Abstract
Cancer drug resistance is a major problem, with the majority of patients with metastatic disease ultimately developing multidrug resistance and succumbing to their disease. Our understanding of molecular events underpinning treatment failure has been enhanced by new genomic technologies and pre‐clinical studies. Intratumour genetic heterogeneity (ITH) is a prominent contributor to therapeutic failure, and it is becoming increasingly apparent that individual tumours may achieve resistance via multiple routes simultaneously – termed polyclonal resistance. Efforts to target single resistance mechanisms to overcome therapeutic failure may therefore yield only limited success. Clinical studies with sequential analysis of tumour material are needed to enhance our understanding of inter‐clonal functional relationships and tumour evolution during therapy, and to improve drug development strategies in cancer medicine.
Keywords: Intratumour heterogeneity, Drug resistance, Genomic instability, Cancer evolution
Abbreviations
- WES
whole exome sequencing
- WGS
whole genome sequencing
- ITH
intratumour heterogeneity
- RNAseq
whole transcriptome sequencing
- CGH
comparative genomic hybridisation
- ctDNA
circulating tumour DNA
- FISH
fluorescence in-situ hybridisation
- TMZ
temozolomide
- GBM
glioblastoma multiforme
- TKI
tyrosine kinase inhibitor
- PFS
Progression free survival
1. Introduction
In recent years, considerable efforts have been made to personalise cancer therapy, guided by specific characteristics of each patient's cancer. This individualisation of cancer therapy has centred on the development and use of targeted therapies, such as tyrosine kinase inhibitors (e.g. gefitinib, erlotinib, imatinib) and monoclonal antibodies (e.g. trastuzumab, bevacizumab) (Blair et al., 2014). However, both intrinsic (primary) and acquired (secondary) drug resistance are major problems. In a clinical setting, intrinsic resistance refers to a failure of a tumour to respond to therapy at the first interval imaging assessment, while in acquired resistance, an initially responsive tumour subsequently progresses on treatment. At a cellular level, intrinsic resistance arises due to the presence of a resistance mechanism in the tumour cells prior to treatment, whereas in acquired resistance the majority of tumour cells are initially responsive to treatment, and resistance develops over time. The development of targeted agents is highly costly and progression free survival benefits in the advanced disease setting are often limited, frequently failing to translate into overall survival benefits (Blair et al., 2014; Holohan et al., 2013). Hence there is a critical need to understand their limitations and address new opportunities within the context of emerging insights in tumour biology.
Historical knowledge of heterogeneity in cancer, both from a histopathological and genetic perspective (Lengauer et al., 1998; Swanton, 2012), coupled with a large number of recent studies documenting extensive intratumour genetic heterogeneity in a wide range of malignancies (reviewed in Burrell et al., 2013; Kreso and Dick, 2014; Swanton, 2012; Yates and Campbell, 2012), suggest that pre‐existing drug‐resistant subclones might be a substantial contributor to therapeutic resistance in oncology. Tumours are dynamically evolving entities both genetically and epigenetically. Their evolution is also dynamic in spatial organisation both locally and throughout the body, as well as temporally throughout the disease course. Sampling different parts of the same tumour may therefore reveal striking differences in the genetic and epigenetic make up of the cancer cells at each site (Bashashati et al., 2013, 2010, 2014, 2012, 2013, 2013). Similarly, sampling the tumour at different time‐points, for example after disease progression, might reveal genetic evolution or differences in the clonal composition of the tumour as the disease progresses (Keats et al., 2012; Landau et al., 2013).
Cancer therapy constitutes a uniquely defined, often stringent, directional selection pressure in the evolution of a tumour (Almendro et al., 2014; Merlo et al., 2006), and on this basis, the molecular mechanisms for resistance will vary with the therapeutic agent in question. Furthermore, for any given therapy there may be multiple mechanisms of resistance (reviewed elsewhere (Blair et al., 2014; Holohan et al., 2013) and in other reviews in this issue of Molecular Oncology).
In this review, we will examine studies that have monitored tumour evolution and subclonal tumour architecture over the course of treatment. We will review the evidence that acquired drug resistance can frequently be attributed to the selection of pre‐existing intrinsically resistant subclones, and consider the possibility of de novo generation of resistance‐conferring mutations. The genetic and molecular mechanisms of resistance to targeted therapies are particularly well described, and will be used in this review to illustrate how intratumour genetic heterogeneity contributes to drug resistance. We conclude by considering the challenges faced in light of tumour heterogeneity in devising therapeutic strategies, as well as in the interpretation of clinical trial data.
2. Sources of heterogeneity in tumours
Intratumour heterogeneity can be observed at many different levels, and may be attributable to a number of different factors. Heterogeneity occurs first at the cellular level (intercellular heterogeneity), but with the selective outgrowth of any given cell clone, varying degrees of clonal heterogeneity may arise. Subclones may expand and evolve in a sequential linear fashion, or else may continue to diverge, following branched evolutionary trajectories (Figure 1) (Burrell and Swanton, 2014). There are many recent reviews summarising the evidence for intratumour heterogeneity and different modes of cancer evolution that are beyond the scope of this article (Burrell et al., 2013; Greaves and Maley, 2012; Marusyk et al., 2012; Merlo et al., 2006; Swanton, 2012; Yates and Campbell, 2012) and we will present only a brief summary here.
Figure 1.
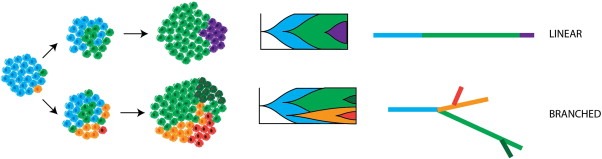
Linear and Branched Cancer Evolution. Schematic illustrating different patterns of cancer evolution. Intercellular heterogeneity followed by clonal selection leads to outgrowth of one or more subclones. If the emerging subclone outcompetes the rest of the tumour cell population, this is described as a clonal sweep, and the subclonal genotype has ‘fixed’ in the population. In linear evolution, subclones arise sequentially (top panel), while if divergent subclones emerge independently then evolution is branched (bottom panel). Incomplete clonal sweeps will generate clonal heterogeneity, which can arise in both linear and branch evolutionary trajectories. Subclonal genotypes allow the monitoring of tumour evolution over time.
Diversity in tumours is evident at the genetic, epigenetic, transcriptomic and proteomic level (Burrell et al., 2013; Marusyk et al., 2012). Genomic instability can affect DNA sequence, chromosome structure and chromosome number, with some forms of instability compromising genome integrity at multiple levels simultaneously (Burrell et al., 2013). Genomic instability, a feature of a high proportion of solid tumours, generates a high level of intercellular genetic heterogeneity (Lengauer et al., 1998) and has been linked with both drug resistance and poor prognosis in cancer (Holohan et al., 2013; Lee et al., 2011; Swanton et al., 2009). Epigenetic, transcriptomic and proteomic heterogeneity may arise due to underlying genotypic variation, but can also reflect cell cycle stage, stochastic variation between cells, or hierarchical organisation of cells according to the cancer stem cell theory (Arora et al., 2013; Gupta et al., 2011; Kreso et al., 2013; Meacham and Morrison, 2013; Nathanson et al., 2014).
In addition, diverse phenotypes can result from extrinsic factors such as pH, hypoxia, and paracrine signalling interactions with stromal and other tumour cells (Gatenby et al., 2010; Junttila and de Sauvage, 2013). Extrinsic factors can directly generate phenotypic diversity, through the modulation of cellular signalling, but also act as selection pressures, supporting clonal expansion of those cells that proliferate efficiently in a given micro‐environmental context. The effects of extrinsic factors, including exogenous carcinogens such as tobacco, may be evident both at a limited local level and more widely, as reflected by apparent organ‐specific features of metastatic tumour lesions (Campbell et al., 2010; Yachida et al., 2010; Yates and Campbell, 2012).
Clusters of mutations defining the genotypes of individual subclonal populations within a tumour can be tracked over time, giving insights into how tumours evolve in the face of different selection pressures. This is perhaps most pertinent to those pressures acting at metastatic sites, and those imposed by treatment. Even if genotype is not always directly responsible for the expansion of a given subclone, genotype tracking enables the monitoring of subclonal architecture over time. Clonal heterogeneity may be evident within single samples (biopsies for solid tumours, blood samples for haematopoietic malignancies) (Anderson et al., 2011; Landau et al., 2013; Lohr et al., 2014; Nik‐Zainal et al., 2012; Snuderl et al., 2011; Szerlip et al., 2012), and can also be observed between different tumour regions within the same primary or between primary and metastatic sites (regional heterogeneity) (Figure 2) (Bashashati et al., 2013, 2010, 2014, 2012, 2009, 2013, 2012, 2010). In a cohort of ten renal cell carcinomas, subclones appeared to be relatively spatially separated and, in general, more heterogeneity and more driver events were discovered as the number of biopsies increased (Gerlinger et al., 2014; Ricketts and Linehan, 2014). Examining temporally separated samples, mostly in haematopoietic cancers, has revealed that clonal heterogeneity can vary substantially over time, likely influenced by multiple factors including genetic drift, drug treatment and the acquisition of new driver mutations (Figure 2) (Bolli et al., 2014; Haffner et al., 2013; Keats et al., 2012; Landau et al., 2013; Lohr et al., 2014; Schuh et al., 2012; Swanton, 2012).
Figure 2.
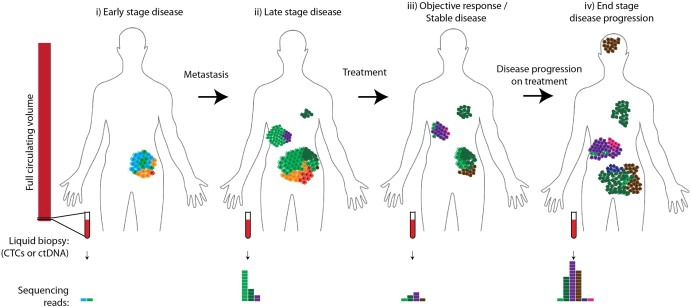
Temporal and Spatial Heterogeneity in Cancer Progression. i) The primary tumour is composed of multiple subclones; biopsies of any one part would therefore be subject to sampling bias. ii) Multiple subclones (light and dark green clones) have seeded metastases, and metastatic clones have continued to evolve (emergence of purple subclone). In the primary tumour the ancestral blue subclone is now extinct. iii) Following treatment, differential clonal response has occurred, resulting in an objective clinical response or stable disease. Orange, red and light green clones have all responded either partially or completely to treatment, while pre‐existing resistant clones (dark green, purple, brown) have continued to expand during treatment. A resistant clone (pink) has developed de novo during treatment. iv) Clinical disease progression has occurred – purple, pink, dark green and brown clones have all expanded, and a new brain metastasis has been seeded (brown). A further new drug resistant clone has emerged (blue). Biopsies taken at different timepoints in disease progression or from different sites of disease would be subject to sampling bias. Assaying circulating tumour DNA (ctDNA) may help minimise this bias, and can be performed longitudinally (bottom panel). ctDNA concentration increases with disease burden and is higher in metastatic than localised disease (Bettegowda et al., 2014). However, as only a small fraction of the circulating volume is sampled, individual subclones may not be detected (this sampling bias also affects analysis of haematopoetic malignancies) and furthermore it is not clear how uniformly different subclones shed DNA into the circulation.
Together these studies indicate that even with ultra‐deep‐sequencing and collecting multiple samples over both time and space, we are likely to be grossly underestimating the degree of intratumour genetic heterogeneity in some cases, where only a small fraction of the total tumour burden is subject to deep sequencing analysis (Figure 2). The impact of such heterogeneity upon outcome and drug response is not yet fully understood, but an increasing number of studies are describing dynamic, and sometimes unexpected, changes in the subclonal architecture during treatment, with reports of multiple distinct resistant clones arising within the same tumour.
3. Monitoring clonal evolution during cancer therapy
Therapy represents a very defined and stringent selection pressure during the evolution of a cancer (Greaves and Maley, 2012; Merlo et al., 2006), and several studies have now traced the clonal evolution of tumours during the course of treatment. Due to the comparative ease of longitudinal sampling, these studies have, to date, mostly been in haematopoietic malignancies such as chronic myeloid leukaemia and multiple myeloma (Bolli et al., 2014; Ding et al., 2012; Keats et al., 2012; Lohr et al., 2014; Mullighan et al., 2008; Schuh et al., 2012). There have been only a limited number of studies of the clonal evolution of solid tumours pre‐ and post‐treatment (Bashashati et al., 2013; Castellarin et al., 2013; Johnson et al., 2014; Shi et al., 2014). The relationship between different modes of tumour evolution and patient outcome is as yet unclear (Bolli et al., 2014; Landau et al., 2013). In view of this, a large clinical study has been set up that aims to track the subclonal dynamics of primary non‐small cell lung cancer through the disease course in patients (TRACERx – clinicaltrials.gov NCT01888601). If the evolutionary trajectories of cancers are affected by treatment in a relatively predictable manner, it might be possible to take advantage of this in devising the optimal therapeutic strategy to prolong relapse free survival, exploiting evolutionary dead‐ends.
3.1. Acute lymphoblastic leukaemia (ALL)
An early study of 61 cases of ALL (Mullighan et al., 2008), found that 58 per cent of relapsed disease samples lacked some copy number alterations detected in the diagnostic sample, implying that the relapsed clone was ancestral to the diagnostic clone. Indeed backtracking analyses revealed that the relapsed ancestral clone was often present as a minor subpopulation at diagnosis. This indicates branched evolution of ALL, and demonstrates the influence of treatment upon the evolutionary trajectory of ALL, by removing the incumbent clone. Branched evolution and shifts in clonal architecture between diagnosis and relapse have subsequently been demonstrated in an independent ALL study (Anderson et al., 2011). In this study, which used multiplex fluorescence in‐situ hybridisation (FISH) to examine 30 cases of ALL, five cases had paired pre‐ and post‐treatment samples. Clonal architecture at relapse was distinct to that observed at diagnosis, with relapse deriving from either major or minor subclones. There was some evidence to suggest that relapse could be driven by more than one subclone, and in addition that the dominant clone at relapse continues to evolve, acquiring new genetic lesions.
3.2. Acute myeloid leukaemia (AML)
Whole‐genome sequencing (WGS) was used to compare diagnostic and relapse samples from 8 AML patients (Ding et al., 2012). Two patterns of clonal evolution were observed at relapse after chemotherapy: either a new mutation was found to have fixed in the original founding clone at relapse, or else a subclonal population of the founding clone was identified with new mutations. Chemotherapy always failed to eradicate the founding clone. An increased frequency of DNA base transversions at relapse versus diagnosis was suggestive of therapy‐induced mutagenesis, raising the possibility that therapy influences cancer evolution through the direct induction of mutations.
3.3. Chronic lymphocytic leukaemia (CLL)
At least two studies have now been published that have longitudinally examined the evolution of CLL through the disease course, including treatment (Landau et al., 2013; Schuh et al., 2012). CLL is a slow‐growing malignancy, with a very variable disease course, that may not require treatment for several years. In the earlier of these two studies, WGS of multiple samples from three patients revealed fluctuating subclonal architecture with therapy, to differing extents at different points in the disease course (Schuh et al., 2012).
The second study (Landau et al., 2013) used whole‐exome sequencing (WES) and copy number analysis to examine 18 CLL patients with two temporally separated samples (12 patients were treated, 6 untreated). Ten out of the 12 treated cases underwent clonal evolution, in contrast to 1 of the 6 untreated cases. 5 of the cases undergoing clonal evolution displayed branched evolution, while the remaining 5 underwent linear clonal evolution (see Figure 1), illustrating the selection of one or more subclones during treatment (2, 3). Interestingly, when comparing unpaired treated and untreated samples across a larger cohort of 160 cases, there was enrichment for subclonal driver mutations in treated versus untreated tumours, and an apparent increase in subclonal complexity (at the level of larger, expanded subclones). The presence of subclonal driver mutations was also an independent risk factor for disease progression. Together with observations of clonal evolution during treatment, this suggests that treatment might select for more aggressive subclones (harbouring driver mutations), potentially through relieving interclonal competition following therapeutic targeting of the dominant clone (Landau et al., 2013).
Figure 3.
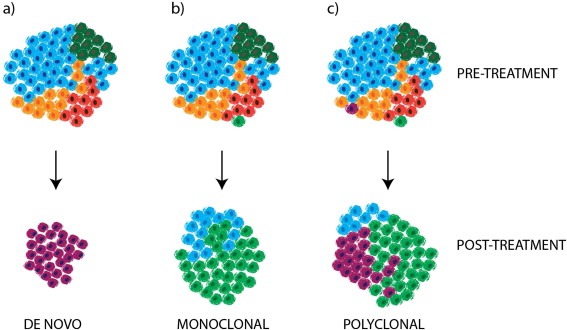
Clonal Evolution and Drug Resistance. Acquired drug resistance may emerge as a consequence of the selective expansion of: a) the de novo development of resistance in a previously sensitive clone (e.g. as a consequence of treatment induced mutagenesis, or tumour‐intrinsic genomic instability) b) a pre‐existing resistant minor subclone or c) multiple pre‐existing or de novo resistant subclones, which may be present across one or multiple sites of disease.
3.4. Multiple myeloma
The complex and dynamic nature of multiple myeloma subclonal architecture has now been examined in multiple studies (Bolli et al., 2014; Keats et al., 2012; Lohr et al., 2014). The earliest of these studies performed serial genomic analyses using comparative genomic hybridisation (CGH), and described a particularly striking example of dynamic subclonal architecture, with one case that was sampled at seven different timepoints showing alternating clonal dominance between two subclones during therapy, followed by dramatic linear evolution of a subclone that had been barely detectable at diagnosis, but which dominated the disease at death (Keats et al., 2012). In a cohort of 15 cases, various different patterns of clonal evolution were observed during treatment (Bolli et al., 2014). In some cases new subclones emerged through either linear (2/15) or branched evolution (4/15), while in four other cases existing subclones displayed differential (and occasionally alternating) responses, consistent with the observations of Keats and colleagues (Keats et al., 2012). An interesting subset of 5 patients displayed stable clonal architecture over time, even though they had been treated and some had shown a substantial therapeutic response. This suggests all subclones were equivalently affected by therapy in these cases. Interestingly there was no apparent relationship between the mode of evolution and treatment response or survival, although the cohort was small so this conclusion warrants validation in a larger cohort. There was, however, evidence for selection of clones bearing driver mutations over time, consistent with observations in CLL (Landau et al., 2013). Similarly, a third study found that mutations which were significantly recurrent across 203 patients were more commonly clonal in previously treated versus untreated cases, implying that treatment might select for driver mutations that were initially present subclonally (Lohr et al., 2014).
3.5. Longitudinal studies of clonal evolution during treatment in solid tumours
Relatively few studies of clonal evolution during treatment have been undertaken in solid tumours. This is likely to be due to difficulties in sampling; biopsies of metastases may not be clinically indicated or immediately accessible and there may be multiple metastases, precluding full sampling of the tumour's genomic landscape (Figure 2). In many cases, only specific resistance mutations have been examined with the expectation that the drug resistance mutation identified is responsible for therapeutic failure at all sites of disease and in all subclones, limiting the capacity to draw conclusions about modes of clonal evolution.
Clonal evolution has been analysed by WES in BRAF‐mutant melanoma patients at disease progression on BRAF inhibitors, for whom multiple geographically and/or temporally separated biopsies were available (Shi et al., 2014). In depth analysis of one patient (for whom two baseline and nine progressive tumour samples were available) revealed that all nine progressive sites of disease had followed branched rather than linear evolutionary trajectories. Each progressive lesion harboured unique mutations not observed elsewhere or at baseline, and different lesions also harboured distinct drug resistance mechanisms. Branched evolution at relapse after treatment was also demonstrated in three further patients, again revealing distinct drug resistance mechanisms in different lesions in some patients. Interestingly, expression of the proliferation marker Ki67 was dramatically higher in progressive disease, in keeping with selection for fitter, highly proliferative disease during treatment.
In ovarian cancer a small number of tumours have been analysed over the course of treatment (Bashashati et al., 2013; Castellarin et al., 2013). In three patients with high‐grade serous ovarian carcinoma (HGSC), tumour cells harvested from ascites at the primary presentation and two subsequent, post‐chemotherapy recurrences were analysed by WES (Castellarin et al., 2013). Clustering mutations according to mutant allele frequencies revealed that all three tumours harboured multiple subclones in the primary sample (Castellarin et al., 2013), consistent with a second study that sampled multiple regions from five HGSCs (Bashashati et al., 2013). One case displayed complex clonal dynamics, with one clone increasing in frequency between primary and first relapse sample, while another disappeared (Castellarin et al., 2013). Two tumours had relatively stable clonal architecture over time, although in both cases some mutations decreased or increased in frequency between primary and relapse samples, implying some degree of clonal evolution (Castellarin et al., 2013). Stable clonal architecture was also observed in a longitudinally sampled tumour in a separate study (Bashashati et al., 2013). Across both studies, in all four cases, chemotherapy failed to destroy the major clones, inferred by the disappearance of fewer than 10 per cent of mutations after treatment (Bashashati et al., 2013; Castellarin et al., 2013). This is similar to findings in AML (Ding et al., 2012). Perhaps surprisingly, given the amount of DNA damaging chemotherapy all four patients received, relatively few new mutations were observed. This suggests that resistance was derived from outgrowth of a pre‐existing resistant subclone, or else selective persistence of existing clones (Figure 3). An alternative possibility is that therapy‐induced or intrinsic genomic instability can generate chromosomal rearrangements, rather than point mutations, which drive the emergence of resistant disease. It remains to be seen whether the patterns of evolution observed in this small number of patients will be representative of HGSC evolution through treatment more generally.
A recent study of recurrent gliomas found that they are often ancestral to the dominant clone at surgical excision of the primary tumour, with frequent branched evolution during adjuvant therapy (Johnson et al., 2014). In contrast to the above cases of ovarian cancer, and similar to observations in AML (Ding et al., 2012), treatment has been found to profoundly alter the course of genome evolution in glioma (Hunter et al., 2006; Johnson et al., 2014). The alkylating agent temozolomide (TMZ) is often given as adjuvant therapy following surgical excision of low‐grade gliomas, but disease recurrence is common. Recurrent TMZ‐treated gliomas have frequently been found to have a hypermutation phenotype, associated with inactivation of the mismatch repair pathway, and display a mutational profile consistent with induction by alkylating agents (Hunter et al., 2006; Johnson et al., 2014). Six recurrent gliomas that harboured TMZ‐induced hypermutation all underwent malignant progression to glioblastoma multiforme (GBM), and in each case TMZ was implicated in mutations affecting genes in the RB and Akt‐mTOR signalling pathways (Johnson et al., 2014). Non‐hypermutated recurrent tumours that progressed to GBM also acquired mutations in these pathways, but through alternative mechanisms, supporting the notion that derangement of these pathways is key in malignant progression, and that TMZ may drive this process in some tumours (Figure 3).
4. Drug resistance and cancer evolution
There are many factors that influence drug response. However, pharmacokinetic factors, such as drug absorption in the gut, while generally important in drug response are unlikely to be a major factor explaining any relationship between ITH and drug resistance, as it seems likely that intratumour heterogeneity affects variation in drug response predominantly at the cellular level. An exception to this cell‐intrinsic mode of resistance would be the role of tumour cells in determining stromal architecture and vascularisation in the immediate local environment, since this may influence drug delivery at a regional level within tumours, and such micro‐environmental heterogeneity may also generate phenotypic diversity among tumour cells.
At the level of the tumour cell, there are multiple different routes through which drug resistance might be achieved: drugs may not be taken up efficiently, efflux may be upregulated, or drugs may be readily inactivated or metabolised within the cell. Alternatively, cells may alter the drug target itself, or rewire cellular signalling so as to negate the effect of the drug, which is particularly relevant to targeted therapies (Blair et al., 2014; Holohan et al., 2013). This can be achieved in a number of different ways, such as epigenetic modulation, point mutation, amplification, deletion or down‐regulation of the target itself or another gene, often in the same or a parallel signalling pathway (Blair et al., 2014; Holohan et al., 2013).
Mechanisms of resistance to targeted therapies have been particularly well studied in this regard, since acquired resistance to these drugs is almost universally observed in advanced disease and they act upon a genetically defined target and signalling pathway (Blair et al., 2014). Figure 4 summarises some of the described mechanisms of resistance to some targeted therapies. Some chemotherapy agents, such as the anthracyclines and anti‐metabolites, also target specific enzymes, and in vitro studies suggest that resistance to these may be also achieved through target alteration, although it is not known whether this occurs in tumours (Bugg et al., 1991; Sugimoto et al., 1990). In contrast, other chemotherapy drugs, such as cisplatin and mitomycin C, directly generate extensive DNA damage. Resistance to such agents cannot therefore be achieved by modifying the drug target, but must depend instead on mechanisms of resistance acting up‐ or down‐stream of the DNA damage, for example reducing cellular uptake of drug or modulating DNA repair (Edwards et al., 2008; Holohan et al., 2013; Lord and Ashworth, 2012).
Figure 4.
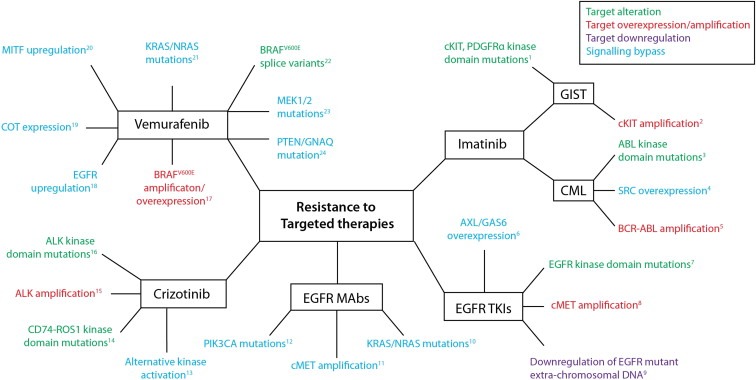
Mechanisms of resistance to targeted therapies. Schematic summarising some of the described mechanisms of resistance to a selection of targeted therapies: Vemurafenib (BRAFV600E inhibitor), Imatinib (BCR‐ABL/cKIT/PDGFRA), EGFR tyrosine kinase inhibitors (e.g. gefitinib, erlotinib), EGFR targeted monoclonal antibodies (e.g. cetuximab, panitumumab), Crizotinib (ALK, ROS1 inhibitor). 1. (Heinrich et al., 2006; Liegl et al., 2008; Lim et al., 2008; Wardelmann et al., 2006) 2. (Debiec‐Rychter et al., 2005; Heinrich et al., 2006) 3. (Shah et al., 2002) 4. (Mahon et al., 2008) 5. (Gorre et al., 2001) 6. (Zhang et al., 2012) 7. (Inukai et al., 2006; Kosaka et al., 2006; Maheswaran et al., 2008) 8. (Turke et al., 2010) 9. (Nathanson et al., 2014) 10. (Amado et al., 2008; Bardelli et al., 2013; Diaz et al., 2012; Misale et al., 2012) 11. (Bardelli et al., 2013; Engelman et al., 2007) 12. (Sartore‐Bianchi et al., 2009) 13. (Katayama et al., 2012) 14. (Awad et al., 2013) 15. (Katayama et al., 2012) 16. (Choi et al., 2010; Katayama et al., 2012) 17. (Shi et al., 2014; Shi et al., 2012; Van Allen et al., 2014) 18. (Prahallad et al., 2012) 19. (Johannessen et al., 2010) 20. (Johannessen et al., 2013; Van Allen et al., 2014) 21. (Nazarian et al., 2010; Shi et al., 2014; Van Allen et al., 2014) 22. (Poulikakos et al., 2011; Shi et al., 2012) 23. (Shi et al., 2014; Van Allen et al., 2014) 24. (Turajlic et al., 2014).
An important question in understanding cancer evolution during treatment is whether the drug resistant cells that lead to disease progression are 1) present prior to treatment 2) generated directly by treatment; or 3) generated during (but independently of) treatment and finally 4) whether the resistant cells are homogeneous (clonal) within individual patients (Figure 3).
4.1. Selection of pre‐existing resistant subclones in heterogeneous tumours
It is thought that acquired resistance often reflects positive selection of pre‐existing subclones harbouring resistance‐conferring mutations (2, 3) (Diaz et al., 2012; Gerlinger and Swanton, 2010). Direct evidence for such minor populations comes from a limited number of studies in various tumour types including lung (Inukai et al., 2006; Maheswaran et al., 2008; Su et al., 2012; Turke et al., 2010), melanoma (Van Allen et al., 2014) and chronic myeloid leukaemia (CML) (Roche‐Lestienne et al., 2002; Shah et al., 2002), probably due to the challenges of detecting such low frequency resistant cell populations. Far more commonly, the identified resistance mutation(s) is not detected in the pre‐treatment sample (Bettegowda et al., 2014; Diaz et al., 2012; Inukai et al., 2006; Kosaka et al., 2006; Liegl et al., 2008; Misale et al., 2014; Shi et al., 2014; Wagle et al., 2014), although in many cases, it has been proposed that these relapse‐specific mutations were nevertheless likely to have been present in small populations of cells pre‐treatment, a view that has been supported by mathematical modelling (Diaz et al., 2012).
Imatinib is commonly used for the treatment of CML expressing the fusion protein BCR‐ABL, and in the treatment of gastro‐intestinal stromal tumours (GIST) harbouring mutant cKIT and PDGFRA. In CML, the most common mechanism of resistance is the acquisition of secondary mutations in ABL (Blair et al., 2014; Gorre et al., 2001; Shah et al., 2002), with 29 of 32 CML patients who relapsed after an initial response to imatinib harbouring kinase domain mutations (Figure 4) (Shah et al., 2002). In two studies, cytogenetic non‐responders (who had initial haematological responses) were found to have pre‐treatment mutations (Roche‐Lestienne et al., 2002; Shah et al., 2002), which were apparently clonally selected during treatment. Two of four patients in blast crisis that failed to respond to imatinib were also found to have pre‐existing ABL kinase domain mutations, supporting the notion that primary and secondary resistance mechanisms are largely similar (Shah et al., 2002). In GIST, secondary mutations in the cKIT kinase domain have been identified in a high proportion of tumours relapsing after treatment with imatinib or sunitinib (a multi‐target kinase inhibitor, with activity against both cKIT and PDGFRA) (Figure 4) (Liegl et al., 2008; Lim et al., 2008; Wardelmann et al., 2006), which were not detected in the pre‐treatment sample tumour.
The selection of resistant minor subclones has been observed in the context of EGFR (epithelial growth factor receptor) tyrosine kinase inhibitor treatment in lung cancer (gefitinib, erlotinib), where the most common mechanism of resistance is the acquisition of a secondary gatekeeper mutation in the ATP binding pocket of the tyrosine kinase domain of EGFR (T790M) (Figure 4) (Inukai et al., 2006; Kosaka et al., 2006; Maheswaran et al., 2008). T790M mutations have been detected in untreated samples using sequencing approaches that enrich for the mutation (Inukai et al., 2006; Maheswaran et al., 2008; Su et al., 2012), and the detection of a pre‐treatment T790M mutation has been found to be associated with reduced progression‐free survival (Maheswaran et al., 2008; Su et al., 2012). Rare pre‐treatment cells (<1%) harbouring MET amplification have also been identified in four lung cancers that went on to develop resistance to EGFR inhibitors through MET amplification (Turke et al., 2010), as well as in a lung cancer cell line that developed resistance via MET amplification. Interestingly, in experimental systems, the selective outgrowth of rare MET‐amplified cells seemed to be enhanced by hepatocyte growth factor (HGF) exposure (Turke et al., 2010).
In glioblastoma, tumours have been identified that contain mixed populations of cells harbouring either PDGFR, MET or EGFR amplification (Snuderl et al., 2011; Szerlip et al., 2012). In vitro, cell lines derived from co‐amplified tumours required inhibition of both EGFR and PDGFR in order to inhibit signalling via the PI3K‐AKT‐mTOR pathway (Szerlip et al., 2012). This suggests that PDGFR‐amplified cells in co‐amplified glioblastomas might contribute to resistance to EGFR inhibition, although this awaits clinical validation.
Also in the context of EGFR blockade, but this time with monoclonal antibodies in colon cancer, KRAS mutations (Figure 4) were identified in circulating tumour DNA from sera of 9/24 patients that developed resistance. Mathematical modelling, based on the time taken for emergence of drug resistance, suggested that KRAS mutant subclones were present in tumours prior to treatment (Diaz et al., 2012).
4.2. De novo acquisition of resistance mutations
As described above, there are relatively few studies that have identified defined resistance‐conferring mutations in pre‐treatment samples. This raises the possibility that at least in some cases, these mutations may arise de novo during treatment (Figure 3). However, it is challenging to definitively ascertain the extent to which resistant subclones are generated during treatment. Even in cases where pre‐treatment samples were sequenced to very high depth (Bolli et al., 2014; Wagle et al., 2014) and still failed to detect the emerging subclone, it is still entirely plausible that the resistant subclone was present and was not sampled (either due to regional heterogeneity in solid tumours or limited sample collection size for haematopoietic malignancies) or alternatively that the subclone was present, but not sufficiently expanded to be detectable by population‐based methodologies such as sequencing (Figure 2). Further development of single‐cell sequencing approaches could improve the sensitivity of detection of resistant subclones within samples (although it will not eliminate potential problems of sampling bias between spatially separated regions of the same tumour), but at this point, the technology is not sufficiently high throughput and robust.
The contributions of genomic instability and mutagenic therapies to the ongoing evolution of tumours should not be overlooked, however (Burrell et al., 2013; Cahill et al., 1999; Ding et al., 2012; Gerlinger and Swanton, 2010; Johnson et al., 2014). As described in Sections 3.2 and 3.5, chemotherapy has been found to contribute to mutagenesis in AML and glioma (Ding et al., 2012; Johnson et al., 2014). In glioma, mutations induced through a combination of mutagenic TMZ therapy and acquired genomic instability (mismatch repair deficiency) appear to affect key pathways involved in the progression from glioma to GBM (Johnson et al., 2014). However, in other studies tumours displayed limited evidence of new mutations following treatment with mutagenic therapy (Bashashati et al., 2013; Castellarin et al., 2013). Nevertheless, this provides support in principle for the notion that mutations acquired through therapy could lead to therapeutic resistance (Figure 3).
In turn, this suggests that de novo mutations arising during therapy as a consequence of tumour‐intrinsic (as opposed to therapy‐induced) genomic instability could contribute to drug resistance (Cahill et al., 1999; Gerlinger and Swanton, 2010; Nowell, 1976). Coupled with the increased likelihood of pre‐existing resistant clones in the context of genomic instability, this could explain the relationship between chromosomal instability (possibly the commonest form of genomic instability) and both drug resistance and poor outcome (Lee et al., 2011; McGranahan et al., 2012). Based on experimental studies of evolution in microbial populations, the increased mutation rate conferred by genomic instability is likely to be particularly advantageous in small populations, by decreasing the wait time for a beneficial mutation to be acquired (Arjan et al., 1999; Burrell and Swanton, 2014; Sprouffske et al., 2012). The contribution of genomic instability to drug resistance could therefore be particularly pertinent in the adjuvant setting following surgery for localised disease, given the relatively small size of the tumour cell population at this point in the disease course.
4.3. Selection versus rapid adaptation
Selection of resistant subclones, whether pre‐existing or generated de novo, is likely to be a major, if not the major, contributor to therapeutic acquired resistance. However, it is important to note that there are alternative dynamic routes to resistance that do not require such selection. Feedback re‐wiring of cellular signalling networks in response to targeted therapies, in the absence of any requirement for new mutations, is an example of such adaptive change (Arora et al., 2013; Blair et al., 2014; Prahallad et al., 2012). For example, it has recently been shown that glucocorticoid receptor expression can lead to resistance to androgen receptor inhibition in prostate cancer cells (Arora et al., 2013). Androgen receptor signalling normally mediates repression of glucocorticoid receptor expression, and the relief of this repression upon treatment leads to induction of glucocorticoid receptor expression in a subset of cells. This has potential implications for the use of steroids in the management of prostate cancer (Arora et al., 2013).
In another example of adaptive change in the face of treatment, it has recently been found that EGFR mutant glioblastoma cells harbour copies of mutant EGFR in reservoirs of extra‐chromosomal DNA (Nathanson et al., 2014). Upon treatment with EGFR inhibitors, EGFR mutant extra‐chromosomal DNA was essentially eliminated, reducing the level of expression and achieving resistance to the effects of therapy. This effect was mirrored by the reduction in the percentage of tumour cells exhibiting high EGFR expression following treatment with EGFR inhibitors. Upon removal of EGFR inhibition, the extra‐chromosomal copies of mutant EGFR re‐emerged. In this instance, therefore, impressive genomic plasticity enables drug resistance to be rapidly achieved. Comparable to this effect, a transient drug‐resistant state has been observed in cultured cells treated with a range of different agents, which appeared to be mediated via IGF‐1 receptor signalling and an altered chromatin state. The drug tolerant population of cells could be selectively targeted with IGF‐1 receptor inhibitors or chromatin‐modifying agents (Sharma et al., 2010). Similarly, a recent pre‐clinical study in ALL identified a key role for chromatin modulation in resistance to NOTCH1 inhibitors, which could be overcome by inhibiting the chromatin regulator BRD4 (Knoechel et al., 2014). The ability to modulate adaptive drug resistance through targeting chromatin regulators lends hope to novel combination therapies.
5. Polyclonal evolution of drug resistance
Given that cancers have been shown to evolve both in a branched and linear fashion (Figure 1), it is reasonable to suppose that this will also be reflected in the emergence of drug resistance, with multiple distinct resistant subclones arising in a patient's tumour. The observation that recurrent disease after chemotherapy is composed of multiple subclones in AML, ALL, CLL, multiple myeloma, glioma and ovarian cancer, suggests that polyclonal resistance might be a common mode of resistance to chemotherapy (see Section 3, above). The specific genetic lesions involved in resistance in these cases remains unclear.
In the case of many targeted therapies, our knowledge of molecular and genetic mechanisms of acquired resistance is quite detailed (Figure 4), particularly given the relatively short time for which these agents have been in clinical use. This understanding has been gleaned through our knowledge of the targeted pathways, as well as from pre‐clinical models, and the ability to use rationally designed, targeted sequencing approaches to identify resistance mechanisms. This has enabled studies in which pre‐ and post‐treatment resistant samples are assayed for a range of different resistance mechanisms. In an increasing number of cases, this is revealing polyclonal resistance, whereby multiple distinct resistance‐conferring mutations are identified in the same tumour (examples are summarised in Table 1). The simultaneous emergence of multiple resistance mechanisms indicates branched tumour evolution during treatment (1, 3).
Table Table 1.
Clinical studies identifying polyclonal genetic mechanisms of drug resistance.
| Inhibitor | Target | Tumour type | Resistance mechanisms identified | Description – Patients with multiple resistance mechanisms | Study |
|---|---|---|---|---|---|
| Imatinib | BCR‐ABL | Chronic Myeloid Leukaemia | BCR‐ABL secondary mutations | Multiple resistant clones identified in 12/32 patients | (Shah et al., 2002) |
| cKIT/PDGFRA | Gastro‐intestinal stromal tumours | Secondary KIT mutations | 4 patients, multiple different secondary KIT mutations | (Wardelmann et al., 2006) | |
| 2 patients with different KIT mutations at different progression sites | (Lim et al., 2008) | ||||
| 6/11 KIT mutant GISTs had 2‐5 secondary mutations in different lesions3/11 had 2 secondary KIT mutations in same lesion | (Liegl et al., 2008) | ||||
| Cetuximab/Panitumumab | EGFR | Colon | KRAS mutations | 3 of 9 patients that developed resistance had >1 mutation in KRAS (2 with 2 mutations, 1 with 4 mutations). | (Diaz et al., 2012) |
| KRAS, NRAS, BRAF, EGFR mutations | Average number of mutations detected per patient = 2.9 (range 0–12) | (Bettegowda et al., 2014) | |||
| KRAS, NRAS mutations | 2 cases – multiple KRAS variants2 cases – NRAS and KRAS variants | (Misale et al., 2014) | |||
| Vemurafenib/Dabrafenib | BRAFV600E | Melanoma | Mutations: NRAS, KRAS, MAP2K1 Other: BRAF amplificationBRAF splice variants | 9/44 patients with at least 2 mechanisms of resistanceIn 16 patients with multiple samples 13/16 had multiple mechanisms of resistance | (Shi et al., 2014) |
| NRAS, MEK1, MEK2 mutations.BRAF amp | 3 of 23 patients with known resistance mutations identified showed multiple alterations | (Van Allen et al., 2014) | |||
| Crizotinib | ALK | Lung | ALK secondary mutations | Patient with 2 independent subclones with different ALK secondary mutations | (Choi et al., 2010) |
| ALK secondary mutations, amplification.KIT amplification, EGFR activation | 3 patients with multiple mechanisms | (Katayama et al., 2012) |
5.1. Polyclonal resistance to targeted therapies
An early appreciation of the potential complexity of resistance to targeted agents came with the report of CML patients harbouring multiple distinct mutations in the ABL kinase domain following disease progression on imatinib treatment (Shah et al., 2002). In 12 of 32 patients developing resistance to imatinib, more than one resistant clone was identified, with 2–4 clones identified per patient. BCR‐ABL amplification was also observed in 2 of these polyclonally resistant patients (Figure 4). This has been mirrored in GISTs treated with imatinib (Liegl et al., 2008; Lim et al., 2008; Wardelmann et al., 2006). At recurrence, patients with GISTs often undergo debulking surgery for symptom relief, due to the growth of multiple tumour lesions within the abdomen. This enables the sampling of multiple sites of disease. In three studies, patients have been identified that have distinct secondary mutations in KIT at different sites of disease (Liegl et al., 2008; Lim et al., 2008; Wardelmann et al., 2006) (Table 1), as well as within the same lesion (Liegl et al., 2008). Some cases of recurrent disease did not harbour secondary KIT mutations, suggesting further heterogeneity with alternative mechanisms driving resistance. In lung cancer patients with tumours bearing ALK rearrangements, resistance after crizotinib treatment appeared to be driven by multiple mechanisms within the same patient, including secondary ALK kinase domain mutations, as well as amplification of the mutant fusion gene or KIT (Table 1) (Choi et al., 2010; Katayama et al., 2012).
There have also been reports of polyclonal resistance to EGFR blockade with monoclonal antibodies (Bettegowda et al., 2014, 2012, 2014, 2012) in colorectal cancer (Table 1). Three of these studies assessed disease recurrence by sequencing circulating tumour DNA (ctDNA) (Bettegowda et al., 2014; Diaz et al., 2012; Misale et al., 2014). In samples analysed specifically for mutations in KRAS codons 12 and 13 (associated with resistance to EGFR inhibition (Amado et al., 2008)), 3 of 9 patients that developed KRAS mutations in their sera following panitumumab treatment harboured more than one KRAS mutation (Diaz et al., 2012). Two simultaneous distinct KRAS mutations were also identified at low allele frequencies in a metastatic tumour sample (Misale et al., 2012). In more recent studies in which multiple different resistance mechanisms, including NRAS, BRAF and EGFR mutations in addition to KRAS, were assayed, as many as 12 co‐occurring mutations were identified in a single patient (Bettegowda et al., 2014; Misale et al., 2014). Taken together, these data suggest that accurately assessing the frequency of polyclonal drug resistance is limited by the number of resistance mechanisms assessed, the depth of sequencing and the quantity of tumour DNA assessed in resistant samples.
The importance of the effects of both investigational and sampling bias is reinforced by recent studies in RAF inhibitor resistant BRAF‐mutant melanoma (Shi et al., 2014; Van Allen et al., 2014). In a cohort of 16 patients from whom multiple samples were available at disease progression, 13 (81%) harboured multiple mechanisms of resistance (Shi et al., 2014). This was in contrast to 9 of 44 patients (20%) when patients for whom only one recurrence sample was available were also included. While this indicates heterogeneity between different sites of disease, multiple mechanisms of resistance have also been identified within the same biopsy in melanomas (Romano et al., 2013; Shi et al., 2014; Van Allen et al., 2014; Wilmott et al., 2012). Detailed analysis (WES) of the phylogeny of nine progressive tumours relative to two baseline biopsies for one patient revealed that all nine progressive tumours underwent branched evolution (Shi et al., 2014). Four distinct characterised resistance mechanisms (and probably at least one further mechanism) contributed to relapse in this case. A single biopsy would have substantially underestimated the complexity of resistance in this case, and in other case reports of RAF inhibitor resistant melanoma (Romano et al., 2013; Wilmott et al., 2012). In a recently described case of metastatic melanoma, resistance was conferred by two mutations in PTEN and GNAQ, found ubiquitously throughout all sites of disease (Turajlic et al., 2014). This illustrates a general theme of drug resistance studies: intrinsic resistance may be driven by ubiquitous or ‘truncal’ mutations, while acquired drug resistance can be driven by subclonal mutations, often located on the branches of tumour phylogenetic trees, resulting in frequent polyclonality of resistance mechanisms (Figure 5).
Figure 5.
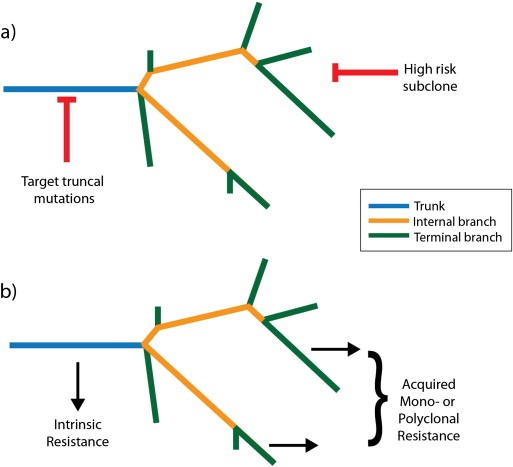
Strategies for targeting heterogeneous tumours. A) Effective therapeutic strategies for targeting heterogeneous tumours could include targeting clonal, or truncal mutations, or else targeting high‐risk subclones – this might be particularly efficacious in the context of adjuvant therapy. B) Intrinsic resistance is likely to be driven by truncal mutations, while acquired resistance is likely to be driven by one or more subclonal mutations (branched mutations in branched evolution, nested subclones in linear evolution – see Figure 1).
In summary, there are now multiple reported cases, in various malignancies, of drug resistance being driven by more than clone. At present we are probably underestimating the true frequency of polyclonal drug resistance, due to studies being confined either to the interrogation of a limited number of resistance mechanisms, or else a limited number of samples. Furthermore, non‐genetic mechanisms of resistance are frequently not assessed, for example the generation of BRAF splice variants or gene overexpression (Figure 4). Thus whether those cases in which only one resistance mechanism is identified are truly monoclonal requires further investigation. One additional possibility is that more than one clone harbours the same resistance mutation, creating an illusion of monoclonality when only this mutation is assayed in resistant samples.
5.2. Phenotypic convergence of polyclonal resistance mechanisms
One common theme of the above studies is that the same resistance mechanisms are detected recurrently across patients, even if multiple different mechanisms are identified in the same patient. This suggests that, at least in the case of targeted therapies, there might be a limited armoury of resistance mechanisms that cancer cells can draw upon, indicating significant constraints to tumour evolution and drug resistance. In addition, these mechanisms often converge upon similar phenotypic outcomes, for example reactivation of MAPK and ERK signalling upon development of resistance to BRAF or EGFR inhibitors (Blair et al., 2014). Such convergence is observed across different tumour types, between patients, and within individual patients between subclones present at the same or multiple sites of disease, and is perhaps unsurprising given the relatively defined selection pressure of drug treatment, particularly targeted therapies.
This is consistent with reports of parallel evolution of distinct tumour subclones bearing mutations in the same pathways that are expected to result in some degree of phenotypic convergence, now observed in multiple tumours (Bolli et al., 2014, 2014, 2012, 2014, 2014). Such phenotypic convergence could lend hope to strategies that aim to target the effect of the various resistance mechanisms, rather than specific alterations themselves (Misale et al., 2014). However, in general relapse samples have been assessed for a focussed set of mutations, rather than being subject to relatively unbiased WGS or WES approaches, and it remains possible that there are still many unknown mechanisms of resistance, including non‐genetic mechanisms. It is also possible that the end point of the convergence of these disparate resistance lesions will be non‐specific attributes (i.e. the hallmarks of cancer (Hanahan and Weinberg, 2011)) that are difficult to target, other than with chemotherapy.
6. The challenges of treating heterogeneous tumours
Inter‐ and intra‐tumour heterogeneity pose significant challenges to cancer therapy, both to our ability to personalise therapy, but also in the development of drug resistance (Fedele et al., 2014; Gerlinger and Swanton, 2010; Yap et al., 2012). In addition, genomic instability and mutagenic therapy contribute to the on‐going dynamic evolution of tumours (Almendro et al., 2014; Hunter et al., 2006; Johnson et al., 2014), conceivably enabling constant shifting of the therapeutic goalposts (Burrell et al., 2013; Burrell and Swanton, 2014; Cahill et al., 1999).
6.1. Identifying actionable mutations – when to treat?
Efforts to target therapy to specific genetic lesions depend upon the identification of these aberrations in biopsies prior to treatment. In the face of heterogeneity, detection of a given ‘actionable’ mutation may be confounded by sampling bias (Gerlinger et al., 2014, 2012), such that detection in one sample does not mean that the mutation is ubiquitously found in every tumour subclone throughout all geographical sites of the tumour, potentially limiting the efficacy of treatment. Conversely absence of a lesion does not mean that it is not found in other, un‐sampled, subclones (Figure 2).
It has been proposed by our group and others that targeting ubiquitous mutations, present on the trunk of the tumour's evolutionary tree, might be the optimal therapeutic strategy (Figure 5) (Gerlinger et al., 2012; Yap et al., 2012). Nevertheless, as discussed above, the likelihood of (potentially multiple) subclonal resistance‐conferring mutations being present is high, rendering even the targeting of clonally dominant lesions challenging (Figure 5). Patients with subclonal actionable mutations are also likely to derive some benefit from therapy, especially if the targeted subclone is aggressive. This might be particularly relevant in combination with surgery, whereby adjuvant targeting of potentially lethal tumour cell populations (whether clonal or subclonal in the primary tumour) leads to improved outcome (Figure 5) (Burrell et al., 2010). In some situations, targeting one subclone could disrupt synergistic functional interactions between genotypically distinct clonal populations, such as have been reported in glioblastoma cells (Inda et al., 2010). Liquid biopsies, which assay mutation status from either circulating tumour cells or circulating tumour DNA may help in the detection of subclonal mutations (Bettegowda et al., 2014; Maheswaran et al., 2008; Murtaza et al., 2013) without the need for multiple biopsies (Figure 2).
However, it is also possible that targeting a subclonal driver mutation could have a negative impact upon patient outcome, by accelerating the growth of other clones. BRAF inhibitors paradoxically activate ERK signalling in BRAF wild‐type cells (Poulikakos et al., 2010), and this effect may be even more pronounced in cells bearing KRAS or NRAS mutations (Lohr et al., 2014). The observation of squamous cell carcinomas and new primary melanomas forming during BRAF inhibitor treatment underscores the strength of this effect (Sosman et al., 2012; Zimmer et al., 2012). Since BRAF, KRAS and NRAS mutations may be subclonal in the same patient in multiple myeloma (Lohr et al., 2014), this sounds a note of caution for the use of BRAF inhibitors in this setting, and underscores the need to understand the effects of targeted therapies in different genetic backgrounds.
Clinical trials will now be needed to assess the potential benefit of treating both clonal (or truncal) and subclonal (or branched) mutations. Combination therapy may provide some benefit in tumours composed of multiple subclones. However, it must be borne in mind that parallels with anti‐microbial or anti‐retroviral therapy, where polychemotherapy regimens are often the norm, cannot necessarily be extended to cancer. In the former examples, drug therapy is aimed at targeting processes within the microbe or virus that have diverged from humans in evolutionary terms, billions of years ago. Cancers have diverged from the host within only years, with somatic aberrations leading to relatively subtle cellular phenotypic changes, resulting in narrower therapeutic indices and rendering multiple targeting strategies more prone to side effects. There is therefore a limit to the number of drugs that can be taken simultaneously for toxicity reasons. In addition, frequently subclones do not harbour currently ‘actionable’ mutations. It is likely that the combinations of subclonal drivers within the same tumour will be distinct from patient to patient, rendering conventional drug development strategies to assess efficacy of combination approaches challenging. Sampling bias and intratumour heterogeneity may further complicate decisions based on the presence or absence of actionable or resistance‐conferring mutations.
6.2. Pre‐empting resistance – a tractable approach?
The elucidation of common mechanisms of resistance to targeted therapies has led to the consideration of pre‐emptively treating tumours in order to prevent the development of resistance. For example, treating melanomas with both a BRAF inhibitor and a MEK inhibitor (since many mechanisms of resistance converge upon ERK/MEK re‐activation) has been shown to lead to a modest improvement in progression free survival compared to BRAF inhibitor monotherapy (Flaherty et al., 2012). However, patients still progress, so resistance evidently develops to combination therapy, albeit later than to monotherapy. Thus while this strategy may offer improvement, it is unlikely to be a solution in most cases. Resistance mechanisms to this particular combination therapy nevertheless do appear to converge upon reactivation of MAPK signalling (Wagle et al., 2014), suggesting there is scope for optimising this strategy.
However, it is conceivable that prospectively drugging common resistance mechanisms might not improve overall survival, even if progression free survival is lengthened in the short term (Cunningham et al., 2011; Gillies et al., 2012). Taking the example of T790M mutations in EGFR mutant lung cancer, which are now druggable (Walter et al., 2013), one option might be to treat with inhibitors that can target the T790M kinase from the outset, to prevent this clone from expanding. However, learning lessons from existing studies of the clonal evolution of tumours during drug treatment, and the many examples of polyclonal resistance to targeted agents, it seems likely that resistance via another route will then emerge. Since patients with subclonal T790M mutations do derive some benefit from EGFR inhibitor treatment, even though their time to progression is shorter (Maheswaran et al., 2008; Su et al., 2012), treating sequentially, or potentially in an alternating fashion, might confer longer progression free survival intervals overall. The goal of treatment would then be to maintain the tumour in a ‘treatable’ state for as long as possible, rather than seeking dramatic initial responses, selecting for subclones for which no therapeutic modality exists.
Anticipating both the development of resistance, and the likely mechanism (coupled with knowledge of how to treat the resistant disease) might therefore be superior to treating the drug resistance mechanism pre‐emptively, although this hypothesis requires formal assessment. Needless to say, there are complexities in predicting resistance based simply on the presence of resistance mutations pre‐treatment. For example, pre‐treatment MEK1 mutations have been associated with rapid progression in BRAF‐mutant melanoma, and have emerged in drug resistant samples, yet other patients with pre‐treatment MEK1 mutations have derived benefit from RAF inhibition (Van Allen et al., 2014). It has been suggested that dynamic regulation of MEK1 expression could contribute to this variability (Van Allen et al., 2014), while it is also possible that epistasis plays a role in this and other similar contexts.
Further prospective trials are therefore required to examine the best strategy for employing our knowledge of polyclonal drug resistance mechanisms in the presence of intratumour heterogeneity to targeted therapies.
6.3. Can treatment exacerbate cancer evolution and drug resistance?
It is possible that in some cases, cancer therapy does not merely shape cancer evolution, but actually accelerates cancer progression. Perhaps the clearest example of treatment exacerbating disease progression to date is in TMZ‐treated glioma, where mutations in key pathways for malignant progression can be attributed, with a reasonable confidence, to the mutagenic effect of therapy (Johnson et al., 2014). The mutagenic effect of therapy or of acquired genomic instability, might drive further divergence between tumour subclones, generating greater intratumour heterogeneity, and further increasing the likelihood of resistance to future therapies.
Studies of CLL and multiple myeloma have also provided evidence for the hypothesis that therapy might select for more aggressive disease, with driver mutations present sub‐clonally prior to treatment appearing to be selected during therapy (Bolli et al., 2014; Landau et al., 2013; Lohr et al., 2014). In CLL, it was suggested that this could be due to the relief of interclonal competition when the dominant disease clone is removed by treatment (Landau et al., 2013). Studies of microbial evolution suggest that interclonal competition (or clonal interference) is a significant factor in shaping the evolution of exponentially growing populations (Lang et al., 2013; Sprouffske et al., 2012). Thus targeting individual subclones, or even the dominant clone, could lead to the emergence of more aggressive populations of tumour cells, previously restrained by clonal interference, and raises the question of whether targeting the dominant clone is always the most sensible therapeutic strategy from the point of view of tumour evolution (Cunningham et al., 2011, 2009, 2010).
7. Future perspectives
The increasing number of studies revealing polyclonal resistance to both targeted therapies and chemotherapy presents a substantial challenge to our current models for dealing with drug resistance, and suggests that efforts to target single mechanisms of drug resistance with second generation inhibitors may have limited success. However, phenotypic convergence of multiple mechanisms of resistance upon one or a few signalling pathways lends hope to the development of strategies to manage intratumour heterogeneity during the acquisition of drug resistance. More unbiased studies using WES or WGS of samples pre‐ and post‐treatment will improve our estimate of the frequency of such polyclonal resistance. Given that chemotherapy is still the mainstay of treatment for many tumours, there is a striking lack of studies examining resistance to these agents since the advent of next generation sequencing, and our understanding of the most common molecular mechanisms of resistance to these agents remains poor.
There is also clearly a need to better understand the evolutionary dynamics of tumours during treatment with longitudinal studies such as TRACERx, such that the optimal therapeutic strategies for life‐prolonging therapy can be devised. Mathematical modelling approaches have suggested that these may not be the most obvious strategies (Zhao et al., 2014), or those therapies that lead to dramatic (but short term) therapeutic responses, but rather those that achieve evolutionary equilibrium. Liquid biopsies of ctDNA or circulating tumour cells will be key in facilitating the monitoring of solid tumours with inaccessible metastases over time, not least because they represent an acceptable test that could be used clinically in the future. However, given sensitivity issues with ctDNA in some tumour types (Bettegowda et al., 2014) and our lack of knowledge as to how representative circulating tumour DNA is of heterogeneous tumours, direct biopsies of progressive lesions should be taken where possible. Warm autopsy studies are also likely to be key in this regard.
Resistance is an inevitable part of treatment with any agent in most advanced solid tumours, and therefore clinical trials of new and existing agents should have tissue collection protocols built in to expedite the study of resistance mechanisms and reduce the lag phase between the identification of a potentially successful therapy and the understanding of resistance, such that second generation inhibitors can be developed. Pre‐clinical research has been essential to our understanding of drug resistance mechanisms, and should ideally be integrated into clinical trial set‐up to ensure efficient transfer of knowledge from bench to bedside. The development of better pre‐clinical models, recapitulating intratumour heterogeneity of human tumours (McFadden et al., 2014), will improve the contribution of these studies to our understanding of drug resistance and cancer progression.
Acknowledgements
C.S. is a senior Cancer Research UK (Grant codes A19310 and A17786) clinical research fellow and is funded by Cancer Research UK, the Rosetrees Trust, EU FP7 (projects PREDICT and RESPONSIFY, ID:259303), the Prostate Cancer Foundation, the European Research Council and the Breast Cancer Research Foundation. Research is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Burrella Rebecca A., Swantona Charles, (2014), Tumour heterogeneity and the evolution of polyclonal drug resistance, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.06.005.
Contributor Information
Rebecca A. Burrell, Email: r.burrell@ucl.ac.uk
Charles Swanton, Email: charles.swanton@cancer.org.uk.
References
- Almendro, V. , Cheng, Y.K. , Randles, A. , Itzkovitz, S. , Marusyk, A. , Ametller, E. , Gonzalez-Farre, X. , Munoz, M. , Russnes, H.G. , Helland, A. , 2014. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep.. 6, 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado, R.G. , Wolf, M. , Peeters, M. , Van Cutsem, E. , Siena, S. , Freeman, D.J. , Juan, T. , Sikorski, R. , Suggs, S. , Radinsky, R. , 2008. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol.: Official Journal of the American Society of Clinical Oncology. 26, 1626–1634. [DOI] [PubMed] [Google Scholar]
- Anderson, K. , Lutz, C. , van Delft, F.W. , Bateman, C.M. , Guo, Y. , Colman, S.M. , Kempski, H. , Moorman, A.V. , Titley, I. , Swansbury, J. , 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 469, 356–361. [DOI] [PubMed] [Google Scholar]
- Arjan, J.A. , Visser, M. , Zeyl, C.W. , Gerrish, P.J. , Blanchard, J.L. , Lenski, R.E. , 1999. Diminishing returns from mutation supply rate in asexual populations. Science. 283, 404–406. [DOI] [PubMed] [Google Scholar]
- Arora, V.K. , Schenkein, E. , Murali, R. , Subudhi, S.K. , Wongvipat, J. , Balbas, M.D. , Shah, N. , Cai, L. , Efstathiou, E. , Logothetis, C. , 2013. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 155, 1309–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad, M.M. , Katayama, R. , McTigue, M. , Liu, W. , Deng, Y.L. , Brooun, A. , Friboulet, L. , Huang, D. , Falk, M.D. , Timofeevski, S. , 2013. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N. Engl. J. Med.. 368, 2395–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli, A. , Corso, S. , Bertotti, A. , Hobor, S. , Valtorta, E. , Siravegna, G. , Sartore-Bianchi, A. , Scala, E. , Cassingena, A. , Zecchin, D. , 2013. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov.. 3, 658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashashati, A. , Ha, G. , Tone, A. , Ding, J. , Prentice, L.M. , Roth, A. , Rosner, J. , Shumansky, K. , Kalloger, S. , Senz, J. , 2013. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J. Pathol.. 231, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda, C. , Sausen, M. , Leary, R.J. , Kinde, I. , Wang, Y. , Agrawal, N. , Bartlett, B.R. , Wang, H. , Luber, B. , Alani, R.M. , 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med.. 6, 224ra24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, B.G. , Bardelli, A. , Park, B.H. , 2014. Somatic alterations as the basis for resistance to targeted therapies. J. Pathol.. 232, 244–254. [DOI] [PubMed] [Google Scholar]
- Bolli, N. , Avet-Loiseau, H. , Wedge, D.C. , Van Loo, P. , Alexandrov, L.B. , Martincorena, I. , Dawson, K.J. , Iorio, F. , Nik-Zainal, S. , Bignell, G.R. , 2014. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun.. 5, 2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg, B.Y. , Danks, M.K. , Beck, W.T. , Suttle, D.P. , 1991. Expression of a mutant DNA topoisomerase II in CCRF-CEM human leukemic cells selected for resistance to teniposide. Proc. Natl. Acad. Sci. U. S. A.. 88, 7654–7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell, R.A. , Juul, N. , Johnston, S.R. , Reis-Filho, J.S. , Szallasi, Z. , Swanton, C. , 2010. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J. Cell. Biochem.. 111, 782–790. [DOI] [PubMed] [Google Scholar]
- Burrell, R.A. , McGranahan, N. , Bartek, J. , Swanton, C. , 2013. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 501, 338–345. [DOI] [PubMed] [Google Scholar]
- Burrell, R.A. , Swanton, C. , 2014. The evolution of the unstable cancer genome. Curr. Opin. Genet. Dev.. 24, 61–67. [DOI] [PubMed] [Google Scholar]
- Cahill, D.P. , Kinzler, K.W. , Vogelstein, B. , Lengauer, C. , 1999. Genetic instability and Darwinian selection in tumours. Trends Cell Biol.. 9, M57–M60. [PubMed] [Google Scholar]
- Campbell, P.J. , Yachida, S. , Mudie, L.J. , Stephens, P.J. , Pleasance, E.D. , Stebbings, L.A. , Morsberger, L.A. , Latimer, C. , McLaren, S. , Lin, M.L. , 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 467, 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin, M. , Milne, K. , Zeng, T. , Tse, K. , Mayo, M. , Zhao, Y. , Webb, J.R. , Watson, P.H. , Nelson, B.H. , Holt, R.A. , 2013. Clonal evolution of high-grade serous ovarian carcinoma from primary to recurrent disease. J. Pathol.. 229, 515–524. [DOI] [PubMed] [Google Scholar]
- Choi, Y.L. , Soda, M. , Yamashita, Y. , Ueno, T. , Takashima, J. , Nakajima, T. , Yatabe, Y. , Takeuchi, K. , Hamada, T. , Haruta, H. , 2010. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med.. 363, 1734–1739. [DOI] [PubMed] [Google Scholar]
- Cunningham, J.J. , Gatenby, R.A. , Brown, J.S. , 2011. Evolutionary dynamics in cancer therapy. Mol. Pharm.. 8, 2094–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debiec-Rychter, M. , Cools, J. , Dumez, H. , Sciot, R. , Stul, M. , Mentens, N. , Vranckx, H. , Wasag, B. , Prenen, H. , Roesel, J. , 2005. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology. 128, 270–279. [DOI] [PubMed] [Google Scholar]
- Diaz, L.A. , Williams, R.T. , Wu, J. , Kinde, I. , Hecht, J.R. , Berlin, J. , Allen, B. , Bozic, I. , Reiter, J.G. , Nowak, M.A. , 2012. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 486, 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L. , Ley, T.J. , Larson, D.E. , Miller, C.A. , Koboldt, D.C. , Welch, J.S. , Ritchey, J.K. , Young, M.A. , Lamprecht, T. , McLellan, M.D. , 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 481, 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, S.L. , Brough, R. , Lord, C.J. , Natrajan, R. , Vatcheva, R. , Levine, D.A. , Boyd, J. , Reis-Filho, J.S. , Ashworth, A. , 2008. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 451, 1111–1115. [DOI] [PubMed] [Google Scholar]
- Engelman, J.A. , Zejnullahu, K. , Mitsudomi, T. , Song, Y. , Hyland, C. , Park, J.O. , Lindeman, N. , Gale, C.M. , Zhao, X. , Christensen, J. , 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 316, 1039–1043. [DOI] [PubMed] [Google Scholar]
- Fedele, C. , Tothill, R.W. , McArthur, G.A. , 2014. Navigating the challenge of tumor heterogeneity in cancer therapy. Cancer Discov.. 4, 146–148. [DOI] [PubMed] [Google Scholar]
- Flaherty, K.T. , Infante, J.R. , Daud, A. , Gonzalez, R. , Kefford, R.F. , Sosman, J. , Hamid, O. , Schuchter, L. , Cebon, J. , Ibrahim, N. , 2012. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med.. 367, 1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby, R.A. , Brown, J. , Vincent, T. , 2009. Lessons from applied ecology: cancer control using an evolutionary double bind. Cancer Res.. 69, 7499–7502. [DOI] [PubMed] [Google Scholar]
- Gatenby, R.A. , Gillies, R.J. , Brown, J.S. , 2010. Evolutionary dynamics of cancer prevention. Nat. Rev. Cancer. 10, 526–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger, M. , Horswell, S. , Larkin, J. , Rowan, A.J. , Salm, M.P. , Varela, I. , Fisher, R. , McGranahan, N. , Matthews, N. , Santos, C.R. , 2014. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet.. 46, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger, M. , Rowan, A.J. , Horswell, S. , Larkin, J. , Endesfelder, D. , Gronroos, E. , Martinez, P. , Matthews, N. , Stewart, A. , Tarpey, P. , 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med.. 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger, M. , Swanton, C. , 2010. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br. J. Cancer. 103, 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies, R.J. , Verduzco, D. , Gatenby, R.A. , 2012. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer. 12, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre, M.E. , Mohammed, M. , Ellwood, K. , Hsu, N. , Paquette, R. , Rao, P.N. , Sawyers, C.L. , 2001. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 293, 876–880. [DOI] [PubMed] [Google Scholar]
- Greaves, M. , Maley, C.C. , 2012. Clonal evolution in cancer. Nature. 481, 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P.B. , Fillmore, C.M. , Jiang, G. , Shapira, S.D. , Tao, K. , Kuperwasser, C. , Lander, E.S. , 2011. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 146, 633–644. [DOI] [PubMed] [Google Scholar]
- Haffner, M.C. , Mosbruger, T. , Esopi, D.M. , Fedor, H. , Heaphy, C.M. , Walker, D.A. , Adejola, N. , Gurel, M. , Hicks, J. , Meeker, A.K. , 2013. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest.. 123, 4918–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. , Weinberg, R.A. , 2011. Hallmarks of cancer: the next generation. Cell. 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Heinrich, M.C. , Corless, C.L. , Blanke, C.D. , Demetri, G.D. , Joensuu, H. , Roberts, P.J. , Eisenberg, B.L. , von Mehren, M. , Fletcher, C.D. , Sandau, K. , 2006. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J. Clin. Oncol.: Official Journal of the American Society of Clinical Oncology. 24, 4764–4774. [DOI] [PubMed] [Google Scholar]
- Holohan, C. , Van Schaeybroeck, S. , Longley, D.B. , Johnston, P.G. , 2013. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 13, 714–726. [DOI] [PubMed] [Google Scholar]
- Hunter, C. , Smith, R. , Cahill, D.P. , Stephens, P. , Stevens, C. , Teague, J. , Greenman, C. , Edkins, S. , Bignell, G. , Davies, H. , 2006. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res.. 66, 3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda, M.M. , Bonavia, R. , Mukasa, A. , Narita, Y. , Sah, D.W. , Vandenberg, S. , Brennan, C. , Johns, T.G. , Bachoo, R. , Hadwiger, P. , 2010. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev.. 24, 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai, M. , Toyooka, S. , Ito, S. , Asano, H. , Ichihara, S. , Soh, J. , Suehisa, H. , Ouchida, M. , Aoe, K. , Aoe, M. , 2006. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res.. 66, 7854–7858. [DOI] [PubMed] [Google Scholar]
- Johannessen, C.M. , Boehm, J.S. , Kim, S.Y. , Thomas, S.R. , Wardwell, L. , Johnson, L.A. , Emery, C.M. , Stransky, N. , Cogdill, A.P. , Barretina, J. , 2010. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 468, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen, C.M. , Johnson, L.A. , Piccioni, F. , Townes, A. , Frederick, D.T. , Donahue, M.K. , Narayan, R. , Flaherty, K.T. , Wargo, J.A. , Root, D.E. , 2013. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 504, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B.E. , Mazor, T. , Hong, C. , Barnes, M. , Aihara, K. , McLean, C.Y. , Fouse, S.D. , Yamamoto, S. , Ueda, H. , Tatsuno, K. , 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 343, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila, M.R. , de Sauvage, F.J. , 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 501, 346–354. [DOI] [PubMed] [Google Scholar]
- Katayama, R. , Shaw, A.T. , Khan, T.M. , Mino-Kenudson, M. , Solomon, B.J. , Halmos, B. , Jessop, N.A. , Wain, J.C. , Yeo, A.T. , Benes, C. , 2012. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci. Transl. Med.. 4, 120ra17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats, J.J. , Chesi, M. , Egan, J.B. , Garbitt, V.M. , Palmer, S.E. , Braggio, E. , Van Wier, S. , Blackburn, P.R. , Baker, A.S. , Dispenzieri, A. , 2012. Clonal competition with alternating dominance in multiple myeloma. Blood. 120, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoechel, B. , Roderick, J.E. , Williamson, K.E. , Zhu, J. , Lohr, J.G. , Cotton, M.J. , Gillespie, S.M. , Fernandez, D. , Ku, M. , Wang, H. , 2014. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet.. 46, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka, T. , Yatabe, Y. , Endoh, H. , Yoshida, K. , Hida, T. , Tsuboi, M. , Tada, H. , Kuwano, H. , Mitsudomi, T. , 2006. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin. Cancer Res.. 12, 5764–5769. [DOI] [PubMed] [Google Scholar]
- Kreso, A. , Dick, J.E. , 2014. Evolution of the cancer stem cell model. Cell Stem Cell. 14, 275–291. [DOI] [PubMed] [Google Scholar]
- Kreso, A. , O'Brien, C.A. , van Galen, P. , Gan, O.I. , Notta, F. , Brown, A.M. , Ng, K. , Ma, J. , Wienholds, E. , Dunant, C. , 2013. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 339, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau, D.A. , Carter, S.L. , Stojanov, P. , McKenna, A. , Stevenson, K. , Lawrence, M.S. , Sougnez, C. , Stewart, C. , Sivachenko, A. , Wang, L. , 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 152, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, G.I. , Rice, D.P. , Hickman, M.J. , Sodergren, E. , Weinstock, G.M. , Botstein, D. , Desai, M.M. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 500, 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.J. , Endesfelder, D. , Rowan, A.J. , Walther, A. , Birkbak, N.J. , Futreal, P.A. , Downward, J. , Szallasi, Z. , Tomlinson, I.P. , Howell, M. , 2011. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res.. 71, 1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer, C. , Kinzler, K.W. , Vogelstein, B. , 1998. Genetic instabilities in human cancers. Nature. 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Liegl, B. , Kepten, I. , Le, C. , Zhu, M. , Demetri, G.D. , Heinrich, M.C. , Fletcher, C.D. , Corless, C.L. , Fletcher, J.A. , 2008. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J. Pathol.. 216, 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K.H. , Huang, M.J. , Chen, L.T. , Wang, T.E. , Liu, C.L. , Chang, C.S. , Liu, M.C. , Hsieh, R.K. , Tzen, C.Y. , 2008. Molecular analysis of secondary kinase mutations in imatinib-resistant gastrointestinal stromal tumors. Med. Oncol.. 25, 207–213. [DOI] [PubMed] [Google Scholar]
- Lohr, J.G. , Stojanov, P. , Carter, S.L. , Cruz-Gordillo, P. , Lawrence, M.S. , Auclair, D. , Sougnez, C. , Knoechel, B. , Gould, J. , Saksena, G. , 2014. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 25, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.J. , Ashworth, A. , 2012. The DNA damage response and cancer therapy. Nature. 481, 287–294. [DOI] [PubMed] [Google Scholar]
- Maheswaran, S. , Sequist, L.V. , Nagrath, S. , Ulkus, L. , Brannigan, B. , Collura, C.V. , Inserra, E. , Diederichs, S. , Iafrate, A.J. , Bell, D.W. , 2008. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med.. 359, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon, F.X. , Hayette, S. , Lagarde, V. , Belloc, F. , Turcq, B. , Nicolini, F. , Belanger, C. , Manley, P.W. , Leroy, C. , Etienne, G. , 2008. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res.. 68, 9809–9816. [DOI] [PubMed] [Google Scholar]
- Marusyk, A. , Almendro, V. , Polyak, K. , 2012. Intra-tumour heterogeneity: a looking glass for cancer?. Nat. Rev. Cancer. 12, 323–334. [DOI] [PubMed] [Google Scholar]
- McFadden, D.G. , Papagiannakopoulos, T. , Taylor-Weiner, A. , Stewart, C. , Carter, S.L. , Cibulskis, K. , Bhutkar, A. , McKenna, A. , Dooley, A. , Vernon, A. , 2014. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 156, 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan, N. , Burrell, R.A. , Endesfelder, D. , Novelli, M.R. , Swanton, C. , 2012. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep.. 13, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham, C.E. , Morrison, S.J. , 2013. Tumour heterogeneity and cancer cell plasticity. Nature. 501, 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo, L.M. , Pepper, J.W. , Reid, B.J. , Maley, C.C. , 2006. Cancer as an evolutionary and ecological process. Nat. Rev. Cancer. 6, 924–935. [DOI] [PubMed] [Google Scholar]
- Misale, S. , Arena, S. , Lamba, S. , Siravegna, G. , Lallo, A. , Hobor, S. , Russo, M. , Buscarino, M. , Lazzari, L. , Sartore-Bianchi, A. , 2014. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal Cancer. Sci. Transl. Med.. 6, 224 ra226 [DOI] [PubMed] [Google Scholar]
- Misale, S. , Yaeger, R. , Hobor, S. , Scala, E. , Janakiraman, M. , Liska, D. , Valtorta, E. , Schiavo, R. , Buscarino, M. , Siravegna, G. , 2012. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 486, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan, C.G. , Phillips, L.A. , Su, X. , Ma, J. , Miller, C.B. , Shurtleff, S.A. , Downing, J.R. , 2008. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 322, 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza, M. , Dawson, S.J. , Tsui, D.W. , Gale, D. , Forshew, T. , Piskorz, A.M. , Parkinson, C. , Chin, S.F. , Kingsbury, Z. , Wong, A.S. , 2013. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 497, 108–112. [DOI] [PubMed] [Google Scholar]
- Nathanson, D.A. , Gini, B. , Mottahedeh, J. , Visnyei, K. , Koga, T. , Gomez, G. , Eskin, A. , Hwang, K. , Wang, J. , Masui, K. , 2014. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 343, 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian, R. , Shi, H. , Wang, Q. , Kong, X. , Koya, R.C. , Lee, H. , Chen, Z. , Lee, M.K. , Attar, N. , Sazegar, H. , 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 468, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S. , Van Loo, P. , Wedge, D.C. , Alexandrov, L.B. , Greenman, C.D. , Lau, K.W. , Raine, K. , Jones, D. , Marshall, J. , Ramakrishna, M. , 2012. The life history of 21 breast cancers. Cell. 149, 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell, P.C. , 1976. The clonal evolution of tumor cell populations. Science. 194, 23–28. [DOI] [PubMed] [Google Scholar]
- Poulikakos, P.I. , Persaud, Y. , Janakiraman, M. , Kong, X. , Ng, C. , Moriceau, G. , Shi, H. , Atefi, M. , Titz, B. , Gabay, M.T. , 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 480, 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos, P.I. , Zhang, C. , Bollag, G. , Shokat, K.M. , Rosen, N. , 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 464, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad, A. , Sun, C. , Huang, S. , Di Nicolantonio, F. , Salazar, R. , Zecchin, D. , Beijersbergen, R.L. , Bardelli, A. , Bernards, R. , 2012. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 483, 100–103. [DOI] [PubMed] [Google Scholar]
- Ricketts, C.J. , Linehan, W.M. , 2014. Intratumoral heterogeneity in kidney cancer. Nat. Genet.. 46, 214–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche-Lestienne, C. , Soenen-Cornu, V. , Grardel-Duflos, N. , Lai, J.L. , Philippe, N. , Facon, T. , Fenaux, P. , Preudhomme, C. , 2002. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 100, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Romano, E. , Pradervand, S. , Paillusson, A. , Weber, J. , Harshman, K. , Muehlethaler, K. , Speiser, D. , Peters, S. , Rimoldi, D. , Michielin, O. , 2013. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin. Cancer Res.. 19, 5749–5757. [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi, A. , Martini, M. , Molinari, F. , Veronese, S. , Nichelatti, M. , Artale, S. , Di Nicolantonio, F. , Saletti, P. , De Dosso, S. , Mazzucchelli, L. , 2009. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res.. 69, 1851–1857. [DOI] [PubMed] [Google Scholar]
- Schuh, A. , Becq, J. , Humphray, S. , Alexa, A. , Burns, A. , Clifford, R. , Feller, S.M. , Grocock, R. , Henderson, S. , Khrebtukova, I. , 2012. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 120, 4191–4196. [DOI] [PubMed] [Google Scholar]
- Shah, N.P. , Nicoll, J.M. , Nagar, B. , Gorre, M.E. , Paquette, R.L. , Kuriyan, J. , Sawyers, C.L. , 2002. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2, 117–125. [DOI] [PubMed] [Google Scholar]
- Shah, S.P. , Morin, R.D. , Khattra, J. , Prentice, L. , Pugh, T. , Burleigh, A. , Delaney, A. , Gelmon, K. , Guliany, R. , Senz, J. , 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 461, 809–813. [DOI] [PubMed] [Google Scholar]
- Sharma, S.V. , Lee, D.Y. , Li, B. , Quinlan, M.P. , Takahashi, F. , Maheswaran, S. , McDermott, U. , Azizian, N. , Zou, L. , Fischbach, M.A. , 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 141, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Hugo, W. , Kong, X. , Hong, A. , Koya, R.C. , Moriceau, G. , Chodon, T. , Guo, R. , Johnson, D.B. , Dahlman, K.B. , 2014. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov.. 4, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Moriceau, G. , Kong, X. , Lee, M.K. , Lee, H. , Koya, R.C. , Ng, C. , Chodon, T. , Scolyer, R.A. , Dahlman, K.B. , 2012. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun.. 3, 724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl, M. , Fazlollahi, L. , Le, L.P. , Nitta, M. , Zhelyazkova, B.H. , Davidson, C.J. , Akhavanfard, S. , Cahill, D.P. , Aldape, K.D. , Betensky, R.A. , 2011. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 20, 810–817. [DOI] [PubMed] [Google Scholar]
- Sosman, J.A. , Kim, K.B. , Schuchter, L. , Gonzalez, R. , Pavlick, A.C. , Weber, J.S. , McArthur, G.A. , Hutson, T.E. , Moschos, S.J. , Flaherty, K.T. , 2012. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med.. 366, 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva, A. , Spiteri, I. , Piccirillo, S.G. , Touloumis, A. , Collins, V.P. , Marioni, J.C. , Curtis, C. , Watts, C. , Tavaré, S. , 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A.. 110, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske, K. , Merlo, L.M. , Gerrish, P.J. , Maley, C.C. , Sniegowski, P.D. , 2012. Cancer in light of experimental evolution. Curr. Biol.. 22, R762–R771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, K.Y. , Chen, H.Y. , Li, K.C. , Kuo, M.L. , Yang, J.C. , Chan, W.K. , Ho, B.C. , Chang, G.C. , Shih, J.Y. , Yu, S.L. , 2012. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J. Clin. Oncol.: Official Journal of the American Society of Clinical Oncology. 30, 433–440. [DOI] [PubMed] [Google Scholar]
- Sugimoto, Y. , Tsukahara, S. , Oh-hara, T. , Isoe, T. , Tsuruo, T. , 1990. Decreased expression of DNA topoisomerase I in camptothecin-resistant tumor cell lines as determined by a monoclonal antibody. Cancer Res.. 50, 6925–6930. [PubMed] [Google Scholar]
- Swanton, C. , 2012. Intratumor heterogeneity: evolution through space and time. Cancer Res.. 72, 4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton, C. , Nicke, B. , Schuett, M. , Eklund, A.C. , Ng, C. , Li, Q. , Hardcastle, T. , Lee, A. , Roy, R. , East, P. , 2009. Chromosomal instability determines taxane response. Proc. Natl. Acad. Sci. U. S. A.. 106, 8671–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip, N.J. , Pedraza, A. , Chakravarty, D. , Azim, M. , McGuire, J. , Fang, Y. , Ozawa, T. , Holland, E.C. , Huse, J.T. , Jhanwar, S. , 2012. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. U. S. A.. 109, 3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic, S. , Furney, S.J. , Stamp, G. , Rana, S. , Ricken, G. , Oduko, Y. , Saturno, G. , Springer, C. , Hayes, A. , Gore, M. , 2014. Whole genome sequencing reveals complex mechanisms of intrinsic resistance to BRAF inhibition. Ann. Oncol.: Official Journal of the European Society for Medical Oncology/ESMO. 25, 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turke, A.B. , Zejnullahu, K. , Wu, Y.L. , Song, Y. , Dias-Santagata, D. , Lifshits, E. , Toschi, L. , Rogers, A. , Mok, T. , Sequist, L. , 2010. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 17, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen, E.M. , Wagle, N. , Sucker, A. , Treacy, D.J. , Johannessen, C.M. , Goetz, E.M. , Place, C.S. , Taylor-Weiner, A. , Whittaker, S. , Kryukov, G.V. , 2014. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov.. 4, 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle, N. , Van Allen, E.M. , Treacy, D.J. , Frederick, D.T. , Cooper, Z.A. , Taylor-Weiner, A. , Rosenberg, M. , Goetz, E.M. , Sullivan, R.J. , Farlow, D.N. , 2014. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov.. 4, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, A.O. , Sjin, R.T. , Haringsma, H.J. , Ohashi, K. , Sun, J. , Lee, K. , Dubrovskiy, A. , Labenski, M. , Zhu, Z. , Wang, Z. , 2013. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov.. 3, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardelmann, E. , Merkelbach-Bruse, S. , Pauls, K. , Thomas, N. , Schildhaus, H.U. , Heinicke, T. , Speidel, N. , Pietsch, T. , Buettner, R. , Pink, D. , 2006. Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin. Cancer Res.. 12, 1743–1749. [DOI] [PubMed] [Google Scholar]
- Wilmott, J.S. , Tembe, V. , Howle, J.R. , Sharma, R. , Thompson, J.F. , Rizos, H. , Lo, R.S. , Kefford, R.F. , Scolyer, R.A. , Long, G.V. , 2012. Intratumoral molecular heterogeneity in a BRAF-mutant, BRAF inhibitor-resistant melanoma: a case illustrating the challenges for personalized medicine. Mol. Cancer Ther.. 11, 2704–2708. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Northcott, P.A. , Dubuc, A. , Dupuy, A.J. , Shih, D.J. , Witt, H. , Croul, S. , Bouffet, E. , Fults, D.W. , Eberhart, C.G. , 2012. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 482, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida, S. , Jones, S. , Bozic, I. , Antal, T. , Leary, R. , Fu, B. , Kamiyama, M. , Hruban, R.H. , Eshleman, J.R. , Nowak, M.A. , 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467, 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, T.A. , Gerlinger, M. , Futreal, P.A. , Pusztai, L. , Swanton, C. , 2012. Intratumor heterogeneity: seeing the wood for the trees. Sci. Transl. Med.. 4, 127ps10 [DOI] [PubMed] [Google Scholar]
- Yates, L.R. , Campbell, P.J. , 2012. Evolution of the cancer genome. Nat. Rev. Genet.. 13, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Lee, J.C. , Lin, L. , Olivas, V. , Au, V. , LaFramboise, T. , Abdel-Rahman, M. , Wang, X. , Levine, A.D. , Rho, J.K. , 2012. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet.. 44, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Pritchard, J.R. , Lauffenburger, D.A. , Hemann, M.T. , 2014. Addressing genetic tumor heterogeneity through computationally predictive combination therapy. Cancer Discov.. 4, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, L. , Hillen, U. , Livingstone, E. , Lacouture, M.E. , Busam, K. , Carvajal, R.D. , Egberts, F. , Hauschild, A. , Kashani-Sabet, M. , Goldinger, S.M. , 2012. Atypical melanocytic proliferations and new primary melanomas in patients with advanced melanoma undergoing selective BRAF inhibition. J. Clin. Oncol.: Official Journal of the American Society of Clinical Oncology. 30, 2375–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]


