Abstract
Despite the advancements in the cancer therapeutics, gastric cancer ranks as the second most common cancers with high global mortality rate. Integrative functional genomic investigation is a powerful approach to understand the major dysregulations and to identify the potential targets toward the development of targeted therapeutics for various cancers. Intestinal and diffuse type gastric tumors remain the major subtypes and the molecular determinants and drivers of these distinct subtypes remain unidentified. In this investigation, by exploring the network of gene coexpression association in gastric tumors, mRNA expressions of 20,318 genes across 200 gastric tumors were categorized into 21 modules. The genes and the hub genes of the modules show gastric cancer subtype specific expression. The expression patterns of the modules were correlated with intestinal and diffuse subtypes as well as with the differentiation status of gastric tumors. Among these, G1 module has been identified as a major driving force of diffuse type gastric tumors with the features of (i) enriched mesenchymal, mesenchymal stem cell like, and mesenchymal derived multiple lineages, (ii) elevated OCT1 mediated transcription, (iii) involvement of Notch activation, and (iv) reduced polycomb mediated epigenetic repression. G13 module has been identified as key factor in intestinal type gastric tumors and found to have the characteristic features of (i) involvement of embryonic stem cell like properties, (ii) Wnt, MYC and E2F mediated transcription programs, and (iii) involvement of polycomb mediated repression. Thus the differential transcription programs, differential epigenetic regulation and varying stem cell features involved in two major subtypes of gastric cancer were delineated by exploring the gene coexpression network. The identified subtype specific dysregulations could be optimally employed in developing subtype specific therapeutic targeting strategies for gastric cancer.
Keywords: Gastric cancer, Intestinal subtype, Diffuse subtype, Mesenchymal stem cells, Embryonic stem cells, Notch signaling, Polycomb repression, Mesenchymal stem cell differentiation
Highlights
Derived 21 modules from gastric tumor transcriptome by coexpression networking.
Driving hallmarks of diffuse and intestinal gastric tumors were identified.
Mesenchymal stemness and reduced polycomb repression confer diffuse type tumors.
Embryonic stem cell features with Wnt‐MYC‐E2F transcription drive intestinal type.
Contrasting stem cell features and dysregulations of cancer subtypes delineated.
1. Introduction
Despite the advancements in the cancer research and developments in cancer therapeutics, gastric cancer remains one of the cancers with highest annual mortality rate (Nagini, 2012). Potential therapeutic targets and targeted therapeutics still remain to be identified and developed to tackle different types of gastric cancers. Heterogeneities and complexities remain the major limiting factors in identifying the realistic therapeutic targets (Sehn, 2012; Chen et al., 2003). Stratification of gastric cancer patients based on the dysregulations and identifying the therapeutic targets for the different subgroups of patients are the current needs in the development of next generation diagnostics and therapeutics. Genome‐wide profiling of cancers and integrative functional genomics approaches are found useful in delineating the complexities and heterogeneities in various cancers and molecular stratification of cancer patients (TCGA, 2012; Schroeder et al., 2013).
With the advent of genomic investigations, several advancements have been made in understanding the biology of gastric cancer. Identification of key cancer genes, molecular stratification of tumors, and identification of predominant pathways involved in gastric cancer are the major outcomes (Cho et al., 2011; Ooi et al., 2009; Zang et al., 2011). Interactions among the genes of a single cell type and cross talks among various types of cells in the tumor microenvironment contribute tremendously in cancers and needs to be investigated (Hanahan and Weinberg, 2011). However, these interactions could not be investigated by conventional genomics which often merely involves screening the genes differentially altered in tumors. Integrative and multi‐dimensional analysis of microarray data, particularly the transcriptomics data is capable of yielding system level information (Barabasi and Oltvai, 2004; Stuart et al., 2003).
Gene coexpression network based gene pattern analysis from the transcriptomics data by applying the weighted gene coexpression network analysis (WGCNA) organizes the whole genome expression data into functional clusters of coexpressed genes. This is useful to investigate the gene expression profile in functional contexts and to infer the expression pattern of gene sets and their regulations (Horvath and Dong, 2008). Earlier studies involving the construction of coexpression network in gastric cancer had identified coexpressed modular genes and pathways. PLAG2A, a prognostic marker, its coexpression with EPHB2 receptor, association with Wnt/β‐catenin pathway and its β‐catenin mediated regulation were established from the network of gastric cancer transcriptome (Aggarwal et al., 2006; Ganesan et al., 2008). Another recent network from gastric tumors has revealed the enrichment of the stromal cells in diffuse gastric tumors (Wu et al., 2013). Apart from the mass of proliferating cancer cells, tumors are composed of multiple distinct cell types and the aggressiveness of the cancer is influenced by heterotypic interactions among these cells; in particular, the stromal cells and stem cells contribute to the development and progression of cancers upon differentiation and were inferred from mRNA network (Ben‐Porath et al., 2008; Wu et al., 2013; Zhao et al., 2010).
Though multiple networks have been constructed in gastric and many other cancer types, each of these networks have their unique potential in identifying novel system level information in understanding the biology of cancers. In this study, weighted gene coexpression based network analysis of the global transcriptome of gastric tumors was performed to infer the global gene interactions and thus the functional processes playing crucial role in gastric carcinogenesis. It was aimed to connect the gene modules with clinical traits and to understand the gene interactions involved in specific clinical phenotypes. From the coexpression pattern of genes, the major molecular cellular factors involved in two different major subtypes of gastric cancer were identified. Involvements of heterogenous categories of stem cells, varying transcription programs, and different epigenetic dysregulations have been identified as hallmarks of gastric cancer subtypes.
2. Materials and methods
2.1. Microarray data preprocessing
Gene expression profile of gastric tumors was collected from the microarray database Gene expression omnibus (GEO). Since the aim of the study is to obtain the gene network, where each node represents the gene, the MAS 5.0 normalized mRNA profile data was matched with the gene symbol and gene description provided in the corresponding platform file. Gene duplicates were removed by considering the average expression value of multiple probes of genes. The processed profile data was used for the network construction.
2.2. Coexpression network construction
Gastric cancer mRNA profile datasets were collected from Gene expression omnibus (GSE15459, GSE22377) (Forster et al., 2011; Ooi et al., 2009). The first network was constructed from the GSE15459 dataset having the mRNA profile of 200 gastric tumor samples. A weighted coexpression network of the selected 20,318 genes from the gastric cancer transcriptome was constructed by applying the algorithms of WGCNA (Langfelder and Horvath, 2008; Zhang and Horvath, 2005). The correlations among gene expressions were measured based on the Pearson correlation coefficients of all pairs of the genes. The soft thresholding function of WGCNA was used to derive the continuous value of the gene coexpression from Pearson correlation values. The correlation coefficients were further converted into adjacency matrix, using the power adjacency function. The details and scripts used for the construction of coexpression network are provided in Supplemental Method 1.
2.3. Module detection
The adjacency value of each gene along with its degree of shared correlation within the weighted network was transformed into topological overlap, based on which the dissimilarity measure of the topological overlap matrix of the highly coexpressed genes were calculated and the genes were hierarchically clustered into different modules. To sum up the entire module gene expression and to know the strength of the module connections in the network, various network concepts such as maximum adjacency ratio (MAR), heterogeneity, and connectivity of the module genes were calculated by applying the function network concept in WGCNA (Horvath and Dong, 2008). Modules in the network were developed as network model using the visualization tool VisANT (Hu et al., 2008).
2.4. Clustering and analysis of the modules
The genes in the different modules were sorted based on the MAR value and 25 genes with high MAR value were selected as hub genes. The unsupervised clustering of the top 25 hub genes of each of the 21 modules across the gastric tumor mRNA profiles GSE15459 and GSE22377 were performed in dChip (Li and Wong, 2001). Distance of the clustering was calculated based on the 1‐correlation value (Eisen et al., 1998; Golub et al., 1999). Similarly, the expression pattern of genes of the modules and genes in different signatures were analyzed in dChip.
2.5. Screening of the module genes across stem cells
The mRNA profile of mesenchymal stem cells (GSE28974) and embryonic stem cells (GSE29625) were collected from GEO. These two different profiles were combined by median normalization and the combined data was then quantile normalized (Bolstad et al., 2003; Qin et al., 2012). The quantile normalized data was screened for the expression of individual module genes. The mean expression of the gene sets in the samples and subgroups were calculated and the values were plotted as boxplot by applying the function boxplot in R package (Murrell et al., 2005). The fold difference and the significance values were calculated by applying the difference between the mean expression of the genes in MSC and ESC, and the significance was calculated by two‐tailed t‐test.
2.6. Classification of the gastric cancer cell lines
The gastric cancer cell line profile data GSE22183 was collected from the database GEO. The average mean expressions of the duplicate genes were calculated and the duplicates were merged. The intrinsic subtypes of gastric cancer cell lines based on the molecular classification were obtained from the information provided in the article (Tan et al., 2011). The expression levels of the G1 module genes in intestinal and diffuse class of cell lines were measured based on Z score metric. The mean expression level of genes in G1 module was considered as X sg1 in sample S. X s be the mean expression of all the genes in profile data and the σ s is the standard deviation of the X s. Z score was derived by applying the formula, (Levine et al., 2006).
2.7. In‐vitro reporter assay
In‐vitro Notch/Myc/Wnt/E2F reporter assays was performed in the specified cell lines in 24 well cell culture plates. Upon reaching 70% confluency, the pathway specific firefly luciferase reporter plasmid along with CMV‐renilla reporter plasmid (SA Biosciences) were transfected in the ratio of 100:1 using Fugene transfection reagent (Promega). Upon 24 h of incubation, the cells were harvested and the pathway/transcription factor specific transcriptional activity was measured by dual luciferase assay (Parsons et al., 2000). The basal un‐induced pathway/transcriptional activity of cell lines were calculated in folds by considering the folds of measurement from negative control firefly (firefly reporter without enhancer) transfected sample as one. For SAHA (Suberoylanilide hydroxamic acid) treatment, 24 h after luciferase plasmid transfection, the cells were treated with SAHA and vehicle control. Upon 24 h of incubation, the cells were harvested and the Notch transcriptional activity was measured by dual luciferase assay. For siRNA experiments, pooled siRNA from Dhramacon were used. The cells were transfected with siRNA using oligofectamine (Invitrogen). After 24 h, the cells were transfected with reporter plasmids and further harvested at 24 h after the plasmid transfection. In all the experiments, the necessary controls were used and the assays were performed twice in duplicates or triplicates.
2.8. SAHA treatment, RT‐PCR, and Western blotting
AGS cells seeded in 6 well culture plate were treated with increasing concentration of SAHA and incubated for 48 h. Subsequently, total RNA was extracted using Trizol (Invitrogen) and cDNA was synthesized from the isolated RNA using Reverse Transcriptase (Invitrogen) as per the standard protocol. The cDNA was used for further semi‐quantitative RT‐PCR experiments to analyze the relative gene expression using gene specific primers. Total cell lysate from the specified gastric cancer cell lines were used for the analysis of Myc protein expression by Western blotting. Myc (Santa Cruz) and β‐actin (Sigma Aldrich) antibodies were used for the detection of cellular levels of protein. The secondary antibodies used were anti‐mouse IgG‐HRP (GE Healthcare) and the blots were developed using ECL Prime reagent (GE Healthcare).
3. Results
3.1. Identification of gene coexpression network modules playing crucial role in intestinal and diffuse type gastric tumors
Whole genome expression profiles have unlimited dimensions of information and often remain unexplored. Analysis of the genes differentially expressed in tumors is the very basic outcome of the mRNA profiles. In order to understand the complex transcriptional regulations in gastric cancers and to identify potential pathological and therapeutic leads, mRNA profile of gastric tumors was investigated. The gastric cancer transcriptome dataset (GSE15459) comprising the mRNA profiles of 200 gastric tumor samples was selected for the investigation. Identification of the genes and gene sets having correlated expression pattern across cancer samples is an intriguing approach in identifying the gene interactions, biological processes and global dysregulations in cancers. A weighted coexpression network (WGCNA) of 20,318 genes in the gastric cancer mRNA profile data was constructed by applying the soft thresholding and topological overlap matrix (TOM) algorithms of WGCNA (Langfelder and Horvath, 2008; Zhang and Horvath, 2005) (Supplemental Method 1). Gene–gene associations among the genes were measured based on the Pearson correlation coefficients of all pairs of the genes in the mRNA profile. In order to identify the weight of the correlations throughout the transcriptome, the correlation coefficients of all pairs of the genes were converted into adjacency matrix. The adjacency value of each gene along with its degree of shared correlation within the weighted network was transformed into topological overlap. Based on this, highly coexpressed genes were hierarchically clustered into 21 different modules (Figure 1A and Supplemental Table 1). The highly coexpressed genes were clustered based on dissimilarity measure of TOM (1 ‐ TOM). The genes in the modules and their coexpressed neighbors were viewed as network models in the visualization tool VisANT (Hu et al., 2008). Individual modules with top connections are shown in Figure 1B and Supplemental Data 1. The 21 modules were named from G1 to G21. The numbers of genes across the modules were varying between 3999 and 22. With the selected criteria mentioned above, this is the grouping pattern of highly coexpressed genes in the analyzed gastric tumor mRNA profile. This clustering also means that the transcriptome of gastric tumors comprise 21 distinct or related functional processes and is worth investigating in order to understand the driving forces and molecular patho‐biological processes in gastric cancers.
Figure 1.
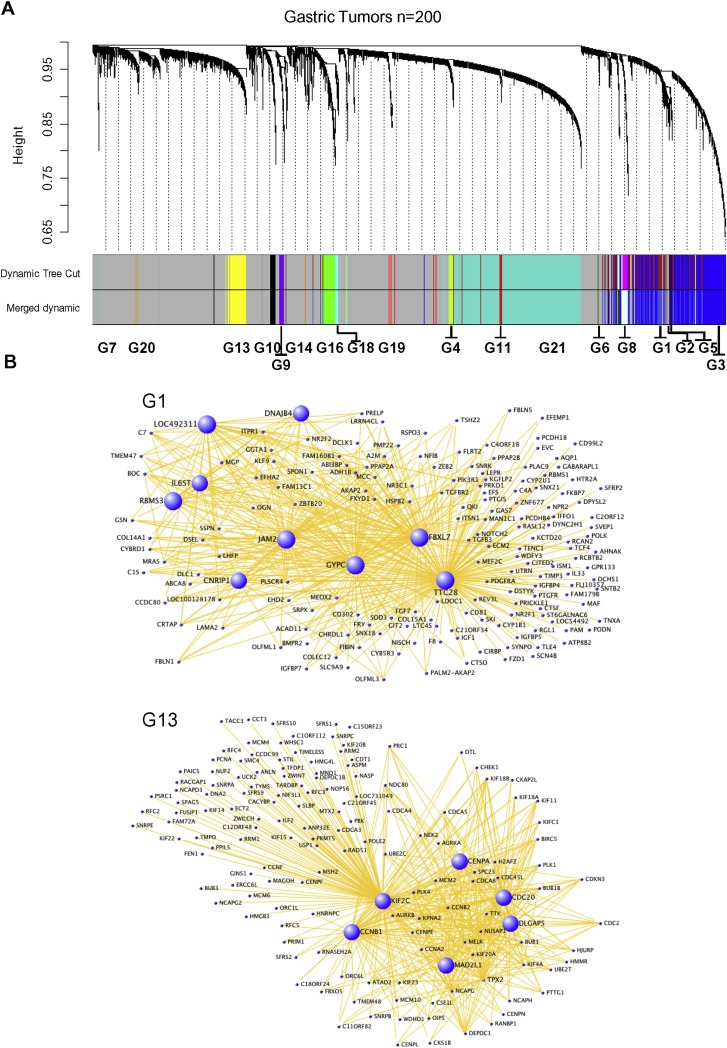
Coexpressed mRNA network of gastric tumors is defined as 21 modules. (A) Highly correlated genes are grouped into modules of coexpressed genes by hierarchical clustering of the TOM dissimilarity measure (1‐TOM). The topological overlap dissimilarities among the genes are represented as height. Each branch of the dendrogram represents a module, and assigned a color at the bottom for clear view. Genes (20,318) are grouped into 21 modules of different colors. Genes that does not fall into any modules are represented in grey. Few closely related modules were merged into single module by merge dynamics. (B) Visualization of highly connected genes of the selected modules G1 and G13 is shown as network. Each dot represents a gene and hub genes are highlighted in blue color and in larger size.
To investigate the modular expression of genes in the clinical contexts of gastric tumors, the expression pattern of different modules were probed in association with the clinical phenotypes. The hub genes are the key genes of the modules connected with majority of the genes in the module and hence the expression of top 25 hub genes of each module (Supplemental Table 2) were analyzed in the gastric tumor mRNA profile GSE15459, comprising 200 gastric tumors. The analysis revealed a clustered expression of different modules in gastric cancer subtypes based on their differentiation status (Figure 2A). Unsupervised clustering of top 25 hub genes of each module classifies gastric tumors into their subtypes. Among the 21 modules, the hub genes of G1, G2, G3, G5, G6, G16 and G18 modules are highly expressed in poorly differentiated tumors than moderately differentiated samples. On the other hand, the hub genes of G13 module are expressed in moderately differentiated tumors. These modular hub genes also show consistent clustering and expression pattern in an independent patient cohort of gastric tumor mRNA profile (GSE22377) with intestinal and diffuse type gastric tumor information (Figure 2B). Some of the other modular genes are partially consistent between two mRNA profiles. In large, the hub genes of the modules are capable of classifying the gastric tumors based on their differentiation status and also into intestinal and diffuse types with significant p values.
Figure 2.
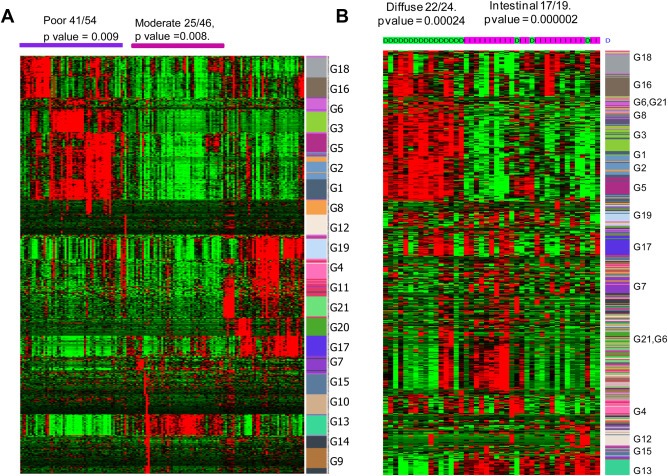
The modular hub genes are differentially expressed in gastric cancer subtypes. (A) Two way hierarchical clustering of the expression of top 25 hub genes of all modules in the gastric tumor mRNA profile, GSE15459 shows the elevated expression of G1, G2, G3, and G5 modules in poorly differentiated tumors. G13 module is expressed in moderately differentiated gastric tumors. In all the heat maps highest to lowest expression is represented by red to green. (B) Expression and clustering pattern of modular genes in an independent cohort of gastric tumors (GSE22377) indicate the expression of G1, G2, G3, and G5 modules in diffuse type gastric tumors and G13 in intestinal type.
The observed striking expression pattern of the modular genes in correlation with the differentiation status of gastric tumors gave the motivation to statistically probe the connections between all modules and the differentiation status. The cumulative expression of complete genes of each of all the modules were computed and correlated with the differentiation status of gastric tumors based on Pearson correlations as described earlier (Ghazalpour et al., 2006). This analysis ranked the modules based on their association with the differentiation status (Figure 3A). G1 and G13 modules were found to be highly associated with the clinical trait, the histopathological status of the tumors, also in an independent gastric tumor mRNA profile (GSE22377). Also, owing to the number of genes present in G1 and G13 modules, they were further investigated. The whole G1 and G13 modular genes show very distinct expression in intestinal and diffuse type gastric tumors in two different cohorts (Figure 3B and C, and Supplemental Data 2). This expression pattern confirms that the genes and the molecular processes conferred by G1 module plays major role in poorly differentiated diffuse type gastric tumors. Similarly, the genes and functions brought out by G13 module are involved in moderately differentiated and intestinal subtype gastric tumors.
Figure 3.
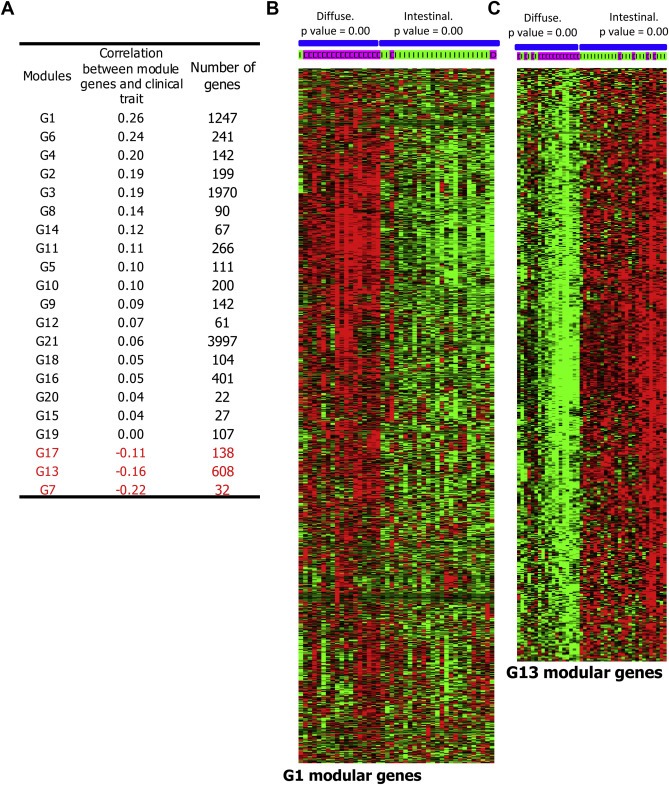
G1 and G13 modules play major role in diffuse and intestinal subtype gastric tumors, respectively. (A) Correlation between the cumulative expressions of modular genes with clinical trait (poor differentiation status) of the samples is listed. Cumulative expression was computed by applying the principal component (module eigengene value) and the correlation was calculated by Pearson correlation. This was performed in the gastric tumor mRNA profile, GSE15459. (B–C) Analysis of the mRNA expression pattern of G1 and G13 modular genes in the profile GSE22377 shows higher level expression of G1 genes in diffuse type tumors (B). G13 modular genes are expressed in intestinal gastric tumors (C).
3.2. Identification of the involvement of different types of stem cells in different subtypes of gastric cancer
To understand the molecular and cellular functional features conferred by the selected subtype specific modules, the G1 and G13 modular genes were analyzed for gene set enrichment. In addition to the gene ontology (GO) database, curated gene sets of MsigDB and gene sets collected from multiple publications and online pathway databases (Liberzon et al., 2011; Subramanian et al., 2005) also were probed for this gene set enrichment analysis. Curated gene set analysis in MsigDB shows the enrichment of mesenchymal stem cell features with G1 module genes. On the other side, G13 module is enriched with embryonic stem cell property (Supplemental Table 3). The gene ontology analysis in DAVID database (Huang da et al., 2009) shows the involvement of extracellular matrix, angiogenesis, cell cycle, immune response related features among the genes in G1 and few of its related modules. Analysis of the transcription factor binding sites in the promoter region of G1 and G13 modular genes using the tool Dire (Gotea and Ovcharenko, 2008) showed the enrichment of OCT1 binding sites in G1 module and the enrichment of MYC and E2F transcription factor binding sites in G13 module (Supplemental Table 4).
Since both of the contrasting G1 and G13 modules are enriched for varying types of stem cell features, these modular genes were further extensively screened for the differing stem cell properties by investigating with multiple stem cell signatures established to date. Gene signatures in GenesigDb database (Culhane et al., 2010) and from publications (Ben‐Porath et al., 2008; Mizuno et al., 2010) were used for this analysis. In total, 184 stem cell signatures were collected and the G1 and G13 modules were investigated (Supplemental Table 5). To find out the profound biological dissimilarities between these contrasting modules, the stem cell features that are distinctly enriched in G1 and not in G13 and vice‐versa were explored (Supplemental Table 6). This analysis reveals that G1 module is enriched with mesenchymal stem cell (MSC) features and G13 module with embryonic stem cell (ESC) features. Further, G1 modular genes found enriched with the features of hMSC, chondrogeneic markers, endothelial differentiation, and hypoxia induced genes (Supplemental Table 6). Indeed these are the known common features of mesenchymal stem cell maintenance and differentiation (Abdollahi et al., 2011). On the other side, the G13 genes are enriched for the ontological features such as hESC, hypoxia repressed genes, and MYC target genes. This observation compelled us to hypothesize that two varying stem cell types are enriched and involved in intestinal and diffuse subtypes of gastric cancer in an exclusive manner.
In order to verify the involvement of MSC like and ESC like features embedded in G1 and G13 modules, the available mRNA profiles of mesenchymal stem cells and embryonic stem cells were analyzed. The mRNA profiles (GSE28974, GSE29625) from two different studies (Giritharan et al., 2011; Kuijjer et al., 2012) were quantile normalized, compiled and used for investigating the expression pattern of G1 and G13 genes. The mean expressions of the module genes were analyzed. The mean expression of the G1 modular genes show elevated expression in mesenchymal stem cells. Similarly, the G13 modular genes show higher expression in embryonic stem cells with significant p value and fold difference (Figure 4 A–D). Analysis of the expression patterns of G1 modular genes in multiple mRNA profiles of ESC and MSC (GSE28974, GSE29625 and GSE2248) also showed their enriched expression in MSC. Interestingly, G1 and G13 genes were found capable of classifying the mRNA profile comprising the samples of ESC and MSC origin even in unsupervised clustering (Figure 4E–H). Thus the expression pattern and enrichment screening of G1 and G13 modules across different stem cell derived expression profiles reveal the MSC related features conferred by G1 modular genes and their involvement in diffuse type gastric tumors. In contrast, G13 module has the features resembling ESC and is involved in the carcinogenesis associated with intestinal type gastric cancer.
Figure 4.
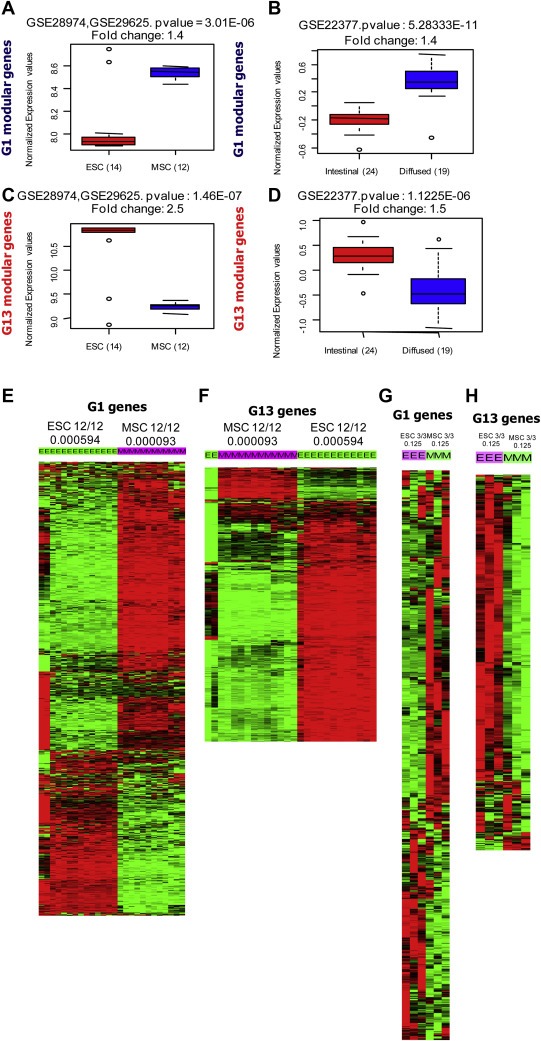
G1 and G13 module genes show the characteristics of varying stem cell features. (A) Box plot showing the mean expression of G1 modular genes in mesenchymal stem cells (MSC) and embryonic stem cells (ESC). (B) The mean expression of G1 modular genes in diffuse and intestinal subtype gastric tumors. (C–D) G13 genes are expressed in ESC (C) and intestinal subtype gastric tumors (D). (A–D) The GEO profile used for the analysis, p value and fold changes are shown on the top of box plots. The sample numbers of different groups are shown in parenthesis. (E–H) Majority of the G1 genes are highly expressed in MSC (E and G) and G13 genes are highly expressed in ESC (F and H) in multiple mRNA profiles. The stem cell profiles used are: MSC – GSE28974 and ESC – GSE29625, MSC and ESC – GSE2248. The number of samples in different subgroups and p value for enriched genes in sample cluster are shown on the top of heat maps.
We further investigated the selective expression of previously defined (i) mesenchymal stem cell specific markers, and (ii) mesenchymal migration factors (Spaeth et al., 2008) in G1 modules and diffuse type gastric tumors and observed the elevated expression of mesenchymal markers in diffuse type tumors (Figure 5A). Normally, these migration factors favor the MSC localization to the site of tissue damage as well as tumor microenvironment (Karnoub et al., 2007). Similarly, chemokine genes also were found over expressed in diffuse type tumors (Figure 5B). Altogether, expression of mesenchymal genes, expression of MSC markers, and migration factors including chemokine/receptor genes are the characteristic feature of diffuse type gastric cancer samples and was identified through genome‐wide transcriptional networking.
Figure 5.
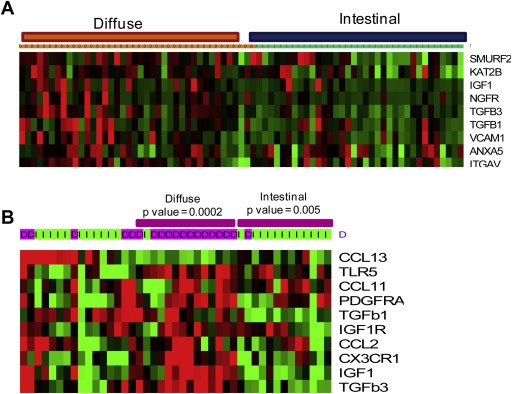
Mesenchymal specific markers and mesenchymal stem cell migration inducing cytokines are components of G1 module and expressed in diffuse type gastric tumors. (A) A sub‐set of G1 modular genes are mesenchymal markers and show elevated expression in diffuse type gastric tumors. (B) MSC migration inducing cytokines show significant higher expression in diffuse gastric tumors. This analysis is from the mRNA profile GSE22377.
3.3. Mesenchymal stem cell differentiation rich cell niche is the hallmark of diffuse type gastric tumors
The expression pattern of G1 modular genes in a unique mRNA profile comprising multiple cell lineages derived from mesenchymal stem cells (GSE9451) shows that the genes in the G1 module are heterogeneously expressed in multiple lineages such as fibroblasts, adipocytes, chondrocytes, and osteoblasts differentiated from mesenchymal stem cells (Figure 6A). Since the G1 modular genes are predominantly expressed in diffuse type gastric tumors, occurrence and involvement of these multiple mesenchymal stem cell derived cell lineages and likely the differentiation stems from the MSC seems the hallmark of diffuse gastric tumors. Transcription factor binding site enrichment analysis among G1 modular genes also supports the occurrence of transcription factors associated with these lineages. Specifically, the binding sequences of the transcription factors like OCT1, GATA1, and CEBPB which are known to induce the endothelial and osteoblast differentiation (Bhasin et al., 2010; Qi et al., 2003; Stains and Civitelli, 2003) occur at higher frequency among G1 genes (Supplemental Table 4). Similarly, the binding sites of the transcription factors PPARG and SOX5 known to involve in the induction of adipocyte and chondrocyte differentiation (Akiyama et al., 2002) are enriched in the promoters of G1 genes (Figure 6B). This shows that stromal cell rich tumor microenvironment stemming from mesenchymal stem like cell differentiation is the key molecular process in diffuse type gastric tumors. From this, it can be inferred that occurrence and differentiation of MSC into chondrocyte, osteoblast, fibroblast, endothelial, and adipocyte lineage (Figure 6C) like cells are obviously responsible for the aggressiveness of the diffuse type gastric tumors. However, since this observation is from mRNA profiles of gastric tumors, the number of cell types present in diffuse type gastric tumor microenvironment is not clear and needs to be identified.
Figure 6.
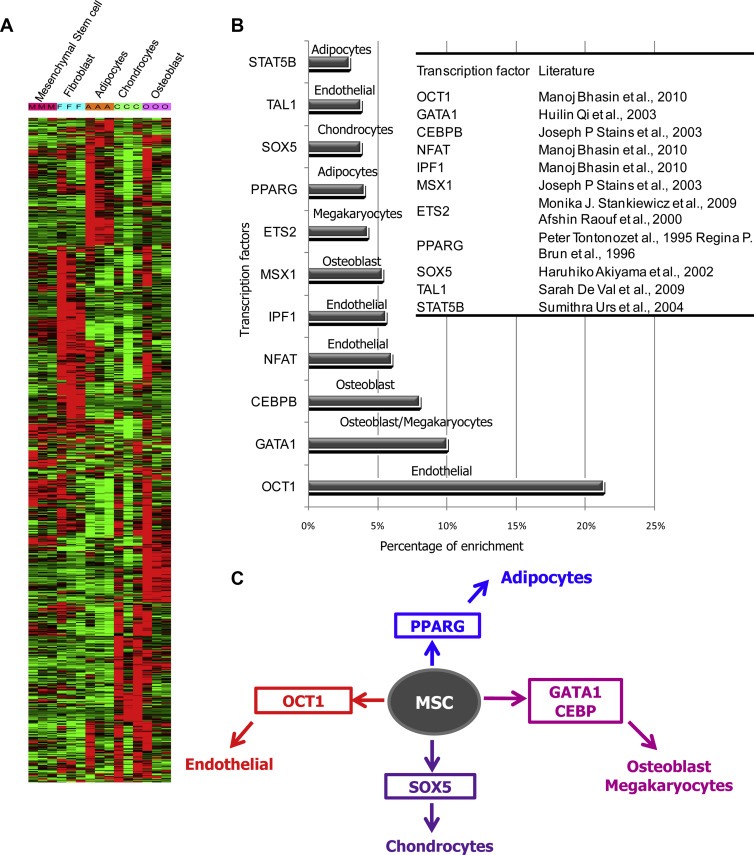
Expression pattern of G1 modular genes across mesenchymal stem cell lineages. (A) Expression of G1 genes across different mesenchymal stem cell derived and differentiated lineages (GSE 9451). G1 genes are differentially expressed in different lineages derived from mesenchymal stem cells (Fibroblast – F, Adipocytes – A, Chondrocytes – C, Osteoblast – O, Mesenchymal stem cell – M). (B) The binding sites of mesenchymal lineage specific differentiation inducing transcription factors OCT1, GATA1, CEBPB, NFAT, IPF1, MSX1, etc., are enriched in the promoters of G1 genes. The table shows the references for the lineage specificity associated with the indicated transcription factors. (C) Scheme showing the occurrence of mesenchymal stem cell (MSC) derived lineages/properties which is the characteristic feature of G1 module and the hallmark of tumor milieu in diffuse tumors.
3.4. Involvement of epigenetic and Myc mediated dysregulations in diffuse and intestinal tumors
Upon analyzing the G1 and G13 modular genes for their overlap in a massive panel of 184 stem cell signatures collected from GeneSigDb and literature (Supplemental Table 5), multiple epigenetic and polycomb repression related gene sets were found enriched in G1 modular genes. Notably, the genes which are targets of polycomb repressive complex 2 (PRC2), EED, EZH2 and SUZ12 are found enriched among G1 modular genes (Figure 7A). In addition to the polycomb repressed genes, the genes with H3K27 trimethylation marks in their promoter are also enriched among G1 genes. PRC2, EED, EZH2 and SUZ12 target genes present in G1 module were analyzed for their expression across gastric tumors and found to express highly in diffuse type gastric tumors (Figure 7B). It is worth mentioning that the G1 genes show elevated expression in poorly differentiated gastric tumor and the constructed network is exclusively with the positive associations among the genes. The redundant and independent identification of four different epigenetic signatures (i) SUZ12 repressed genes, (ii) EZH2 repressed genes, (iii) EED repressed genes, and (iv) genes with H3K27 trimethylation marks in their promoter, which all indicate the reduced polycomb target gene repression as the characteristic dysregulation of the G1 module. Since five signatures point toward the same phenomenon, this observation could be considered very authentic. Moreover, analysis of the mean mRNA expression of PRC2 subunit genes SUZ12, EED, and EZH2 reveals their under‐expression in diffuse type gastric tumors (Figure 7C). This shows that epigenetic dysregulation due to reduced polycomb (PRC2), EED and SUZ12 mediated suppression of G1 genes and the concomitant loss of H3K27 trimethylation in the promoters of target genes as the features of diffuse type gastric tumors. This implies that these are the hallmarks of poorly differentiated and diffuse subtype gastric tumors. This observation also independently supports the major involvement of epigenetic regulation in mesenchymal stem cell differentiation which yields multiple lineages and confers poorly differentiated and aggressive tumor microenvironment.
Figure 7.
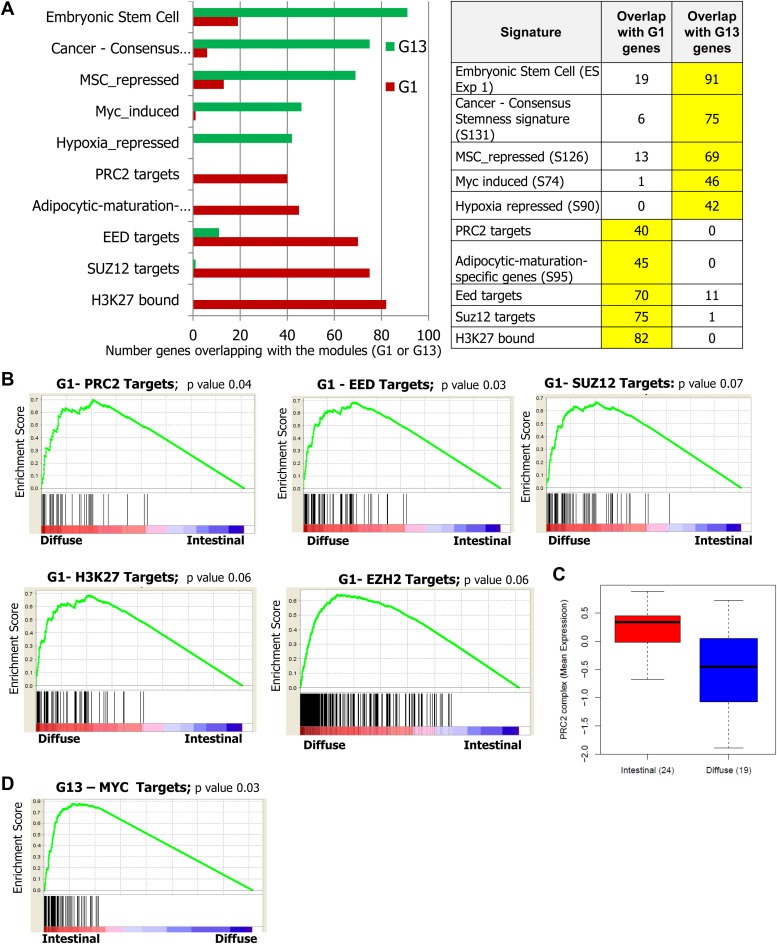
Identification of reduced polycomb mediated repression as major dysregulation among G1 modular genes. (A) Screening of G1 and G13 modules for gene set enrichment against 184 stem cell signatures shows that the G1 genes enriched with the targets of SUZ12, EED, PRC2, and H3K27. Myc target genes are found enriched in G13 module. The list of the signatures is shown in Supplemental Table 5. The numbers of genes in the modules (G1 and G13) that overlap with the selected top ranking signatures are shown in the graph (left) and table (right). (B) Gene set enrichment analysis of gastric tumors for PRC2, EED, SUZ12, EZH2 and H3K27 target genes across gastric tumors indicate their elevated expression in diffuse type samples with the enrichment score of ∼0.7 and significance. This analysis was performed in the samples in GSE22377 profile. (C) While PRC2 targets show elevated expression in diffuse gastric tumors, the PRC2 genes EED, EZH2 and SUZ12 show reduced expression in diffuse type tumors. (D) Gene set enrichment analysis of gastric tumors for MYC target genes across gastric tumors indicate their elevated expression in intestinal type samples.
On the other hand, the G13 module is enriched with the target genes of MYC (Figure 7A). This shows the MYC mediated transcription program as the key dysregulation driving the G13 modular gene expression. Accordingly, these Myc target genes in G13 module are found highly expressed in intestinal type gastric tumors (Figure 7D). Thus the key driving dysregulations of G1 and G13 modules or diffuse and intestinal subtypes of gastric cancer were identified.
3.5. Suppression of polycomb mediated repression is associated with elevated notch signaling
As mentioned earlier, G1 and G13 modular genes are capable of stratifying the gastric tumors into intestinal and diffuse types. Molecular stratification of gastric tumor samples into intestinal and diffuse subtypes based on the expression of G1 modular genes was also found reproducible in a panel of gastric cancer cell lines. The mRNA profile of 37 gastric cancer cell lines (GSE22183) (Tan et al., 2011) was investigated and the combined expression pattern of all G1 genes across cell lines were measured based on the Z score metric (Levine et al., 2006). The analysis of the expression of G1 genes in 37 gastric cancer cell lines classified the cell lines into intestinal and diffuse types (Figure 8A). G1 genes are highly expressed in diffuse type gastric cancer cell lines Hs746T, MKN1, YCC20, RerfGC1B, and YCC16, whereas found relatively downregulated in intestinal type cell lines YCC3, and IM95. Similarly, the expression pattern of G13 genes also was analyzed. However, G13 genes were not capable of stratifying the cell lines based on their histological types (Supplemental Data 3). As mentioned earlier, G1 genes were found to be good classifiers. Upon comparing the specific and exclusive expression of G1 and G13 genes, YCC16 was identified as a good representative model cell line expressing G1 modular genes. On the other side, YCC3, and IM95 were identified to express lesser levels of G1 genes and more of G13 genes (Figure 8A–B). The most commonly used gastric cancer cell line model AGS was found to be an intermediate.
Figure 8.

Reduced expression of polycomb component genes and elevated Notch mediated transcription are the features of G1 modular genes. (A) Overall expression of G1 modular genes across 37 gastric cancer cell lines shows the predominant expression in diffuse type gastric tumors. The combined expression of G1 genes was calculated based on Z score from the mRNA profile GSE22183. (B) Upon comparing the exclusive and selective expression of G1 and G13 genes, YCC16 was identified as good representative model for G1 modular expression and, IM95 and YCC3 as models for G13 expression. (C) Basal and un‐induced Notch in‐vitro transcription factor reporter activity measured across selected gastric cancer cell lines. Reporter activity was measured by dual luciferase assay and expressed in folds of reporter activity in comparison to the firefly reporter without enhancer. (D) RNAi mediated silencing of PRC2 components EED and EZH2 results in activation of Notch reporter/transcriptional activity in AGS cells. (E) EED silencing results in elevated Notch2 gene expression in AGS cells. (F) HDAC inhibitor SAHA treatment results in a reduction in the expression of polycomb component genes EED, EZH2, and SUZ12 in AGS cells. (G) Modest elevation in Notch reporter/transcriptional activity in AGS cells upon treatment with HDAC inhibitor SAHA.
Upon identifying the representative cell line models for the modules, it was further attempted to explore and evaluate the major signaling pathway dysregulations playing key role in the G1 and G13 modules. Recently, quantitative analysis of the in‐vitro activities of 45 transcription factors/signaling pathways in 8 gastric cancer cell lines was achieved (Periasamy et al., 2014) and comparison of all these transcription factor activities in the cell lines in accordance with the expression of G1 genes showed relative higher level Notch transcriptional activity in YCC16 cell line while compared to YCC3 and IM95 (Figure 8C). This shows that Notch might be a key pathway involved in regulating G1 modular genes and thus also being a critical pathway involved in diffuse type gastric cancers. Interestingly, one of the Notch family genes, NOTCH2, was found among G1 modular genes. On the other hand, the polycomb genes EZH2, EED and SUZ12 were found to have reduced expression in diffuse gastric tumors (Figure 7C). Therefore we hypothesized the suppression of the expression of polycomb repressor genes (EZH2, EED, SUZ12) as the cause for activated Notch signaling in diffuse type gastric tumors. In AGS cells, one of the cell lines with diminished Notch transcriptional activity, siRNA mediated silencing of EED and EZH2 resulted in the elevation of Notch transcriptional activity (Figure 8D). AGS cells were transfected with EED or EZH2 siRNA separately and 24 h after transfection, the cells were transfected with the Notch reporter plasmid (Notch enhancer driven firefly luciferase reporter construct, SA Biosciences) and CMV‐renilla luciferase reporter plasmid. The firefly and renilla luciferase reporter activities were measured by dual luciferase assay at 24 h after reporter plasmid transfection. The normalized Notch enhancer driven firefly reporter activity was measured as Notch transcriptional activity and found elevated at 48 h after EED and EZH2 siRNA transfection. Concordantly, one of the notch family genes NOTCH2 was found elevated at mRNA level upon EED silencing in AGS cells (Figure 8E). Here, in AGS cells, at 48 h after EED siRNA transfection, RT‐PCR was performed and the elevated NOTCH2 gene expression due to EED silencing was inferred.
Histone deacetylase (HDAC) inhibitors are also known to deplete polycomb repressive complex 2 (PRC2) proteins EED, EZH2 and SUZ12 (Fiskus et al., 2006). Therefore, the same gastric cancer cell line AGS was treated with SAHA, one of the HDAC inhibitors. Upon SAHA treatment, the impact on the expression of the PRC2 complex genes EZH2, SUZ12, and EED genes were analyzed by RT‐PCR and found diminished (Figure 8F). Subsequently, the impact of the SAHA treatment on Notch transcriptional activity was assessed by Notch reporter activity. HDAC inhibitor treatment which has been identified to mediate the repression of the expression of PRC2 complex genes EZH2, SUZ12, and EED results in the elevation of Notch transcriptional program in AGS cells (Figure 8G). Thus both siRNA mediated genetic silencing and chemical mediated inhibition of PRC2 genes result in the elevation of Notch transcriptional activity in AGS gastric cancer cells, a cell line with basal non‐activated Notch transcriptional activity. This shows that the expression of polycomb genes and the histone deacetylation are capable of repressing Notch activity. Reduction in polycomb gene expression results in the elevated Notch activity. In diffuse gastric tumors, the G1 modular genes are highly expressed and are indicative of reduced polycomb repression and elevated Notch activity. Therefore, PRC2 complex gene expression and elevated Notch transcriptional activity are potential therapeutic targets for diffuse type gastric tumors or patients with elevated G1 modular genes, and hence modulation of PRC2 complex or Notch activation is the possible targeted therapeutic approach for diffuse type gastric cancer patients in large and more specifically to the gastric tumors with elevated G1 modular genes.
While Notch activation and reduced polycomb gene expression were identified as major dysregulations in diffuse type gastric tumors, the signaling dysregulations in intestinal type gastric tumors was investigated using the G13 modular genes and G13 modular gene expressing cell lines. Myc and E2F binding sites are enriched among the G13 modular genes (Supplemental Table 4). This shows Myc and E2F mediated transcriptional programs as critical drivers of G13 module and intestinal type gastric tumors. Upon referring the mRNA profile of gastric cancer cell lines (GSE22183), the quantity of Myc mRNA was found higher in the G13 model cell lines IM95 and YCC3 in contrast to the G1 model cell line YCC16 (Figure 9A). Myc protein also was found to express highly in IM95 and YCC3 cells (Figure 9B). AGS cells, the non‐G1 and non‐G13 cell line was found to highly express Myc at protein level. Though there is discordance between relative Myc expression at mRNA and protein levels in AGS cells, this is known for many genes and might be due to post‐transcriptional or post‐translational stabilization. Similarly, YCC16 also shows more Myc transcriptional activity but with less Myc mRNA and deserves a separate investigation. However, G13 modular model cell lines IM95 and YCC3, along with the Myc expressing AGS cells are already known for their inherent Wnt signaling pathway activation (Periasamy et al., 2014). Accordingly, TCF/LEF mediated Wnt transcriptional activity is high in YCC3, IM95, AGS cells and found less in YCC16 (Figure 9C). Interestingly, the basal Wnt transcriptional activity of the cell line panel is very well correlating with the Myc expression pattern. It is known that Myc and E2F transcription factors are regulated by Wnt/β‐catenin mediated regulation (Calvisi et al., 2005). Further, in AGS cells, silencing of β‐catenin (CTNNB1) resulted in the down regulation of Myc and E2F mediated transcriptional activity (Figure 9D–E). As described earlier, Myc and E2F transcriptional activity were measured using pMyc‐Luc and pE2F‐Luc reporter plasmids (SA Biosciences) by dual luciferase assay. Moreover, it is known that Wnt is often activated in intestinal type gastric tumors (Ogasawara et al., 2006) and also in embryonic stem cells (Van Camp et al., 2013). This shows Wnt/β‐catenin mediated transcriptional regulation involving Myc and E2F mediated transcription program is the major dysregulation in intestinal gastric tumors and has been identified from the features of G13 module of gastric cancer coexpression network. Wnt‐Myc‐E2F signaling cascade is the potential therapeutic target for intestinal gastric tumors and more precisely for gastric tumors with elevated G13 modular gene expression.
Figure 9.
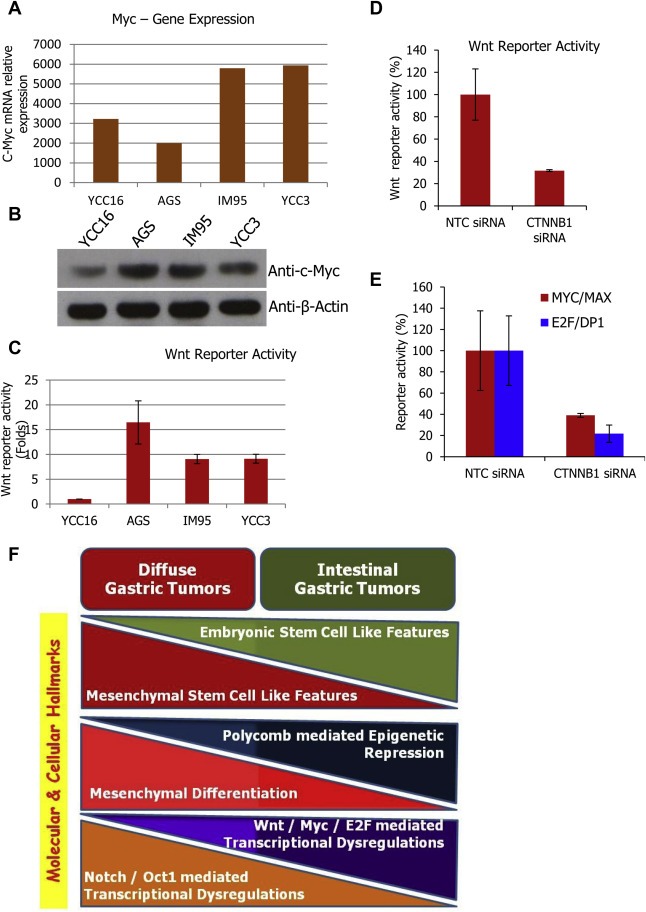
Wnt‐Myc‐E2F mediated transcription program is activated in intestinal type gastric cancer cells. (A) Relative quantity of c‐Myc mRNA across selected low level G13 modular gene expressing (YCC16 and AGS) and high level G13 modular gene expressing (IM95 and YCC3) gastric cancer cell lines. This expression values are from the genome‐wide mRNA profile of GSE22183. (B) Immunoblot showing relative c‐Myc protein quantity in the same cell line panel. (C) Basal and un‐induced Wnt/TCF in‐vitro transcription factor reporter activity measured across selected gastric cancer cell lines. Reporter activity was measured by dual luciferase assay and expressed in folds of reporter activity in comparison to the firefly reporter without enhancer. (D) Confirmation of the repressed Wnt transcriptional activity upon silencing the CTNNB1 (β‐catenin) gene in AGS cells (Wnt hyperactive cell line) by transfecting the CTNNB1 siRNA. (E) RNAi mediated silencing of CTNNB1 results in the reduction in MYC & E2F reporter/transcriptional activity in AGS cells. (F) Model summarizing all the differential and contrasting features identified from this investigation for their association and involvement in intestinal and diffuse subtypes of gastric tumors. These are the novel molecular and cellular hallmarks of gastric cancer subtypes identified from the current investigation.
4. Discussion
Integrative functional genomic analysis of the transcriptomes of tumors improves our understanding of the molecular heterogeneity and complexity of cancers (Gerlinger et al., 2012). Network modeling based on coexpression pattern analysis of genes have been employed in various cancers to understand the biology of cancers, cancer microenvironment and to gain clinical insights (Cho et al., 2012; Doig et al., 2013). Cancer progression is correlated with series of changes in gene expression patterns, gene interactions, and changes in cells and their microenvironment. While compared to gene focused studies, gene set and gene signature focused functional genomic investigations seem much rewarding in gaining functional insights (Ponomarev et al., 2012). The concept of network has led to the discovery of novel functions and functional interactions of BRCA1 in breast cancer (Pujana et al., 2007), novel pathways involved in cancer subtypes (Teschendorff et al., 2010) and heterogeneity within breast cancer subtypes (Fredlund et al., 2012). The known biomarkers have been expanded to prognostic biomarker panels in Chronic lymphocytic leukemia (CLL) based on coexpressed genes of a reported biomarker ZAP70 (Zhang et al., 2010). Coexpression network has identified the involvement of epigenetic components in alcohol induced changes in brain gene expression (Ponomarev et al., 2012).
Recently, network based coexpression analysis of gastric cancer transcriptome has resulted in the identification of enriched stromal cell population in diffuse gastric tumors (Wu et al., 2013). However, the heterogeneities among the stem cell types and factors associated with stromal cell enrichment were not explored. In the current investigation, weighted coexpression network was constructed to investigate the transcriptome of gastric cancer. From the clustering pattern of 20,000 genes, 21 coexpressed modules were derived. Like histopathology based clinical classification of gastric cancer subtypes, the 21 network modules and the selected hub genes from the modules could classify gastric cancer into subtypes in unsupervised manner. Thus, from the network, set of modules clearly associated with diffuse subtype of gastric tumors and modules related to intestinal subtype of gastric tumors were identified. Lauren's classification of gastric tumors as intestinal and diffuse subtypes is the current classification and the intestinal type has the characteristics of cohesive, moderately differentiated, glandular cells with precursor lesions. In contrast, the diffuse type has the features of non‐cohesive, highly infiltrating, poorly differentiated cells, without glandular and precursor lesions (Hamilton and Meltzer, 2006; Lauren, 1965). Apart from these extensively studied features of gastric cancer subtypes, other published gene expression analysis based molecular classifications did recapitulate the histological subtypes with additional sub grouping possibilities. In these studies, tumors were stratified into tumorigenic, reactive, and gastric‐like subtypes independent of Lauren's classification. Also gastric cancer cell lines were subgrouped into proximal, distal gastric cancers‐intestinal type, and diffuse/signet cell subtype based on cancer location in association with the Lauren's classification (Shah et al., 2011; Tay et al., 2003). However, the current investigation was aimed to characterize the gene sets toward identifying their molecular dysregulations. Nevertheless the selected modules show close association with Lauren's classification. More specifically, the analyses led to the identification of new molecular characteristics such as differential stem cell properties among these known gastric cancer subtypes.
Among the modules, G1 modular genes are highly expressed in poorly differentiated/diffuse type tumors and of G13 modules are expressed in well and moderately differentiated tumors. G1 module shows the features of mesenchymal and mesenchymal stem cells with the signs for mesenchymal stem cell differentiation. G1 modular genes also comprise some of the MSC markers. Thus the involvement of these features in diffuse type gastric tumors has been identified. Apart from the enriched representation of mesenchymal markers and mesenchymal lineage specific genes, G1 modular genes reveal the presence of mesenchymal stem cells and mesenchymal stem cell derived multiple cell lineages or cell types as the hallmark of diffuse gastric tumors. For the first time, mRNA expression revealing the occurrence of different cell types in diffuse type gastric tumors and specifically, the occurrence of the diversified mesenchymal stem cell derived cell types has been identified. The mesenchymal stem cell derived fibroblasts and endothelial cells in tumors indicate the kind of tumor microenvironment that favors differentiation, invasions and cancer progression. G1 genes such as IL‐6, IGF1, FGF, and ACVR1 are known for their expression in stromal cell type (Deng et al., 2008; Sung et al., 2007). G13 module shows some of the features of embryonic stem cells with an elevated expression in intestinal tumors. This module shows the features of high proliferation driven by E2F and Myc transcription factors. Altogether, the differential and varying stem cell properties observed in different gastric cancer subtypes is a notable and novel observation in gastric cancer and also in any cancers.
Using molecular signatures, occurrence of stem cells across breast, glioma, bladder and other cancers have been extensively demonstrated (Ben‐Porath et al., 2008). The stemness associated with cancer cells and its association with poor differentiation status of cancers are well established (Sell, 2004; Zhang et al., 2013). However, it has not been described beyond that (Ben‐Porath et al., 2008; Wong et al., 2008). There are reports stating the existence of multipotent stem cell property in gastric stem cells, the Lgr5+ mutation in stem cells leading to the intestinal gastric cancer, tissue stem cells in the resident organ acting as candidate of cancer stem cells, involvement of bone marrow derived stem cells in epithelial cancers and increased metastasis through CCL5 secretion (Milne et al., 2009). Recent in‐vitro and in‐vivo studies reveal the involvement of normal stem cells, mesenchymal cells, and cancer stem cells in gastric cancer and tumor growth (Kitadai, 2010; Lei et al., 2013; Saikawa et al., 2010; Singh, 2013; Yang et al., 2011). Possible targeting of the stromal interactions also has been suggested as cancer therapeutic strategy (Yashiro and Hirakawa, 2010). However, this is the first report for the occurrence of two distinct stem cell types and their varied expression in cancer subtypes. Specifically, the differential occurrence and involvement of embryonic stem cell like cells and mesenchymal stem cell like cells in intestinal and diffuse type gastric tumors has been identified with molecular biological approaches (Figure 9F). This tremendously adds to the developing concepts in stem cell research and in delineating the molecular processes involved gastric carcinogenesis.
Mesenchymal stem cells are the essential and fundamental components in developing the tumor microenvironment and the presence of the scattered population of cells like cancer associated fibroblast (CAF), endothelial cells, immune cells, and other stromal cells in tumor microenvironment are emerging (Alphonso and Alahari, 2009). The involvement of CAF in angiogenesis through the production of factors like VEGF, FGF and the role of endothelial cells in forming new vessels with these factors have been reported in gastric cancer (Augsten et al., 2010). In addition, our analysis reveals the possible presence of few other cell types like osteoblast and chondrocytes in tumor microenvironment. Since this inference is from the global mRNA expression pattern, the involvement of osteoblast like and chondrocyte like cellular features is very clear. However, whether this is due to the expression of these genes by other types of cells or the different types of cells occurring in the diffuse gastric tumor is not known. From the current data, this could not be discriminated and needs to be investigated.
The regulatory mechanisms of the stromal cells are under exploration (Tuxhorn et al., 2002). Till now, involvement of histone modification in regulating tumor microenvironment has not been defined (Dey, 2011). The current study shows the involvement of epigenetic regulatory mechanism in the identified mesenchymal stem cell derived cellular differentiation involving the occurrence of endothelial like, osteoblast like and chondrocyte like features/cells in diffuse type gastric tumors. Reduced expression of polycomb repressive complex genes was found associated with the tumor microenvironment of diffuse type gastric cancer. Silencing of tumor suppressor genes by epigenetic regulation is known to be involved in carcinogenesis and cancer progression (Ron‐Bigger et al., 2010). The polycomb repressive complex is regulating the genes involved in maintaining the stem cells and genes playing crucial role in lineage specific differentiation (Aranda et al., 2009). The major regulators of the complex SUZ12, EED, and EZH2, all found suppressed in diffuse type gastric tumors and has been identified as the features of G1 module. Loss of H3K27 trimethylation (H3K27me3) repression mark in the promoters of the target genes is known in multiple cancers and low level of H3K27me3 in promoters show poor survival (Ellinger et al., 2012). Among the investigated 184 gene signatures, a gene set indicative of H3K27me3 repression marks in their promoter is found upregulated among G1 modular genes (MAN1C1, SPON1, PTGFR, PODN, PDGFRA, FBLN5, and SMAD11). Strikingly, multiple signatures point toward the dysregulated polycomb repression as the feature of G1 module and diffuse type gastric tumors.
Among the gastric cancer cell lines, YCC16 is the cell line with the elevated expression of G1 modular genes and the reduced expression of polycomb genes (EED, EZH2). This further supports the loss or reduced expression of the polycomb genes SUZ12, EED, and EZH2 is the major dysregulation in diffuse type gastric tumors. However, whether this is the primary cause for carcinogenesis or the consequence of some other dysregulation is a key question in cancer genetics and needs to be determined. Our experiments in gastric cancer cell lines show the elevated level of Notch signaling upon the loss of polycomb repression.
The degree of epigenetic silencing of genes varies across different cell types and in stem cells the differentiation inducing genes are repressed by H3K27 trimethylation repression mark (Aranda et al., 2009). Recently, it has been found that the cell adhesion and cell communication genes in endothelial cells favor angiogenesis upon EZH2 silencing (Dreger et al., 2012). Studies on the epigenetic regulation of stem cells reveal that embryonic stem cells exert high repression marks and repress the differentiation genes whereas in mesenchymal stem cells, there is a reduction in repression marks and hence the differentiation genes are active (Aranda et al., 2009). This supports the identified lower level expression of PRC2 complex genes in diffuse type gastric tumors enriched with mesenchymal stem cell like cells while compared to intestinal type tumors with embryonic stem cell like cells. Thus the under‐expression of the factors inducing epigenetic repression switch the mesenchymal stem cells toward the differentiation of endothelial cells and favors the cell–cell communication, invasion, metastasis and angiogenesis in diffuse type gastric cancers. This deserves a sequential and extensive investigation in animal models.
Mesenchymal markers IGF1 and TGFB1 were reported to be involved in epithelial to mesenchymal transition (EMT) in prostate cancer (Graham et al., 2008), and VCAM1 is known to confer high invasiveness to gastric cancer (Ding et al., 2003). Notably, IGF1, TGFB1 and VCAM1 all being the G1 modular genes and supporting the aggressive features associated with diffuse type gastric tumors. All these imply that selected G1 genes may be developed as molecular markers for the stratification of gastric tumors.
5. Conclusion
Upon investigating gene coexpression pattern in the gastric tumor mRNA profile, the key and contrasting stem cell features and dysregulations involved in intestinal and diffuse subtypes of gastric tumors have been identified. Investigation of G1 and G13 modules across stem cell factors has extended the cancer stem cell research with a new paradigm that different gastric cancer subtypes have different stem cell properties. Diffuse tumors are found to have the features of mesenchymal stem cells, their differentiation to multiple cell types along with the dysregulations involving Notch and polycomb repressors. On the other hand, embryonic stem cell like features with Wnt‐Myc‐E2F mediated transcriptional dysregulations have been found to be the driving factors in intestinal type gastric tumors.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgment
UGC‐Meritorious fellowship support to Kalaivani Kalamohan is acknowledged. The experiments were performed with the research grant support from Department of Biotechnology (DBT), Govt. of India, through the research grant BT/PR11625/MED/30/155/2008 to Kumaresan Ganesan, Madurai Kamaraj University. Instrumentation supports of UGC‐CEGS, DBT‐IPLS, DST‐PURSE, UGC‐NRCBS, and UGC‐CAS programme supported central facilities of SBS, MKU are acknowledged.
Supplementary data 1.
Supplementary data related to this article can be found at doi:10.1016/j.molonc.2014.04.005.
Kalamohan Kalaivani, Periasamy Jayaprakash, Bhaskar Rao Divya, Barnabas Georgina D., Ponnaiyan Srigayatri, Ganesan Kumaresan, (2014), Transcriptional coexpression network reveals the involvement of varying stem cell features with different dysregulations in different gastric cancer subtypes, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.04.005.
References
- Abdollahi, H. , Harris, L.J. , Zhang, P. , McIlhenny, S. , Srinivas, V. , Tulenko, T. , DiMuzio, P.J. , 2011. The role of hypoxia in stem cell differentiation and therapeutics. J. Surg. Res. 165, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal, A. , Guo, D.L. , Hoshida, Y. , Yuen, S.T. , Chu, K.M. , So, S. , Boussioutas, A. , Chen, X. , Bowtell, D. , Aburatani, H. , Leung, S.Y. , Tan, P. , 2006. Topological and functional discovery in a gene coexpression meta-network of gastric cancer. Cancer Res. 66, 232–241. [DOI] [PubMed] [Google Scholar]
- Akiyama, H. , Chaboissier, M.C. , Martin, J.F. , Schedl, A. , de Crombrugghe, B. , 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphonso, A. , Alahari, S.K. , 2009. Stromal cells and integrins: conforming to the needs of the tumor microenvironment. Neoplasia. 11, 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda, P. , Agirre, X. , Ballestar, E. , Andreu, E.J. , Roman-Gomez, J. , Prieto, I. , Martin-Subero, J.I. , Cigudosa, J.C. , Siebert, R. , Esteller, M. , Prosper, F. , 2009. Epigenetic signatures associated with different levels of differentiation potential in human stem cells. PLoS One. 4, e7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten, M. , Hagglof, C. , Pena, C. , Ostman, A. , 2010. A digest on the role of the tumor microenvironment in gastrointestinal cancers. Cancer Microenviron. 3, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi, A.L. , Oltvai, Z.N. , 2004. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113. [DOI] [PubMed] [Google Scholar]
- Ben-Porath, I. , Thomson, M.W. , Carey, V.J. , Ge, R. , Bell, G.W. , Regev, A. , Weinberg, R.A. , 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin, M. , Yuan, L. , Keskin, D.B. , Otu, H.H. , Libermann, T.A. , Oettgen, P. , 2010. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genomics. 11, 342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad, B.M. , Irizarry, R.A. , Astrand, M. , Speed, T.P. , 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Calvisi, D.F. , Conner, E.A. , Ladu, S. , Lemmer, E.R. , Factor, V.M. , Thorgeirsson, S.S. , 2005. Activation of the canonical Wnt/beta-catenin pathway confers growth advantages in c-Myc/E2F1 transgenic mouse model of liver cancer. J. Hepatol. 42, 842–849. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Leung, S.Y. , Yuen, S.T. , Chu, K.M. , Ji, J. , Li, R. , Chan, A.S. , Law, S. , Troyanskaya, O.G. , Wong, J. , So, S. , Botstein, D. , Brown, P.O. , 2003. Variation in gene expression patterns in human gastric cancers. Mol. Biol. Cell. 14, 3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J.Y. , Lim, J.Y. , Cheong, J.H. , Park, Y.Y. , Yoon, S.L. , Kim, S.M. , Kim, S.B. , Kim, H. , Hong, S.W. , Park, Y.N. , Noh, S.H. , Park, E.S. , Chu, I.S. , Hong, W.K. , Ajani, J.A. , Lee, J.S. , 2011. Gene expression signature-based prognostic risk score in gastric cancer. Clin. Cancer Res. 17, 1850–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, D.Y. , Kim, Y.A. , Przytycka, T.M. , 2012. Chapter 5: network biology approach to complex diseases. Plos Comput. Biol. 8, e1002820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane, A.C. , Schwarzl, T. , Sultana, R. , Picard, K.C. , Picard, S.C. , Lu, T.H. , Franklin, K.R. , French, S.J. , Papenhausen, G. , Correll, M. , Quackenbush, J. , 2010. GeneSigDB – a curated database of gene expression signatures. Nucleic Acids Res. 38, D716–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Z.L. , Sharff, K.A. , Tang, N. , Song, W.X. , Luo, J. , Luo, X. , Chen, J. , Bennett, E. , Reid, R. , Manning, D. , Xue, A. , Montag, A.G. , Luu, H.H. , Haydon, R.C. , He, T.C. , 2008. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 13, 2001–2021. [DOI] [PubMed] [Google Scholar]
- Dey, P. , 2011. Epigenetic changes in tumor microenvironment. Indian J. Cancer. 48, 507–512. [DOI] [PubMed] [Google Scholar]
- Ding, Y.B. , Chen, G.Y. , Xia, J.G. , Zang, X.W. , Yang, H.Y. , Yang, L. , 2003. Association of VCAM-1 overexpression with oncogenesis, tumor angiogenesis and metastasis of gastric carcinoma. World J. Gastroenterol. 9, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig, T.N. , Hume, D.A. , Theocharidis, T. , Goodlad, J.R. , Gregory, C.D. , Freeman, T.C. , 2013. Coexpression analysis of large cancer datasets provides insight into the cellular phenotypes of the tumour microenvironment. BMC Genomics. 14, 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger, H. , Ludwig, A. , Weller, A. , Stangl, V. , Baumann, G. , Meiners, S. , Stangl, K. , 2012. Epigenetic regulation of cell adhesion and communication by enhancer of zeste homolog 2 in human endothelial cells. Hypertension. 60, 1176–1183. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B. , Spellman, P.T. , Brown, P.O. , Botstein, D. , 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U S A. 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger, J. , Kahl, P. , von der Gathen, J. , Heukamp, L.C. , Gutgemann, I. , Walter, B. , Hofstadter, F. , Bastian, P.J. , von Ruecker, A. , Muller, S.C. , Rogenhofer, S. , 2012. Global histone H3K27 methylation levels are different in localized and metastatic prostate cancer. Cancer Invest. 30, 92–97. [DOI] [PubMed] [Google Scholar]
- Fiskus, W. , Pranpat, M. , Balasis, M. , Herger, B. , Rao, R. , Chinnaiyan, A. , Atadja, P. , Bhalla, K. , 2006. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol. Cancer Ther. 5, 3096–3104. [DOI] [PubMed] [Google Scholar]
- Forster, S. , Gretschel, S. , Jons, T. , Yashiro, M. , Kemmner, W. , 2011. THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Mod. Pathol. 24, 1390–1403. [DOI] [PubMed] [Google Scholar]
- Fredlund, E. , Staaf, J. , Rantala, J.K. , Kallioniemi, O. , Borg, A. , Ringner, M. , 2012. The gene expression landscape of breast cancer is shaped by tumor protein p53 status and epithelial-mesenchymal transition. Breast Cancer Res. 14, R113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan, K. , Ivanova, T. , Wu, Y. , Rajasegaran, V. , Wu, J. , Lee, M.H. , Yu, K. , Rha, S.Y. , Chung, H.C. , Ylstra, B. , Meijer, G. , Lian, K.O. , Grabsch, H. , Tan, P. , 2008. Inhibition of gastric cancer invasion and metastasis by PLA2G2A, a novel beta-catenin/TCF target gene. Cancer Res. 68, 4277–4286. [DOI] [PubMed] [Google Scholar]
- Gerlinger, M. , Rowan, A.J. , Horswell, S. , Larkin, J. , Endesfelder, D. , Gronroos, E. , Martinez, P. , Matthews, N. , Stewart, A. , Tarpey, P. , Varela, I. , Phillimore, B. , Begum, S. , McDonald, N.Q. , Butler, A. , Jones, D. , Raine, K. , Latimer, C. , Santos, C.R. , Nohadani, M. , Eklund, A.C. , Spencer-Dene, B. , Clark, G. , Pickering, L. , Stamp, G. , Gore, M. , Szallasi, Z. , Downward, J. , Futreal, P.A. , Swanton, C. , 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour, A. , Doss, S. , Zhang, B. , Wang, S. , Plaisier, C. , Castellanos, R. , Brozell, A. , Schadt, E.E. , Drake, T.A. , Lusis, A.J. , Horvath, S. , 2006. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2, e130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritharan, G. , Ilic, D. , Gormley, M. , Krtolica, A. , 2011. Human embryonic stem cells derived from embryos at different stages of development share similar transcription profiles. PLoS One. 6, e26570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub, T.R. , Slonim, D.K. , Tamayo, P. , Huard, C. , Gaasenbeek, M. , Mesirov, J.P. , Coller, H. , Loh, M.L. , Downing, J.R. , Caligiuri, M.A. , Bloomfield, C.D. , Lander, E.S. , 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 286, 531–537. [DOI] [PubMed] [Google Scholar]
- Gotea, V. , Ovcharenko, I. , 2008. DiRE: identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res. 36, W133–W139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, T.R. , Zhau, H.E. , Odero-Marah, V.A. , Osunkoya, A.O. , Kimbro, K.S. , Tighiouart, M. , Liu, T. , Simons, J.W. , O'Regan, R.M. , 2008. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 68, 2479–2488. [DOI] [PubMed] [Google Scholar]
- Hamilton, J.P. , Meltzer, S.J. , 2006. A review of the genomics of gastric cancer. Clin. Gastroenterol. Hepatol. 4, 416–425. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. , Weinberg, R.A. , 2011. Hallmarks of cancer: the next generation. Cell. 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Horvath, S. , Dong, J. , 2008. Geometric interpretation of gene coexpression network analysis. Plos Comput. Biol. 4, e1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Snitkin, E.S. , DeLisi, C. , 2008. VisANT: an integrative framework for networks in systems biology. Brief Bioinform. 9, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da, W. , Sherman, B.T. , Lempicki, R.A. , 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Karnoub, A.E. , Dash, A.B. , Vo, A.P. , Sullivan, A. , Brooks, M.W. , Bell, G.W. , Richardson, A.L. , Polyak, K. , Tubo, R. , Weinberg, R.A. , 2007. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 449, 557–563. [DOI] [PubMed] [Google Scholar]
- Kitadai, Y. , 2010. Cancer-stromal cell interaction and tumor angiogenesis in gastric cancer. Cancer Microenviron. 3, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijjer, M.L. , Rydbeck, H. , Kresse, S.H. , Buddingh, E.P. , Lid, A.B. , Roelofs, H. , Burger, H. , Myklebost, O. , Hogendoorn, P.C. , Meza-Zepeda, L.A. , Cleton-Jansen, A.M. , 2012. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer. 51, 696–706. [DOI] [PubMed] [Google Scholar]
- Langfelder, P. , Horvath, S. , 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9, 559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauren, P. , 1965. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 64, 31–49. [DOI] [PubMed] [Google Scholar]
- Lei, Z. , Tan, I.B. , Das, K. , Deng, N. , Zouridis, H. , Pattison, S. , Chua, C. , Feng, Z. , Guan, Y.K. , Ooi, C.H. , Ivanova, T. , Zhang, S. , Lee, M. , Wu, J. , Ngo, A. , Manesh, S. , Tan, E. , Teh, B.T. , So, J.B. , Goh, L.K. , Boussioutas, A. , Lim, T.K. , Flotow, H. , Tan, P. , Rozen, S.G. , 2013. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 145, 554–565. [DOI] [PubMed] [Google Scholar]
- Levine, D.M. , Haynor, D.R. , Castle, J.C. , Stepaniants, S.B. , Pellegrini, M. , Mao, M. , Johnson, J.M. , 2006. Pathway and gene-set activation measurement from mRNA expression data: the tissue distribution of human pathways. Genome Biol. 7, R93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Wong, W.H. , 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U S A. 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon, A. , Subramanian, A. , Pinchback, R. , Thorvaldsdottir, H. , Tamayo, P. , Mesirov, J.P. , 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, A.N. , Carneiro, F. , O'Morain, C. , Offerhaus, G.J. , 2009. Nature meets nurture: molecular genetics of gastric cancer. Hum. Genet. 126, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, H. , Spike, B.T. , Wahl, G.M. , Levine, A.J. , 2010. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc. Natl. Acad. Sci. U S A. 107, 22745–22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagini, S. , 2012. Carcinoma of the stomach: a review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J. Gastrointest. Oncol. 4, 156–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara, N. , Tsukamoto, T. , Mizoshita, T. , Inada, K. , Cao, X. , Takenaka, Y. , Joh, T. , Tatematsu, M. , 2006. Mutations and nuclear accumulation of beta-catenin correlate with intestinal phenotypic expression in human gastric cancer. Histopathology. 49, 612–621. [DOI] [PubMed] [Google Scholar]
- Ooi, C.H. , Ivanova, T. , Wu, J. , Lee, M. , Tan, I.B. , Tao, J. , Ward, L. , Koo, J.H. , Gopalakrishnan, V. , Zhu, Y. , Cheng, L.L. , Lee, J. , Rha, S.Y. , Chung, H.C. , Ganesan, K. , So, J. , Soo, K.C. , Lim, D. , Chan, W.H. , Wong, W.K. , Bowtell, D. , Yeoh, K.G. , Grabsch, H. , Boussioutas, A. , Tan, P. , 2009. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. Plos Genet. 5, e1000676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, S.J. , Rhodes, S.A. , Connor, H.E. , Rees, S. , Brown, J. , Giles, H. , 2000. Use of a dual firefly and Renilla luciferase reporter gene assay to simultaneously determine drug selectivity at human corticotrophin releasing hormone 1 and 2 receptors. Anal. Biochemistry. 281, 187–192. [DOI] [PubMed] [Google Scholar]
- Periasamy, J. , Muthuswami, M. , Rao, D.B. , Tan, P. , Ganesan, K. , 2014. Stratification and delineation of gastric cancer signaling by in vitro transcription factor activity profiling and integrative genomics. Cell Signalling. 26, 880–894. [DOI] [PubMed] [Google Scholar]
- Ponomarev, I. , Wang, S. , Zhang, L. , Harris, R.A. , Mayfield, R.D. , 2012. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. 32, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana, M.A. , Han, J.D. , Starita, L.M. , Stevens, K.N. , Tewari, M. , Ahn, J.S. , Rennert, G. , Moreno, V. , Kirchhoff, T. , Gold, B. , Assmann, V. , Elshamy, W.M. , Rual, J.F. , Levine, D. , Rozek, L.S. , Gelman, R.S. , Gunsalus, K.C. , Greenberg, R.A. , Sobhian, B. , Bertin, N. , Venkatesan, K. , Ayivi-Guedehoussou, N. , Sole, X. , Hernandez, P. , Lazaro, C. , Nathanson, K.L. , Weber, B.L. , Cusick, M.E. , Hill, D.E. , Offit, K. , Livingston, D.M. , Gruber, S.B. , Parvin, J.D. , Vidal, M. , 2007. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 39, 1338–1349. [DOI] [PubMed] [Google Scholar]
- Qi, H. , Aguiar, D.J. , Williams, S.M. , La Pean, A. , Pan, W. , Verfaillie, C.M. , 2003. Identification of genes responsible for osteoblast differentiation from human mesodermal progenitor cells. Proc. Natl. Acad. Sci. U S A. 100, 3305–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, S. , Kim, J. , Arafat, D. , Gibson, G. , 2012. Effect of normalization on statistical and biological interpretation of gene expression profiles. Front Genet. 3, 160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Bigger, S. , Bar-Nur, O. , Isaac, S. , Bocker, M. , Lyko, F. , Eden, A. , 2010. Aberrant epigenetic silencing of tumor suppressor genes is reversed by direct reprogramming. Stem Cells. 28, 1349–1354. [DOI] [PubMed] [Google Scholar]
- Saikawa, Y. , Fukuda, K. , Takahashi, T. , Nakamura, R. , Takeuchi, H. , Kitagawa, Y. , 2010. Gastric carcinogenesis and the cancer stem cell hypothesis. Gastric Cancer. 13, 11–24. [DOI] [PubMed] [Google Scholar]
- Schroeder, M.P. , Gonzalez-Perez, A. , Lopez-Bigas, N. , 2013. Visualizing multidimensional cancer genomics data. Genome Med. 5, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehn, L.H. , 2012. Paramount prognostic factors that guide therapeutic strategies in diffuse large B-cell lymphoma. Hematology Am. Soc. Hematol. Educ. Program. 1, 402–409. [DOI] [PubMed] [Google Scholar]
- Sell, S. , 2004. Stem cell origin of cancer and differentiation therapy. Crit. Rev. Oncol. Hematol. 51, 1–28. [DOI] [PubMed] [Google Scholar]
- Shah, M.A. , Khanin, R. , Tang, L. , Janjigian, Y.Y. , Klimstra, D.S. , Gerdes, H. , Kelsen, D.P. , 2011. Molecular classification of gastric cancer: a new paradigm. Clin. Cancer Res. 17, 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.R. , 2013. Gastric cancer stem cells: a novel therapeutic target. Cancer Lett. 338, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth, E. , Klopp, A. , Dembinski, J. , Andreeff, M. , Marini, F. , 2008. Inflammation and tumor microenvironments: defining the migratory itinerary of mesenchymal stem cells. Gene Ther. 15, 730–738. [DOI] [PubMed] [Google Scholar]
- Stains, J.P. , Civitelli, R. , 2003. Genomic approaches to identifying transcriptional regulators of osteoblast differentiation. Genome Biol. 4, 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, J.M. , Segal, E. , Koller, D. , Kim, S.K. , 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science. 302, 249–255. [DOI] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo, P. , Mootha, V.K. , Mukherjee, S. , Ebert, B.L. , Gillette, M.A. , Paulovich, A. , Pomeroy, S.L. , Golub, T.R. , Lander, E.S. , Mesirov, J.P. , 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S.Y. , Hsieh, C.L. , Wu, D. , Chung, L.W. , Johnstone, P.A. , 2007. Tumor microenvironment promotes cancer progression, metastasis, and therapeutic resistance. Curr. Probl. Cancer. 31, 36–100. [DOI] [PubMed] [Google Scholar]
- Tan, I.B. , Ivanova, T. , Lim, K.H. , Ong, C.W. , Deng, N. , Lee, J. , Tan, S.H. , Wu, J. , Lee, M.H. , Ooi, C.H. , Rha, S.Y. , Wong, W.K. , Boussioutas, A. , Yeoh, K.G. , So, J. , Yong, W.P. , Tsuburaya, A. , Grabsch, H. , Toh, H.C. , Rozen, S. , Cheong, J.H. , Noh, S.H. , Wan, W.K. , Ajani, J.A. , Lee, J.S. , Tellez, M.S. , Tan, P. , 2011. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 141, 476–485. 485 e471–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, S.T. , Leong, S.H. , Yu, K. , Aggarwal, A. , Tan, S.Y. , Lee, C.H. , Wong, K. , Visvanathan, J. , Lim, D. , Wong, W.K. , Soo, K.C. , Kon, O.L. , Tan, P. , 2003. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 63, 3309–3316. [PubMed] [Google Scholar]
- Teschendorff, A.E. , Gomez, S. , Arenas, A. , El-Ashry, D. , Schmidt, M. , Gehrmann, M. , Caldas, C. , 2010. Improved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modules. BMC Cancer. 10, 604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas, 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxhorn, J.A. , Ayala, G.E. , Smith, M.J. , Smith, V.C. , Dang, T.D. , Rowley, D.R. , 2002. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin. Cancer Res. 8, 2912–2923. [PubMed] [Google Scholar]
- Wong, D.J. , Liu, H. , Ridky, T.W. , Cassarino, D. , Segal, E. , Chang, H.Y. , 2008. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Grabsch, H. , Ivanova, T. , Tan, I.B. , Murray, J. , Ooi, C.H. , Wright, A.I. , West, N.P. , Hutchins, G.G. , Wu, J. , Lee, M. , Lee, J. , Koo, J.H. , Yeoh, K.G. , van Grieken, N. , Ylstra, B. , Rha, S.Y. , Ajani, J.A. , Cheong, J.H. , Noh, S.H. , Lim, K.H. , Boussioutas, A. , Lee, J.S. , Tan, P. , 2013. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut. 62, 1100–1111. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Ping, Y.F. , Yu, X. , Qian, F. , Guo, Z.J. , Qian, C. , Cui, Y.H. , Bian, X.W. , 2011. Gastric cancer stem-like cells possess higher capability of invasion and metastasis in association with a mesenchymal transition phenotype. Cancer Lett. 310, 46–52. [DOI] [PubMed] [Google Scholar]
- Yashiro, M. , Hirakawa, K. , 2010. Cancer-stromal interactions in scirrhous gastric carcinoma. Cancer Microenviron. 3, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, Z.J. , Ong, C.K. , Cutcutache, I. , Yu, W. , Zhang, S.L. , Huang, D. , Ler, L.D. , Dykema, K. , Gan, A. , Tao, J. , Lim, S. , Liu, Y. , Futreal, P.A. , Grabsch, H. , Furge, K.A. , Goh, L.K. , Rozen, S. , Teh, B.T. , Tan, P. , 2011. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer Res. 71, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Horvath, S. , 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article 17 [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Xiang, Y. , Ding, L. , Keen-Circle, K. , Borlawsky, T.B. , Ozer, H.G. , Jin, R. , Payne, P. , Huang, K. , 2010. Using gene co-expression network analysis to predict biomarkers for chronic lymphocytic leukemia. BMC Bioinformatics. 11, (Suppl. 9) S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Zhou, Y. , Qian, H. , Shao, G. , Lu, X. , Chen, Q. , Sun, X. , Chen, D. , Yin, R. , Zhu, H. , Shao, Q. , Xu, W. , 2013. Stemness and inducing differentiation of small cell lung cancer NCI-H446 cells. Cell Death Dis. 4, e633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Dumur, C.I. , Holt, S.E. , Beckman, M.J. , Elmore, L.W. , 2010. Multipotent adipose stromal cells and breast cancer development: think globally, act locally. Mol. Carcinog. 49, 923–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


