Abstract
Patient derived xenografts (PDXs) are increasingly appreciated models in cancer research, particularly for preclinical testing, as they reflect the patient's tumor biology more accurately than cancer cell lines. We have established a collection of 20 breast PDXs and characterized their biological and clinical features, as well as their genetic stability. While most PDXs originated from triple negative breast cancers (70%), our collection comprised five ER + cases (25%). Remarkably, the tumors that produced PDXs derived from a subset of aggressive breast cancers with a high proportion of grade 3 tumors and reduced recurrence‐free survival. Consistent with this, we found significant differences between the transcriptomic signatures of tumors that produced a PDX (Take) and those that did not (No Take). The PDXs faithfully recapitulate the histological features of their primary tumors, and retain an excellent conservation of molecular classification assignment and Copy Number Change (CNC). Furthermore, the CNC profiles of different PDXs established from the same tumor overlap significantly. However, a small fraction of CNCs in the primary tumor that correspond to oligoclonal events were gradually lost during sequential passaging, suggesting that the PDXs' genetic structure eventually stabilizes around a dominant clone present in the tumor of origin. Finally, de novo occurring genetic events covering up to 9% of the genome were found in only a minority of the PDXs, showing that PDXs have limited genetic instability. These data show that breast cancer PDXs represent a subset of aggressive tumors prone to relapse, and that despite of an excellent conservation of original features, they remain genetically dynamic elements.
Keywords: Breast cancer, Patient derived xenografts, Genetic stability, Adverse prognosis
Highlights
A collection of breast cancer PDXs is described.
PDXs represent a subset of clinically aggressive breast tumors.
Most PDXs derive from a dominant genetic subclone present in the tumor of origin.
PDXs are genetically dynamic and susceptible to slow drift.
Abbreviations
- PDX
patient derived xenografts
- CNC
copy number changes
- CGH
comparative genomic hybridization
- RFS
recurrence free survival
1. Introduction
Considerable progress has been made in understanding the molecular mechanisms underlying breast cancer development. A number of new therapeutic strategies are currently being devised aiming at targeting specific oncogenic pathways. However, a significant gap still exists between the experimental work and the development of effective clinical treatments. To validate the anti‐tumor potential of candidate drugs, preclinical studies based on model systems must be performed, and it is paramount that these models accurately reproduce the biological features of the cancer in the patients (Hait, 2010).
Preclinical research traditionally relies on cell lines established from tumor specimens. Cancer cell lines have yielded highly valuable and informative data, and remain indispensable models for molecular genetics and biochemistry studies (Kao et al., 2009). However, cancer cell lines only partially recapitulate the biology of the original tumors, because they propagate as adherent monolayers in vitro, in conditions that differ drastically from those in the tumor. Furthermore, a number of cell lines used routinely have been established decades ago and passed along from one laboratory to another, thus encountering changes in culture conditions that may lead to the derivation of subsets with genetic and phenotypic differences (Nugoli et al., 2003). Finally, patient tumor material is available in small amounts, which limits the number of experiments that can be performed with primary cultures.
Human tumors propagated in immunocompromised animals, called patient derived xenografts (PDXs) or tumor grafts, could be interesting alternatives to cancer cell lines. Research on colorectal and breast cancer has shown that grafts from primary tumors faithfully reproduce the histology and morphology of the tumor they stemmed from (Fiebig et al., 2004; Marangoni et al., 2007). These pioneer studies were the proof of principle that PDXs could be an excellent bridge between clinical material and cancer cell lines, and recent research shows that established PDXs are permanent sources of tumor material that can be subjected to treatment and monitored for response (Landis et al., 2013). Furthermore, breast cancer PDXs faithfully reproduce tumor pathology, growth and metastasis of breast cancer (DeRose et al., 2011), and also seem to maintain the genetic characteristics of their original tumors (Petrillo et al., 2012; Reyal et al., 2012; Zhang et al., 2013). Together these studies demonstrate the power of PDX models for cancer research.
We describe here a collection of 20 established breast cancer PDXs that resulted from engraftment of 130 primary tumors. To investigate what differentiates tumors that produce PDXs from those that do not, we have carefully analyzed the determinants of graft take in breast cancer, and characterized the primary tumors and their resulting PDXs by array‐CGH and transcriptome analysis. We show that tumor grafts from breast cancer display remarkable conservation of the morphological and genetic features of the tumor they stem from. CNC profiles remained stable over several passages, but a small fraction of CNCs in the primary tumor that corresponded to oligoclonal events were gradually lost during sequential passaging. As expected, grade 3 and ER‐ breast tumors were positively selected in the grafting process (Landis et al., 2013; Petrillo et al., 2012; Zhang et al., 2013). Finally, we show that breast tumors that give rise to stabilized PDXs derive from a subset of aggressive cancer prone to relapse and have a transcriptomic signature clearly distinct from tumors that fail to produce PDXs.
2. Materials and methods
2.1. Patients and tumor material
A total of 130 fresh breast cancer samples were collected from the pathology department upon macroscopic dissections, transferred to the animal facility and implanted within a maximum of 60 min after surgical removal. Full description of all the tumors grafted is provided in Supplementary Table 1. This study was reviewed and approved by the Montpellier Cancer Center – Val d'Aurelle Institutional Review Board and informed consent was obtained from all patients. Samples were systematically anonymized. Mean follow‐up time was of 34 months.
2.2. Grafting, passaging and preservation of PDXs
This project was first reviewed and approved by an internal animal ethics committee, and then by the University of Montpellier animal ethics committee. There was always a minimum of three mice per experimental group. We implanted a single fragment of fresh or frozen tumor (∼8 mm3) into the inter‐scapular fat pads of 3 to 4‐week‐old female Swiss‐nude mice. For ER+ tumors, mice were supplemented with estrogen by weekly application on the skin of 10 μl of a 15 mg/ml estrogen (Sigma‐Aldricht, St Louis, MI, USA) solution in ethanol. Tumor growth was measured weekly using calipers. Tumors were passaged onto a further cohort of mice once grafts reached a maximum of 2000 mm3. At each passage, fragments were (1) frozen at −180 °C in L15 culture (Sigma‐Aldricht) medium, with 40% FCS serum, 10% DMSO, for eventual retransplantation, (2) frozen dry at −80 °C for DNA, RNA or protein extraction, (3) fixed into 4% buffered formaldehyde and embedded in paraffin.
2.3. Histological analysis
Histology was assessed on hematoxylin‐eosin stained sections. Immunochemistry was performed on standard sections, deparaffinized, rehydrated and treated for epitope retrieval in boiling EDTA (pH 9). Neutralization of endogenous peroxidase was done with H2O2. Sections were incubated for 20 min at room temperature with anti‐ERa (clone 6F11, 1:100, LeicaBiosystems, St Germain en Laye, France), anti‐PR (clone PgR636, 1:400, Dako, Glostrup, Denmark) mouse monoclonal antibodies. Antibody binding was amplified using the EnVision™ FLEX detection system (Dako). Endogeneous biotin and avidin were saturated with a blocking kit (Vector Laboratories‐CliniSciences, Nanterre, France). Sections were then incubated with primary antibodies or mouse isotype control (Mouse IgG1κ, Sigma–Aldrich) overnight at 4 °C. Sections were then washed 3X in 0.1% Tween20‐PBS solution at 25 °C for 3 h and incubated 10 min with the secondary antibody. Amplification of the signal was done using the ABC kit (Vector Laboratories‐CliniSciences). Sections were washed as above. Binding was visualized by incubating with DAB (3,3‐diaminobenzidine) substrate. The sections were then counterstained with haematoxylin, dehydrated and permanently mounted. Slides were scanned with a digital slide scanner NanoZoomer (Hamamatsu). Pictures were visualized and exported using NDP.view 1 software. Slides were read by a pathologist (F.B.). The presence of invasive carcinoma was assessed and histology of the tumors and corresponding PDX was compared. Immunohistochemistry results were evaluated according to standard practice.
2.4. DNA and RNA extraction
DNA and RNA were isolated from frozen tissues using the QIAmp DNA Mini kit and Rneasy Mini Kit (Qiagen S.A. France, Courtaboeuf, France). Each DNA sample was quantified by nanospectrophotometry (NanoView, GE Healthcare, Orsay, France) and qualified by 0.8% agarose electrophoresis. Qualification of mRNA was performed using a Bioanalyser (Agilent, Santa Clara, CA, USA).
2.5. Array‐CGH
The genomic profiles were obtained by array comparative genomic hybridization (array‐CGH) using HG18 CGH 385K Whole Genome v2.0 array (Roche NimbleGen, Madison, WI, USA). DNA from a pool of 20 normal females was used as reference. For hybridization, 1 μg of genomic DNA and reference DNA were labeled using NimbleGen Dual‐Color DNA Labeling Kit (Roche Diagnostics, Meylan, France). Labeling products were precipitated with isopropanol and resuspended in water. Test (Cy3) and reference (Cy5) samples were combined in 40 μl of NimbleGen Hybridization buffer. Hybridization was performed in a NimbleGen Hybridization system 4 for 48 h at 42 °C with agitation mode B and washed using NimbleGen Wash Buffer kit according to manufacturer's instructions. Arrays were scanned at 5 μm resolution using the GenePix4000B scanner (Axon Instruments, Molecular Devices Corp., Sunnyvale, CA). Data were extracted from scanned images using NimbleScan 2.5 extraction software (Roche NimbleGen, Madison, WI, USA), which allows automated grid alignment, extraction, normalization, and export of data files. Normalized files were used as input for the Nexus 6.1 Software (Biodiscovery, El Segundo, CA, USA). Analysis settings for data segmentation and calling were the following: significant threshold for FASTST2 Segmentation algorithm: 1.0E‐7, Max Continuous Probe Spacing: 1000, Min number of probes per segment: 10, high level gain: 0.485, gain: 0.17, loss: −0.2, homozygous copy loss: −0.485. Hierarchical clustering was done using Nexus 6.1 using average linkage setting.
2.6. Expression profiling and molecular classification
Expression profiling was performed on Affymetrix Human Genome GeneChip U133Plus2 and Biotinylated cRNA were prepared according to the Affymetrix IVT Express protocol from 100 or 200 ng total RNA and hybridization was done as follows. CRNA were fragmented, 12 μg hybridized for 16 h at 45 °C, washed and stained in the Affymetrix Fluidics Station 450 with Hybridization Wash & Stain kit. GeneChips were scanned using the Affymetrix GeneChip Scanner 3000 7G. Raw feature data were normalized using Robust Multi‐array Average (RMA) method (R package affy). All subsequent analyses were performed on normalized datasets. For the CIT/Guedj classification we applied a classical distance‐to‐centroid approach, implemented in the citbcmst R package available at http://cran.rproject.org/web/packages/citbcmst/index.html). For further details see (Guedj et al., 2012). For the Hu, PAM 50 and Sorlie classifications we implemented the script published by Alan Mackay available at http://rock.icr.ac.uk/collaborations/Mackay/centroid.correlations.Eset/ExpressionSetNearest Centroid Correlations.pdf). For the BasalA/B classification we used the gene centroid list as in (Neve et al., 2006). Pearson metrics was employed for all classifications except for Sorlie (Spearman metric). For the VEGF and IL8 signatures the function sig.score from the R program package Genefu was used to compute a signature score after MAS5 normalization of the data (R package affy). Probes list are included in the IL8 (Hu et al., 2009), VEGF (Waugh and Wilson, 2008) and proliferation (Rody et al., 2011) lists are shown in Supplementary Table 2.
2.7. Survival analyses
On grafted tumors we performed Univariate Kaplan–Meier analysis with a log‐rank (cox‐Mantel‐Haenzel) test using the GenePattern package. The CIT cohort dataset was dowloaded from Arrayexpress (E‐MTAB‐365) and the 428 samples stratified with survival follow‐up data as ER + or basal‐like subtype. Univariate Kaplan–Meier was performed with a log‐rank (cox‐Mantel‐Haenzel) test using GenePattern package with the VEGF signature.
2.8. Exome sequencing
High‐throughput exome sequencing was performed by Integragen (Evry, France) using the Human All Exon v4 −70 Mb kit (Agilent) followed by 75 base end‐sequencing on an Illumina HiSEQ 2000 on 6 samples at a mean 60X depth from three primary/PDXs pairs (Gnirke et al., 2009). Bioinformatic analysis used the pipeline provided by Illumina (CASAVA1.8) on the build37 of the human genome and with ELANDv2e alignment algorithm. Annotation includes gene (Refseq), polymorphism (dbsnp132, 1000 genomes) and frequency of mutation in Integragen database (IGdb) of 150 expected to be normal Exome sequenced at Integragen facilities. For each mutation, localization (exon, intron) as well as effect (missense, no‐sense, synonymous) was annotated. Variant detection was done by pair (Primary/PDX) with CASAVA1.8 (Theta = 0.01). This is the standard pipeline for output delivered by our contractor (Integragen SARL, Evry). Due to the absence of Normal blood DNA sequencing from the patient the filtering was done according to a stringent strategy in three steps as described below. (1) Mutations present in the 1000 genomes database or in IGdb were excluded from analysis as they represent polymorphisms or sequencing errors. (2) Only missense or nonsense mutations were considered. (3) Only the 518 genes previously found to be mutated twice in (Banerji et al., 2012) or in the Significantly mutated genes list of (Koboldt et al., 2012) were included. This filtering process in three steps resulted in the approval of 35 mutations for B3977 samples and 28 mutations for B3029 and 5 for the B3921 case (SupTable 3). Due to this low number of mutations in B3921, we considered this sample as unsuitable to determine a general trend in the comparison between primary and PDX. Frequency of the mutated allele was calculated in the primary and the PDX on the basis of respective read numbers.
2.9. Statistical analysis
Anova or t‐test were performed when appropriate using the Rcmdr R package. Multivariate logistic regression analysis was performed to identify independent predictors of the “PDX take”.
3. Results
3.1. Establishment of the breast tumor PDX collection
With the aim to establish stabilized breast cancer PDXs, we grafted a total of 130 fresh breast tumor specimens into the inter‐scapular fat pad of Swiss‐Nude mice within an hour of surgical resection. We chose to graft into the inter‐scapular fat pad rather than the mammary gland because of quicker and easier handling, as our strategy was to graft rapidly a large number of tumors within a limited time span. We rarely observed macroscopic metastases to the lung and never to the liver. All grafted breast tumors were primary tumors, except one tumor that was a local recurrence from a primary breast cancer treated three years earlier in our institution. We observed take (tumor regrowth at first graft) in 39 (30%) cases and stabilization (after at least three serial passages) of 20 (15%) PDX lines. Thus, 19 grafts that initially took were not sustained beyond one or two passages. The majority of these grafts (15/19) were ER + tumors with a slow growth rate. Indeed, while established PDXs required on average 130–150 days of growth between each passage (and 63 days for a group of fast growers; Figure 1), non‐established grafts took about 15 months to become palpable. These slow‐growing grafts were therefore at increased risk of being lost because of premature death of the recipient animal. We found a significant correlation between PDX take and/or establishment, and steroid receptor levels and SBR grade (Table 1). Indeed, a majority of PDXs were of the ER/PR/HER2 triple negative (TN) phenotype (14/20) and predominantly mutated at the TP53 gene (65%), while only five PDXs were ER+ and three were HER2+ (two ER+/HER2+ and one ER−/HER2+) (Figure 2, Supplementary Figures 1B and C).
Figure 1.
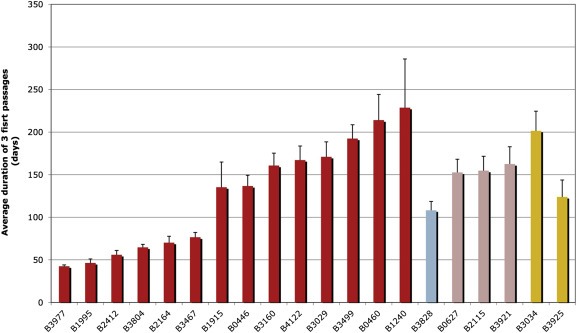
Growth kinetics of established breast PDXs. The average duration of the 3 first passage was calculated for each PDX and used as an estimate of their growth rate. Time is presented in days. Each bar represents a PDX. Color of the bar indicates the molecular subtype to which the PDX was assigned. As shown in Figure 2 red = Bas‐L, light blue = Lum‐B, pink = Lum‐C, orange = m‐Apo.
Table 1.
Clinical and pathological characteristics that differentiate breast tumors that give rise to a PDX from those which fail to do so. Distribution of characteristics showing significant differences are presented for tumors that gave rise to primary take (passage 1) and tumors that reached passage 3 (a stage at which the PDXs were stabilized). Tumor Grade was determined according to the EE‐SBR guide lines. Estrogen and progesterone receptor status was assessed by immunohistochemistry and scored as defined in the Materials and Methods section.
| Passage 1 Nb (%) | No passage 1 Nb (%) | Fisher‐test p‐value | Passage 3 Nb (%) | No passage 3 Nb (%) | Fisher‐test p‐value | |
|---|---|---|---|---|---|---|
| Steroid receptors | 0.0003 | 0.000001 | ||||
| ER + PR+ | 10 (15%) | 55 (85%) | 2 (3%) | 63 (97%) | ||
| ER + PR− | 12 (35%) | 22 (65%) | 4 (12%) | 30 (88%) | ||
| ER−PR− | 16 (53%) | 14 (47%) | 14 (47%) | 16 (53%) | ||
| HER2 | N.S. | N.S | ||||
| Negative | 31 (31%) | 70 (69%) | 16 (16%) | 85 (84%) | ||
| Positive | 7 (25%) | 21 (75%) | 3 (11%) | 25 (89%) | ||
| GRADE | 0.0002 | 0.00005 | ||||
| Grade I | 1 (11%) | 8 (89%) | 0 (0%) | 9 (100%) | ||
| Grade II | 10 (16%) | 53 (84%) | 2 (3%) | 61 (97%) | ||
| Grade III | 28 (48%) | 30 (52%) | 18 (31%) | 40 (69%) | ||
| Size (pT) | 0.01 | 0.034 | ||||
| 1 | 6 (15%) | 35 (85%) | 2 (5%) | 39 (95%) | ||
| 2 and more | 33 (38%) | 55 (62%) | 18 (21%) | 70 (79%) | ||
| pN | N.S. | N.S. | ||||
| 0 | 17 (31%) | 38 (69%) | 10 (18%) | 45 (82%) | ||
| 1 to3 | 19 (30%) | 45 (70%) | 8 (13%) | 56 (87%) | ||
| Relapse | 0.01 | 0.0001 | ||||
| Yes | 13 (62%) | 8 (38%) | 10 (48%) | 11 (52%) | ||
| No | 26 (24%) | 83 (76%) | 10 (9%) | 99 (91%) |
Figure 2.
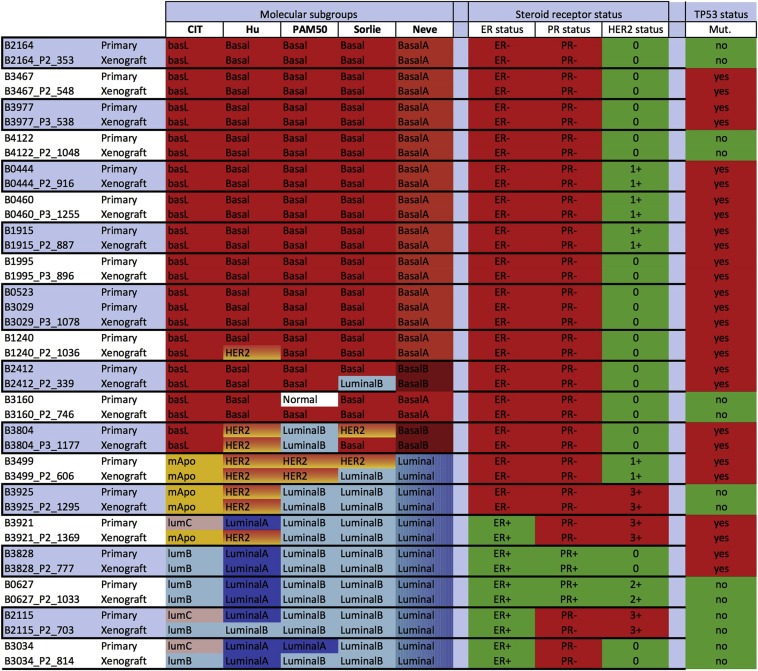
Molecular classification of PDX and tumors of origin are highly concordant. Identical assignment were found in 87% of the cases with at least 3/5 identical classifications for each PDX. Divergences were restricted to the luminal subtypes. Transcriptomes of PDX and corresponding tumor of origin were classified using the Guedj, Hu, PAM 50, Sorlie and Neve breast cancer classifiers and assignments were indicated by a color code. Red: Basal or Bas‐L, Orange: M‐Apo (ER−/HER2+), Pink: Lum‐C (ER+/PR−), light blue: Lum‐B (ER+/PR+/HER2−), dark blue: Luminal A. ER, PR status determined by IHC is presented in red for ER−, in green for ER+. HER2 status was noted negative in green when scored <2+, positive in red score = 3+. TP53 mutation status determined by DNA sequencing, mutated in red, wt in green.
3.2. PDXs reproduce the phenotype of the tumors they originate from
We next investigated whether the PDXs retained the phenotypic features of the tumors of origin. We found that indeed they showed remarkable conservation of histological and morphological features (Supplementary Figure 1A). Furthermore, immunohistochemical staining in PDXs originating from ER + tumors showed that ER and PR expression was stably conserved during passages (Supplementary Figure 1B). We also detected matching HER2 overexpression patterns in HER2+ primary tumors and their corresponding PDXs (Supplementary Figure 1C).
To further assess the phenotypic stability of the PDXs in reference to the primaries, we examined their respective assignments into molecular subtypes. We established expression profiles for the primary tumors and PDXs on Affymetrix U133 Plus 2 GeneChips, and then classified them according to the CIT/Guedj classification (Guedj et al., 2012). Next, we compared these results with four other nearest centroid classifiers (Figure 2). Of the 20 tumors that gave rise to grafts, 13 were assigned to the Bas‐L (in coherence with their TN phenotype), two to the ER‐ mApo (one had been diagnosed HER2 3+, while the second was determined HER2 1+) and five to ER + Lum‐B or Lum‐C molecular subgroups of the CIT/Guedj classification (Guedj et al., 2012). Subgroup assignments using these classifiers were globally concordant, thus confirming the conservation of the expression characteristics between primaries and PDXs (Figure 2). Together these results show that breast cancer PDXs faithfully reproduce the phenotype of the tumor of origin.
3.3. PDXs preserve copy number change profiles of the original tumor
To assess the genomic stability of the PDXs, we established copy number change (CNC) profiles of primary tumors and their PDXs by array‐CGH on high density oligo‐chips. Similar patterns of genomic aberrations in PDXs and the primary tumors they originated from suggest that CNC profiles evolve in parallel in primary tumors and their grafts (Supplementary Figure 2A). However, in a number of cases the CNC profile of the primary tumor was less accentuated than the corresponding PDX profile, which resulted in a higher number of aberration call in the grafts (Supplementary Figure 2A). This effect could be explained by variable contingents of contaminating normal stromal cells in the primary tumors, resulting in the dilution and subsequent attenuation of the CNC signal. In the PDXs, contamination by normal tissue should not occur because stromal cells do not interfere with the tumor DNA signal, as they have murine origin and there is limited homology on our arrays. We estimated the fraction of normal cells present in each primary tumor and then inferred the level of dilution of tumor DNA by normal DNA in the CNC profiles of primary tumors. CNC profiles of primary tumors and PDXs were then normalized taking this dilution factor into account. Indeed, normalization led to a significant decrease in the divergence between CNC profiles from primary tumors and their corresponding PDXs (Supplementary Figure 2B, C). Furthermore, clustering analysis performed on corrected profiles revealed that PDXs and their cognate primaries were systematically co‐clustered, thus showing a high level of correlation (Figure 3).
Figure 3.
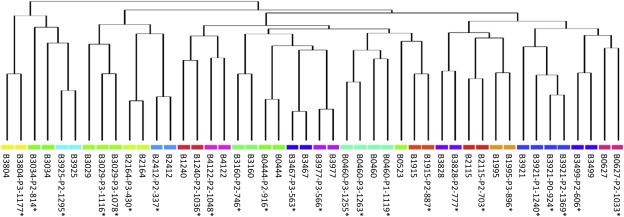
Breast tumors of origin and corresponding PDXs show elevated similarity at the genomic level. CNC profiles of all PDXs (* indicates corrected profiles) and breast tumors of origin were determined by array‐CGH and convergence tested by clustering analysis. Breast tumors and their cognate PDXs were systematically clustered together (pairs, triplet or quadriplet according to profile availability).
Next, we analyzed the genetic stability of the PDXs during passages. Visual inspection revealed that CNC profiles of different grafts obtained from the same tumor were almost interchangeable (Figure 4C). Consistent with this, when compared to the primary tumor, the concordance ratio of CNC profiles of PDXs from passages 0, 1 or 2 suggests that changes occurred mainly during the initial graft (Figure 4B). These data indicate that propagation of breast tumors as xenografts on immunocompromised mice preserves preexisting genomic features, and that these genomic features change little during passages.
Figure 4.
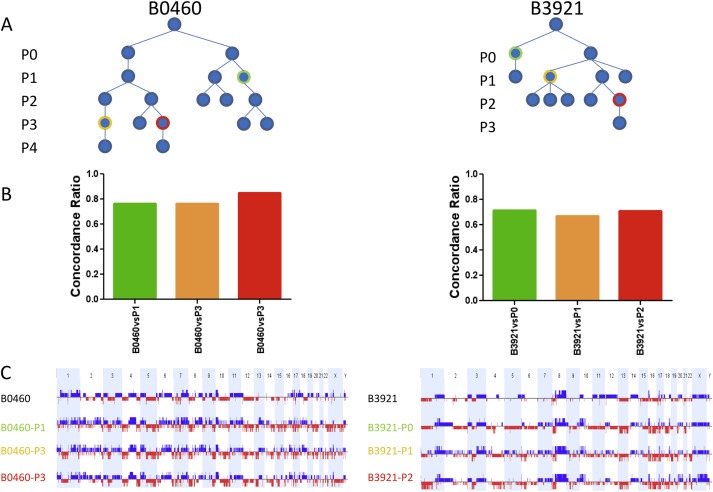
CNC profiles of different grafts from the same primary tumor remain remarkably stable .A: Graft tree indicating the position of the analyzed PDXs which have been highlighted by a color code. B: Histograms showing the fraction of overlapping events in the tumor of origin and the corresponding PDX which are identified by color codes indicated in the graft tree. C: whole genome CNC profiles, gains are shown in blue, losses in red, chromosomes by alternating light blue and white bars. Samples are identified as shown in the tree.
3.4. PDXs uncover limited levels of oligoclonality in breast tumors
Despite the genomic conservation of the PDXs and their tumors of origin, a detailed analysis of the CGH‐profiles revealed that a fraction of the CNCs showed divergence at specific loci (examples are shown in Supplementary Figure 3A, B). We were interested in CNCs present in the tumors of origin but absent in the PDXs, which were clear signs of oligoclonality in the primary tumor. According to the tumor analyzed, these events represented up to 6% of the human genome (Figure 5A) and reached at least 2% in about half of the tumors. These findings suggest that a limited but sizeable level of oligoclonality is present in our breast cancer set. We next determined the precise fraction of de novo occurring anomalies in the PDXs when compared to the original tumors, as these de novo events are representative of ongoing genetic instability. We compared the CGH profiles of the PDXs and their cognate primary tumors after correction for normal stromal cells contamination (Supplementary Figure 2). Similarly to oligoclonal anomalies, de novo events fluctuated from one PDX line to another, representing 0.3–9% of the human genome, with a quarter of the lines encompassing at least 4% of the genome (Figure 5B). Importantly, we observed no correlation between elevated oligoclonality and high occurrence of de novo CNCs. Results obtained from CGH profiles were confirmed by exome sequencing performed on a subset of three primary tumor/PDX pairs that we selected because of their CNC profile differences. However, since we could not obtain normal tissue from the patients to use as reference, our determination of somatic mutations could not be definitive. To circumvent this pitfall, we restricted our analysis to a subset of genes with mutations and indels described in previous high‐throughput sequencing studies on breast cancer (Banerji et al., 2012; Koboldt et al., 2012). We were able to identify in each of the three breast tumors analyzed 5, 28 and 32 genes with deleterious mutations, respectively (Supplementary Table 3), and to compare the mutation frequency in the primary tumor vs the PDX (Supplementary Figure 4). In the primary tumor/PDX pairs with sufficient numbers to perform a statistical analysis, we found that mutation frequencies in the PDXs and in their original tumors were highly correlated (R 2 coefficient 0.65 and 0.80). In particular, we observed a general trend toward an enrichment of mutation frequencies in the PDXs, probably as a consequence of the elimination of normal stromal cells. While a minority of the mutations (possibly representing founder or essential events such as TP53) was selected to homogeneity in the PDXs, a large fraction of the mutations remained at an intermediary frequency rate (0; 400; 0.25–0.7) indicating that they were not subjected to a drastic selection process. Mutations restricted to the primary tumor (and lost in the PDX) represented 5.7% and 17% for B3977 and B3029 respectively. These results confirm our CNC data showing sizable levels of oligoclonality in these tumors. A limited set of de novo events (seven events in two cases) were also found. Together these data show that there are differences in clonality in our breast cancer PDX collection, but that there is a general tendency for the gradual emergence of a dominant clone in the PDXs after three passages. Limited genetic drift could also be observed.
Figure 5.
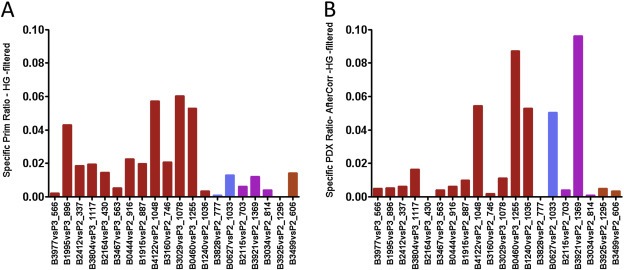
Quantification of CNC specifically found in each primary tumors (A) or specific of each PDX (B). Events specific to the tumors give an insight on tumor heteroclonality, whereas those found only in the PDX represent events acquired de novo during propagation in the mice. A: Each bar represents the percentage of the genome interrogated by the array involved in primary tumor specific CNC. B: the fraction of the genome corresponding to CNC occurring de novo in the PDX. CGH profiles of the PDXs and cognate primary tumors were analyzed after correction for the contamination by normal stromal cells.
3.5. Breast tumors that take correspond to a subset of aggressive breast cancer
The prevalence of grade 3 and TP53‐mutated tumors in our PDX collection suggested that stable grafts could represent a subset of aggressive breast cancers. A multivariate regression logistics analysis showing that PDX take was significantly associated to Grade 3, ER‐ and metastatic disease further supported this model (Supplementary Table 4). To investigate the link between PDX take and aggressive breast cancer, we compared recurrence‐free survival (RFS) in the breast cancer patients. Indeed, we found that patients whose primary tumor gave rise to a PDX had a lower RFS than patients whose tumor did not take (Supplementary Figure 5A). RFS may reflect known differences in outcome between TN basal cancers (predominant in the Take group) and ER + luminal cancers (overrepresented in the No Take group). However, when we stratified the patients in TN and ER + tumors, we still found a significant difference between RFS in the Take and the No Take groups in ER + tumors, and an observable trend in TN breast cancers (Supplementary Figure 5B, C).
The contrast between outcome in patients from Take and No Take groups raised the possibility that tumors that produce PDXs have distinct gene expression signatures from tumors that do not take. Our first attempt to identify a set of genes differentially expressed in the breast primary tumors using a supervised approach did not succeed because of limited sample size and insufficient statistical power. We therefore selected 16 expression signatures with reported prognostic significance in breast cancer to test for significant transcriptomic differences in the primary tumors used in our set. Five of these expression signatures showed a significant increase in the Take group, namely GGI‐grade (Sotiriou et al., 2006), proliferation (Rody et al., 2011), wound healing (Chang et al., 2004), VEGF (Hu et al., 2009) and IL8 (Waugh and Wilson, 2008)(Figure 6). To avoid a confounding effect due to compositional differences, we classified the tumors according to molecular subtypes (namely Bas‐L and Lum‐B/Lum‐C for the two subtypes with a sufficiently large sample size) and repeated the signature analysis. Remarkably, GGI‐grade was significantly increased in the Take vs the No Take groups in both Bas‐L and Lum tumors, while three signatures showed a restricted pattern. The wound healing signature was significantly increased in the Take group in Lum‐B/Lum‐C tumors, whereas VEGF and IL8 signatures were restricted to the Take group in Bas‐L tumors. Interestingly, the proliferation signature showed differential expression in Lum tumors but did not reach significance, likely due to small sample size. Taken together, these results show that tumors that give rise to PDXs correspond to a distinct subgroup of aggressive breast cancers irrespective of the molecular subtype they belong to.
Figure 6.
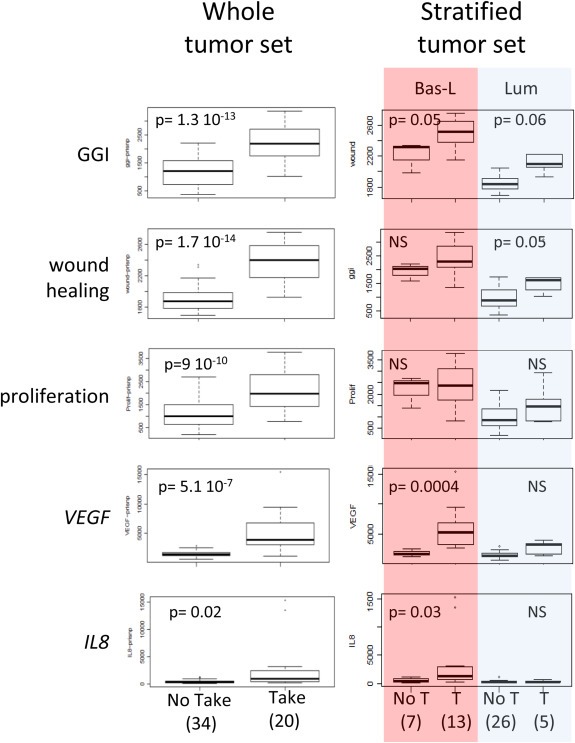
Expression differences between primary tumors that give rise to PDX (Take) and those that did not (No Take). Number of cases in each category are indicated in brackets. We present 5 expression signatures with known prognostic significance that showed significant expression differences. The tumor set was stratified according to molecular subtypes to verify whether these differences were not due to differences in composition of the Take and the No Take groups. GGI identifies the molecular grade signature.
4. Discussion
4.1. Grafting efficiency
In comparison to other cancers, such as colorectal cancer, breast cancer PDXs are characterized by a relatively low take rate upon grafting on immunocompromised mice. In particular, several studies have shown that high grade and ER‐negative tumors are overrepresented in breast PDXs (DeRose et al., 2011; Landis et al., 2013; Marangoni et al., 2007; Zhang et al., 2013). Our work is consistent with these findings, given that from the 20 stabilized PDXs we established, 14 stemmed from TN breast cancers and 18 were grade 3. Furthermore, the number of takes at first graft (39 take out of 130 grafts) was almost double the number of stabilized PDXs (at least three passages). Grafts that we could not stabilize were predominantly slow‐growing ER + tumors. It therefore appears that despite estradiol supplementation and grafting on female mice, ER + breast tumors (predominantly of grade 2 in our series) are negatively selected in this system.
Research on melanoma shows that some xenografts may require severely immunosuppressed animals, such as NOD‐SCID gamma‐delta mice, and it has therefore been proposed that the level of immunosuppression of recipient mice is a determinant factor for graft stabilization (Quintana et al., 2008). However, our attempt to graft on severely immunocompromised mice did not lead to a clear breakthrough, consistent with what others reported for breast cancer grafts (DeRose et al., 2011; Landis et al., 2013). It has also been suggested that the graft site could make a difference in graft take rates. For instance, breast cancer orthotopic grafting in cleared mammary fat pads has been preferred by several groups, but did not lead to a clear increase of ER + tumor take rate (DeRose et al., 2011; Zhang et al., 2013). Another alternative could be to graft into the renal capsule (Cutz et al., 2006), however, this is a complex procedure and it is therefore not very commonly used. Hence, primary breast tumor grafting is still open for improvement and co‐grafting of mesenchymal feeder cells or engineered mice expressing cocktails of cytokines could represent interesting leads.
4.2. PDXs represent a subset of aggressive and recurrence‐prone breast tumors
In our collection the permanently stabilized PDXs corresponded to a subset of aggressive breast cancers associated to a lower RFS. This correlation between graft stabilization and cancer aggressiveness is in line with reports showing that metastatic lesions, such as pleural effusions, have a high take rate (Landis et al., 2013). We found that the potential to take and produce xenografts is inherent to the primary tumors, as demonstrated by the clear differences at the RNA expression level between tumors that took and those that did not. Overall, tumors producing stable PDXs showed increased expression of several gene signatures associated with adverse prognosis, namely GGI, wound healing, proliferation, IL8 and VEGF (Chang et al., 2004; Hu et al., 2009; Rody et al., 2011; Sotiriou et al., 2006; Waugh and Wilson, 2008). The core of the first three signatures is built around genes governing cell cycle progression, DNA synthesis and repair genes, which suggests there is increased proliferation in the tumors of the Take group. Elevated expression of IL8 and VEGF is characteristic of aggressive breast tumors. In particular, elevated VEGF expression is known as a marker of adverse prognosis in breast cancer and is associated with hypoxia and increased vasculature (Dhakal et al., 2012; Gasparini, 2000) (Supplementary Figure 6). Furthermore, increased IL8 signaling in breast cancer has been shown to favor cancer stem cell maintenance and propagation (Ginestier et al., 2010; Fernando et al., 2011), by way of cross‐connections with PI3K/AKT and SRC (Hartman et al., 2013; Singh et al., 2013). It is reasonable to assume that elevated production of IL8 and/or VEGF by tumor cells (Hu et al., 2009; Rody et al., 2011; Waugh and Wilson, 2008) could be critical for the grafting process and thus contribute to the higher take rate of basal‐like breast tumors, for instance, by favoring cancer stem cell renewal and the angiogenic shift necessary to the long‐term establishment of tumors of large size (Waugh and Wilson, 2008).
4.3. PDXs reproduce the phenotypic and genetic characteristics of the grafted tumor fragment
An excellent conservation of the histological features of the tumor of origin in breast cancer PDXs was first shown by Marangoni and colleagues (Marangoni et al., 2007), and then repeatedly observed by others (DeRose et al., 2011; Landis et al., 2013). Our results are in agreement with these observations, thus confirming that PDXs perfectly recapitulate the morphological characteristics of primary tumors. Because sporadic loss of ER expression in PDXs established from ER + breast tumors has been reported (Bergamaschi et al., 2009), we carefully checked ER expression in our PDXs. However, we could not observe any reversion of ER expression in our ER + PDXs (Supplementary Figure 1B). Importantly, these findings were corroborated by transcriptome analysis in the primary tumors and corresponding PDXs, which show very similar expression profiles and molecular classification assignments, in concordance with recent research (Petrillo et al., 2012).
Besides transcriptomic stability, the PDXs show remarkable stability at the genomic level, as clearly demonstrated by the excellent conservation of the CNC profiles of the tumors of origin. These results differ slightly from that of Reyal and coworkers, however, who observed a global increase in the number of CNCs in the PDXs (Reyal et al., 2012). Our analysis suggests that this discrepancy can be attributed to the presence of large contingents of contaminating stromal cells in the primary tumors, which dilutes out the tumors' DNA signal. Interestingly, we found that the CNC profiles in different PDXs established in parallel from the same tumor are nearly identical. These findings are very reassuring concerning the genetic stability of breast PDXs and their reliability as cancer models. We also found that oligoclonal CNC differences represent 0.2–6% of the genome, depending on the primary tumor, which is significantly less than what others reported for some breast tumors (Nik‐Zainal et al., 2012). It is possible that our assessment of oligoclonality levels based on CNC profile differences is an underestimation, when compared to the levels detected by whole genome sequencing. Nevertheless, our data are consistent with findings by Nik‐Zainal et al. (2012) showing that oligoclonality varies appreciably from one breast tumor to another, and that the genetic structure of each tumor stabilizes around a dominant clone (Nik‐Zainal et al., 2012). The analysis of different PDXs established from the same tumor revealed that there is gradual loss of the minor clones over sequential passages. This result is however not surprising, as grafting onto immunocompromised mice represents a change of environment and selective pressure. In addition, gradual loss of minor clones could also result from the outgrowth of de novo occurring mutations.
In summary, our results show that PDXs are a faithful representation of the tumor of origin, but sequential passages are associated with gradual selection of a dominant genetic clone and some models can show limited genetic drift. These data suggest that PDXs are dynamic biologic elements that should therefore be regularly checked for genetic and phenotypic stability.
4.4. PDXs as cancer models
Grafts established from clinical tumor material are increasingly appreciated models for preclinical testing because they are closer representations of the disease than cancer cell lines. PDXs are particularly well suited to test targeted therapy, as they can be comprehensively characterized at the molecular level. PDXs also allow detection of the oncogenic cascades that fuel the tumor. One could therefore expect that grafts will be increasingly used for the definition of therapeutic combinations, and they should play an important role in the current therapeutic evolution toward personalized medicine. Furthermore, PDXs could be key for unraveling the causes and the mechanisms underlying treatment failure, and for testing alternative therapies to overcome tumor resistance. Indeed, resistant and sensitive sublines can be derived from tumors with a clinically documented response to therapy, and sublines with variable levels of treatment sensitivity can be further established in vivo (Landis et al., 2013). Hence, a precise assessment of the tumor's level of oligoclonality and/or genetic plasticity could be of great value. Combined with short term ex vivo culture, it is foreseeable that PDXs will give rise to families of genetically modified models expressing shRNA, specific target genes or reporter genes, which will open the way to functional assays similar to those currently performed in cancer cell lines. For instance, fluorescent or luminescent models will be of special interest to monitor metastatic dissemination using in vivo imaging and to test compounds for the reduction of the tumor burden.
In summary, we have established a valuable collection of breast cancer PDXs that faithfully reproduce the tumors of origin. We have shown that breast PDXs retain transcriptomic and genomic stability over sequential passages, with gradual selection of a dominant genetic clone. Our results show that breast tumors producing stabilized PDXs derive from a subset of aggressive cancers associated with a poor clinical outcome, and that despite of an excellent conservation of original features, they remain genetically dynamic elements. We conclude that our PDX collection could be an important model for breast cancer research and preclinical testing.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Acknowledgements
This work was supported by grants from INCa MOPRECLI and INCa “Role of cancer stem cells during metastatic progression of breast cancer” as well as Ligue Contre le Cancer comité du Haut‐Rhin grant 2010 and Ligue Contre le Cancer Comité de l'Hérault grant 2011. The authors wish to thank the personnel of the IRCM animal facility team, the histology (RHEM) platform, the Affymetrix platform of Montpellier and Dr Caroline Mollevi from the Biostatistics platform at ICM for their help in this project. The constant support of ICM and SIRIC Montpellier‐Cancer is gratefully acknowledged.
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.11.010
du Manoir Stanislas, Orsetti Béatrice, Bras-Gonçalves Rui, Nguyen Tien-Tuan, Lasorsa Laurence, Boissière Florence, Massemin Blandine, Colombo Pierre-Emmanuel, Bibeau Frédéric, Jacot William and Theillet Charles, (2014), Breast tumor PDXs are genetically plastic and correspond to a subset of aggressive cancers prone to relapse, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.11.010.
References
- Banerji, S. , Cibulskis, K. , Rangel-Escareno, C. , Brown, K.K. , Carter, S.L. , Frederick, A.M. , Lawrence, M.S. , Sivachenko, A.Y. , Sougnez, C. , Zou, L. , Cortes, M.L. , Fernandez-Lopez, J.C. , Peng, S. , Ardlie, K.G. , Auclair, D. , Bautista-Piña, V. , Duke, F. , Francis, J. , Jung, J. , Maffuz-Aziz, A. , Onofrio, R.C. , Parkin, M. , Pho, N.H. , Quintanar-Jurado, V. , Ramos, A.H. , Rebollar-Vega, R. , Rodriguez-Cuevas, S. , Romero-Cordoba, S.L. , Schumacher, S.E. , Stransky, N. , Thompson, K.M. , Uribe-Figueroa, L. , Baselga, J. , Beroukhim, R. , Polyak, K. , Sgroi, D.C. , Richardson, A.L. , Jimenez-Sanchez, G. , Lander, E.S. , Gabriel, S.B. , Garraway, L.A. , Golub, T.R. , Melendez-Zajgla, J. , Toker, A. , Getz, G. , Hidalgo-Miranda, A. , Meyerson, M. , 2012. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 486, 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi, A. , Hjortland, G.O. , Triulzi, T. , Sørlie, T. , Johnsen, H. , Ree, A.H. , Russnes, H.G. , Tronnes, S. , Maelandsmo, G.M. , Fodstad, O. , Borresen-Dale, A.-L. , Engebraaten, O. , 2009. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol. Oncol.. 3, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.Y. , Sneddon, J.B. , Alizadeh, A.A. , Sood, R. , West, R.B. , Montgomery, K. , Chi, J.-T. , van de Rijn, M. , Botstein, D. , Brown, P.O. , 2004. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol.. 2, E7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutz, J.C. , Guan, J. , Bayani, J. , Yoshimoto, M. , Xue, H. , Sutcliffe, M. , English, J. , Flint, J. , LeRiche, J. , Yee, J. , Squire, J.A. , Gout, P.W. , Lam, S. , Wang, Y.Z. , 2006. Establishment in severe combined immunodeficiency mice of subrenal capsule xenografts and transplantable tumor lines from a variety of primary human lung cancers: potential models for studying tumor progression-related changes. Clin. Cancer Res.. 12, 4043–4054. [DOI] [PubMed] [Google Scholar]
- DeRose, Y.S. , Wang, G. , Lin, Y.-C. , Bernard, P.S. , Buys, S.S. , Ebbert, M.T.W. , Factor, R. , Matsen, C. , Milash, B.A. , Nelson, E. , Neumayer, L. , Randall, R.L. , Stijleman, I.J. , Welm, B.E. , Welm, A.L. , 2011. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med.. 17, 1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal, H.P. , Naume, B. , Synnestvedt, M. , Borgen, E. , Kaaresen, R. , Schlichting, E. , Wiedswang, G. , Bassarova, A. , Holm, R. , Giercksky, K.-E. , Nesland, J.M. , 2012. Expression of vascular endothelial growth factor and vascular endothelial growth factor receptors 1 and 2 in invasive breast carcinoma: prognostic significance and relationship with markers for aggressiveness. Histopathology. 61, 350–364. [DOI] [PubMed] [Google Scholar]
- Fernando, R.I. , Castillo, M.D. , Litzinger, M. , Hamilton, D.H. , Palena, C. , 2011. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res.. 71, 5296–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig, H.H. , Maier, A. , Burger, A.M. , 2004. Clonogenic assay with established human tumour xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur. J. Cancer. 40, 802–820. [DOI] [PubMed] [Google Scholar]
- Gasparini, G. , 2000. Prognostic value of vascular endothelial growth factor in breast cancer. Oncologist. 5, (Suppl. 1) 37–44. [DOI] [PubMed] [Google Scholar]
- Ginestier, C. , Liu, S. , Diebel, M.E. , Korkaya, H. , Luo, M. , Brown, M. , Wicinski, J. , Cabaud, O. , Charafe-Jauffret, E. , Birnbaum, D. , Guan, J.-L. , Dontu, G. , Wicha, M.S. , 2010. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J. Clin. Invest.. 120, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke, A. , Melnikov, A. , Maguire, J. , Rogov, P. , LeProust, E.M. , Brockman, W. , Fennell, T. , Giannoukos, G. , Fisher, S. , Russ, C. , Gabriel, S. , Jaffe, D.B. , Lander, E.S. , Nusbaum, C. , 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol.. 27, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj, M. , Marisa, L. , de Reynies, A. , Orsetti, B. , Schiappa, R. , Bibeau, F. , Macgrogan, G. , Lerebours, F. , Finetti, P. , Longy, M. , Bertheau, P. , Bertrand, F. , Bonnet, F. , Martin, A.L. , Feugeas, J.P. , Bièche, I. , Lehmann-Che, J. , Lidereau, R. , Birnbaum, D. , Bertucci, F. , de Thé, H. , Theillet, C. , 2012. A refined molecular taxonomy of breast cancer. Oncogene. 31, 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait, W.N. , 2010. Anticancer drug development: the grand challenges. Nat. Rev. Drug Discov.. 9, 253–254. [DOI] [PubMed] [Google Scholar]
- Hartman, Z.C. , Poage, G.M. , Hollander, den, P. , Tsimelzon, A. , Hill, J. , Panupinthu, N. , Zhang, Y. , Mazumdar, A. , Hilsenbeck, S.G. , Mills, G.B. , Brown, P.H. , 2013. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res.. 73, 3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Fan, C. , Livasy, C. , He, X. , Oh, D.S. , Ewend, M.G. , Carey, L.A. , Subramanian, S. , West, R. , Ikpatt, F. , Olopade, O.I. , van de Rijn, M. , Perou, C.M. , 2009. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med.. 7, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, J. , Salari, K. , Bocanegra, M. , Choi, Y.-L. , Girard, L. , Gandhi, J. , Kwei, K.A. , Hernandez-Boussard, T. , Wang, P. , Gazdar, A.F. , Minna, J.D. , Pollack, J.R. , 2009. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE. 4, e6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt, D.C. , Fulton, R.S. , McLellan, M.D. , Schmidt, H. , Kalicki-Veizer, J. , Mcmichael, J.F. , Fulton, L.L. , Dooling, D.J. , Ding, L. , Mardis, E.R. , Wilson, R.K. , Ally, A. , Balasundaram, M. , Butterfield, Y.S.N. , Carlsen, R. , Carter, C. , Chu, A. , Chuah, E. , Chun, H.-J.E. , Coope, R.J.N. , Dhalla, N. , Guin, R. , Hirst, C. , Hirst, M. , Holt, R.A. , Lee, D. , Li, H.I. , Mayo, M. , Moore, R.A. , Mungall, A.J. , Pleasance, E. , Gordon Robertson, A. , Schein, J.E. , Shafiei, A. , Sipahimalani, P. , Slobodan, J.R. , Stoll, D. , Tam, A. , Thiessen, N. , Varhol, R.J. , Wye, N. , Zeng, T. , Zhao, Y. , Birol, I. , Jones, S.J.M. , Marra, M.A. , Cherniack, A.D. , Saksena, G. , Onofrio, R.C. , Pho, N.H. , Carter, S.L. , Schumacher, S.E. , Tabak, B. , Hernandez, B. , Gentry, J. , Nguyen, H. , Crenshaw, A. , Ardlie, K. , Beroukhim, R. , Winckler, W. , Getz, G. , Gabriel, S.B. , Meyerson, M. , Chin, L. , Park, P.J. , Kucherlapati, R. , Hoadley, K.A. , Todd Auman, J. , Fan, C. , Turman, Y.J. , Shi, Y. , Li, L. , Topal, M.D. , He, X. , Chao, H.-H. , Prat, A. , Silva, G.O. , Iglesia, M.D. , Zhao, W. , Usary, J. , Berg, J.S. , Adams, M. , Booker, J. , Wu, J. , Gulabani, A. , Bodenheimer, T. , Hoyle, A.P. , Simons, J.V. , Soloway, M.G. , Mose, L.E. , Jefferys, S.R. , Balu, S. , Parker, J.S. , Neil Hayes, D. , Perou, C.M. , Malik, S. , Mahurkar, S. , Shen, H. , Weisenberger, D.J. , Triche, T. , Lai, P.H. , Bootwalla, M.S. , Maglinte, D.T. , Berman, B.P. , Van Den Berg, D.J. , Baylin, S.B. , Laird, P.W. , Creighton, C.J. , Donehower, L.A. , Getz, G. , Noble, M. , Voet, D. , Saksena, G. , Gehlenborg, N. , DiCara, D. , Zhang, J. , Zhang, H. , Wu, C.-J. , Yingchun Liu, S. , Lawrence, M.S. , Zou, L. , Sivachenko, A. , Lin, P. , Stojanov, P. , Jing, R. , Cho, J. , Sinha, R. , Park, R.W. , Nazaire, M.-D. , Robinson, J. , Thorvaldsdottir, H. , Mesirov, J. , Park, P.J. , Chin, L. , Reynolds, S. , Kreisberg, R.B. , Bernard, B. , Bressler, R. , Erkkila, T. , Lin, J. , Thorsson, V. , Zhang, W. , Shmulevich, I. , Ciriello, G. , Weinhold, N. , Schultz, N. , Gao, J. , Cerami, E. , Gross, B. , Jacobsen, A. , Sinha, R. , Arman Aksoy, B. , Antipin, Y. , Reva, B. , Shen, R. , Taylor, B.S. , Ladanyi, M. , Sander, C. , Anur, P. , Spellman, P.T. , Lu, Y. , Liu, W. , Verhaak, R.R.G. , Mills, G.B. , Akbani, R. , Zhang, N. , Broom, B.M. , Casasent, T.D. , Wakefield, C. , Unruh, A.K. , Baggerly, K. , Coombes, K. , Weinstein, J.N. , Haussler, D. , Benz, C.C. , Stuart, J.M. , Benz, S.C. , Zhu, J. , Szeto, C.C. , Scott, G.K. , Yau, C. , Paull, E.O. , Carlin, D. , Wong, C. , Sokolov, A. , Thusberg, J. , Mooney, S. , Ng, S. , Goldstein, T.C. , Ellrott, K. , Grifford, M. , Wilks, C. , Ma, S. , Craft, B. , Yan, C. , Hu, Y. , Meerzaman, D. , Gastier-Foster, J.M. , Bowen, J. , Ramirez, N.C. , Black, A.D. , Pyatt, R.E. , White, P. , Zmuda, E.J. , Frick, J. , Lichtenberg, T.M. , Brookens, R. , George, M.M. , Gerken, M.A. , Harper, H.A. , Leraas, K.M. , Wise, L.J. , Tabler, T.R. , McAllister, C. , Barr, T. , Hart-Kothari, M. , Tarvin, K. , Saller, C. , Sandusky, G. , Mitchell, C. , Iacocca, M.V. , Brown, J. , Rabeno, B. , Czerwinski, C. , Petrelli, N. , Dolzhansky, O. , Abramov, M. , Voronina, O. , Potapova, O. , Marks, J.R. , Suchorska, W.M. , Murawa, D. , Kycler, W. , Ibbs, M. , Korski, K. , Spychała, A. , Murawa, P. , Brzeziński, J.J. , Perz, H. , Łaźniak, R. , Teresiak, M. , Tatka, H. , Leporowska, E. , Bogusz-Czerniewicz, M. , Malicki, J. , Mackiewicz, A. , Wiznerowicz, M. , Van Le, X. , Kohl, B. , Viet Tien, N. , Thorp, R. , Van Bang, N. , Sussman, H. , Duc Phu, B. , Hajek, R. , Phi Hung, N. , Viet The Phuong, T. , Quyet Thang, H. , Zaki Khan, K. , Penny, R. , Mallery, D. , Curley, E. , Shelton, C. , Yena, P. , Ingle, J.N. , Couch, F.J. , Lingle, W.L. , King, T.A. , Maria Gonzalez-Angulo, A. , Mills, G.B. , Dyer, M.D. , Liu, S. , Meng, X. , Patangan, M. , Waldman, F. , Stöppler, H. , Kimryn Rathmell, W. , Thorne, L. , Huang, M. , Boice, L. , Hill, A. , Morrison, C. , Gaudioso, C. , Bshara, W. , Daily, K. , Egea, S.C. , Pegram, M.D. , Gomez-Fernandez, C. , Dhir, R. , Bhargava, R. , Brufsky, A. , Shriver, C.D. , Hooke, J.A. , Leigh Campbell, J. , Mural, R.J. , Hu, H. , Somiari, S. , Larson, C. , Deyarmin, B. , Kvecher, L. , Kovatich, A.J. , Ellis, M.J. , King, T.A. , Hu, H. , Couch, F.J. , Mural, R.J. , Stricker, T. , White, K. , Olopade, O. , Ingle, J.N. , Luo, C. , Chen, Y. , Marks, J.R. , Waldman, F. , Wiznerowicz, M. , Bose, R. , Chang, L.-W. , Beck, A.H. , Maria Gonzalez-Angulo, A. , Pihl, T. , Jensen, M. , Sfeir, R. , Kahn, A. , Chu, A. , Kothiyal, P. , Wang, Z. , Snyder, E. , Pontius, J. , Ayala, B. , Backus, M. , Walton, J. , Baboud, J. , Berton, D. , Nicholls, M. , Srinivasan, D. , Raman, R. , Girshik, S. , Kigonya, P. , Alonso, S. , Sanbhadti, R. , Barletta, S. , Pot, D. , Sheth, M. , Demchok, J.A. , Mills Shaw, K.R. , Yang, L. , Eley, G. , Ferguson, M.L. , Tarnuzzer, R.W. , Zhang, J. , Dillon, L.A.L. , Buetow, K. , Fielding, P. , Ozenberger, B.A. , Guyer, M.S. , Sofia, H.J. , Palchik, J.D. , 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis, M.D. , Lehmann, B.D. , Pietenpol, J.A. , Chang, J.C. , 2013. Patient-derived breast tumor xenografts facilitating personalized cancer therapy. Breast Cancer Res.. 15, 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni, E. , Vincent-Salomon, A. , Auger, N. , Degeorges, A. , Assayag, F. , de Cremoux, P. , de Plater, L. , Guyader, C. , De Pinieux, G. , Judde, J.G. , Rebucci, M. , Tran-Perennou, C. , Sastre-Garau, X. , Sigal-Zafrani, B. , Delattre, O. , Dieras, V. , Poupon, M.-F. , 2007. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res.. 13, 3989–3998. [DOI] [PubMed] [Google Scholar]
- Neve, R.M. , Chin, K. , Fridlyand, J. , Yeh, J. , Baehner, F.L. , Fevr, T. , Clark, L. , Bayani, N. , Coppe, J.-P. , Tong, F. , Speed, T. , Spellman, P.T. , DeVries, S. , Lapuk, A. , Wang, N.J. , Kuo, W.-L. , Stilwell, J.L. , Pinkel, D. , Albertson, D.G. , Waldman, F.M. , McCormick, F. , Dickson, R.B. , Johnson, M.D. , Lippman, M. , Ethier, S. , Gazdar, A. , Gray, J.W. , 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S. , Van Loo, P. , Wedge, D.C. , Alexandrov, L.B. , Greenman, C.D. , Lau, K.W. , Raine, K. , Jones, D. , Marshall, J. , Ramakrishna, M. , Shlien, A. , Cooke, S.L. , Hinton, J. , Menzies, A. , Stebbings, L.A. , Leroy, C. , Jia, M. , Rance, R. , Mudie, L.J. , Gamble, S.J. , Stephens, P.J. , Mclaren, S. , Tarpey, P.S. , Papaemmanuil, E. , Davies, H.R. , Varela, I. , McBride, D.J. , Bignell, G.R. , Leung, K. , Butler, A.P. , Teague, J.W. , Martin, S. , Jönsson, G. , Mariani, O. , Boyault, S. , Miron, P. , Fatima, A. , Langerød, A. , Aparicio, S.A.J.R. , Tutt, A. , Sieuwerts, A.M. , Borg, A. , Thomas, G. , Salomon, A.V. , Richardson, A.L. , Børresen-Dale, A.-L. , Futreal, P.A. , Stratton, M.R. , Campbell, P.J. , Breast Cancer Working Group of the International Cancer Genome Consortium, 2012. The life history of 21 breast cancers. Cell. 149, 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugoli, M. , Chuchana, P. , Vendrell, J. , Orsetti, B. , Ursule, L. , Nguyen, C. , Birnbaum, D. , Douzery, E.J.P. , Cohen, P. , Theillet, C. , 2003. Genetic variability in MCF-7 sublines: evidence of rapid genomic and RNA expression profile modifications. BMC Cancer. 3, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo, L.A. , Wolf, D.M. , Kapoun, A.M. , Wang, N.J. , Barczak, A. , Xiao, Y. , Korkaya, H. , Baehner, F. , Lewicki, J. , Wicha, M.S. , Spellman, P.T. , Gray, J.W. , van't Veer, L.J. , Esserman, L.J. , 2012. Xenografts faithfully recapitulate breast cancer-specific gene expression patterns of parent primary breast tumors. Breast Cancer Res. Treat.. 135, 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana, E. , Shackleton, M. , Sabel, M.S. , Fullen, D.R. , Johnson, T.M. , Morrison, S.J. , 2008. Efficient tumour formation by single human melanoma cells. Nature. 456, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyal, F. , Guyader, C. , Decraene, C. , Lucchesi, C. , Auger, N. , Assayag, F. , De Plater, L. , Gentien, D. , Poupon, M.-F. , Cottu, P. , de Cremoux, P. , Gestraud, P. , Vincent-Salomon, A. , Fontaine, J.-J. , Roman-Roman, S. , Delattre, O. , Decaudin, D. , Marangoni, E. , 2012. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res.. 14, R11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rody, A. , Karn, T. , Liedtke, C. , Pusztai, L. , Ruckhaeberle, E. , Hanker, L. , Gaetje, R. , Solbach, C. , Ahr, A. , Metzler, D. , Schmidt, M. , Müller, V. , Holtrich, U. , Kaufmann, M. , 2011. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res.. 13, R97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, J.K. , Farnie, G. , Bundred, N.J. , Simoes, B.M. , Shergill, A. , Landberg, G. , Howell, S.J. , Clarke, R.B. , 2013. Targeting CXCR1/2 significantly reduces breast cancer stem cell activity and increases the efficacy of inhibiting HER2 via HER2-dependent and – Independent mechanisms. Clin. Cancer Res.. 19, 643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou, C. , Wirapati, P. , Loi, S. , Harris, A. , Fox, S. , Smeds, J. , Nordgren, H. , Farmer, P. , Praz, V. , Haibe-Kains, B. , Desmedt, C. , Larsimont, D. , Cardoso, F. , Peterse, H. , Nuyten, D. , Buyse, M. , van de Vijver, M.J. , Bergh, J. , Piccart, M. , Delorenzi, M. , 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst.. 98, 262–272. [DOI] [PubMed] [Google Scholar]
- Waugh, D.J.J. , Wilson, C. , 2008. The interleukin-8 pathway in cancer. Clin. Cancer Res.. 14, 6735–6741. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Claerhout, S. , Prat, A. , Dobrolecki, L. , Petrovic, I. , Lai, Q. , Landis, M. , Wiechmann, L. , Schiff, R. , Giuliano, M. , Wong, H. , Fuqua, S. , Contreras, A. , Gutierrez, C. , Huang, J. , Mao, S. , Pavlick, A. , Froehlich, A.M. , Wu, M.F. , Tsimelzon, A. , Hilsenbeck, S.G. , Chen, E. , Zuloaga, P. , Shaw, C. , Rimawi, M.F. , Perou, C.M. , Mills, G.B. , Chang, J.C. , Lewis, M.T. , 2013. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenografts. Cancer Res.. 73, 4885–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data


