Abstract
In our previous study, we identified 1241 loci with somatic copy number alterations in human hepatocellular carcinoma (HCC) using Affymetrix SNP 6.0 arrays, and a putative cancer gene SERPINA5 was uncovered in a novel chromosomal region with recurrent copy number loss at 14q31.1–32.13. The SERPINA5 was reported to be deregulated in renal, breast, prostate and ovarian cancers. However, the roles of SERPINA5 in cancer remain greatly elusive. In this study, we found that the DNA dosage and expression level of the SERPINA5 gene were significantly decreased in HCC by quantitative real‐time PCR. Notably, the expression levels of SERPINA5 negatively correlated with malignant progression of HCC. The SERPINA5 gene was further observed to reduce in vitro and in vivo metastatic potential of HCC cells. Moreover, secreted SERPINA5 protein also could inhibit the metastatic ability of HCC cells. Finally, we discovered that one of the mechanisms explaining SERPINA5 inhibition of HCC metastasis is through direct interaction with fibronectin and disruption of the fibronectin–integrin signaling pathway. These findings highlight an important role of SERPINA5 in the regulation of migratory and metastatic potentials of HCC and suggest a potential application of SERPINA5 in cancer treatment.
Keywords: SERPINA5, Migration, Metastasis, Hepatocellular carcinoma
Highlights
Downregulation of SERPINA5 negatively correlates with malignant progression of HCC.
SERPINA5 reduces in vitro and in vivo metastatic potential of HCC cells.
SERPINA5 inhibits HCC cell migration by directly interacting with fibronectin.
SERPINA5 disrupts the fibronectin–integrin β1 signaling pathway.
1. Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and the third leading cause of cancer death worldwide (Yang and Roberts, 2010). The identification and characterization of genes within amplified and deleted chromosomal loci can provide new insights into the pathogenesis of cancer and also lead to new approaches for diagnosis and therapy (Speleman et al., 2008). To identify novel cancer‐related genes, we previously identified 1241 loci with somatic copy number alterations (CNA) in human HCC using Affymetrix genome‐wide SNP 6.0 arrays (Jia et al., 2011). Importantly, a number of new CNAs were also found, including a novel recurrent region of copy number loss at 14q31.1–32.13 with a frequency of 8.6% (5/58) in HCC (Jia et al., 2011).
Copy number loss of 14q has been reported in several other cancers, including renal cell carcinoma (Alimov et al., 2004), adenocarcinoma of the gastroesophageal junction (GEJ) (Fortenberry et al., 2010; van Dekken et al., 1999), head and neck squamous cell carcinoma (HNSCC) (Pehlivan et al., 2008), colorectal adenocarcinoma (Thorstensen et al., 2001), and gastric cancer (Buffart et al., 2012). Moreover, loss of 14q was also correlated with an early age of onset, advanced stage, high grade, and poor disease outcome (Alimov et al., 2004; Buffart et al., 2012; Fortenberry et al., 2010; Pehlivan et al., 2008; Thorstensen et al., 2001). The focal region has been isolated to 14q31.1–32.13 in adenocarcinoma of the GEJ and renal cell carcinoma, suggesting that this region encodes a putative tumor suppressor gene involved in disease progression (Alimov et al., 2004; Fortenberry et al., 2010; van Dekken et al., 1999). Therefore, the identification of the genes at locus 14q31.1–32.13 responsible for copy number loss is imperative to understand the molecular mechanisms of cancer initiation and progression in many solid tumors, including HCC. To identify the potential driver genes located in this region, we previously focused on differentially expressed genes at this locus with integrated copy number analysis and expression profiling, from which one candidate gene, SERPINA5, was derived (Jia et al., 2011).
SERPINA5 (also called Protein C Inhibitor, PCI) belongs to the serine protease inhibitor superfamily which is a multi‐functional protein, including preventing metastasis and anti‐angiogenesis in tumor cells (Sil et al., 2011). SERPINA5 was first isolated from human plasma. Human plasma SERPINA5 is mainly synthesized in the liver (Francis and Thomas, 1984), but it is also produced in kidneys and reproductive organs, including testes, seminal vesicles and ovaries (Laurell et al., 1992). Recently, SERPINA5 expression has been shown to be decreased in renal, breast, prostate and ovarian cancers (Asanuma et al., 2007; Bijsmans et al., 2011; Cao et al., 2003; Wakita et al., 2004). However, the expression status, biological function and molecular mechanisms of SERPINA5 in HCC are largely unknown.
In this study, we demonstrated that SERPINA5 is pathologically downregulated in HCC specimens. Ectopic expression of SERPINA5 could inhibit the metastatic abilities of HCC cell lines in vitro and in vivo. Moreover, we found that one of the mechanisms by which SERPINA5 contributes to these malignant feature was explored to through disrupting the fibronectin–integrin signaling pathway. Together, our findings not only advance the molecular understanding of tumor metastasis, but also provide a novel therapeutic target for the treatment of metastatic HCC.
2. Materials and methods
2.1. Cell lines and cell culture
Seven liver cancer cell lines were used in this study: HUH‐7, HepG2, SMMC‐7721, Hep3B, MHCC‐97H, HCCLM3 and SNU‐449. The SMMC‐7721 cells were cultured at 37 °C with a 5% CO2 atmosphere in DMEM supplemented with 10% newborn calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin. The other six cancer cell lines and HEK‐293T cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were regularly examined to ensure that they were free of mycoplasma contamination.
2.2. Antibodies, plasmids and other reagents
Specific antibodies against integrin β1, FAK, phospho‐FAK (Y397), Src and phospho‐Src (Y416) were purchased from CST (Danvers, MA, USA). Specific antibody against integrin β1 (Y788/789) were purchased from Invitrogen (Grand Island, NY, USA). The antibody against β‐actin was purchased from Sigma (St. Louis, MO, USA). The SERPINA5 antibody used for Western blot was purchased from Abcam (Hong Kong, China), and the antibody for Co‐immunoprecipitation was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The SERPINA5 construct was previously described (Jia et al., 2011). Lentiviral shRNA vectors targeting SERPINA5 and scrambled control shRNA vectors were purchased from Open Biosystems (Thermo scientific). siRNAs targeting integrin β1 and negative control siRNA were ordered from SMARTpool (Thermo scientific). The shRNA targeting fibronectin was constructed as previously reported (Jia et al., 2010). Human plasma‐derived fibronectin was purchased from Millipore (Billerica, MA, USA). DMEM without serum or phenol red was purchased from Invitrogen (Grand Island, NY, USA). Recombinant human SERPINA5 was ordered from R&D (Minneapolis, MN). Chaperone competent cell BL21 were purchased from Takara (Dalian, China).
2.3. Lentiviral vector construction, packaging and infection
The experiments were performed as previously described (Jia et al., 2011). The entire coding sequence of the target cDNAs was amplified and cloned into the pWPXL vector, which was obtained from Addgene. Lentivirus production and transduction were performed according to instructions supplied by Addgene (http://www.addgene.org).
2.4. HCC specimens and clinical data
HCC primary tumors and the adjacent non‐tumor liver tissues (3 cm from the tumor) were obtained from the surgical specimen archives of the Qidong Liver Cancer Institute, Jiangsu Province, China. Participants that these samples were obtained from provided their written informed consent to participate in the study, and the Ethical Review Committee of the WHO Collaborating Center for Research in Human Production authorized by the Shanghai Municipal Government approved this study as well as the consent procedure.
Genomic DNA was extracted from 125 HCC primary tumors and adjacent non‐tumor tissues. Total RNA was extracted from 130 HCC primary tumors and adjacent non‐tumor tissues. Forty‐six HCC specimens with genomic DNA and total RNA were used to analyze the correlation between DNA dosage and mRNA expression of the SERPINA5 gene. Among the 130 paired HCC specimens with cDNA, fifty‐eight HCC specimens with detailed clinical information were used to analyze the correlation between mRNA expression of the SERPINA5 gene and the clinical features of HCC. Additionally, we mainly employed the Edmondson and Steiner grading system (EGS) to determine the histopathological grade of HCC, and two categories were considered (low grade, EGS I–II; high grade, EGS III–IV).
2.5. Quantitative real‐time PCR
Genomic DNA dosage and mRNA expression levels were quantified using a 7900 Real‐Time PCR System with SDS 2.3 software (Applied Biosystems) according to the manufacturer's instructions. In brief, total genomic DNA and mRNA were extracted from tumor tissues. First‐strand cDNA synthesis and amplification were performed using Reverse Transcription Reagents (Takara) according to the manufacturer's instructions. cDNA templates were mixed with SYBR Green premix with Rox (Takara) to perform quantitative PCR reactions. A repetitive element (Line‐1) and β‐actin were used as endogenous controls for the DNA and mRNA levels, respectively. Details of the primers sequences are provided in the Table S1.
2.6. Cell proliferation assays
Cells were seeded at a density of 2000 cells per well and incubated with complete medium or conditioned medium collected from stable cell lines and supplemented with 10% fetal bovine serum in 96‐well plates. Culturing medium was removed and aliquots (10 μl) from the Cell‐Counting Kit (CCK‐8, Dojindo, Kumamoto, Japan) and 90 μl complete medium were added to the wells and incubated for 2 h. After incubation, the absorbance was measured at 450 nm to calculate the number of viable cells in each well. Each measurement was performed in triplicate, and the experiments were repeated twice.
2.7. Trans‐well migration assays
Cell migration assays were performed using 6.5‐mm trans‐well chambers (8 μm pore size, BD) as described previously with some modifications (Jia et al., 2010). Cells were seeded into trans‐well chambers at a density of 40,000 cells per well, and serum (10% FBS in DMEM) was used as the chemoattractant in lower chamber. The wells were washed with D‐PBS after 16–24 h. The cells that migrated to the basal side of the membrane were fixed and stained with crystal violet, visualized and photographed with a CKX41 microscope (Olympus, Japan) at 200× magnification. Images of three random fields from three replicate wells were obtained, and the cells that migrated were counted. Chambers for migration on purified fibronectin assay were coated with 10 μg/ml fibronectin overnight at 4 °C and blocked with 2% BSA at 37 °C for 2 h. 1 × 105 cells in 200 μl serum‐free DMEM were seeded to upper chamber. After 4 h, cells in the upper side were removed and cells in the underside were fixed, stained and counted as described above.
2.8. Secreted protein preparation
Cells were incubated in a 6‐cm dish overnight at the same density. When the confluent reached 60–70%, cells were washed three times with D‐PBS to get rid of dead cells, serum and phenol red. Cells were washed with DMEM without serum or phenol red (conditioned medium, CM) once, and then 3 mL of CM was added to the plate. After 24–48 h incubation, CM was collected, centrifuged and filtered with a 0.22 μm sterile filter (Millipore). Then conditioned medium containing secreted protein can be used to treat cells for following up experiments.
2.9. Secreted SERPINA5, human plasma‐derived fibronectin and purified recombinant SERPINA5 treatments
One day before treatment, cells were incubated in 6‐cm dishes. When the confluent was 50%–60%, cells were washed with D‐PBS three times, and with DMEM once to get rid of dead cells and serum. Three milliliter of conditioned medium collected from SERPINA5‐overexpressing or knockdown cells was added. 100 μg/ml fibronectin was supplied in designated experiments and D‐PBS was used as negative control. After 24–36 h incubation in designated culturing conditions, cells were digested and subjected to transwell assays. For recombinant SERPINA5 treatment, cells were seeded and washed as described above, and then treated with 3 mL serum‐free DMEM containing different concentrations (0.02 μg/ml, 0.1 μg/ml and 1 μg/ml) recombinant SERPINA5. D‐PBS was used as negative control. Cells were harvested for trans‐well migration assays after 24 h.
2.10. Secreted protein concentration
To concentrate proteins, Amicon® Ultra‐15 Centrifugal Filter Units (10 kDa) and Ultra‐4 Centrifugal Filter Units (10 kDa) (Millipore) were used. The collected conditioned medium was added to the upper chamber of the centrifugal filter units. The units were centrifuged using a horizontal rotor at 4000 rpm or angled rotor at 12,000 rcf for 15–30 min until the volume of liquid in upper chamber reached about 200 μl for Ultra‐15 and 50 μl for Ultra‐4. The residue in the upper chamber was measured for protein content using the Bradford assay. Same amount of protein was applied for western blot and immunoprecipitation. Concentrated CM was tested for α‐tubulin by immunoblotting to ensure cellular protein was not present.
2.11. Membrane and peripheral protein extraction
The membrane and peripheral protein extraction experiments were performed by using the ProteoExtract® transmembrane protein extraction kit (Novagen) according to the manufacturer's instructions. Both integral membrane and peripheral associated proteins were extracted from HCCLM3‐SERPINA5 cells. Protein content was measured by Bradford assay, and same amount of protein was applied for Co‐IP.
2.12. Co‐immunoprecipitation (Co‐IP)
Co‐IP experiments were performed with the Pierce® Co‐immunoprecipitation (Co‐IP) Kit (Thermo Scientific) according to the manufacturer's instructions. Whole cell lysate, conditioned medium and membrane and associated proteins were collected from the specified cells and subjected to co‐IP analysis.
2.13. Western blot analysis
For western blot analysis, the protein concentration was determined by the Bradford assay. Cell lysates, secreted proteins and immunoprecipitated proteins were separated by SDS‐PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% non‐fat milk in TBST and incubated with specific primary antibodies. Detection was performed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) followed by exposure to X‐ray film. According to the abundance of target proteins in different HCC cells, we shortened or extended the exposure time to get clearly presented bands on X‐ray film. Therefore, the density of bands represents a relatively low or high level of target proteins.
2.14. Animal experiments and histological analysis
Animal experiments were performed as previously described (Jia et al., 2010). Five‐ to six‐week‐old, male, congenitally immune‐deficient nude mice were maintained under specific pathogen‐free (SPF) conditions. The mice were manipulated and housed according to protocols approved by the Shanghai Medical Experimental Animal Care Commission. For experimental metastasis assays, HCCLM3‐SERPINA5 and HCCLM3‐Vector cells (2 × 106 per mouse) were injected into the tail veins of nude mice. Six weeks later, the mice were euthanized, and the organs, including the lung and liver, were removed and processed for standard histological study. For histological analysis, the primary tumors and mouse organs were harvested at necropsy and fixed in 10% formalin. The fixed samples were then embedded in paraffin, and three non‐sequential serial sections were obtained from each animal. The sections were stained with hematoxylin and eosin (H&E) and analyzed for the presence of metastases.
2.15. GST pull down and in vitro binding assay
Chaperone competent cell BL21 was used to expression GST‐SERPINA5 fusion protein. 1 mM IPTG was added to induce protein expression in 30 °C for 5 h. The cell lysis and binding steps were performed according to instructions of MagneGST™ Pull‐Down System (Promega). 100 ng human purified fibronectin (Millipore) was incubated with MagneGST™ Particles carrying GST‐SERPINA5 fusion protein or GST in room temperature for 1 h. MagneGST™ Particles were washed with binding buffer and eluted with 1× SDS loading buffer. An aliquot of MagneGST™ Particles carrying GST‐SERPINA5 fusion protein or GST and human purified fibronectin were boiled in 1× SDS loading buffer as input. All samples were analyzed by Western blotting.
2.16. Statistics
The statistical analysis and graphical depiction of data were performed using GraphPad Prism 5.0. The results are presented as mean ± s.e.m and were evaluated with Student's t‐test (two‐tailed; P < 0.05 was considered significant), unless otherwise specified (paired t‐test, Pearson's correlation). Additionally, certain statistical calculations were performed using SPSS (Statistical Package for the Social Sciences), version 19.0 for Windows. The chi‐square (χ 2) test was used to evaluate the association between SERPINA5 expression and the clinicopathological parameters of the HCC specimens. P < 0.05 was considered significant.
3. Results
3.1. Downregulation of SERPINA5 in HCC specimens
To determine the role of SERPINA5 in the initiation and progression of HCC, the DNA dosage and expression level of this gene were examined in HCC tumor and paired adjacent non‐tumor tissues, respectively. By quantitative real‐time PCR (q‐PCR), we showed that both of the gene dosage and expression levels of SERPINA5 are significantly reduced in HCC specimens, compared with those of adjacent non‐tumor tissues (Figure 1A and B). Furthermore, the positive correlation between the DNA dosage and expression levels of SERPINA5 was also confirmed (Figure 1C). Consistent with aberrant expression of SERPINA5 reported in various tumors, Oncomine expression analysis revealed lower SERPINA5 expression in HCC tissues than in normal liver tissues in two independent cohorts of HCC specimens (Figure S1). Additionally, the decreased protein level of SERPINA5 in HCC tissues was also confirmed by immunoblotting (Figure 1D). Therefore, these data suggest that the decreased SERPINA5 expression was a frequent event in human HCC tissues, which may have functional roles in HCC progression.
Figure 1.
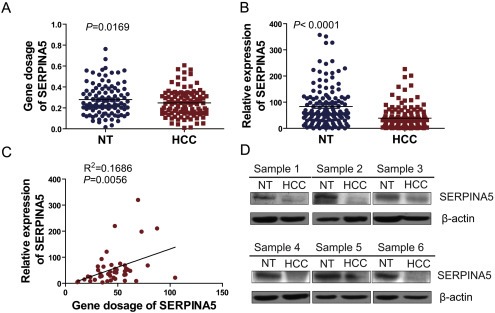
Downregulation of SERPINA5 gene in HCC. (A) Determination of DNA dosages of the SERPINA5 gene by quantitative real‐time PCR (q‐PCR) in an independent cohort of 125 paired HCCs and adjacent non‐tumor tissues (NT) with genomic DNA (paired Student's t‐test). (B) Determination of expression levels of the SERPINA5 gene by q‐PCR in an independent cohort of 130 paired HCCs and adjacent non‐tumor tissues (NT) with complementary DNA (cDNA) (paired Student's t‐test). (C) Correlation analysis of DNA dosage and mRNA expression for the SERPINA5 gene in 46 HCC primary tissues with both genomic DNA and cDNA. The correlation was analyzed with a two‐tailed Pearson Correlation Test. (D) The SERPINA5 protein levels decreased in the HCC primary tumors as opposed to the adjacent non‐tumor tissues. The detection of the relative protein levels of SERPINA5 in six pairs of HCC primary tumors and adjacent non‐tumor tissues by immunoblotting (HCC, Primary tumor; NT, non‐tumor tissue).
3.2. Loss of SERPINA5 expression is associated with malignant progression of HCC
Based on the relative expression levels of the SERPINA5 gene in 58 paired HCC primary tumors and adjacent non‐tumor tissues derived from q‐PCR experiments, we analyzed the clinical significance of SERPINA5 expression in HCC. By comparing the relative SERPINA5 expression levels between the primary tumor and paired adjacent non‐tumor tissue, we found that the SERPINA5 expression was predominantly downregulated in HCC specimens with a frequency of 64% (Figure 2A). Importantly, correlation analysis showed that the downregulation of SERPINA5 negatively correlates with the tumor grade and intrahepatic metastasis of HCC specimens, with lower expression of SERPINA5 in HCC specimens with high grade and intrahepatic metastasis (Figure 2B–C).
Figure 2.
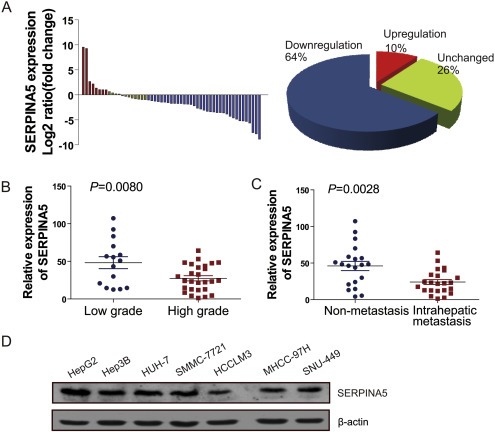
Loss of SERPINA5 expression correlates with malignant features of HCC. (A) The expression level of SERPINA5 gene in 58 paired HCC and matched non‐tumor tissues were determined by q‐PCR. The data are expressed as the log2 fold change (ΔCt [HCC/Non.]). Significant downregulation of SERPINA5 in paired HCC/non‐tumor samples was defined as a log2 fold change <1 (i.e., two‐fold). The pie chart shows the proportions of HCC samples showing upregulation (red), downregulation (blue), and no change (green). (B) Downregulation of SERPINA5 in primary HCCs in high grade (grade III–IV) compared with those in low grade (grade I–II) according to the results of q‐PCR. (C) Downregulation of SERPINA5 in primary HCCs with intrahepatic metastasis compared with those without intrahepatic metastasis according to the results of q‐PCR. Statistical analysis of the difference between groups was performed with an unpaired Student's t‐test. (D) Relative protein levels of SERPINA5 in seven HCC cell lines analyzed by immunoblotting.
Furthermore, the relative expression levels of SERPINA5 in seven HCC cell lines were examined by q‐PCR and immunoblotting. Specifically, the expression levels of SERPINA5 were relatively high in Hep3B, HepG2 and HUH‐7 cells, which have no or low metastatic potential (Cui et al., 2006; Genda et al., 1999; Lee et al., 2006), whereas the expression level of SERPINA5 was relatively low in SNU‐449, HCCLM3, and MHCC‐97H cells (Figure 2D and Figure S2), which have high metastatic potential (Fuchs et al., 2008; Lee et al., 2006; Li et al., 2004). These results imply that the loss of SERPINA5 might be associated with the malignant progression of HCC, providing clues to explore its biological function in HCC progression.
3.3. Targeted knockdown of SERPINA5 promotes HCC cell migration
To investigate the potential functions of SERPINA5 in HCC, we stably knocked down the expression of this gene using lentiviral infection in HCC cell lines. The knockdown efficiencies in Hep3B and HUH‐7 were validated by q‐PCR and immunoblotting (Figure 3A–B). Subsequently, the effect of SERPINA5 knockdown on proliferation was determined by CCK‐8 assays. We observed that disruption of SERPINA5 gene expression had no significant effect on the proliferation of HCC cells (Figure S3B). However, we found that knockdown of SERPINA5 expression could promote the migratory ability of HCC cells in trans‐well assays (Figure 3C). Briefly, these results suggest that SERPINA5 may play an important role in HCC cell migration.
Figure 3.
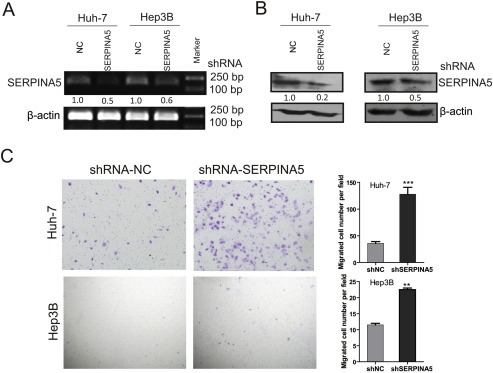
Targeted downregulation of SERPINA5 promotes HCC cell migration. (A) Detection of shRNA‐mediated knockdown of SERPINA5 gene in HUH‐7 and Hep3B cells by RT‐PCR. (B) Detection of shRNA‐mediated knockdown of SERPINA5 in HUH‐7 and Hep3B cells by immunoblotting. (C) Representative results of the trans‐well migration assay after SERPINA5 knockdown in HUH‐7 and Hep3B cells. Statistical analysis of the difference between groups was performed with an unpaired Student's t‐test. The results are shown as mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.4. Ectopic expression of SERPINA5 inhibits the migration and metastatic potential of HCC cells
The SERPINA5 gene was further overexpressed by lentiviral infection in MHCC‐97H, SNU‐449 and HCCLM3 cells and confirmed by immunoblotting (Figure 4A). Subsequently, we determined the effect of SERPINA5 overexpression on HCC cell proliferation, which showed that exogenous overexpression of SERPINA5, has no significant effect on HCC cell proliferation (Figure S3A), which was consistent with the above results. Furthermore, we examined the effect of SERPINA5 overexpression on the migratory and invasive abilities of HCC cells using trans‐well migration assays, and found that ectopic expression of SERPINA5 significantly inhibit the in vitro migration and invasion of HCC cells (Figure 4B–C). To further investigate whether SERPINA5 overexpression can inhibit tumor metastasis in vivo, by using of mouse models, HCCLM3‐SERPINA5 and HCCLM3‐VECTOR cells were injected into nude mice via tail vein and the lung metastasis was analyzed after 6 weeks. The incidence of lung metastasis was significantly inhibited in the SERPINA5 group of mouse models (Figure 4D). Moreover, hematoxylin and eosin (H&E) staining of lungs showed that the number of micrometastases was significantly lower in the SERPINA5 group than those in the VECTOR group (Figure 4E). These results indicate that ectopic expression of the SERPINA5 gene can inhibit the metastatic potentials of HCC cells.
Figure 4.
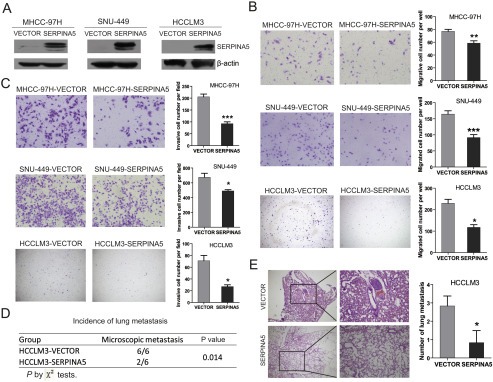
Overexpression of SERPINA5 inhibits HCC cell migration and metastasis. (A) The detection of lentiviral‐mediated overexpression of SERPINA5 in MHCC‐97H, SNU‐449 and HCCLM3 cells by immunoblotting. (B) Representative result of the trans‐well migration assays for the effects of SERPINA5 on the in vitro migratory ability of MHCC‐97H, SNU‐449 and HCCLM3 cells (unpaired Student's t‐test, mean ± s.e.m). (C) Representative result of the trans‐well invasion assays for the effects of SERPINA5 on the in vitro invasive ability of MHCC‐97H, SNU‐449 and HCCLM3 cells (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (D and E) The effects of SERPINA5 on the in vivo metastatic ability of HCCLM3 cells in xenograft models of nude mice (n = 6) determined by examination of mouse lungs for microscopic nodules. The incidence of lung metastasis in the two groups of mouse model (D), and representative results of the histological examination of mouse lungs for metastatic nodules from two groups, HCCLM3‐VECTOR and HCCLM3‐SERPINA5 (E) were presented. Statistical analysis of the difference between groups was performed by a chi‐square (χ2) test and an unpaired Students' t‐test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.5. Secreted SERPINA5 protein inhibits the migratory ability of HCC cells
SERPINA5 was originally identified in human plasma as an APC inhibitor (Suzuki et al., 1983), and its concentration in plasma is ∼100 nM (Geiger, 2007). To investigate whether HCC cells secrete SERPINA5, the levels of secreted SERPINA5 in conditioned medium were examined in the seven HCC cell lines. Secreted SERPINA5 levels were relatively high in three HCC cell lines (HepG2, Hep3B and HUH‐7) with low metastatic potential compared to the HCC cell lines with high metastatic potential (Figure 5A). Subsequently, three HCC cell lines (SNU‐449, HCCLM3 and MHCC‐97H) that secrete low SERPINA5 were stably transfected to express full‐length SERPINA5. Conditioned medium (CM) from these SERPINA5‐overexpressing cells were collected, and SERPINA5 was detected by immunoblotting. The results showed that SERPINA5 protein can be detected in the conditioned medium of SERPINA5‐expressing cells (Figure 5B).
Figure 5.
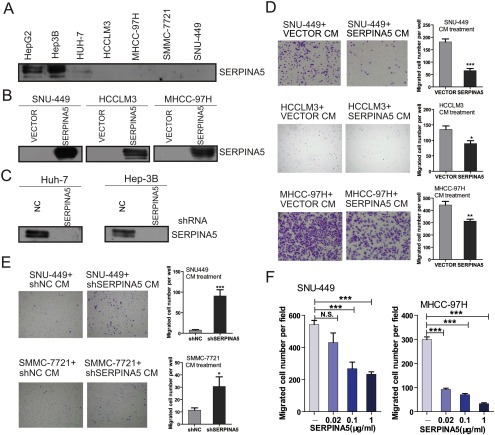
Secreted SERPINA5 inhibits the migration of HCC cells. (A) Detection of the secreted SERPINA5 protein levels in seven HCC cell lines by immunoblotting. (B) Detection of the secreted SERPINA5 protein levels in MHCC‐97H, SNU‐449 and HCCLM3 cells overexpressing ectopic SERPINA5 gene by immunoblotting. (C) Detection of the secreted SERPINA5 protein levels in HUH‐7 and Hep3B cells expressing shRNA targeting the SERPINA5 gene using immunoblotting. (D) Representative result of the trans‐well migration assays examining the effect of conditioned medium from SERPINA5‐overexpressing cells on the in vitro migration of SNU‐449, HCCLM3 and MHCC‐97H cells (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (E) Representative result of the trans‐well migration assays examining the effect of conditioned medium from HUH‐7‐shSERPINA5 cells on the in vitro migration of SNU‐449 and SMMC‐7721 cells (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (F) The recombinant human SERPINA5 reduces the migratory abilities of SNU‐449 and HCCLM3 cells in trans‐well migration assays (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001).
To further determine whether secreted SERPINA5 produces an inhibitory effect on HCC cell migration, we collected conditioned media from the SERPINA5‐overexpressing cells and SERPINA5‐knockdown cells, respectively (Figure 5B and C). The conditioned media collected from three SERPINA5‐overexpressing cell lines (SNU‐449‐SERPINA5, HCCLM3‐SERPINA5 and MHCC‐97H‐SERPINA5) were used to treat SNU‐449, HCCLM3 and MHCC‐97H cells, respectively. The conditioned medium from SERPINA5‐knockdown cells (HUH‐7‐shSERPINA5) was used to treat SNU‐449 and SMMC‐7721 cells. We performed CCK‐8 assays and trans‐well migration assays to determine the effect of secreted SERPINA5 on the proliferation and migration of HCC cells. We found that the secreted SERPINA5 has no significant effect on the proliferation of HCC cells (Figure S3C–D). However, the secreted SERPINA5 significantly inhibited the migratory potential of HCC cells (Figure 5D and E), which was consistent with the above results. Additionally, we also employed purified recombinant human SERPINA5 to treat HCC cells, and found that the recombinant protein can reduce the migration of HCC cells (Figure 5F). Together, these findings suggest that the secreted SERPINA5 protein has an inhibitory effect on HCC cell migration.
3.6. SERPINA5 inhibits HCC cell migration by interacting with fibronectin
During cancer progression, the extracellular matrix (ECM) of the tissue in which the tumor grows is extensively remodeled (Kaspar et al., 2006). It has been shown that SERPINA5 can bind to glycosaminoglycans, such as heparin and certain phospholipids, which modulate SERPINA5 inhibitory activity (Huntington and Li, 2009; Shirk et al., 1994). To explore potential SERPINA5 interaction partners leading to inhibition of HCC cell migration and tumor metastasis, we used LC‐MS/MS to analyze immunoprecipitated HCCLM3‐SERPINA5 membrane and peripheral proteins with SERPINA5 antibody and identified an ECM component, fibronectin (Figure 6A and Table S2). Fibronectin (FN) is a high molecular weight adhesive glycoprotein that plays a prominent role in mediating ECM function because it is present in high abundance and interacts with cellular components (Kaspar et al., 2006).
Figure 6.
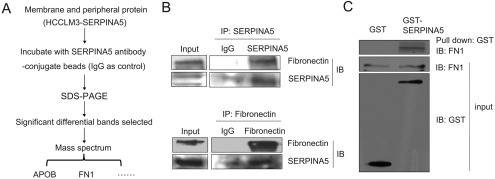
The direct interaction of SERPINA5 with fibronectin. (A) Schematic model of the identification of SERPINA5‐binding proteins using LC‐MS/MS to analyze immunoprecipitated proteins, by use of antibody against the SERPINA5 in membrane and peripheral proteins of HCCLM3‐SERPINA5, and separated by SDS‐PAGE. (B) Co‐immunoprecipitation (IP) analysis of SERPINA5 and fibronectin association in human HepG2 cells expressing endogenous SERPINA5. The proteins isolated by immunoprecipitation with an antibody against SERPINA5 were employed to detect the fibronectin protein, whereas the proteins isolated by immunoprecipitation with an antibody against fibronectin were subjected to detect the SERPINA5 protein using immunoblotting. (C) Purified SERPINA5 protein binds to fibronectin in vitro by GST pull‐down assays. The protein binding to MagneGST™ Particles was analyzed by immunoblotting (upper). An aliquot of MagneGST™ Particles (lower) and fibronectin (middle) were used as input.
To further verify the binding of SERPINA5 to fibronectin, co‐immunoprecipitation was performed using HepG2 whole cell lysate and conditioned medium with antibodies against either SERPINA5 or fibronectin, respectively. We identified an interaction between SERPINA5 and fibronectin in whole cell lysate (Figure 6B) but not in conditioned medium, which may be caused by the domain modification caused by glycosylation (Figure S4). In addition, we demonstrated that purified SERPINA5 protein can bind to fibronectin in vitro by GST pull‐down assays (Figure 6C). Together, these results suggest that SERPINA5 can directly bind to fibronectin and might participate in the fibronectin signaling pathway.
3.7. SERPINA5 inhibits HCC cell migration partially by disrupting the fibronectin/integrin β1 signaling pathway
We subsequently determined the effect of exogenously applied human plasma‐derived fibronectin on the migration of HCC cells after treatment with conditioned medium. Treatment of exogenous fibronectin reversed the inhibitory effect of SERPINA5 on the migration of SNU‐449 and HCCLM3 cells (Figure 7A). Furthermore, we found that the secreted SERPINA5 inhibits the migratory potential of HCC cells on chambers coated with fibronectin (Figure 7B). In addition, we determined that stable knockdown of the SERPINA5 gene enhanced the migration of HUH‐7 cells, whereas shRNA‐mediated knockdown of the fibronectin gene significantly attenuated the migration of HUH‐7‐shSERPINA5 cells (Figure 7C). Our findings suggest that SERPINA5 binds to the glycoprotein fibronectin, which in turn modulates SERPINA5 inhibition of migration.
Figure 7.
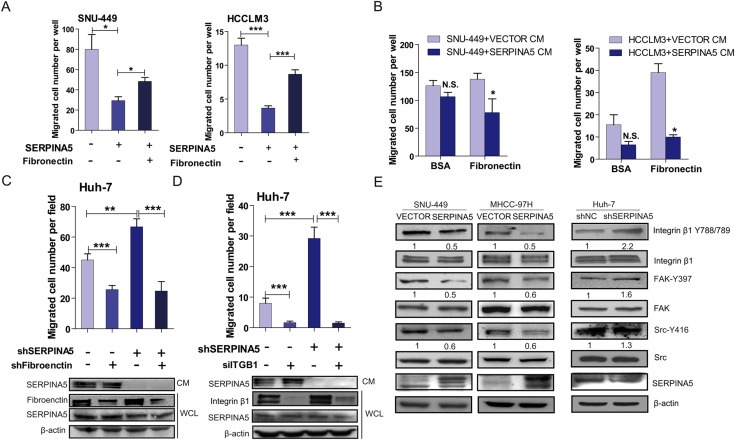
SERPINA5 inhibits HCC cell migration by disrupting the fibronectin/integrin β1 signaling pathway. (A) Treatment of SERPINA5 significantly inhibits the migration of SNU‐449 and HCCLM3 cells, which can be restored by exogenous fibronectin (100 μg/ml) addition (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) The secreted SERPINA5 inhibits the migratory potential of SNU‐449 and HCCLM3 cells on chambers coated with fibronectin (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (C) Knockdown of SERPINA5 promotes the migration of HUH‐7 cells, whereas targeted downregulation of fibronectin significantly attenuates cell migration induced by loss of SERPINA5 (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (D) Knockdown of SERPINA5 promotes the migration of HUH‐7 cells, whereas the targeted downregulation of integrin β1 (ITGB1) significantly attenuates cell migration induced by loss of SERPINA5 (unpaired Student's t‐test, mean ± s.e.m, *, P < 0.05; **, P < 0.01; ***, P < 0.001). (E) Ectopic expression of SERPINA5 leads to decreased phosphorylation levels of multiple pro‐metastatic proteins. The overexpression of SERPINA5 in SNU‐449 and MHCC‐97H cells decreases the phosphorylation of integrin β1 (Y788/789), FAK (Y397) and Src (Y416). Knockdown of SERPINA5 in HUH‐7 cells increases the phosphorylation of integrin β1 (Y788/789), FAK (Y397) and Src (Y416) proteins.
In human tumors, fibronectin expression is positively associated with tumor metastatic potential. Fibronectin binds to membrane‐spanning integrin receptors such as integrin β1 and plays an important role in cell signaling, defining cellular shape, and regulating cell mobility (Sengupta et al., 2010). We first demonstrated that stable knockdown of the SERPINA5 gene enhanced the migration of HUH‐7 cells, whereas siRNA‐mediated knockdown of the integrin β1 gene significantly attenuated the migration of HUH‐7‐shSERPINA5 cells (Figure 7D). Furthermore, we analyzed the downstream signaling targets of integrin β1 and demonstrated reduced phosphorylation of integrin β1 (Y788/789), FAK (Y397) and Src kinase (Y416) upon overexpression of SERPINA5 in SNU‐449 and MHCC‐97H cells, whereas targeted knockdown of the SERPINA5 gene leads to increased phosphorylation of these proteins in HUH‐7 cells (Figure 7E). Briefly, these findings indicate that one mechanism by which SERPINA5 inhibits HCC cell migration is through disruption of the fibronectin–integrin β1 signaling pathway.
4. Discussion
An effective way to identify driver genes with causal roles in carcinogenesis is the detection of genomic regions that undergo frequent alterations in cancers. To identify recurrent regions of CNAs in hepatocellular carcinoma (HCC), we recently performed a genome‐wide copy number analysis of 58 HCC primary tumors and paired non‐tumor tissues using the Affymetrix SNP 6.0 array, from which a total of 1241 regions of somatic CNAs were derived (Jia et al., 2011). Importantly, a number of new regions with somatic CNAs have also been uncovered in HCC, including a recurrent region of copy number loss at 14q31.1–32.13 occurring at a frequency of 8.6% (5/58). By using integrated analysis of copy number alteration and gene expression profiling, a putative tumor suppressor genes (SERPINA5) was uncovered in the recurrent region of copy number loss, 14q31.1–32.13, in HCC.
SERPINA5 is a member of the superfamily of serine protease inhibitors (Suzuki and Hayashi, 2007). SERPINA5 functions as an inhibitor of several plasma proteases involved in blood coagulation and is a potent inhibitor of uPA (Stief et al., 1987). Given both the role of uPA in tumor cell metastasis and its inhibition by SERPINA5, the physiological relevance of SERPINA5 in tumor metastasis is apparent (Jackson et al., 1997). This relevance was substantiated by a report on prostate cancer in which loss of SERPINA5 expression was found in high‐grade tumors (Cao et al., 2003). Moreover, overexpression of SERPINA5 results in decreased invasion, metastatic potential and angiogenesis in breast cancer, and elevated levels of SERPINA5 protein have been reported to be associated with increased survival in primary breast carcinomas (Asanuma et al., 2007; Castello et al., 2007). Additionally, the expression of SERPINA5 has been shown to be decreased in renal, prostate and ovarian cancers (Alimov et al., 2004; Bijsmans et al., 2011; Cao et al., 2003). However, the expression status and functional roles of the SERPINA5 gene in HCC are largely unknown. In this study, we followed up on our previous study in which the SERPINA5 gene was identified in a genomic region of copy number loss at 14q31.1–32.13, and found the decreased level of SERPINA5 in HCC tissues. Importantly, the downregulation of SERPINA5 negatively correlated with the malignant progression of HCC specimens. Moreover, SERPINA5 was observed to inhibit the in vitro and in vivo metastatic potential of HCC cells.
To further explore the underlying mechanism by which SERPINA5 inhibits cell migration and tumor metastasis, we found that SERPINA5 can bind to fibronectin, a high‐molecular weight extracellular matrix glycoprotein that can be secreted by tumor cells (Pankov and Yamada, 2002). Fibronectin, a ligand for the integrin β1 receptor, binds to membrane‐spanning integrin receptors and plays an important role in cell signaling, defining cellular shape, mobility, and regulating cell proliferation and cell survival (Sengupta et al., 2010). Moreover, fibronectin plays an important role in cell signaling pertinent to tumor progression and is important for the proliferation of different types of cancers, including HCC (Han et al., 2006; Ohnishi et al., 1998; Sherbet et al., 1982). In this study, we found that shRNA‐mediated knockdown of fibronectin expression attenuated the increased migration of HCC cells caused by targeted knockdown of SERPINA5. Moreover, the migratory potential of HCC cells on chambers coated with fibronectin was suppressed by secreted SERPINA5. The binding of integrins to their ligands triggers downstream signal transduction cascades. The fibronectin receptor (α5β1‐integrin) primarily binds to fibronectin, leading to cell anchoring, and subsequently activates the non‐receptor tyrosine kinases FAK and Src, which play an important role in tumorigenesis by promoting the proliferation and invasion of cancer cells (Caswell et al., 2007; Mitra and Schlaepfer, 2006). In this study, we found that overexpression of SERPINA5 decreases the phosphorylation levels of FAK and Src kinase, whereas inhibition of integrin β1 expression inhibits increased migration of HCC cells caused by targeted knockdown of SERPINA5.
It has been reported that fibronectin–integrin β1 is an important regulator of MMP‐2 and MMP‐9 activity (Milner et al., 2007; Sil et al., 2011). Therefore, we employed gelatin zymography assay to determine whether the binding of SERPINA5 to fibronectin–integrin modulates the activity of MMP‐2 and MMP‐9. However, we found that overexpression of SERPINA5 has no significant effect on the activity of MMPs in HCC cells (Figure S5). This finding implies that alternative downstream effectors rather than the activation of MMP‐2 and MMP‐9 are involved in the fibronectin–integrin β1 pathway, which deserves further research in next study. In addition, the proteolytic activity of SERPINA5 on the migratory ability of HCC cells was further determined by constructing two SERPINA5 mutants (R354A and T341R) without proteolytic activity (Rehault et al., 2005). The results showed that both of the two mutants inhibit the migratory ability of HCC cells (Figure S6). Overall, this study demonstrated that SERPINA5 might regulate tumor migration and metastasis independent of protease inhibitory activity by interacting with fibronectin and disrupting fibronectin–integrin signaling in HCC. This evidence suggests that SERPINA5 may function as a fibronectin antagonist.
In conclusion, we have, for the first time, demonstrated that SERPINA5 is a novel metastasis suppressor gene in HCC. Molecular studies revealed that the metastasis‐suppressing function of SERPINA5 in HCC might be associated with its role in decreasing cell migration by directly interacting with fibronectin and disrupting the fibronectin–integrin signaling pathway. Our findings suggest that SERPINA5 is a target gene within the 14q31.1–32.13 deletion region that plays a pivotal role in HCC metastasis.
Disclosure
There are no potential conflicts of interest with regard to this paper.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgments
We are most grateful for the pWPXL, psPAX2 and pMD2.G lentivirus plasmids provided by Professor Didier Trono from School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, 1015 Lausanne, Switzerland. This work was partially supported by grants from The National 973 Key Basic Research Program (2013CB910500); The National Natural Science Foundation of China (81125016, 81071637, and 91029728).
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.12.003.
Jing Ying, Jia Deshui, Wong Chun-Ming, Oi-Lin Ng Irene, Zhang Zhenfeng, Liu Li, Wang Qifeng, Zhao Fangyu, Li Jinjun, Yao Ming, Wu Xingzhong and He Xianghuo, (2014), SERPINA5 inhibits tumor cell migration by modulating the fibronectin‐integrin β1 signaling pathway in hepatocellular carcinoma, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.12.003.
Contributor Information
Xingzhong Wu, Email: xz_wu@shmu.edu.cn.
Xianghuo He, Email: xhhe@shsci.org.
References
- Alimov, A. , Sundelin, B. , Wang, N. , Larsson, C. , Bergerheim, U. , 2004. Loss of 14q31-q32.2 in renal cell carcinoma is associated with high malignancy grade and poor survival. Int. J. Oncol.. 25, 179–185. [PubMed] [Google Scholar]
- Asanuma, K. , Yoshikawa, T. , Hayashi, T. , Akita, N. , Nakagawa, N. , Hamada, Y. , Nishioka, J. , Kamada, H. , Gabazza, E.C. , Ido, M. , Uchida, A. , Suzuki, K. , 2007. Protein C inhibitor inhibits breast cancer cell growth, metastasis and angiogenesis independently of its protease inhibitory activity. Int. J. Cancer. 121, 955–965. [DOI] [PubMed] [Google Scholar]
- Bijsmans, I.T. , Smits, K.M. , de Graeff, P. , Wisman, G.B. , van der Zee, A.G. , Slangen, B.F. , de Bruine, A.P. , van Engeland, M. , Sieben, N.L. , Van de Vijver, K.K. , 2011. Loss of SerpinA5 protein expression is associated with advanced-stage serous ovarian tumors. Mod. Pathol.. 24, 463–470. [DOI] [PubMed] [Google Scholar]
- Buffart, T.E. , Carvalho, B. , van Grieken, N.C. , van Wieringen, W.N. , Tijssen, M. , Kranenbarg, E.M. , Verheul, H.M. , Grabsch, H.I. , Ylstra, B. , van de Velde, C.J. , Meijer, G.A. , 2012. Losses of chromosome 5q and 14q are associated with favorable clinical outcome of patients with gastric cancer. Oncologist. 17, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Becker, C. , Lundwall, A. , Christensson, A. , Gadaleanu, V. , Lilja, H. , Bjartell, A. , 2003. Expression of protein C inhibitor (PCI) in benign and malignant prostatic tissues. Prostate. 57, 196–204. [DOI] [PubMed] [Google Scholar]
- Castello, R. , Landete, J.M. , Espana, F. , Vazquez, C. , Fuster, C. , Almenar, S.M. , Ramon, L.A. , Radtke, K.P. , Estelles, A. , 2007. Expression of plasminogen activator inhibitors type 1 and type 3 and urokinase plasminogen activator protein and mRNA in breast cancer. Thromb. Res.. 120, 753–762. [DOI] [PubMed] [Google Scholar]
- Caswell, P.T. , Spence, H.J. , Parsons, M. , White, D.P. , Clark, K. , Cheng, K.W. , Mills, G.B. , Humphries, M.J. , Messent, A.J. , Anderson, K.I. , McCaffrey, M.W. , Ozanne, B.W. , Norman, J.C. , 2007. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell. 13, 496–510. [DOI] [PubMed] [Google Scholar]
- Cui, J.F. , Liu, Y.K. , Zhang, L.J. , Shen, H.L. , Song, H.Y. , Dai, Z. , Yu, Y.L. , Zhang, Y. , Sun, R.X. , Chen, J. , Tang, Z.Y. , Yang, P.Y. , 2006. Identification of metastasis candidate proteins among HCC cell lines by comparative proteome and biological function analysis of S100A4 in metastasis in vitro. Proteomics. 6, 5953–5961. [DOI] [PubMed] [Google Scholar]
- Fortenberry, Y.M. , Brandal, S. , Bialas, R.C. , Church, F.C. , 2010. Protein C inhibitor regulates both cathepsin L activity and cell-mediated tumor cell migration. Biochim. Biophys. Acta. 1800, 580–590. [DOI] [PubMed] [Google Scholar]
- Francis, R.B. , Thomas, W. , 1984. Behaviour of protein C inhibitor in intravascular coagulation and liver disease. Thromb. Haemost.. 52, 71–74. [PubMed] [Google Scholar]
- Fuchs, B.C. , Fujii, T. , Dorfman, J.D. , Goodwin, J.M. , Zhu, A.X. , Lanuti, M. , Tanabe, K.K. , 2008. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res.. 68, 2391–2399. [DOI] [PubMed] [Google Scholar]
- Geiger, M. , 2007. Protein C inhibitor, a serpin with functions in- and outside vascular biology. Thromb. Haemost.. 97, 343–347. [PubMed] [Google Scholar]
- Genda, T. , Sakamoto, M. , Ichida, T. , Asakura, H. , Kojiro, M. , Narumiya, S. , Hirohashi, S. , 1999. Cell motility mediated by rho and Rho-associated protein kinase plays a critical role in intrahepatic metastasis of human hepatocellular carcinoma. Hepatology. 30, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Han, S. , Khuri, F.R. , Roman, J. , 2006. Fibronectin stimulates non-small cell lung carcinoma cell growth through activation of Akt/mammalian target of rapamycin/S6 kinase and inactivation of LKB1/AMP-activated protein kinase signal pathways. Cancer Res.. 66, 315–323. [DOI] [PubMed] [Google Scholar]
- Huntington, J.A. , Li, W. , 2009. Structural insights into the multiple functions of protein C inhibitor. Cell Mol. Life Sci.. 66, 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, T.P. , Cooper, S.T. , Church, F.C. , 1997. Assessment of the interaction between urokinase and reactive site mutants of protein C inhibitor. J. Protein Chem.. 16, 819–828. [DOI] [PubMed] [Google Scholar]
- Jia, D. , Wei, L. , Guo, W. , Zha, R. , Bao, M. , Chen, Z. , Zhao, Y. , Ge, C. , Zhao, F. , Chen, T. , Yao, M. , Li, J. , Wang, H. , Gu, J. , He, X. , 2011. Genome-wide copy number analyses identified novel cancer genes in hepatocellular carcinoma. Hepatology. 54, 1227–1236. [DOI] [PubMed] [Google Scholar]
- Jia, D. , Yan, M. , Wang, X. , Hao, X. , Liang, L. , Liu, L. , Kong, H. , He, X. , Li, J. , Yao, M. , 2010. Development of a highly metastatic model that reveals a crucial role of fibronectin in lung cancer cell migration and invasion. BMC Cancer. 10, 364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar, M. , Zardi, L. , Neri, D. , 2006. Fibronectin as target for tumor therapy. Int. J. Cancer. 118, 1331–1339. [DOI] [PubMed] [Google Scholar]
- Laurell, M. , Christensson, A. , Abrahamsson, P.A. , Stenflo, J. , Lilja, H. , 1992. Protein C inhibitor in human body fluids. Seminal plasma is rich in inhibitor antigen deriving from cells throughout the male reproductive system. J. Clin. Invest.. 89, 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.K. , Poon, R.T. , Yuen, A.P. , Ling, M.T. , Kwok, W.K. , Wang, X.H. , Wong, Y.C. , Guan, X.Y. , Man, K. , Chau, K.L. , Fan, S.T. , 2006. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin. Cancer Res.. 12, 5369–5376. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Tian, B. , Yang, J. , Zhao, L. , Wu, X. , Ye, S.L. , Liu, Y.K. , Tang, Z.Y. , 2004. Stepwise metastatic human hepatocellular carcinoma cell model system with multiple metastatic potentials established through consecutive in vivo selection and studies on metastatic characteristics. J. Cancer Res. Clin. Oncol.. 130, 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, R. , Crocker, S.J. , Hung, S. , Wang, X. , Frausto, R.F. , del Zoppo, G.J. , 2007. Fibronectin- and vitronectin-induced microglial activation and matrix metalloproteinase-9 expression is mediated by integrins alpha5beta1 and alphavbeta5. J. Immunol.. 178, 8158–8167. [DOI] [PubMed] [Google Scholar]
- Mitra, S.K. , Schlaepfer, D.D. , 2006. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol.. 18, 516–523. [DOI] [PubMed] [Google Scholar]
- Ohnishi, T. , Hiraga, S. , Izumoto, S. , Matsumura, H. , Kanemura, Y. , Arita, N. , Hayakawa, T. , 1998. Role of fibronectin-stimulated tumor cell migration in glioma invasion in vivo: clinical significance of fibronectin and fibronectin receptor expressed in human glioma tissues. Clin. Exp. Metastasis. 16, 729–741. [DOI] [PubMed] [Google Scholar]
- Pankov, R. , Yamada, K.M. , 2002. Fibronectin at a glance. J. Cell Sci.. 115, 3861–3863. [DOI] [PubMed] [Google Scholar]
- Pehlivan, D. , Gunduz, E. , Gunduz, M. , Nagatsuka, H. , Beder, L.B. , Cengiz, B. , Rivera, R.S. , Fukushima, K. , Palanduz, S. , Ozturk, S. , Yamanaka, N. , Shimizu, K. , 2008. Loss of heterozygosity at chromosome 14q is associated with poor prognosis in head and neck squamous cell carcinomas. J. Cancer Res. Clin. Oncol.. 134, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehault, S.M. , Zechmeister-Machhart, M. , Fortenberry, Y.M. , Malleier, J. , Binz, N.M. , Cooper, S.T. , Geiger, M. , Church, F.C. , 2005. Characterization of recombinant human protein C inhibitor expressed in Escherichia coli . Biochim. Biophys. Acta. 1748, 57–65. [DOI] [PubMed] [Google Scholar]
- Sengupta, S. , Nandi, S. , Hindi, E.S. , Wainwright, D.A. , Han, Y. , Lesniak, M.S. , 2010. Short hairpin RNA-mediated fibronectin knockdown delays tumor growth in a mouse glioma model. Neoplasia. 12, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherbet, G.V. , Tindle, M.E. , Stidolph, S. , 1982. Fibronectin in human astrocytomas grown in tissue culture. Anticancer Res.. 2, 251–254. [PubMed] [Google Scholar]
- Shirk, R.A. , Elisen, M.G. , Meijers, J.C. , Church, F.C. , 1994. Role of the H helix in heparin binding to protein C inhibitor. J. Biol. Chem.. 269, 28690–28695. [PubMed] [Google Scholar]
- Sil, H. , Sen, T. , Chatterjee, A. , 2011. Fibronectin-integrin (alpha5beta1) modulates migration and invasion of murine melanoma cell line B16F10 by involving MMP-9. Oncol. Res.. 19, 335–348. [DOI] [PubMed] [Google Scholar]
- Speleman, F. , Kumps, C. , Buysse, K. , Poppe, B. , Menten, B. , De Preter, K. , 2008. Copy number alterations and copy number variation in cancer: close encounters of the bad kind. Cytogenet. Genome Res.. 123, 176–182. [DOI] [PubMed] [Google Scholar]
- Stief, T.W. , Radtke, K.P. , Heimburger, N. , 1987. Inhibition of urokinase by protein C-inhibitor (PCI). Evidence for identity of PCI and plasminogen activator inhibitor 3. Biol. Chem. Hoppe Seyler. 368, 1427–1433. [DOI] [PubMed] [Google Scholar]
- Suzuki, K. , Hayashi, T. , 2007. Protein C and its inhibitor in malignancy. Semin. Thromb. Hemost.. 33, 667–672. [DOI] [PubMed] [Google Scholar]
- Suzuki, K. , Nishioka, J. , Hashimoto, S. , 1983. Protein C inhibitor. Purification from human plasma and characterization. J. Biol. Chem.. 258, 163–168. [PubMed] [Google Scholar]
- Thorstensen, L. , Qvist, H. , Nesland, J.M. , Giercksky, K.E. , Lothe, R.A. , 2001. Identification of two potential suppressor gene regions on chromosome arm 14q that are commonly lost in advanced colorectal carcinomas. Scand. J. Gastroenterol.. 36, 1327–1331. [DOI] [PubMed] [Google Scholar]
- van Dekken, H. , Geelen, E. , Dinjens, W.N. , Wijnhoven, B.P. , Tilanus, H.W. , Tanke, H.J. , Rosenberg, C. , 1999. Comparative genomic hybridization of cancer of the gastroesophageal junction: deletion of 14Q31-32.1 discriminates between esophageal (Barrett's) and gastric cardia adenocarcinomas. Cancer Res.. 59, 748–752. [PubMed] [Google Scholar]
- Wakita, T. , Hayashi, T. , Nishioka, J. , Tamaru, H. , Akita, N. , Asanuma, K. , Kamada, H. , Gabazza, E.C. , Ido, M. , Kawamura, J. , Suzuki, K. , 2004. Regulation of carcinoma cell invasion by protein C inhibitor whose expression is decreased in renal cell carcinoma. Int. J. Cancer. 108, 516–523. [DOI] [PubMed] [Google Scholar]
- Yang, J.D. , Roberts, L.R. , 2010. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol.. 7, 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


