [DOI] [PubMed] [Google Scholar]
- Kim, C.H. , Kim, H.K. , Rettig, R.L. , Kim, J. , Lee, E.T. , Aprelikova, O. , Choi, I.J. , Munroe, D.J. , Green, J.E. , 2011. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med. Genomics 4, 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X. , Du, Y. , Wang, G. , Gao, J. , Gong, Y. , Li, L. , Zhang, Z. , Zhu, J. , Jing, Q. , Qin, Y. , Li, Z. , 2011. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig. Dis. Sci. 56, 602–609. [DOI] [PubMed] [Google Scholar]
- Kwak, H.J. , Kim, Y.J. , Chun, K.R. , Woo, Y.M. , Park, S.J. , Jeong, J.A. , Jo, S.H. , Kim, T.H. , Min, H.S. , Chae, J.S. , Choi, E.J. , Kim, G. , Shin, S.H. , Gwak, H.S. , Kim, S.K. , Hong, E.K. , Lee, G.K. , Choi, K.H. , Kim, J.H. , Yoo, H. , Park, J.B. , Lee, S.H. , 2011. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene 30, 2433–2442. [DOI] [PubMed] [Google Scholar]
- LaConti, J.J. , Shivapurkar, N. , Preet, A. , Deslattes Mays, A. , Peran, I. , Kim, S.E. , Marshall, J.L. , Riegel, A.T. , Wellstein, A. , 2011. Tissue and serum microRNAs in the Kras(G12D) transgenic animal model and in patients with pancreatic cancer. PLoS One 6, e20687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie, C.H. , Gal, S. , Dunlop, H.M. , Pushkaran, B. , Liggins, A.P. , Pulford, K. , Banham, A.H. , Pezzella, F. , Boultwood, J. , Wainscoat, J.S. , Hatton, C.S. , Harris, A.L. , 2008. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 141, 672–675. [DOI] [PubMed] [Google Scholar]
- Lee, J.W. , Park, Y.A. , Choi, J.J. , Lee, Y.Y. , Kim, C.J. , Choi, C. , Kim, T.J. , Lee, N.W. , Kim, B.G. , Bae, D.S. , 2011. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol. Oncol. 120, 56–62. [DOI] [PubMed] [Google Scholar]
- Lekanne Deprez, R.H. , Fijnvandraat, A.C. , Ruijter, J.M. , Moorman, A.F. , 2002. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal. Biochem. 307, 63–69. [DOI] [PubMed] [Google Scholar]
- Li, D. , Xie, K. , Wolff, R. , Abbruzzese, J.L. , 2004. Pancreatic cancer. Lancet 363, 1049–1057. [DOI] [PubMed] [Google Scholar]
- Li, J. , Chen, Y. , Zhao, J. , Kong, F. , Zhang, Y. , 2011. miR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett. 304, 52–59. [DOI] [PubMed] [Google Scholar]
- Lim, L.P. , Lau, N.C. , Garrett-Engele, P. , Grimson, A. , Schelter, J.M. , Castle, J. , Bartel, D.P. , Linsley, P.S. , Johnson, J.M. , 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Gao, J. , Du, Y. , Li, Z. , Ren, Y. , Gu, J. , Wang, X. , Gong, Y. , Wang, W. , Kong, X. , 2012. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer 131, 683–691. [DOI] [PubMed] [Google Scholar]
- Liu, R. , Chen, X. , Du, Y. , Yao, W. , Shen, L. , Wang, C. , Hu, Z. , Zhuang, R. , Ning, G. , Zhang, C. , Yuan, Y. , Li, Z. , Zen, K. , Ba, Y. , Zhang, C.Y. , 2012. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin. Chem. 58, 610–618. [DOI] [PubMed] [Google Scholar]
- Mardin, W.A. , Mees, S.T. , 2009. MicroRNAs: novel diagnostic and therapeutic tools for pancreatic ductal adenocarcinoma?. Ann. Surg. Oncol. 16, 3183–3189. [DOI] [PubMed] [Google Scholar]
- Meng, Z. , Garrett, C.R. , Shen, Y. , Liu, L. , Yang, P. , Huo, Y. , Zhao, Q. , Spelman, A.R. , Ng, C.S. , Chang, D.Z. , Cohen, L. , 2012. Prospective randomised evaluation of traditional Chinese medicine combined with chemotherapy: a randomised phase II study of wild toad extract plus gemcitabine in patients with advanced pancreatic adenocarcinomas. Br. J. Cancer 107, 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P.S. , Parkin, R.K. , Kroh, E.M. , Fritz, B.R. , Wyman, S.K. , Pogosova-Agadjanyan, E.L. , Peterson, A. , Noteboom, J. , O'Briant, K.C. , Allen, A. , Lin, D.W. , Urban, N. , Drescher, C.W. , Knudsen, B.S. , Stirewalt, D.L. , Gentleman, R. , Vessella, R.L. , Nelson, P.S. , Martin, D.B. , Tewari, M. , 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U S A 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, T. , Ohuchida, K. , Mizumoto, K. , Yu, J. , Sato, N. , Nabae, T. , Takahata, S. , Toma, H. , Nagai, E. , Tanaka, M. , 2009. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 8, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Nagao, Y. , Hisaoka, M. , Matsuyama, A. , Kanemitsu, S. , Hamada, T. , Fukuyama, T. , Nakano, R. , Uchiyama, A. , Kawamoto, M. , Yamaguchi, K. , Hashimoto, H. , 2011. Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod. Pathol. 25, 112–121. [DOI] [PubMed] [Google Scholar]
- Ng, E.K. , Chong, W.W. , Jin, H. , Lam, E.K. , Shin, V.Y. , Yu, J. , Poon, T.C. , Ng, S.S. , Sung, J.J. , 2009. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58, 1375–1381. [DOI] [PubMed] [Google Scholar]
- Ouyang, H. , Wang, P. , Meng, Z. , Chen, Z. , Yu, E. , Jin, H. , Chang, D.Z. , Liao, Z. , Cohen, L. , Liu, L. , 2011. Multimodality treatment of pancreatic cancer with liver metastases using chemotherapy, radiation therapy, and/or Chinese herbal medicine. Pancreas 40, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed, D. , He, M. , Hong, C. , Gao, S. , Rane, S. , Yang, Z. , Abdellatif, M. , 2010. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J. Biol. Chem. 285, 20281–20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R. , Naishadham, D. , Jemal, A. , 2012. Cancer statistics, 2012. CA Cancer J. Clin. 62, 10–29. [DOI] [PubMed] [Google Scholar]
- Song, J. , Sapi, E. , Brown, W. , Nilsen, J. , Tartaro, K. , Kacinski, B.M. , Craft, J. , Naftolin, F. , Mor, G. , 2000. Roles of Fas and Fas ligand during mammary gland remodeling. J. Clin. Invest. 106, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, B. , Wang, Y. , Xi, Y. , Kudo, K. , Bruheim, S. , Botchkina, G.I. , Gavin, E. , Wan, Y. , Formentini, A. , Kornmann, M. , Fodstad, O. , Ju, J. , 2009. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28, 4065–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, W. , 1990. The clinical utility of the CA 19-9 tumor-associated antigen. Am. J. Gastroenterol. 85, 350–355. [PubMed] [Google Scholar]
- Su, H. , Yang, J.R. , Xu, T. , Huang, J. , Xu, L. , Yuan, Y. , Zhuang, S.M. , 2009. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 69, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Sun, M. , Fink, P.J. , 2007. A new class of reverse signaling costimulators belongs to the TNF family. J. Immunol. 179, 4307–4312. [DOI] [PubMed] [Google Scholar]
- Szafranska, A.E. , Davison, T.S. , John, J. , Cannon, T. , Sipos, B. , Maghnouj, A. , Labourier, E. , Hahn, S.A. , 2007. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26, 4442–4452. [DOI] [PubMed] [Google Scholar]
- Tomimaru, Y. , Eguchi, H. , Nagano, H. , Wada, H. , Tomokuni, A. , Kobayashi, S. , Marubashi, S. , Takeda, Y. , Tanemura, M. , Umeshita, K. , Doki, Y. , Mori, M. , 2010. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br. J. Cancer 103, 1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Li, P.F. , 2010. Foxo3a regulates apoptosis by negatively targeting miR-21. J. Biol. Chem. 285, 16958–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Chen, Z. , Meng, Z.Q. , Fan, J. , Luo, J.M. , Liang, W. , Lin, J.H. , Zhou, Z.H. , Chen, H. , Wang, K. , Shen, Y.H. , Xu, Z.D. , Liu, L.M. , 2009. Dual role of Ski in pancreatic cancer cells: tumor-promoting versus metastasis-suppressive function. Carcinogenesis 30, 1497–1506. [DOI] [PubMed] [Google Scholar]
- Yang, C.H. , Yue, J. , Pfeffer, S.R. , Handorf, C.R. , Pfeffer, L.M. , 2011. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J. Biol. Chem. 286, 39172–39178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Yu, L. , Gao, X. , Hu, J. , Wang, J. , Dai, Z. , Wang, J.F. , Zhang, Z. , Lu, S. , Huang, X. , Wang, Z. , Qiu, S. , Wang, X. , Yang, G. , Sun, H. , Tang, Z. , Wu, Y. , Zhu, H. , Fan, J. , 2011. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 29, 4781–4788. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Yu, F. , Jiao, Y. , Feng, J. , Tang, W. , Yao, H. , Gong, C. , Chen, J. , Su, F. , Zhang, Y. , Song, E. , 2011. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 17, 7105–7115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


