Abstract
Several biomarkers have been proposed as useful parameters to better specify the prognosis or to delineate new target therapy strategies for glioblastoma patients. MicroRNAs could represent putative target molecules, considering their role in tumorigenesis, cancer progression and their specific tissue expression. Although several studies have tried to identify microRNA signature for glioblastoma, a microRNA profile is still far from being well‐defined.
In this work the expression of 19 microRNAs (miR‐7, miR‐9, miR‐9∗, miR‐10a, miR‐10b, miR‐17, miR‐20a, miR‐21, miR‐26a, miR‐27a, miR‐31, miR‐34a, miR‐101, miR‐137, miR‐182, miR‐221, miR‐222, miR‐330, miR‐519d) was evaluated in sixty formalin‐fixed and paraffin‐embedded glioblastoma samples using a locked nucleic acid real‐time PCR. Moreover, a comparison of miRNA expressions was performed between primary brain neoplasias of different grades (grades IV–I).
The analysis of 14 validated miRNA expression in the 60 glioblastomas, using three different non‐neoplastic references as controls, revealed a putative miRNA signature: mir‐10b and miR‐21 were up‐regulated, while miR‐7, miR‐31, miR‐101, miR‐137, miR‐222 and miR‐330 were down‐regulated in glioblastomas. Comparing miRNA expression between glioblastoma group and gliomas of grades I–III, 3 miRNAs (miR‐10b, mir‐34a and miR‐101) showed different regulation statuses between high‐grade and low‐grade tumors. miR‐10b was up‐regulated in high grade and significantly down‐regulated in low‐grade gliomas, suggesting that could be a candidate for a GBM target therapy.
This study provides further data for the identification of a miRNA profile for glioblastoma and suggests that different‐grade neoplasia could be characterized by different expression of specific miRNAs.
Keywords: MicroRNA, Glioblastoma, Brain neoplasia, Low‐grade brain tumors, Real‐time PCR
Highlights
MicroRNA profiles in glioblastoma depend on used non‐neoplastic reference.
Glioblastomas are characterized by a specific miRNA expression pattern.
Low‐grade brain neoplasias show different miRNA expression values than glioblastomas.
Abbreviations
- GBM
Glioblastoma
- miR
MicroRNA
- FFPE
Formalin-Fixed and Paraffin-Embedded
- qRT-LNAPCR
Real-Time Locked Nucleic Acid quantitative PCR
- H&E
Haematoxylin and Eosin
- MS-qLNAPCR
Methylation Sensitive Real-Time Locked Nucleic Acid quantitative PCR
- ASLNAqPCR
Allele-Specific Locked Nucleic Acid quantitative PCR
- FC
Fold Change
- WT
Wild-Type
- MET
Methylated
- UMET
Unmethylated
1. Introduction
Glioblastoma (GBM) is the most frequent and malignant brain tumor in adults. Despite progress in surgical techniques, radiotherapy, chemotherapy and “target therapy” its prognosis remains poor (Brandes et al., 2009; Clarke et al., 2010; Louis et al., 2007). Several biomarkers have been proposed as potentially useful parameters for prognosis, diagnosis or therapy strategies. Among them are microRNAs, or miRNAs, which are small (19–24nt) RNA strands with tissue‐specific expression patterns and which play a functional role in several cellular processes involved in the tumorigenesis and progression of several neoplasia (such as proliferation, invasion, migration and angiogenesis). Due to their role in the regulation of gene expression, in the last few years miRNAs tissue‐specific expression, quantification and functional analysis of miRNAs have been extensively investigated to understand their peculiar involvement in cellular processes. It is now established that each tissue has a characteristic microRNA expression pattern which could be altered in association with a number of different diseases including neoplastic transformation (Galasso et al., 2012; Nana‐Sinkam and Croce, 2013; Zhang et al., 2007). Nowadays, there are more than a hundred ongoing trials incorporating miRNAs as biomarkers (Nana‐Sinkam and Croce, 2013). For all these reasons miRNAs could represent important diagnostic, prognostic or predictive target molecules in the treatment of tumors (Galasso et al., 2012; Lawler and Chiocca, 2009; Nana‐Sinkam and Croce, 2013; Zhang et al., 2007).
In the last few years, several studies have been performed to identify a specific microRNA expression pattern of GBM (Chan et al., 2005; Ciafre et al., 2005; Conti et al., 2009; Dong et al., 2010; Gal et al., 2008; Godlewski et al., 2008; Guan et al., 2010; Hua et al., 2012; Lang et al., 2012; Loftus et al., 2012; Malzkorn et al., 2009; Niyazi et al., 2011; Rao et al., 2010; Sasayama et al., 2009; Silber et al., 2008; Skalsky and Cullen, 2011; Slaby et al., 2010; Srinivasan et al., 2011; Zhang et al., 2013; Zhou et al., 2010). For example, recent works by LeBrun and Li (2011) and Mizoguchi et al. (2012) analyzed the results of many GBM miRNA profiling studies and underlined the adopted techniques, the source of neoplastic specimens and the type of non‐neoplastic reference samples used in each study. Considering all profiling data, several discrepancies have been highlighted between different studies and a specific miRNA expression signature in GBM is far from being well‐defined. These discrepancies could mainly be due to three factors in the experimental design: i) the technique performed to evaluate microRNA expression; ii) the starting material; iii) the choice of reference samples used in the analysis.
In a previous study we reported the feasibility of performing miRNA expression analysis using a real‐time locked nucleic acid quantitative PCR (qRT‐LNAPCR) starting from either fresh/frozen or formalin‐fixed and paraffin‐embedded (FFPE) GBM specimens (de Biase et al., 2012). FFPE‐dissected specimens were chosen as the most reliable material with which to perform a miRNA profiling study. In fact, starting from FFPE samples, a neoplastic cells enrichment step (micro‐ or macro‐dissection) was feasible, FFPE samples could be easily retrieved from the archives of the Department of Pathology and furthermore the obtained results displayed good correlation values between miRNA expression and the corresponding fresh/frozen material (de Biase et al., 2012). We have also shown that it is more advisable to compare miRNA expression data within similar experimental conditions because of the discrepancies observed among GBM miRNA profiles obtained using different non‐neoplastic brain references (Visani et al., 2012).
Keeping in mind both the above considerations and the discrepant results sometimes encountered in literature between GBM miRNAs expression studies, in this study we validated the expression of 19 microRNAs in a group of GBM and described the signature of checked miRNAs, using three different non‐neoplastic brain controls starting from FFPE tumor samples. Furthermore, we evaluated possible correlation between miRNA expression and methylation status of MGMT promoter and the putative targets of deregulated miRNAs. Moreover, we characterized gliomas of grades I–III for the same panel of validated miRNAs to evaluate if some of them could be associated to a specific grade of malignancy.
2. Materials and methods
2.1. Statement of ethics
The study was approved by Ethics Committee of Azienda Sanitaria Locale di Bologna (number of study 08075, protocol number 139/CE of 5th February 2009, Bologna, Italy). All patients signed a written consent for molecular analysis and anonymous data publication for scientific studies, and all information regarding the human material used in this study was managed using anonymous numerical codes.
2.2. Case selection
2.2.1. Neoplastic samples
Sixty cases of primary GBM were collected for miRNA expression analysis from cases retrieved in 2009–2010 at Bellaria (Institute of Anatomic Pathology, Bologna, Italy) and Bufalini (Institute of Anatomic Pathology, Cesena, Italy) Hospitals, within the PERNO (Progetto Emiliano‐Romagnolo di Neuro‐Oncologia) project. Inclusion criteria were: i) primary brain tumors; ii) a cryostat section evaluated by a pathologist (GM or SC); iii) subjects had not undergone neoadjuvant therapy before surgery; iv) subjects resident in Emilia‐Romagna region. The patients included were 34 males and 26 females, aged from 41 to 78 years (mean 62.3 yrs) (Table 1). All 60 samples were diagnosed as GBM according to the 2007 WHO criteria (Louis et al., 2007). Thirty of these 60 cases were previously analyzed for the 19 miRNAs and represented our training set (Visani et al., 2012) (Table 1). The 30 primary GBM belonged to testing set were 20 males and 10 females, aged from 41 to 78 years (mean age 61.7 yrs) (Table 1).
Table 1.
Histological classification, age and sex of the analyzed tumor samples.
| Grade tumor groups | N. Of cases | Age (mean yrs) | Sex | |
|---|---|---|---|---|
| M | F | |||
| Grade IV (GBM) | 60 | 41–78 (62.3) | 34 | 26 |
| Training set | 30 | 42–75 (63.3)a | 14 | 16 |
| Testing set | 30 | 41–78 (61.7)a | 20 | 10 |
| Grade III | 15 | 30–74 (50.7) | 10 | 5 |
| Anaplastic ependymomas | 2 | |||
| Anaplastic oligodendrogliomas | 7 | |||
| Anaplastic astrocytomas | 3 | |||
| Anaplastic oligoastrocytomas | 3 | |||
| Grade II gliomas | 15 | 21–74 (42.8) | 11 | 4 |
| Ependymomas | 2 | |||
| Oligodendrogliomas | 7 | |||
| Pleomorphic xanthoastrocytoma | 1 | |||
| Diffuse astrocytoma | 1 | |||
| Neurocytomas | 4 | |||
| Grade I gliomas | 15 | 2–23 (20.8) | 6 | 9 |
| Pilocytic astrocytomas | 4 | |||
| Gangliogliomas | 11 | |||
M, Male; F, Female.
No statistically significant differences were observed between training and testing set (p= 0.6013, unpaired t‐test).
The following primary brain tumors were also selected: 15 grade III (2 anaplastic ependymomas, 7 anaplastic oligodendrogliomas, 3 anaplastic astrocytomas, 3 anaplastic oligoastrocytomas; 10 males and 5 females, aged 30–74 years, mean 50.7 yrs), 15 grade II (2 ependymomas, 7 oligodendrogliomas, 1 pleomorphic xanthoastrocytoma, 1 diffuse astrocytoma and 4 neurocytomas; 11 males and 4 females, aged 21–74 years, mean 42.8 yrs) and 15 grade I (4 pilocytic astrocytomas and 11 gangliogliomas; 6 males and 9 females, aged 2–23 years, mean 20.8 yrs). All samples were diagnosed according to the 2007 WHO criteria (Louis et al., 2007) (Table 1).
2.2.2. Non‐neoplastic references
Fifteen “normal” tissues, adjacent to the tumor (8 males and 7 females, aged 50–75 years, mean 62.7 yrs), at a distance between 1 and 2 cm from the margin of 15 GBMs, were retrieved. Fifteen polar temporal cortical FFPE specimens, removed in patients submitted to surgery for drug‐resistant epilepsy, were randomly selected from the archives of the Anatomic Pathology division of Bellaria Hospital (7 males and 8 females, aged 25–52 years, mean 39.7 yrs). Additionally, the FirstChoice® Human Brain Reference RNA was utilized (Ambion, Austin, TX, U.S.A).
2.3. Nucleic acid extractions
The haematoxylin and eosin (H&E) sections from FFPE specimens were reviewed by a pathologist (GM) to select the more informative block. One 10 μm‐thick and four 20 μm‐thick sections were cut, followed by one H&E control slide, for DNA and total RNA extractions respectively. The tumor area selected for the analysis was marked on the H&E section to ensure, whenever possible, greater than 90% content of neoplastic cells (avoiding necrosis and lymphocytes). Each section was manually dissected according to the selected area. DNA was extracted using QuickExtract™ FFPE DNA Extraction Kit (Epicentre, Madison, WI, U.S.A.). miRNAs were extracted using RecoverAll Total Nucleic Acid Isolation kit (Ambion, Austin, TX, U.S.A.), according to manufacturer's instructions. SmallRNAs were quantified and cDNA obtained as previously described (de Biase et al., 2012; Visani et al., 2012).
2.4. MicroRNAs analysis
19 miRNAs (miR‐7, miR‐9, miR‐9∗, miR‐10a, miR‐10b, miR‐17, miR‐20a, miR‐21, miR‐26a, miR‐27a, miR‐31, miR‐34a, miR‐101, miR‐137, miR‐182, miR‐221, miR‐222, miR‐330, miR‐519d), were selected for analysis, according to their role in cancer and data published in literature previous to the beginning of the study (de Biase et al., 2012; Visani et al., 2012). Three smallRNAs (RNU49, U54, miR‐103) were used as internal controls (de Biase et al., 2012; Visani et al., 2012). The miRNA analyses were performed using real‐time PCR as previously described (de Biase et al., 2012; Visani et al., 2012). Briefly, RNA was retrotranscribed using the NCode miRNA First‐Strand cDNA Synthesis and qRT‐PCR Kits (Invitrogen, Carlsbad, CA, USA), and miRNA expression was evaluated on the AB7000 machine (Applied Biosystem, Foster City, CA, USA) performing a miRNA‐specific real‐time qPCR using LNA primers (de Biase et al., 2012; Visani et al., 2012).
2.5. Validation of the 19 miRNA expression signature in the testing set
The expression of the 19 miRNAs was evaluated in the testing set (n = 30 GBMs) and the values compared with those obtained in the training group (n = 30 GBMs). In order to define expression values of the analyzed miRNAs in GBM reducing possible biases due to reference selection, only microRNAs with the same regulation status in at least 2 out of the 3 profiles in both groups were considered.
2.6. MGMT methylation and IDH1 mutation analysis
2.6.1. MGMT analysis
At least 50 ng of DNA were treated with bisulfite using the EpiTect Bisulfite kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. A Methylation Sensitive Real‐Time qPCR using LNA modified primers and beacon probes (MS‐qLNAPCR) was performed as previously described (Morandi et al., 2010).
2.6.2. IDH1 analysis
Tumor samples were tested for the IDH1‐R132H mutation using Allele‐Specific Locked Nucleic Acid quantitative PCR (ASLNAqPCR) (Morandi et al., 2012). To put briefly, allele‐specific primers that recognized the IDH1 wild‐type sequence or the R132H mutation were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/). Both forward primers were designed with 3′‐locked nucleic acid (LNA – underlined nucleotides within the primer sequences) substitutions to improve mismatch discrimination (Forward primer WT: 5′‐TTGATCCCCATAAGCATGAC‐3′; Forward primer R132H: 5′‐TTGATCCCCATAAGCATGAT‐3′; Reverse primer: 5′‐GTGGCACGGTCTTCAGAGA‐3′). The analysis was performed using FastStart Universal Probe Master with ROX (Roche Applied Science, Mannheim, Germany) on the AB7000 machine (Applied Biosystem, Foster City, CA, USA) with the following program: 2 min at 50 °C, 4 min at 95 °C followed by 37 cycles of 30 s at 95 °C, 30 s at 58 °C and 30 s at 72 °C with fluorescence measurement. GelStar stain (Lonza Bioscience, Rockland, ME, USA) was used for signal detection.
2.7. Statistical analysis
Expression values and fold changes were obtained by relative quantification and 2−ΔΔCT method (Livak and Schmittgen, 2001) using DataAssist v3.01 Tool (Applied Biosystem, Foster City, CA, USA). The unsupervised hierarchical clustering analysis (Pearson Correlation, average linkage) was performed using the same statistical tool. Statistical analysis of miRNA expression was performed using GraphPad Prism 5.0 software. Correlation analysis between the different groups was performed applying Spearman correlation test. For comparing the expression levels of each miRNA between different groups, Mann–Whitney test was used. Gaussian distribution was evaluated by Shapiro–Wilk Test.
A microRNA was considered as down‐regulated with a median fold change (FC) < −2.0, while a microRNA with a median FC ≥ 2.0 was considered as up‐regulated.
Level of significance was p < 0.05 for all statistical analyses. For microRNA target analyses, a statistical over‐representation test (Cho and Campbell, 2000) was performed using PANTHER web tool (http://www.pantherdb.org/tools/uploadFiles.jsp). The list of target genes was compared to a reference list to statistically determine over‐ or under‐representation of PANTHER classification categories. p‐values were calculated with Bonferroni correction; a cut‐off of 0.05 was considered to estimate if a specific PANTHER category was over‐ or under‐represented in a statistically significant way.
2.8. Bioinformatic prediction of microRNA targets
The experimentally‐validated targets of microRNA which were significantly deregulated in the GBM profile were identified using three different online tools: miRecords (http://mirecords.umn.edu/miRecords/) (Xiao et al., 2009), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) (Hsu et al., 2011) and miRWalk (http://www.umm.uni‐heidelberg.de/apps/zmf/mirwalk) (Dweep et al., 2011). The latest releases of miRecords (http://mirecords.umn.edu/miRecords/download.php, updated on November 25th, 2010) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/download.php, “hsa MTI.xls” file, release 3.5 of November 1st, 2012) were downloaded and only microRNAs of interest were selected. The list of selected microRNAs was uploaded on miRWalk “Validated target(gene) of mirna search” section (http://www.umm.uni‐heidelberg.de/apps/zmf/mirwalk/mirnatargetpub.html, updated on March 11th, 2011) to obtain the list of experimentally‐validated targets relative to each microRNAs of interest. Only targets identified by all 3 prediction tools were then considered. Targets were analyzed and grouped according to their molecular functions, biological process involvement and pathways classification, using the PANTHER web tool (http://www.pantherdb.org/) (Mi et al., 2013).
3. Results
3.1. Validation of the 19 miRNA expression signature in the testing set
Considering that microRNA was considered as down‐regulated with a median fold change (FC) < −2.0, while a microRNA with a median FC ≥ 2.0 was considered as up‐regulated, the expression status of the 19 miRNAs in the testing set (n = 30) was compared with those of training group (Table 2). According to this comparison, 14 miRNAs (miR‐7, miR‐9∗, miR‐10a, miR‐10b, miR‐17, miR‐21, miR‐31, miR‐34a, miR‐101, miR‐137, miR‐182, miR‐221, miR‐222 and miR‐330) showed the same regulation status between the two groups. Only these 14 miRNAs were then considered for further analysis.
Table 2.
Validation of the 19 miRNA expression signature in the testing set.
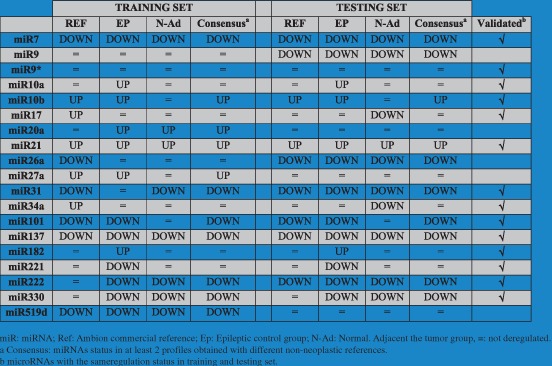
3.2. Expression signature of the 14 validated miRNAs in GBMs
Three different profiles were obtained comparing the 14 validated miRNAs in the entire cohort of 60 GBMs toward the three different non‐neoplastic control groups (normal tissues adjacent to the tumor, epileptic specimens, and a commercial reference). The median expression values of the 14 miRNAs in the GBMs are shown in Suppl. Table 1. 6 miRNAs (miR‐7, miR‐31, miR‐101, miR‐137, miR‐222 and miR‐330) were down‐regulated (Table 3, Suppl. Table 1). All miRNAs with a FC < −2 were down‐regulated in at least 50% of analyzed glioblastoma (Table 3, Suppl. Table 1).
Table 3.
miRNAs up‐ or down‐regulated in GBM in at least 2 profiles obtained with different non‐neoplastic references.
| Status | miRNA | Non‐neoplastic reference | Median fold change of GBM group vs | ||
|---|---|---|---|---|---|
| Refa | Epa | N‐Ada | |||
| Up‐regulated | miR‐10b | Ref, Ep | 2.343 (36/60) | 3.947 (44/60) | = |
| miR‐21 | Ref, Ep, N‐Ad | 6.506 (51/60) | 9.462 (57/60) | 6.415 (52/60) | |
| Down‐regulated | miR‐7 | Ref, Ep, N‐Ad | −23.641 (57/60) | −50.633 (57/60) | −15.723 (53/60) |
| miR‐31 | Ref, N‐Ad | −14.265 (54/60) | = | −3.726 (43/60) | |
| miR‐101 | Ref, Ep | −2.550 (44/60) | −2.750 (46/60) | = | |
| miR‐137 | Ref, Ep, N‐Ad | −10.428 (59/60) | −17.857 (60/60) | −6.215 (54/60) | |
| miR‐222 | Ref, Ep, N‐Ad | −2.186 (33/60) | −14.327 (57/60) | −11.287 (56/60) | |
| miR‐330 | Ep, N‐Ad | = | −5.373 (50/60) | −4.354 (50/60) | |
miR: miRNA; Ref: Ambion commercial reference; EpEpileptic control group; N‐Ad: Normal Adjacent the tumor group, =: not deregulated.
Number of GBMs which shared the same deregulation status are indicated between brackets.
Two microRNAs (miR‐10b and miR‐21) were up‐regulated in GBM group (Table 3, Suppl. Table 1). All miRNAs with a FC ≥ 2 were also up‐regulated in at least 50% of analyzed GBM cases (Table 3, Suppl. Table 1).
Six microRNAs (miR‐9∗, miR‐10a, miR‐17, miR‐34a, miR‐182 and miR‐221) showed expression levels that were within the 2 fold cut‐off and for this reason were considered to not be deregulated (Suppl. Table 1).
The power of the study was calculated considering 70% of GBM deregulated for validated miRNAs and hypothesizing to observe at least 85% of cases with these miRNAs deregulated in our final cohort of 60 GBMs. According to these parameters the power of the study was 0.77.
An unsupervised hierarchical cluster showed that considering these 14 miRNAs, GBM clustered together if compared with to non‐neoplastic references (Suppl. Figure 1).
Correlation between the GBM miRNA signatures obtained with the three non‐neoplastic references were evaluated using the Spearman correlation test (Table 4). A high correlation (>0.65) value (r = 0.859 or r = 0.899) was observed in all the comparisons (Table 4). Expression values obtained using commercial reference and adjacent to the tumor samples as non‐neoplastic controls showed the higher number of miRNAs (n = 11) with the same regulation status (Table 4, Suppl. Table 1).
Table 4.
Spearman correlation ratio between miRNA expression values in GBM obtained using the three different non‐neoplastic references.
| GBM vs N‐Ad | GBM vs Ref | GBM vs Ep | |
|---|---|---|---|
| GBM vs N‐Ad | / | 0.859 (11/14) | 0.899 (8/14) |
| GBM vs Ref | 0.859 (11/14) | / | 0.899 (9/14) |
| GBM vs Ep | 0.899 (8/14) | 0.899 (9/14) | / |
Between brackets the number of miRNAs with the same regulation status. GBM: glioblastoma; Ref: Ambion commercial reference; Ep: Epileptic group; N‐Ad: Normal Adjacent the tumor group. p<0.0001.
3.3. Comparison between MGMT methylated and unmethylated GBM samples
Twenty‐seven out of 59 analyzed cases (45.8%) showed methylation of the MGMT promoter. All cases were wild‐type for IDH1, supporting the classification of “primary GBM” (Yan et al., 2009) (Table 5). In one case, MGMT and IDH1 analysis were not evaluable, probably due to poor quality of extracted DNA.
Table 5.
MGMT and IDH1 status of analyzed GBMs.
| Groups | Methylated (%) | Unmethylated (%) | IDH1 wt (%) | IDH1‐R132H (%) |
|---|---|---|---|---|
| Training (n = 30) | 14 (46.7) | 16 (53.3) | 30 (100) | 0 (0) |
| Testing (n = 29)a | 13 (44.8) | 16 (55.2) | 29 (100) | 0 (0) |
| TOTAL GBM (n = 59)a | 27 (45.8) | 32 (54.2) | 59 (100) | 0 (0) |
In one case MGMT and IDH1 analysis were not evaluable, probably due to poor quality of extracted DNA. No statistical differences were observed between training and testing groups for MGMT methylation status (p=1.000, Fisher's exact test) and IDH1 mutational status (p=1.000, Fisher's exact test).
Considering the variation of expression of the 14 miRNAs validated in GBM profiles, an unsupervised hierarchical clustering analysis was performed to compare MGMT methylated (MET, n = 27) and unmethylated (UMET, n = 32) samples. The results indicated that there was not a significant correlation between methylation status of MGMT promoter and microRNA expression: considering this subset of microRNAs, MET–GBM and UMET–GBM groups did not show a clear clusterization, regardless of the selected non‐neoplastic reference (Suppl. Figure 1). Moreover, the median expression values for each miRNA showed no differences between MET–GBM and UMET–GBM groups irrespective of the control groups used (data not shown). 12 miRNAs showed no differences in regulation status between MET–GBM and UMET–GBM groups. MicroRNA‐182 was up in UMET–GBM samples but not deregulated in MET–GBM; miR‐221 was down‐regulated in UMET–GBM but not deregulated in MET–GBM (data not shown).
3.4. Analysis of putative microRNA targets
To further investigate the biological relevance of the determined GBM miRNA profile, we looked for the known targets of the 8 significantly deregulated microRNAs (Table 3). The analysis was focused on the research of targets experimentally validated in previously published studies. Among available bioinformatics tools online, miRecords, miRTarBase and miRWalk were used. Considering the 8 microRNAs, 81 validated targets on miRecords (http://mirecords.biolead.org/) and 174 on miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) were found, noting that both tools consider manually‐performed experimental interactions based on literature surveying. Using miRWalk (http://www.umm.uni‐heidelberg.de/apps/zmf/mirwalk/), based on automated text‐mining search, 560 targets were found. Combining these results, 46 genes were found in common to the three target prediction databases (Suppl. Table 2). This gene list was entered into the PANTHER database in order to identify significantly over‐ and under‐represented pathways, molecular functions and biological processes, in comparison with a reference list (the default Homo Sapiens Whole Genome list was selected) (Table 6). From the initial list of 46 genes, PANTHER was unable to map the gene KLF4. For this reason all analyses were performed on a total of 45 gene targets. In Table 6, statistically significant (p < 0.05) pathways, molecular function classes and biological processes relevant to the 45 gene targets were listed. In all cases, a statistically significant over‐representation of the number of target genes per category was observed.
Table 6.
PANTHER over‐representation analysis of target genes list.
| PANTHER classification category | Number of genes | Over‐/under‐represented (+/−) | p‐value | % of target listd | ||
|---|---|---|---|---|---|---|
| Reference lista | Target listb | Expectedc | ||||
| Pathways | ||||||
| Insulin/IGF pathway‐MAPKK/MAP kinase cascade | 38 | 4 | 0.09 | + | 3.23E‐04 | 8.89 |
| Angiogenesis | 185 | 6 | 0.42 | + | 6.63E‐04 | 13.33 |
| Interleukin signaling pathway | 112 | 5 | 0.25 | + | 9.88E‐04 | 11.11 |
| Gonadotropin‐releasing hormone receptor pathway | 282 | 6 | 0.63 | + | 7.07E‐03 | 13.33 |
| Insulin/IGF pathway‐PKB signaling cascade | 38 | 3 | 0.09 | + | 1.62E‐02 | 6.67 |
| Molecular functions | ||||||
| Transmembrane receptor protein kinase activity | 120 | 5 | 0.27 | + | 1.16E‐03 | 11.11 |
| Protein kinase activity | 500 | 8 | 1.12 | + | 2.14E‐03 | 17.78 |
| Kinase activity | 664 | 8 | 1.49 | + | 1.58E‐02 | 17.78 |
| Transmembrane receptor protein serine/threonine kinase activity | 52 | 3 | 0.12 | + | 3.42E‐02 | 6.67 |
| Biological Processes | ||||||
| Protein amino acid phosphorylation | 654 | 10 | 1.47 | + | 2.66E‐04 | 22.22 |
| Cellular process | 6072 | 29 | 13.66 | + | 4.24E‐04 | 64.44 |
| Signal transduction | 4019 | 22 | 9.04 | + | 2.49E‐03 | 48.89 |
| Cell communication | 4224 | 22 | 9.50 | + | 5.65E‐03 | 48.89 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | 295 | 6 | 0.66 | + | 8.76E‐03 | 13.33 |
| MAPKKK cascade | 454 | 7 | 1.02 | + | 1.13E‐02 | 15.56 |
| Protein modification process | 1330 | 11 | 2.99 | + | 2.34E‐02 | 24.44 |
| Cell cycle | 1602 | 12 | 3.60 | + | 2.78E‐02 | 26.67 |
Pathways, molecular function classes and biological processes resulted significantly over or under represented by 46 gene targets.
Number of genes in the reference list that map to this PANTHER classification category.
Number of genes in the target genes list that map to this PANTHER classification category.
Expected value is the number of genes that could be expected in target genes list for this PANTHER category based on the reference list.
Percentage of genes in the target list out of the total considered genes in PANTHER (n=45). P‐values are determined by binomial statistics with Bonferroni correction: a p‐value cut‐off of 0.05 was considered.
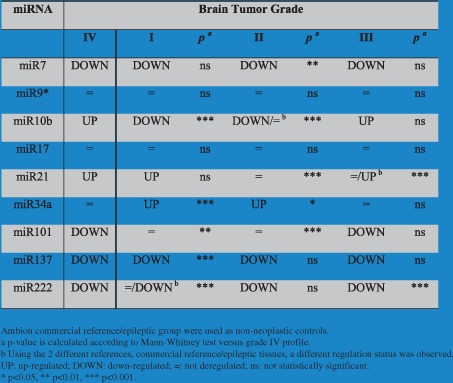
3.5. MicroRNA expression analysis in grades I–III brain tumors
To evaluate possible differences in miRNA expression associated with different grade of malignancy, we further analyzed the miRNA profiles in grades I–III brain tumors (Suppl. Table 3). Considering that normal tissue adjacent to the tumor tissues were not available for grades I–III brain tumors, Ambion and epileptic tissue groups were used for the analysis of miRNAs profile in these samples (Table 7). For this reason, we focused our attention on the 9 miRNAs with the same GBM expression profile with commercial and epileptic control groups (miR‐7, miR‐9∗, miR‐10b, miR‐17, miR‐21, miR‐34a, miR‐101, miR‐137 and miR‐222) (Suppl. Table 1). As happened for GBM analysis, the up‐ (FC ≥ 2) or down‐regulated (FC < −2) miRNAs were deregulated in at least the 50% of analyzed samples (Suppl. Table 3).
Grade I (Table 7, Suppl. Table 3): miR‐7, miR‐10b, miR‐137 and miR‐222 were observed as down‐regulated (miR‐222 only according to the epileptic tissues); miR‐21 and miR‐34a were up‐regulated.
Grade II (Table 7, Suppl. Table 3): miR‐7, miR‐10b, miR‐137 and miR‐222 were observed as down‐regulated (miR‐10b only considering the commercial reference); miR‐34a was up‐regulated.
Grade III (Table 7, Suppl. Table 3): miR‐7, miR‐101, miR‐137 and miR‐22 were down‐regulated; miR‐10b and miR‐21 were up‐regulated (miR‐21 only against epileptic tissues).
Table 7.
MicroRNA expression status in different‐grade tumor groups.
Spearman correlation between the four tumor groups (grades IV–I) showed a high (>0.65) correlation value (r = 0.9167) between high‐grade gliomas (grades IV and III) considering the Ambion commercial reference; using the epileptic tissues as controls the highest values were observed and between high‐grade gliomas (grades IV and III) (r = 0.8833) and between low‐grade gliomas (r = 0.9333) (Suppl. Table 4).
An unsupervised hierarchical clustering analysis showed that there was not a distinct clustering between different‐grade gliomas, both using Ambion commercial reference or epileptic group as controls (data not shown).
3.6. Comparing microRNA expression analysis between grades I–IV brain tumors
The different regulation statuses of miRNAs among the four tumor groups were evaluated. miR‐9∗ and miR‐17 were not deregulated in any analyzed profiles (Table 7).
3 microRNAs (miR‐10b, miR‐34a, miR‐101) shared the same deregulation status in high‐grade gliomas (grades III and IV): miR‐34a was not deregulated in high‐grade gliomas but was up‐regulated in low‐grade (grades I and II) tumors (Figures 1 and 2 and Table 7); miR‐101 was down‐regulated in high grade (III and IV) but not in low‐grade (I and II) gliomas (Figures 1 and 2 and Table 7); miR‐10b was up‐regulated in high‐grade gliomas (III and IV) and meanwhile was significantly down‐regulated in low‐grade (I and II) brain tumors (p < 0.001) (Figures 1 and 2 and Table 7). This last microRNA was down‐regulated or not deregulated in grade II depending on the non‐neoplastic reference (Figures 1 and 2 and Table 7).
Figure 1.
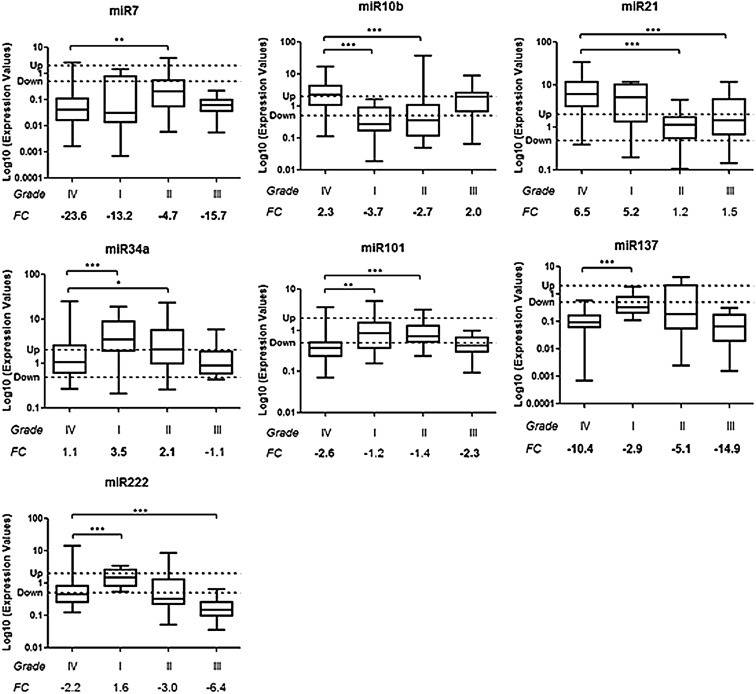
Differences in miRNA expressions among the 4 different‐grade tumor groups evaluated using the Ambion commercial reference as control. Box plots show microRNAs different between GBM Group (grade IV) and the other 3 tumor groups. Y‐axis indicates the microRNA Log10 expression level: “Up” and “Down” lines highlight the cut‐off of 2 fold change used to consider a microRNA as deregulated. In bold the up‐ or down‐regulated median FC values. Bars mean minimum and maximum values of miRNAs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 according to Mann–Whitney test. FC: median Fold Change.
Figure 2.
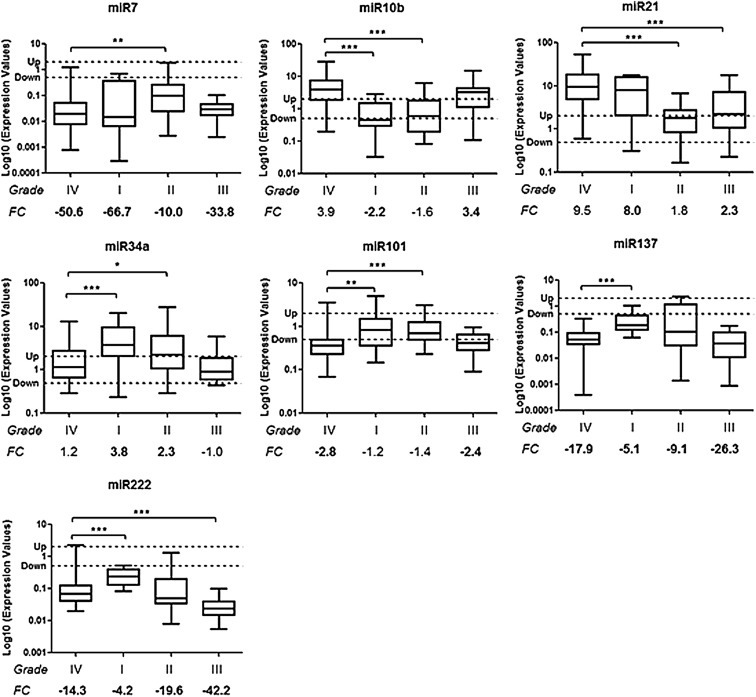
Differences in miRNA expressions among the 4 different‐grade tumor groups was evaluated using the epileptic group as control. Box plots show microRNAs different between GBM Group (grade IV) and the other 3 tumor groups. Y‐axis indicates the microRNA Log10 expression level: “Up” and “Down” lines highlight the cut‐off of 2 fold change used to consider a microRNA as deregulated. In bold are the up‐ or down‐regulated median FC values. Bars mean minimum and maximum values of miRNAs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 according to Mann–Whitney test. FC: median Fold Change.
3 microRNAs (miR‐7, miR‐137 and miR‐222) were down‐regulated in all glioma groups even with observed significant differences in expression levels (e.g. between GBM and grade II for miR‐7 – p < 0.01, or between GBM and grade I for miR‐137 – p < 0.001, Figures 1 and 2 and Table 7). MicroRNA‐222 was down‐regulated or not deregulated in grade I depending on the non‐neoplastic reference (Figures 1 and 2 and Table 7).
miR‐21 was up‐regulated in grade IV and in grade I tumors, but it was not deregulated in those of grade II (Figures 1 and 2 and Table 7). In grade III, miR‐21 showed up‐regulation only when compared with the epileptic reference (Figures 1 and 2 and Table 7).
4. Discussion
Bearing in mind that in GBM the surgery, radio‐ and chemo‐therapies are not enough to combat tumor progression and to ensure a hopeful outcome, the identification of a specific pattern of expression of miRNAs in GBM could provide an important boost in prognosis definition and in new target therapy strategies. Presently, several miRNA profiles are available for GBM (Chan et al., 2005; Ciafre et al., 2005; Conti et al., 2009; Dong et al., 2010; Gal et al., 2008; Godlewski et al., 2008; Guan et al., 2010; Hua et al., 2012; Lang et al., 2012; Loftus et al., 2012; Malzkorn et al., 2009; Niyazi et al., 2011; Rao et al., 2010; Sasayama et al., 2009; Silber et al., 2008; Skalsky and Cullen, 2011; Slaby et al., 2010; Srinivasan et al., 2011; Zhang et al., 2013; Zhou et al., 2010) and a small subset of consistently deregulated miRNAs was further functionally characterized for their activities and downstream targets possibly involved in the tumor (Chen et al., 2008; Huse et al., 2009; Kefas et al., 2008; Li et al., 2011; Sasayama et al., 2009; Smits et al., 2010). However, due to several differences in the study designs, a clear miRNA profile for GBM has not been established yet. Considering the data previously obtained by our group (de Biase et al., 2012; Visani et al., 2012), in this study the expression of 19 miRNAs in GBM was analyzed, starting from FFPE‐dissected samples to ensure a good enrichment in neoplastic cells (de Biase et al., 2012). Three different non‐neoplastic references (Visani et al., 2012) were enrolled for the analysis, to verify if different controls could influence GBM microRNAs expression values. Fourteen out of these 19 miRNAs showed the same deregulation status between training and testing sets and for this reason considered for further analysis.
According to adopted non‐neoplastic references, differences in the median expression values of the 14 validated microRNAs in GBM were observed. Furthermore, different fold‐change values were observed even in those miRNAs with the same regulation status between all three profiles (e.g. miR‐7, miR‐137).
To better define a GBM microRNA signature, only validated microRNAs deregulated in at least 2 out of the 3 profiles obtained were considered. According to our results, 2 miRNAs (miR‐10b and miR‐21) were up‐regulated and 6 miRNAs (miR‐7, miR‐31, miR‐101, miR‐137, miR‐222 and miR‐330) were down‐regulated in group of 60 GBMs.
The microRNA GBM signature found in the present series was in accordance with profiling data previously reported in literature, except for miR‐222 that was down‐regulated in our series. Although miR‐221 and miR‐222 are encoded in the same genomic cluster on the X chromosome (http://www.ncbi.nlm.nih.gov/gene/, Genes ID: 407006 and 407007), miR‐221, differently than miR‐222, was not deregulated. Several studies reported an up‐regulation of miR‐221/222 cluster in GBM (Ciafre et al., 2005; Quintavalle et al., 2012; Zhang et al., 2010). Quintavalle et al. (2012) and Zhang et al. (2010) assessed the up‐regulation of miR‐222 in GBM cell lines in comparison with non‐tumorigenic cell line or the commercial reference. On the contrary, in the present study, we evaluated the regulation of miRNAs in surgical GBM specimens and not in cell lines by using three different non‐neoplastic controls. Ciafre et al. (2005) analyzed miR‐221 in fresh/frozen samples and miR‐222 in cell lines. In these situations, the discrepancies observed with our data could be due to the differences in starting material used for establishing microRNA expression.
The down‐regulation of miR‐221 and miR‐222 status was previously observed by Slaby et al. and Lakomy et al. using stem‐loop RT‐PCR in FFPE‐dissected GBMs (ensuring >90% of tumor cells) in comparison to non‐malignant brain tissues, derived from areas surrounding arteriovenous malformation (AVM) or from commercial reference (Slaby et al., 2010; Lakomy et al., 2011). Slaby et al. (2010) explained this down‐regulation was due to the choice of normal brain samples that could lead to discrepant results with data obtained in other studies.
The differences observed in the expression of miRNAs belonging to the same genomic cluster (miR‐221 and miR‐222) could be due to post‐transcriptional modification, as described, for example, by Davis et al. (2009).
According to our panel, only 2 miRNAs (miR‐182 and miR‐221) showed a different regulation status between methylated and unmethylated samples. MicroRNA‐221 was previously described as mechanism of post‐transcriptional regulation of MGMT (Quintavalle et al., 2013). On the contrary, to the best of our knowledge, a possible correlation between miR‐182 and MGMT methylation status has not been previously described in literature.
Taking into account the 8 microRNAs (miR‐7, miR‐10b, miR‐21, miR‐31, miR‐101, miR‐137, miR‐222 and miR‐330) showing a deregulation in GBMs, a preliminary analysis of their experimentally‐validated targets was performed. Forty‐six targets, shared by three bioinformatics tools, were further analyzed for their molecular function and pathway involvement: interleukin signaling, insulin/IGF pathway‐MAPKK/MAPK cascade, gonadotropin‐releasing hormone receptor (GNRHR), angiogenesis and Insulin/IGF‐PKB pathways were significantly represented by this list of targets and previous studies have demonstrated their decisive role in GBM pathogenesis (Bulnes et al., 2012; de la Iglesia et al., 2008; Ermoian et al., 2002; Low et al., 2008; Montagnani Marelli et al., 2009; Nakada et al., 2011; Trojan et al., 2007; van Groeninghen et al., 1998; Wang et al., 2012).
The expression of the 9 miRNAs (miR‐7, miR‐9∗, miR‐10b, miR‐17, miR‐21, miR‐34a, miR‐101, miR‐137, miR‐222) with consistent regulation status in GBM using Ambion reference or epileptic tissues, was also investigated in lower grade tumors (grades I–III), to evaluate if there were relevant differences associated with different grades of malignancy. Only 3 microRNAs (miR‐7, miR‐137 and miR‐222) showed the same deregulation status in all four tumor grades. Nonetheless, some significant differences in miRNAs expression levels were observed between the groups.
In spite of the power of the study below 0.8 (0.77) and the awareness that a higher number of GBM should be preferable for more accurate results, it should be considered the distinctiveness and uniformity of our cohort, composed of 60 highly selected GBM. In fact all patients were resident in the same geographical region (Emilia‐Romagna) and were surged by the same surgical teams, all diagnoses were obtained with a consensus between neuropathologists and all GBM were primary tumors. All these criteria were chosen for having a homogeneous cohort of specimens and limiting bias of results due to study design.
The unsupervised hierarchical clustering analysis showed that there was no distinct clusterization between different‐grade gliomas. This might suggest that the entire panel of these 9 microRNAs is unsuitable for discriminating gliomas based on the grade of malignancy.
Intriguingly, 3 microRNAs shared the same deregulation status in high‐grade gliomas (grades III and IV): miR‐34a, miR‐101 and miR‐10b. miR‐34a, stable in high‐grade tumors, was on the other hand up‐regulated in low‐grade gliomas (grades I and II); this miRNA was previously reported to be down‐regulated in GBM and its possible role in GBM pathogenesis was investigated (Genovese et al., 2012; Li et al., 2009; Silber et al., 2012). Oncogenes like c‐met, Notch1, Notch2, CDK6, PDGFRA and SMAD4, in TGFβ/SMAD pathway, were validated as targets of miR‐34a and they were effectively over‐expressed in GBM (Genovese et al., 2012; Li et al., 2009; Silber et al., 2012). Although miR‐34a was not deregulated in our high‐grade glioma groups, it showed an up‐regulation in low‐grade tumors, suggesting a possible association between miR‐34a loss of expression and pathogenesis of high‐grade glial neoplasias.
miR‐101 was down‐regulated in high grade but not in low‐grade gliomas, and this result is in accord with previously published studies (Silber et al., 2008; Smits et al., 2010). In particular, we discovered that miR‐101 down‐regulation is present only in association with high‐grade malignancy. Smits et al. (2010) have previously demonstrated one possible role of miR‐101 in GBM progression; they obtained lower levels of miR‐101 in comparison with grades II and III gliomas and they reported the association with over‐expression of miR‐101 target EZH2, which influences proliferation, invasion and angiogenesis.
miR‐10b was up‐regulated in high‐grade gliomas while was significantly down‐regulated in low grade ones. The data concerning GBM is in‐line with results previously published by other studies (Ciafre et al., 2005; Gabriely et al., 2010; Sasayama et al., 2009; Silber et al., 2008). In particular, Gabriely et al. (2010) demonstrated the role of miRNA‐10b in cell proliferation and in cell cycle regulation by targeting Bim (a pro‐apoptotic factor), p16/CDKN2A and p21/CDKN1B. Additionally, the study by Sasayama et al. (2009) reported that levels of invasive factors RhoC and urokinase‐type plasminogen activator receptor (uPAR) were inversely correlated with miR‐10b expression, indicating that it might play a key role in invasion features of gliomas. They reported that miR‐10b expression was associated with glioma grade of malignancy and its expression was significantly lower in low‐grade gliomas compared to high‐grade astrocytic tumors (Sasayama et al., 2009). All of this data has suggested that the silencing of miR‐10b could be a potential therapeutic strategy for GBM treatment. An analysis of miR‐10b expression in primary low‐grade gliomas and in corresponding secondary GBM lesions could be useful to evaluate a possible correlation with tumor progression. It should be considered that the present series of low‐grade gliomas were heterogeneous groups. Further studies are necessary for determining specific differences between GBM and different histotypes of low‐grade brain tumors.
In conclusion, we determined a GBM microRNA profile and performed a comparison of selected miRNAs in different‐grade gliomas. Beyond these results, one of the main conclusions of our study is that GBM microRNA profiling studies should be compared considering similar experimental conditions (i.e. the number of cases analyzed, the type of selected tissue, the non‐neoplastic control chosen and the technique adopted for microRNA expression analysis).
Bearing in mind these considerations, further investigations such as extensive sequencing of miRNAs in a vast group of GBM or the analysis of miRNA expression in low‐grade primary glioma tumors and in their corresponding secondary high‐grade lesions, could be critical to discover new therapeutic strategies and possible correlations with glioma progression.
Statement of author contribution
Conceived and designed the experiments: DdB, MV, AP.
Performed the experiments: DdB, MV.
Cases collection and pathological evaluation: GM, SC, EN.
Analyzed the data: DdB, MV, GM, SC, EN, FA, AB, AP.
Statistical Analysis: DdB, MV, MLBR.
Contributed reagents/materials/analysis tools: AP, AB, PERNO study group.
Wrote the manuscript: DdB, MV, AP.
Conflict of Interest
The authors declare that they have no conflict of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary Figure 1 Unsupervised hierarchical clustering analysis of GBM group for the 14 validated microRNAs. Clustering analysis was performed using the Ambion commercial reference (A), epileptic tissues (B) and normal adjacent to the tumor (C). MET–GBM samples are written in blue, UMET–GBM in pink, non‐neoplastic control samples in green. GBM with methylation status available is reported in red.
Acknowledgment
The authors thank Adriana Giuliani (Indiana University, Indiana, U.S.A.) for her professional revision of the English language. This work has been supported in part by the “Programma di Ricerca Regione‐Università, Regione Emilia‐Romagna” within the “Project of Emilia‐Romagna region on Neuro‐Oncology (PERNO)”.
Supplementary data 1.
Appendix A.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.12.010.
Appendix B.
Steering committee
Baruzzi A. (Chair), Albani F., Calbucci F., D'Alessandro R., Michelucci R. (IRCCS Institute of Neurological Sciences, Bologna, Italy), Brandes A. (Department of Medical Oncology, Bellaria‐Maggiore Hospitals, Bologna, Italy), Eusebi V. (Department of Hematology and Oncological Sciences “L. & A. Seràgnoli”, Section of Anatomic Pathology at Bellaria Hospital, Bologna, Italy), Ceruti S., Fainardi E., Tamarozzi R. (Neuroradiology Unit, Department of Neurosciences and Rehabilitation, S. Anna Hospital, Ferrara, Italy), Emiliani E. (Istituto Oncologico Romagnolo, Department of Medical Oncology, Santa Maria delle Croci Hospital, Ravenna, Italy), Cavallo M. (Division of Neurosurgery, Department of Neurosciences and Rehabilitation, S. Anna Hospital, Ferrara, Italy).
Executive committee
Franceschi E., Tosoni A. (Department of Medical Oncology, Bellaria‐Maggiore Hospitals, Bologna, Italy), Cavallo M. (Division of Neurosurgery, Department of Neurosciences and Rehabilitation, S. Anna Hospital, Ferrara, Italy), Fiorica F. (Department of Radiation Oncology, S. Anna Hospital, Ferrara, Italy), Valentini A. (Division of Neurosurgery, Nuovo Ospedale Civile S. Agostino‐Estense, Baggiovara, Modena, Italy), Depenni R. (Department of Oncology, Policlinico di Modena, Italy), Mucciarini C. (Department of Oncology, Ramazzini Hospital, Carpi, Modena, Italy), Crisi G. (Department of Neuroradiology, Maggiore Hospital, Parma, Italy), Sasso E. (Department of Neurological Sciences, Maggiore Hospital, Parma, Italy), Biasini C., Cavanna L. (Department of Oncology and Hematology, Guglielmo da Saliceto Hospital, Piacenza, Italy), Guidetti D. (Department of Neurology, Guglielmo da Saliceto Hospital, Piacenza, Italy), Marcello N., Pisanello A. (Department of Neurology, Istituto in tecnologie avanzate e modelli assistenziali in oncologia, IRCCS, S. Maria Nuova Hospital, Reggio Emilia, Italy), Cremonini A.M., Guiducci G. (Division of Neurosurgery, M. Bufalini Hospital, Cesena, Italy).
Registry Coordination Office: de Pasqua S., Testoni S. (IRCCS Institute of Neurological Sciences, Bologna, Italy).
Participants
Agati R., Ambrosetto G., Bacci A., Baldin E., Baldrati A., Barbieri E., Bartolini S., Bellavista E., Bisulli F., Bonora E., Bunkheila F., Carelli V., Crisci M., Dall'Occa P., Ferro S., Franceschi C., Frezza G., Grasso V., Leonardi M., Morandi L., Mostacci B., Palandri G., Pasini E., Pastore Trossello M., Poggi R., Riguzzi P., Rinaldi R., Rizzi S., Romeo G., Spagnolli F., Tinuper P., Trocino C. (Bologna), Dall'Agata M., Frattarelli M., Gentili G., Giovannini A., Iorio P., Pasquini U., Galletti G., Guidi C., Neri W., Patuelli A., Strumia S. (Forlì‐Cesena), Faedi M., (IRCCS Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori), Casmiro M., Gamboni A., Rasi F. (Faenza R.A.), Cruciani G. (Lugo, RA), Cenni P., Dazzi C., Guidi A.R., Zumaglini F. (Ravenna), Amadori A., Pasini G., Pasquinelli M., Pasquini E., Polselli A., Ravasio A., Viti B. (Rimini), Sintini M. (Cattolica, RN), Ariatti A., Bertolini F., Bigliardi G., Carpeggiani P., Cavalleri F., Meletti S., Nichelli P., Pettorelli E., Pinna G., Zunarelli E. (Modena), Artioli F., Bernardini I., Costa M., Greco G., Guerzoni R., Stucchi C. (Carpi M.O.), Iaccarino C., Ragazzi M., Rizzi R., Zuccoli G. (Istituto di Ricovero e Cura a Carattere Scientifico, Reggio Emilia), Api P., Cartei F., Fallica E., Granieri E., Latini F., Lelli G., Monetti C., Saletti A., Schivalocchi R., Seraceni S., Tola M.R., Urbini B. (Ferrara), Giorgi C., Montanari E. (Fidenza P.R.), Cerasti D., Crafa P., Dascola I., Florindo I., Giombelli E., Mazza S., Ramponi V., Servadei F., Silini EM., Torelli P. (Parma), Immovilli P., Morelli N., Vanzo C. (Piacenza), Nobile C. (Padova).
Full affiliations and postal addresses of PERNO participants are available at the study website: www.perno.it.
Visani Michela, de Biase Dario, Marucci Gianluca, Cerasoli Serenella, Nigrisoli Evandro, Bacchi Reggiani Maria Letizia, Albani Fiorenzo, Baruzzi Agostino and Pession Annalisa, (2014), Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I–III, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.12.010.
References
- Brandes, A.A. , Tosoni, A. , Franceschi, E. , Sotti, G. , Frezza, G. , Amista, P. , Morandi, L. , Spagnolli, F. , Ermani, M. , 2009. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation with MGMT promoter methylation status. J. Clin. Oncol.. 27, 1275–1279. [DOI] [PubMed] [Google Scholar]
- Bulnes, S. , Bengoetxea, H. , Ortuzar, N. , Argandona, E.G. , Garcia-Blanco, A. , Rico-Barrio, I. , Lafuente, J.V. , 2012. Angiogenic signalling pathways altered in gliomas: selection mechanisms for more aggressive neoplastic subpopulations with invasive phenotype. J. Signal Transduct.. 2012, 597915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J.A. , Krichevsky, A.M. , Kosik, K.S. , 2005. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res.. 65, 6029–6033. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Liu, W. , Chao, T. , Zhang, Y. , Yan, X. , Gong, Y. , Qiang, B. , Yuan, J. , Sun, M. , Peng, X. , 2008. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett.. 272, 197–205. [DOI] [PubMed] [Google Scholar]
- Cho, R.J. , Campbell, M.J. , 2000. Transcription, genomes, function. Trends Genet.. 16, 409–415. [DOI] [PubMed] [Google Scholar]
- Ciafre, S.A. , Galardi, S. , Mangiola, A. , Ferracin, M. , Liu, C.G. , Sabatino, G. , Negrini, M. , Maira, G. , Croce, C.M. , Farace, M.G. , 2005. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun.. 334, 1351–1358. [DOI] [PubMed] [Google Scholar]
- Clarke, J. , Butowski, N. , Chang, S. , 2010. Recent advances in therapy for glioblastoma. Arch. Neurol.. 67, 279–283. [DOI] [PubMed] [Google Scholar]
- Conti, A. , Aguennouz, M. , La Torre, D. , Tomasello, C. , Cardali, S. , Angileri, F.F. , Maio, F. , Cama, A. , Germano, A. , Vita, G. , Tomasello, F. , 2009. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J. Neurooncol.. 93, 325–332. [DOI] [PubMed] [Google Scholar]
- Davis, B.N. , Hilyard, A.C. , Nguyen, P.H. , Lagna, G. , Hata, A. , 2009. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J. Biol. Chem.. 284, 3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Biase, D. , Visani, M. , Morandi, L. , Marucci, G. , Taccioli, C. , Cerasoli, S. , Baruzzi, A. , Pession, A. , 2012. miRNAs expression analysis in paired fresh/frozen and dissected formalin fixed and paraffin embedded glioblastoma using real-time pCR. PLoS ONE. 7, e35596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia, N. , Konopka, G. , Lim, K.L. , Nutt, C.L. , Bromberg, J.F. , Frank, D.A. , Mischel, P.S. , Louis, D.N. , Bonni, A. , 2008. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J. Neurosci.. 28, 5870–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Luo, L. , Hong, S. , Siu, H. , Xiao, Y. , Jin, L. , Chen, R. , Xiong, M. , 2010. Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst. Biol.. 4, 163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep, H. , Sticht, C. , Pandey, P. , Gretz, N. , 2011. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform.. 44, 839–847. [DOI] [PubMed] [Google Scholar]
- Ermoian, R.P. , Furniss, C.S. , Lamborn, K.R. , Basila, D. , Berger, M.S. , Gottschalk, A.R. , Nicholas, M.K. , Stokoe, D. , Haas-Kogan, D.A. , 2002. Dysregulation of PTEN and protein kinase B is associated with glioma histology and patient survival. Clin. Cancer Res.. 8, 1100–1106. [PubMed] [Google Scholar]
- Gabriely, G. , Yi, M. , Narayan, R.S. , Niers, J.M. , Wurdinger, T. , Imitola, J. , Ligon, K.L. , Kesari, S. , Esau, C. , Stephens, R.M. , Tannous, B.A. , Krichevsky, A.M. , 2010. Human glioma growth is controlled by microRNA-10b. Cancer Res.. 71, 3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal, H. , Pandi, G. , Kanner, A.A. , Ram, Z. , Lithwick-Yanai, G. , Amariglio, N. , Rechavi, G. , Givol, D. , 2008. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun.. 376, 86–90. [DOI] [PubMed] [Google Scholar]
- Galasso, M. , Sandhu, S.K. , Volinia, S. , 2012. MicroRNA expression signatures in solid malignancies. Cancer J.. 18, 238–243. [DOI] [PubMed] [Google Scholar]
- Genovese, G. , Ergun, A. , Shukla, S.A. , Campos, B. , Hanna, J. , Ghosh, P. , Quayle, S.N. , Rai, K. , Colla, S. , Ying, H. , Wu, C.J. , Sarkar, S. , Xiao, Y. , Zhang, J. , Zhang, H. , Kwong, L. , Dunn, K. , Wiedemeyer, W.R. , Brennan, C. , Zheng, H. , Rimm, D.L. , Collins, J.J. , Chin, L. , 2012. microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-beta signaling in glioblastoma. Cancer Discov.. 2, 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski, J. , Nowicki, M.O. , Bronisz, A. , Williams, S. , Otsuki, A. , Nuovo, G. , Raychaudhury, A. , Newton, H.B. , Chiocca, E.A. , Lawler, S. , 2008. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res.. 68, 9125–9130. [DOI] [PubMed] [Google Scholar]
- Guan, Y. , Mizoguchi, M. , Yoshimoto, K. , Hata, N. , Shono, T. , Suzuki, S.O. , Araki, Y. , Kuga, D. , Nakamizo, A. , Amano, T. , Ma, X. , Hayashi, K. , Sasaki, T. , 2010. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin. Cancer Res.. 16, 4289–4297. [DOI] [PubMed] [Google Scholar]
- Hsu, S.D. , Lin, F.M. , Wu, W.Y. , Liang, C. , Huang, W.C. , Chan, W.L. , Tsai, W.T. , Chen, G.Z. , Lee, C.J. , Chiu, C.M. , Chien, C.H. , Wu, M.C. , Huang, C.Y. , Tsou, A.P. , Huang, H.D. , 2011. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res.. 39, D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, D. , Mo, F. , Ding, D. , Li, L. , Han, X. , Zhao, N. , Foltz, G. , Lin, B. , Lan, Q. , Huang, Q. , 2012. A catalogue of glioblastoma and brain MicroRNAs identified by deep sequencing. OMICS. 16, 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse, J.T. , Brennan, C. , Hambardzumyan, D. , Wee, B. , Pena, J. , Rouhanifard, S.H. , Sohn-Lee, C. , le Sage, C. , Agami, R. , Tuschl, T. , Holland, E.C. , 2009. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev.. 23, 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas, B. , Godlewski, J. , Comeau, L. , Li, Y. , Abounader, R. , Hawkinson, M. , Lee, J. , Fine, H. , Chiocca, E.A. , Lawler, S. , Purow, B. , 2008. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res.. 68, 3566–3572. [DOI] [PubMed] [Google Scholar]
- Lakomy, R. , Sana, J. , Hankeova, S. , Fadrus, P. , Kren, L. , Lzicarova, E. , Svoboda, M. , Dolezelova, H. , Smrcka, M. , Vyzula, R. , Michalek, J. , Hajduch, M. , Slaby, O. , 2011. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci.. 102, 2186–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, M.F. , Yang, S. , Zhao, C. , Sun, G. , Murai, K. , Wu, X. , Wang, J. , Gao, H. , Brown, C.E. , Liu, X. , Zhou, J. , Peng, L. , Rossi, J.J. , Shi, Y. , 2012. Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS ONE. 7, e36248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler, S. , Chiocca, E.A. , 2009. Emerging functions of microRNAs in glioblastoma. J. Neurooncol.. 92, 297–306. [DOI] [PubMed] [Google Scholar]
- LeBrun, D.G. , Li, M. , 2011. MicroRNAs in glioblastoma multiforme: profiling studies and therapeutic Impacts. Mol. Cell Pharmacol.. 3, 93–105. [Google Scholar]
- Li, Y. , Guessous, F. , Zhang, Y. , Dipierro, C. , Kefas, B. , Johnson, E. , Marcinkiewicz, L. , Jiang, J. , Yang, Y. , Schmittgen, T.D. , Lopes, B. , Schiff, D. , Purow, B. , Abounader, R. , 2009. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res.. 69, 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.B. , Ma, M.W. , Dong, L.J. , Wang, F. , Chen, L.X. , Li, X.R. , 2011. MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol. Ther.. 12, 477–483. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. , Schmittgen, T.D. , 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Loftus, J.C. , Ross, J.T. , Paquette, K.M. , Paulino, V.M. , Nasser, S. , Yang, Z. , Kloss, J. , Kim, S. , Berens, M.E. , Tran, N.L. , 2012. miRNA expression profiling in migrating glioblastoma cells: regulation of cell migration and invasion by miR-23b via targeting of Pyk2. PLoS ONE. 7, e39818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, D.N. , Ohgaki, H. , Wiestler, O.D. , Cavenee, W.K. , 2007. WHO. Classification of Tumours of the Central Nervous System IARC; Lyon: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, S. , Vougioukas, V.I. , Hielscher, T. , Schmidt, U. , Unterberg, A. , Halatsch, M.E. , 2008. Pathogenetic pathways leading to glioblastoma multiforme: association between gene expressions and resistance to erlotinib. Anticancer Res.. 28, 3729–3732. [PubMed] [Google Scholar]
- Malzkorn, B. , Wolter, M. , Liesenberg, F. , Grzendowski, M. , Stuhler, K. , Meyer, H.E. , Reifenberger, G. , 2009. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol.. 20, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. , Thomas, P.D. , 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res.. 41, D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, M. , Guan, Y. , Yoshimoto, K. , Hata, N. , Amano, T. , Nakamizo, A. , Sasaki, T. , 2012. MicroRNAs in human malignant gliomas. J. Oncol.. 2012, 732874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnani Marelli, M. , Moretti, R.M. , Mai, S. , Muller, O. , Van Groeninghen, J.C. , Limonta, P. , 2009. Novel insights into GnRH receptor activity: role in the control of human glioblastoma cell proliferation. Oncol. Rep.. 21, 1277–1282. [DOI] [PubMed] [Google Scholar]
- Morandi, L. , Franceschi, E. , de Biase, D. , Marucci, G. , Tosoni, A. , Ermani, M. , Pession, A. , Tallini, G. , Brandes, A. , 2010. Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer. 10, 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi, L. , de Biase, D. , Visani, M. , Cesari, V. , De Maglio, G. , Pizzolitto, S. , Pession, A. , Tallini, G. , 2012. Allele specific locked nucleic acid quantitative PCR (ASLNAqPCR): an accurate and cost-effective assay to diagnose and quantify KRAS and BRAF mutation. PLoS ONE. 7, e36084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, M. , Kita, D. , Watanabe, T. , Hayashi, Y. , Teng, L. , Pyko, I.V. , Hamada, J. , 2011. Aberrant signaling pathways in glioma. Cancer. 3, 3242–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam, S.P. , Croce, C.M. , 2013. Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther.. 93, 98–104. [DOI] [PubMed] [Google Scholar]
- Niyazi, M. , Zehentmayr, F. , Niemoller, O.M. , Eigenbrod, S. , Kretzschmar, H. , Schulze-Osthoff, K. , Tonn, J.C. , Atkinson, M. , Mortl, S. , Belka, C. , 2011. MiRNA expression patterns predict survival in glioblastoma. Radiat. Oncol.. 6, 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintavalle, C. , Garofalo, M. , Zanca, C. , Romano, G. , Iaboni, M. , del Basso De Caro, M. , Martinez-Montero, J.C. , Incoronato, M. , Nuovo, G. , Croce, C.M. , Condorelli, G. , 2012. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene. 31, 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintavalle, C. , Magnani, D. , Roscino, G. , Romano, G. , Diaz-Lagares, A. , Iaboni, M. , Donnarumma, E. , Fiore, D. , De Marinis, P. , Soini, Y. , Esteller, M. , Condorelli, G. , 2013. miR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS ONE. 8, e74466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, S.A. , Santosh, V. , Somasundaram, K. , 2010. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod. Pathol.. 23, 1404–1417. [DOI] [PubMed] [Google Scholar]
- Sasayama, T. , Nishihara, M. , Kondoh, T. , Hosoda, K. , Kohmura, E. , 2009. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer. 125, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Silber, J. , Lim, D.A. , Petritsch, C. , Persson, A.I. , Maunakea, A.K. , Yu, M. , Vandenberg, S.R. , Ginzinger, D.G. , James, C.D. , Costello, J.F. , Bergers, G. , Weiss, W.A. , Alvarez-Buylla, A. , Hodgson, J.G. , 2008. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med.. 6, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber, J. , Jacobsen, A. , Ozawa, T. , Harinath, G. , Pedraza, A. , Sander, C. , Holland, E.C. , Huse, J.T. , 2012. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS ONE. 7, e33844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky, R.L. , Cullen, B.R. , 2011. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS ONE. 6, e24248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby, O. , Lakomy, R. , Fadrus, P. , Hrstka, R. , Kren, L. , Lzicarova, E. , Smrcka, M. , Svoboda, M. , Dolezalova, H. , Novakova, J. , Valik, D. , Vyzula, R. , Michalek, J. , 2010. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 57, 264–269. [DOI] [PubMed] [Google Scholar]
- Smits, M. , Nilsson, J. , Mir, S.E. , van der Stoop, P.M. , Hulleman, E. , Niers, J.M. , de Witt Hamer, P.C. , Marquez, V.E. , Cloos, J. , Krichevsky, A.M. , Noske, D.P. , Tannous, B.A. , Wurdinger, T. , 2010. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 1, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S. , Patric, I.R. , Somasundaram, K. , 2011. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS ONE. 6, e17438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojan, J. , Cloix, J.F. , Ardourel, M.Y. , Chatel, M. , Anthony, D.D. , 2007. Insulin-like growth factor type I biology and targeting in malignant gliomas. Neuroscience. 145, 795–811. [DOI] [PubMed] [Google Scholar]
- van Groeninghen, J.C. , Kiesel, L. , Winkler, D. , Zwirner, M. , 1998. Effects of luteinising-hormone-releasing hormone on nervous-system tumours. Lancet. 352, 372–373. [DOI] [PubMed] [Google Scholar]
- Visani, M. , de Biase, D. , Marucci, G. , Taccioli, C. , Baruzzi, A. , Pession, A. , 2012. Definition of miRNAs expression profile in glioblastoma samples: the relevance of non-neoplastic brain reference. PLoS ONE. 8, e55314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Liu, Z. , Balivada, S. , Shrestha, T. , Bossmann, S. , Pyle, M. , Pappan, L. , Shi, J. , Troyer, D. , 2012. Interleukin-1beta and transforming growth factor-beta cooperate to induce neurosphere formation and increase tumorigenicity of adherent LN-229 glioma cells. Stem Cell Res. Ther.. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, F. , Zuo, Z. , Cai, G. , Kang, S. , Gao, X. , Li, T. , 2009. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res.. 37, D105–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Parsons, D.W. , Jin, G. , McLendon, R. , Rasheed, B.A. , Yuan, W. , Kos, I. , Batinic-Haberle, I. , Jones, S. , Riggins, G.J. , Friedman, H. , Friedman, A. , Reardon, D. , Herndon, J. , Kinzler, K.W. , Velculescu, V.E. , Vogelstein, B. , Bigner, D.D. , 2009. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med.. 360, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Pan, X. , Cobb, G.P. , Anderson, T.A. , 2007. microRNAs as oncogenes and tumor suppressors. Dev. Biol.. 302, 1–12. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Han, L. , Zhang, A. , Yang, W. , Zhou, X. , Pu, P. , Du, Y. , Zeng, H. , Kang, C. , 2010. Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells. Int. J. Oncol.. 36, 1503–1512. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Zhang, J. , Yan, W. , You, G. , Bao, Z. , Li, S. , Kang, C. , Jiang, C. , You, Y. , Zhang, Y. , Chen, C.C. , Song, S.W. , Jiang, T. , 2013. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer. 119, 814–824. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Ren, Y. , Moore, L. , Mei, M. , You, Y. , Xu, P. , Wang, B. , Wang, G. , Jia, Z. , Pu, P. , Zhang, W. , Kang, C. , 2010. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab. Invest.. 90, 144–155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary Figure 1 Unsupervised hierarchical clustering analysis of GBM group for the 14 validated microRNAs. Clustering analysis was performed using the Ambion commercial reference (A), epileptic tissues (B) and normal adjacent to the tumor (C). MET–GBM samples are written in blue, UMET–GBM in pink, non‐neoplastic control samples in green. GBM with methylation status available is reported in red.


