Abstract
Following the fanfare of initial, often dramatic, success with small molecule inhibitors in the treatment of defined genomic subgroups, it can be argued that the extension of targeted therapeutics to the majority of patients with solid cancers has stalled. Despite encouraging FDA approval rates, the attrition rates of these compounds remains high in early stage clinical studies, with single agent studies repeatedly showing poor efficacy In striking contrast, our understanding of the complexity of solid neoplasms has increased in huge increments, following the publication of large‐scale genomic and transcriptomic datasets from large collaborations such as the International Cancer Genome Consortium (ICGC http://www.icgc.org/) and The Cancer Genome Atlas (TCGA http://cancergenome.nih.gov/). However, there remains a clear disconnect between these rich datasets describing the genomic complexity of cancer, including both intra‐ and inter‐tumour heterogeneity, and what a treating oncologist can consider to be a clinically “actionable” mutation profile. Our understanding of these data is in its infancy and we still find difficulties ascribing characteristics to tumours that consistently predict therapeutic response for the majority of small molecule inhibitors. This article will seek to explore the recent studies of the patterns and impact of mutations in drug resistance, and demonstrate how we may use this data to reshape our thinking about biological pathways, critical dependencies and their therapeutic interruption.
Keywords: Genomics, Therapeutics, Drug resistance
Highlights
Genomic stratification has identified therapeutic biomarkers for some solid tumours.
Resistance to targeted therapies is multifactorial and context specific.
We discuss an overview of recently described clinical resistance mechanisms.
We highlight recent advances in understanding a taxonomy of drug resistance.
We discuss the integration of functional and genomic data to overcome drug resistance.
1.
Following the fanfare of initial, often dramatic, success with small molecule inhibitors in the treatment of defined genomic subgroups, it can be argued that the extension of targeted therapeutics to the majority of patients with solid cancers has stalled. This is despite a drug development program that has prospered with fruitful pipelines producing effective and well tolerated compounds (Rubin EH, 2012). However, despite encouraging FDA approval rates, the attrition rates of these compounds remains high in early stage clinical studies, with single agent studies repeatedly showing poor efficacy (Ivy SP, 2010). In striking contrast, our understanding of the complexity of solid neoplasms has increased in huge increments, following the publication of large‐scale genomic and transcriptomic datasets from large collaborations such as the International Cancer Genome Consortium (ICGC http://www.icgc.org/) and The Cancer Genome Atlas (TCGA http://cancergenome.nih.gov/). In parallel, advances in functional genomics have enabled characterisation of the phenotypic effect of many genes commonly mutated in cancer (Wan PT, 2004; Shalem O 2014).
However, there remains a clear disconnect between these rich datasets describing the genomic complexity of cancer, including both intra‐ and inter‐tumour heterogeneity, and what a treating oncologist can consider to be a clinically “actionable” mutation profile. Our understanding of these data is in its infancy and we still find difficulties ascribing characteristics to tumours that consistently predict therapeutic response for the majority of small molecule inhibitors. This article will seek to explore the recent studies of the patterns and impact of mutations in drug resistance, and demonstrate how we may use this data to reshape our thinking about biological pathways, critical dependencies and their therapeutic interruption.
1. Genomics has opened the door to unexpected therapeutic gains in managing metastatic disease
In trying to appreciate a context for the clinical problem of drug resistance it is important to begin with the notion that sensitivity in treating solid tumours is a relatively novel paradigm. When previously treating metastatic non‐small cell lung cancer or metastatic malignant melanoma, durable responses were the exception to the rule in the setting of cytotoxic chemotherapy (Eigentler TK, 2003; Reck M 2013). The concept of therapeutically relevant genetic subgroups in these elusive diseases has been driven by the discovery and annotation of somatic mutations uncovered by hypothesis‐free genome screens (Stratton MR, 2011). Oncologists treating patients with these same tumour type are now habituated in the use of mutations detected in BRAF, EGFR, or indeed re‐ arrangements in ALK, and more recently ROS or FGFR1, to alter clinical management. This science has changed ideas held by a generation and has been rapidly incorporated into clinical practice (McDermott U NEJM, 2011).
Following on from the success of these examples, the expansion and exploration of large‐scale gene screens by collaborative groups such as TCGA and ICGC has been therapeutically enlightening in many tumour groups. Although not universally acclaimed (Gabor Miklos, 2005) this approach of free exploration, usually of the coding genome, has enabled us a glimpse into the biology of some unusual cancers and often to be surprised by what is uncovered. It could be argued that without this approach we would not be trialling BRAF inhibitors in subtypes of haematological malignancies (hairy cell leukaemia) and in Langerhans cell histiocytosis, both of which harbour activating mutations in BRAF (Haroche J et al., 2013). In addition to providing novel tractable targets in these rare tumours, the large scale “genomic landscape” approach has given us arguably the most complete overview of the heterogeneity and critical signalling dependencies of more common tumours such as breast cancer (Stephens PJ et al., 2012). These findings are now being exploited in gene stratified therapeutic subgroup studies, such as I‐SPY‐2 (http://www.ispy2.org/) and FOCUS4 (http://www.focus4trial.org/), that hope to demonstrate survival benefits in these genetically stratified groups of patients.
Conversely, it is in the use of this technology to explore outlier sensitivities that perhaps more therapeutically relevant biological insights have been gleaned. A perennial question for oncologists is the puzzling phenomenon of the outlier, the patient who responds unusually well to a given therapy. In using genomics to address this question we have learned of examples such as mTOR pathway activating mutations in TSC1 that predispose urothelial tumour patients to unusually durable responses to mTOR inhibition (Iyer G 2012).
Not only has the field explored both common and unusual tumours and found tractable mutations, but the dissection of functional biology surrounding these mutations has enabled progress in therapeutic strategies to exploit the newly identified subgroups. Indeed, in the space of a few years the management of BRAF mutant melanoma has changed so considerably that the focus of debate has now shifted from questioning the complete absence of effective therapeutics, to the most effective manner in which to deploy the multitude of effective MAPK‐directed therapies in order to best delay the emergence of drug resistance (Sullivan RJ, 2013).
2. The emergence of drug resistance is to be universally anticipated with all targeted therapeutics
Despite documented complete remissions and the successful prolongation of both time to progression and overall survival in cancers where pharmacological intervention with targeted therapies is standard of care such as ALK re‐arranged non small cell lung cancer (NSCLC) and BRAF mutant malignant melanoma, resistance to treatment is universal and presents in the form of acquired or intrinsic populations of cancer cells that no longer respond to the therapy (Shi et al., 2014; Wagle et al., 2011). Several recent studies have focussed on these cell populations in order to identify the landscape of genetic and transcriptomic mechanisms of resistance in order to therapeutically circumvent or prevent them emerging.
3. The challenge of the long tail of resistance effectors and clonal heterogeneity
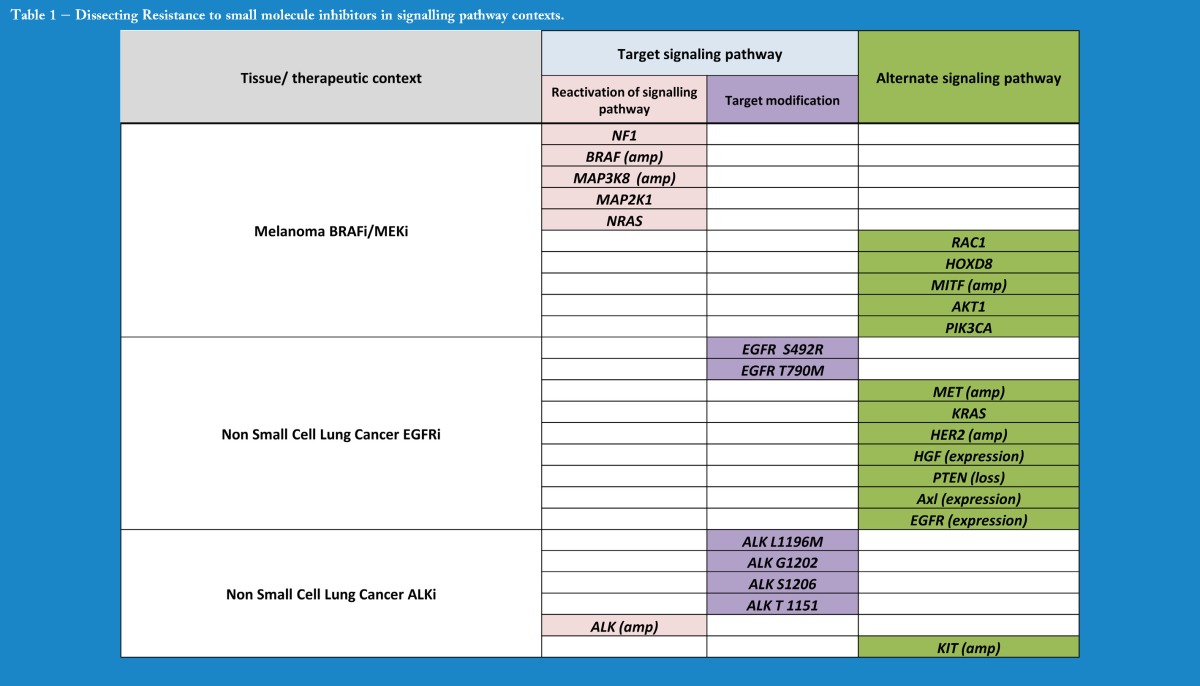
Several pre‐ and post‐therapy biopsy studies have utilised whole exome or whole genome based sequencing approaches to discover resistance effectors in tumour populations that have progressed through therapy. These biologically distinct populations within the tumour retain a proliferative or anti‐apoptotic advantage under the selection pressure of the relevant therapeutic and expand to form the bulk of the progressing tumour, as seen with the emergence of KRAS mutant colorectal cancers in cetuximab treated patients (Misale S 2012). Mutations can serve to either maintain signalling through the preferred oncogenic pathway (despite therapeutic inhibition) or indeed bypass that pathway altogether, and switch dependency to an alternate route (Table 1). In metastatic melanoma, for example, resistance to BRAF inhibitors in the clinic has produced examples that satisfy both of these mechanisms, with mutations in the MAPK pathway that maintain signalling in the face of BRAF inhibition as well as mutations in other pathways that allow the cells to break with their BRAF oncogene addiction (Turajlic S 2014, Van Allen, 2013).
Table 1.
Dissecting Resistance to small molecule inhibitors in signalling pathway contexts.
Such mutations can directly abrogate the function of the target gene itself (Table 1). One of the first studies of this kind in EGFR mutant NSCLC identified “gatekeeper” mutations at residue T790 that change the conformational state of the target kinase pocket, significantly affecting the binding kinetics of the EGFR inhibitor gefitinib (Inukai M 2006). Similar mutations in the ALK kinase domain, or copy number changes in the ALK gene, are seen in acquired resistance to crizotinib in ALK re‐arranged NSCLC (Choi, 2010). Furthermore, mutations or copy number gains that result in high level re‐activation of the target pathway are also seen in BRAF mutant melanoma where COT (MAP3K8), BRAF amplification and NRAS point mutations all result in sustained MAPK activation in the presence of BRAF and/or MEK inhibition (Rizos, 2014; Trunzer K 2013).
This approach has demonstrated conclusively that resistance can be mediated by somatic mutations altering either the pharmacokinetics of the drug itself or modifying the biological consequences of target inhibition. It is presumed that such mutations pre‐date the introduction of therapy and evolve in a Darwinian fashion in the presence of the therapy as an evolutionary bottleneck. Intratumoural genomic heterogeneity has been conclusively demonstrated in clear cell renal carcinoma and presumed to be the substrate for this Darwinian emergence of drug resistant subclones in other tumour types (Gerlinger et al., 2012). Recently, exome screens comparing pre‐ and post‐treatment metastatic tissue in melanoma have confirmed clonal expansion and branched chain evolution of the tumour under BRAF inhibitor selection (Shi H 2014). However, more recent data has shown intriguing evidence that metastatic lesions in ALK re‐arranged NSCLC progressing under crizotinib treatment may indeed be derived from more than one subclone, and that there may be exchanges of tumour cells from non‐contiguous tumour populations – termed “self‐seeding” (Awad et al., 2013).
Such screens have also demonstrated an inherent weakness as they rely heavily on a priori knowledge to identify mutated genes previously annotated in oncogenic pathways. Sample number can be prohibitive in such studies to discover with high confidence novel resistance‐causing variants that present with low frequency. The recent informative study by Van Allen (2014) exploring the clinical landscape to RAF inhibition in melanoma has demonstrated recognisable and recurrent resistance effectors in the majority of the 45 patient cohort, and has highlighted that many tumours fall into the category of harbouring low frequency putative resistance‐causing variants such as HOXD8 and RAC1 that fall outside of the MAPK pathway. This has been termed the “long tail” of resistance effectors, and may explain why 20–30% of these patients have no recognisable resistance‐causing variants, although it is worth noting that somatic mutations are not responsible for alternative splicing of BRAF, or ligand mediated resistance, all of which have been described in this context (Poulikakos et al., 2010; Wilson TR et al., 2012). A similar percentage is quoted in the proportion of crizotinib resistant ALK mutant patients with an unknown resistance mechanism (Doebele RC et al., 2012; Perez CA et al., 2014).
That clinical resistance studies are by necessity underpowered to detect recurrence in infrequently mutated genes poses a specific problem. It has been recently postulated that to detect significant but infrequently mutated genes that recur in under 2% of a tumour, a cohort would require between 900 and 2500 tumour normal pairs depending on the background mutation rate (Garraway and Lander, 2013). This number, while feasible for exploring the genomic architecture of primary tumours, will clearly be challenging to achieve in the context of drug resistance. There will therefore always be a need to have some a priori hypothesis of candidate resistance effector genes to examine in such resistance cohorts, a topic we explore below (Functional approaches exploring modifiers of drug response).
An additional challenge is that posed by recent findings that different drug resistant metastases from within the same patient can harbour not only different mutational mechanisms within the same oncogenic pathway, but also mutations that putatively activate alternate signalling routes. A recent exploration of drug resistant biopsies from a cohort of melanoma patients following treatment with a BRAF inhibitor demonstrated multiple resistance mutations in different genes within separate metastases in the same patient (Shi et al., 2014). In a number of patients mutations activating either MAPK or PI3K signalling were observed in different resistant metastases in the same patient. The implications of these findings are that multiple lesions would require to be biopsied to capture the complete resistance landscape in any single patient, and combinatorial therapy strategies would need to be employed. One potential solution to this problem would be the ability to detect a signal of all of the combined mutations across all metastases by capturing and sequencing circulating cancer DNA (Murtaza et al., 2013). As yet, it remains to be seen whether this is feasible.
4. The challenge of the uncharted genome
Exome based studies are also hampered by the fact that our knowledge of the role the non‐coding genome may play in mediating drug resistance (and which represents 98% of the genome) is still in its infancy. There are recent examples that the parts of the genome that are not explored routinely in drug resistance studies reported to date (largely for reasons of cost) may harbour mutations that are relevant to therapeutics. Two specific nucleotides within the TERT promoter have been shown to be recurrently mutated in over 70% of melanomas and 16% of other tumours producing a novel binding site for the ETS transcription factor (Huang FW et al., 2013). As the number of whole cancer genomes available for analysis world‐wide increases with falling sequencing costs, it is possible that recurrent mutations in these non‐coding regions may attain increased significance in drug resistance cohorts.
Similarly while pleiotropic effects on gene regulation by miRNA have long been thought relevant to drug response, a recent study has specifically shown miRNA effects on the MET oncogene is a factor in resistance to EGFR inhibition in NSCLC (Garofalo et al., 2011). Similarly, miRNA on chromosome 9 that are lost with the tumour suppressor CDKN2A can have a marked effect on response to cisplatin in mesothelioma (Ivanov SV et al., 2010). Our knowledge of the significance of somatic mutations in miRNA genes and target binding sites is still incomplete but already it is clear that germline SNP's in the 3′ UTR binding sites of let‐7a can affect the repression or activity of known oncogenes such as KRAS (Chin LJ, 2008). One can therefore speculate that somatic mutations in non‐coding regions may play an even more complex and multi‐layered role in determining the response to a therapeutic than previously acknowledged.
5. Functional approaches exploring modifiers of drug response
Given the problems and cost associated with uncovering low frequency resistance genes seen in the clinical studies performed to date, novel molecular biological approaches have been utilised to provide an alternative to exploring gene/drug interactions outside the clinical setting.
Forward genetic screens have demonstrated resistance mechanisms that clinical sequencing has not had the ability to detect. Loss of function screens using shRNA have demonstrated EGFR mediated feedback signalling to be a resistance mechanism to BRAF inhibition in BRAF mutant melanoma and colorectal cancer (Prallahad A 2012 Jan 26, 2014 Apr 10). Kinome wide siRNA screens in both melanoma and lung cancer demonstrated that NF1 loss mediates resistance to BRAF inhibition and EGFR inhibition respectively, in both tissues by activating Ras signalling (Whittaker SR, 2013; de Bruin EC et al., 2014). More recently, highly efficient gene knockdown has been achieved using CRISPR/CAS 9 mediated silencing which has independently demonstrated the role of NF1 and NF2 in resistance to BRAF inhibition (Shalem O et al., 2014).
In parallel, advances in gene‐activating open reading frame (ORF) technology has enabled kinome‐ and genome‐wide screens to identify genes whose expression (rather than loss) confers a change in drug response phenotype. One such screen in BRAF mutant melanoma has defined the important role of and MAP3K8 (COT) in mediating resistance to BRAF and MEK inhibition (Johannessen CM et al., 2010). A follow‐up study has defined the role of G‐protein coupled receptors, CREBBP and PKA as part of a lineage specific transcriptional programme in the resistance phenotype for MAPK directed therapies (Johannessen CM et al., 2013).
Such gain‐ and loss of expression screens in cancer have given us valuable insights into resistance mechanisms but are unable to capture specifically those point mutations which account for a number of clinically observed drug resistance mechanisms described to date such as KRAS G12D. To this end the alkylating agent Ethinyl‐nitroso‐urea (ENU) has been used in murine and cell line based phenotype screens by utilising its propensity to cause point mutations stochastically throughout the genome. Crizotinib resistance studies in NSCLC cell lines have demonstrated not only which gatekeeper mutations in the ALK gene enable resistance to crizotinib, but have given an insight into resistance to second generation ALK inhibitors (Zhang S 2011).
The genetic perturbations induced by the means discussed above are susceptible to the criticism that such screens are by their nature artificial and may not reflect phenomena seen in vivo, alongside problems with the reproducibility of certain technologies (Babij C 2011). In this context several large scale cell line based screens seeking to annotate functional characterisation in terms of response to small molecule inhibitors with genomic and transcriptomic biomarkers have been reported recently (Barretina et al., 2012; Garnett MJ et al., 2012) The premise here is that cell autonomous genomic features that modify drug response are so myriad and multi‐layered that one way of gaining insight into such subtleties may be in utilising a great number of features and input cell lines that harbour the same genetic alterations as their parent tumour groups. This effort has been transformative and has identified novel biomarkers as well as known and clinically validated gene–drug interactions (Garnett MJ et al., 2012).
As the technology that underpins such genome‐wide perturbation screens improves, one can imagine a time in the very near future when all targeted agents are routinely tested through such screens to define the putative genetic landscape of drug resistance. Thus, when resistance does occur in the clinic there would be candidate genes to sequence in biopsies obtained upon disease progression and in all likelihood alternate therapeutics identified based on these findings.
6. The future
It is beyond doubt that we have entered the post–genomic age in cancer therapeutics. It is increasingly difficult to consider the care of patients with advanced disease without considering at the same time the molecular taxonomies which have become apparent through next generation sequencing. Classification remains crucial as we continue to move from cumbersome “all comers” late phase clinical studies to rapid moving smaller, enrichment type studies with the knowledge that critical dependencies are indicated by key mutational events and that these likely confer sensitivity to pathway inhibition. Furthermore the introduction of “ basket study” therapeutic trials, such as the recently announced MATCH Trial, where drugs are trialled in a gene rather than tissue specific context is likely to extend the scope of therapeutic intervention outside well established tissue groups (Willyard C 2013).
Beyond this initial stratification however we are learning to utilise these same ideas of a genetic taxonomy to construct a framework for dissecting resistance to treatment. This understanding has already borne fruit in enabling the introduction of second generation drugs effective against the most likely resistance mechanisms likely to occur in a particular context – as seen with Ceritinib in ALK re‐arranged Crizotinib resistant patients (Shaw AT, 2014). Similarly, with the resistance landscape in BRAF mutant melanoma to BRAF inhibitors demonstrating a predilection of MAPK pathway re‐activation, combination MAPK blockade was rapidly trialled improving time to progression and overall survival in BRAF mutant melanoma compared to single agent therapy (Flaherty et al., 2012).
We have arrived at arguably the most exciting time in cancer therapeutics for the last 30 years. Despite those technological advances that now enable one to sequence tumours with greater depth, speed and lower cost than ever before, we are at an inflection point where the philosophy of our approach is changing to reflect the biological questions we deem important. Rather than using genomics in isolation, we suggest that to continue to uncover and respond to resistance mechanisms, we will need to integrate functional approaches to enable the rapid characterisation of phenotype alongside genotype and utilise some of the many approaches discussed above to understand the consequences of somatic mutations in cell lines, animal models and ultimately patients. This will more than ever depend upon our ability to collaborate, and our willingness as a community to share data, resources and expertise.
Alifrangis Constantine C., McDermott Ultan, (2014), Acquired resistance to EGFR‐targeted therapies in colorectal cancer, Molecular Oncolog. 8, doi: 10.1016/j.molonc.2014.05.014.
References
- Awad, M.M. , Engelman, J.A. , Shaw, A.T. , 2013 Sep 19. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N. Engl. J. Med. 369, (12) 1173 http://dx.doi.org/10.1056/NEJMc1309091. [DOI] [PubMed] [Google Scholar]
- Babij, C. , Zhang, Y. , Kurzeja, R.J. , Munzli, A. , Shehabeldin, A. , Fernando, M. , Quon, K. , Kassner, P.D. , Ruefli‐Brasse, A.A. , Watson, V.J. , Fajardo, F. , Jackson, A. , Zondlo, J. , Sun, Y. , Ellison, A.R. , Plewa, C.A. , San, M.T. , Robinson, J. , McCarter, J. , Schwandner, R. , Judd, T. , Carnahan, J. , Dussault, I. , 2011 Sep 1. STK33 kinase activity is nonessential in KRAS‐dependent cancer cells. Cancer Res. 71, (17) 5818–5826. http://dx.doi.org/10.1158/0008-5472.CAN-11-0778. Epub 2011 Jul 8 [DOI] [PubMed] [Google Scholar]
- Barretina, J. , Caponigro, G. , Stransky, N. , Venkatesan, K. , Margolin, A.A. , Kim, S. , Wilson, C.J. , Lehár, J. , Kryukov, G.V. , Sonkin, D. , Reddy, A. , Liu, M. , Murray, L. , Berger, M.F. , Monahan, J.E. , Morais, P. , Meltzer, J. , Korejwa, A. , Jané-Valbuena, J. , Mapa, F.A. , Thibault, J. , Bric-Furlong, E. , Raman, P. , Shipway, A. , Engels, I.H. , Cheng, J. , Yu, G.K. , Yu, J. , Aspesi, P. , de Silva, M. , Jagtap, K. , Jones, M.D. , Wang, L. , Hatton, C. , Palescandolo, E. , Gupta, S. , Mahan, S. , Sougnez, C. , Onofrio, R.C. , Liefeld, T. , MacConaill, L. , Winckler, W. , Reich, M. , Li, N. , Mesirov, J.P. , Gabriel, S.B. , Getz, G. , Ardlie, K. , Chan, V. , Myer, V.E. , Weber, B.L. , Porter, J. , Warmuth, M. , Finan, P. , Harris, J.L. , Meyerson, M. , Golub, T.R. , Morrissey, M.P. , Sellers, W.R. , Schlegel, R. , Garraway, L.A. , 2012 Mar 28. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Natur. 483, (7391) 603–607. http://dx.doi.org/10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, L.J. , Ratner, E. , Leng, S. , Zhai, R. , Nallur, S. , Babar, I. , Muller, R.U. , Straka, E. , Su, L. , Burki, E.A. , Crowell, R.E. , Patel, R. , Kulkarni, T. , Homer, R. , Zelterman, D. , Kidd, K.K. , Zhu, Y. , Christiani, D.C. , Belinsky, S.A. , Slack, F.J. , Weidhaas, J.B. , 2008 Oct 15. A SNP in a let‐7 microRNA complementary site in the KRAS 3' untranslated region increases non‐small cell lung cancer risk. Cancer Res. 68, (20) 8535–8540. http://dx.doi.org/10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.L. , Soda, M. , Yamashita, Y. , Ueno, T. , Takashima, J. , Nakajima, T. , Yatabe, Y. , Takeuchi, K. , Hamada, T. , Haruta, H. , Ishikawa, Y. , Kimura, H. , Mitsudomi, T. , Tanio, Y. , Mano, H. , ALK Lung Cancer Study Group, 2010 Oct 28. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 363, (18) 1734–1739. http://dx.doi.org/10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- de Bruin, E.C. , Cowell, C. , Warne, P.H. , Jiang, M. , Saunders, R.E. , Melnick, M.A. , Gettinger, S. , Walther, Z. , Wurtz, A. , Heynen, G.J. , Heideman, D.A. , Gómez‐Román, J. , García‐Castaño, A. , Gong, Y. , Ladanyi, M. , Varmus, H. , Bernards, R. , Smit, E.F. , Politi, K. , Downward, J. , 2014 May. Reduced NF1 expression confers resistance to EGFR inhibition in lung Cancer. Cancer Discov. 4, (5) 606–619. http://dx.doi.org/10.1158/2159-8290.CD-13-0741. Epub 2014 Feb 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele, R.C. , Pilling, A.B. , Aisner, D.L. , Kutateladze, T.G. , Le, A.T. , Weickhardt, A.J. , Kondo, K.L. , Linderman, D.J. , Heasley, L.E. , Franklin, W.A. , Varella‐Garcia, M. , Camidge, D.R. , 2012 Mar 1. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non‐small cell lung cancer. Clin. Cancer Res. 18, (5) 1472–1482. http://dx.doi.org/10.1158/2159-8290.CD-13-074110.1158/1078-0432.CCR-11-2906. Epub 2012 Jan 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigentler, T.K. , Caroli, U.M. , Radny, P. , Garbe, C. , 2003 Dec. Palliative therapy of disseminated malignant melanoma: a systematic review of 41 randomised clinical trials. Lancet Oncol. 4, (12) 748–759. [DOI] [PubMed] [Google Scholar]
- Flaherty, K.T. , Infante, J.R. , Daud, A. , Gonzalez, R. , Kefford, R.F. , Sosman, J. , Hamid, O. , Schuchter, L. , Cebon, J. , Ibrahim, N. , Kudchadkar, R. , Burris, H.A. , Falchook, G. , Algazi, A. , Lewis, K. , Long, G.V. , Puzanov, I. , Lebowitz, P. , Singh, A. , Little, S. , Sun, P. , Allred, A. , Ouellet, D. , Kim, K.B. , Patel, K. , Weber, J. , 2012 Nov. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 367, (18) 1694–1703. http://dx.doi.org/10.1056/NEJMoa1210093. Epub 2012 Sep. 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor Miklos, G.L. , 2005 May. The human cancer genome project–one more misstep in the war on cancer. Nat. Biotechnol. 23, (5) 535–537. [DOI] [PubMed] [Google Scholar]
- Garnett, M.J. , Edelman, E.J. , Heidorn, S.J. , Greenman, C.D. , Dastur, A. , Lau, K.W. , Greninger, P. , Thompson, I.R. , Luo, X. , Soares, J. , Liu, Q. , Iorio, F. , Surdez, D. , Chen, L. , Milano, R.J. , Bignell, G.R. , Tam, A.T. , Davies, H. , Stevenson, J.A. , Barthorpe, S. , Lutz, S.R. , Kogera, F. , Lawrence, K. , McLaren‐Douglas, A. , Mitropoulos, X. , Mironenko, T. , Thi, H. , Richardson, L. , Zhou, W. , Jewitt, F. , Zhang, T. , O'Brien, P. , Boisvert, J.L. , Price, S. , Hur, W. , Yang, W. , Deng, X. , Butler, A. , Choi, H.G. , Chang, J.W. , Baselga, J. , Stamenkovic, I. , Engelman, J.A. , Sharma, S.V. , Delattre, O. , Saez‐Rodriguez, J. , Gray, N.S. , Settleman, J. , Futreal, P.A. , Haber, D.A. , Stratton, M.R. , Ramaswamy, S. , McDermott, U. , Benes, C.H. , 2012 Mar 28. Systematic identification of genomic markers of drug sensitivity in cancer cells. Natur. 483, (7391) 570–575. http://dx.doi.org/10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo, M. , Romano, G. , Di Leva, G. , Nuovo, G. , Jeon, Y.J. , Ngankeu, A. , Sun, J. , Lovat, F. , Alder, H. , Condorelli, G. , Engelman, J.A. , Ono, M. , Rho, J.K. , Cascione, L. , Volinia, S. , Nephew, K.P. , Croce, C.M. , 2011 Dec 11. EGFR and MET receptor tyrosine kinase‐altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat. Med. 18, (1) 74–82. http://dx.doi.org/10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Garraway, L.A. , Lander, E.S. , 2013 Mar 28. Lessons from the cancer genome. Cel. 153, (1) 17–37. http://dx.doi.org/10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gerlinger, M. , Rowan, A.J. , Horswell, S. , Larkin, J. , Endesfelder, D. , Gronroos, E. , Martinez, P. , Matthews, N. , Stewart, A. , Tarpey, P. , Varela, I. , Phillimore, B. , Begum, S. , McDonald, N.Q. , Butler, A. , Jones, D. , Raine, K. , Latimer, C. , Santos, C.R. , Nohadani, M. , Eklund, A.C. , Spencer‐Dene, B. , Clark, G. , Pickering, L. , Stamp, G. , Gore, M. , Szallasi, Z. , Downward, J. , Futreal, P.A. , Swanton, C. , 2012 Mar 8. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, (10) 883–892. http://dx.doi.org/10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroche, J. , Cohen‐Aubart, F. , Emile, J.F. , Arnaud, L. , Maksud, P. , Charlotte, F. , Cluzel, P. , Drier, A. , Hervier, B. , Benameur, N. , Besnard, S. , Donadieu, J. , Amoura, Z. , Feb 28. 2013. Dramatic efficacy of vemurafenib in both multisystemic and refractory Erdheim‐Chester disease and Langerhans cell histiocytosis harboring the BRAF V600E mutation. Bloo. 121, (9) [DOI] [PubMed] [Google Scholar]
- Huang, F.W. , Hodis, E. , Xu, M.J. , Kryukov, G.V. , Chin, L. , Garraway, L.A. , 2013 Feb 22. Highly recurrent TERT promoter mutations in human melanoma. Scienc. 339, (6122) 957–959. http://dx.doi.org/10.1126/science.1229259. Epub 2013 Jan 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai, M. , Toyooka, S. , Ito, S. , Asano, H. , Ichihara, S. , Soh, J. , Suehisa, H. , Ouchida, M. , Aoe, K. , Aoe, M. , Kiura, K. , Shimizu, N. , Date, H. , 2006 Aug 15. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non‐small cell lung cancer. Cancer Res. 66, (16) 7854–7858. [DOI] [PubMed] [Google Scholar]
- Ivanov, S.V. , Goparaju, C.M. , Lopez, P. , Zavadil, J. , Toren‐Haritan, G. , Rosenwald, S. , Hoshen, M. , Chajut, A. , Cohen, D. , Pass, H.I. , 2010 Jul 23. Pro‐tumorigenic effects of miR‐31 loss in mesothelioma. J. Biol. Chem. 285, (30) 22809–22817. http://dx.doi.org/10.1074/jbc.M110.100354. Epub 2010 May 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy, S.P. , Siu, L.L. , Garrett-Mayer, E. , Rubinstein, L. , 2010 Mar 15. Approaches to phase 1 clinical trial design focused on safety, efficiency, and selected patient populations: a report from the clinical trial design task force of the national cancer institute investigational drug steering committee. Clin. Cancer Res. 16, (6) 1726–1736. http://dx.doi.org/10.1158/1078-0432.CCR-09-1961. Epub 2010 Mar 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, G. , Hanrahan, A.J. , Milowsky, M.I. , Al‐Ahmadie, H. , Scott, S.N. , Janakiraman, M. , Pirun, M. , Sander, C. , Socci, N.D. , Ostrovnaya, I. , Viale, A. , Heguy, A. , Peng, L. , Chan, T.A. , Bochner, B. , Bajorin, D.F. , Berger, M.F. , Taylor, B.S. , Solit, D.B. , 2012 Oct 12. Genome sequencing identifies a basis for everolimus sensitivity. Scienc. 338, (6104) 221 http://dx.doi.org/10.1126/science.1226344. Epub 2012 Aug 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen, C.M. , Boehm, J.S. , Kim, S.Y. , Thomas, S.R. , Wardwell, L. , Johnson, L.A. , Emery, C.M. , Stransky, N. , Cogdill, A.P. , Barretina, J. , Caponigro, G. , Hieronymus, H. , Murray, R.R. , Salehi‐Ashtiani, K. , Hill, D.E. , Vidal, M. , Zhao, J.J. , Yang, X. , Alkan, O. , Kim, S. , Harris, J.L. , Wilson, C.J. , Myer, V.E. , Finan, P.M. , Root, D.E. , Roberts, T.M. , Golub, T. , Flaherty, K.T. , Dummer, R. , Weber, B.L. , Sellers, W.R. , Schlegel, R. , Wargo, J.A. , Hahn, W.C. , Garraway, L.A. , 2010 Dec 16. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Natur. 468, (7326) 968–972. http://dx.doi.org/10.1038/nature09627. Epub 2010 Nov 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen, C.M. , Johnson, L.A. , Piccioni, F. , Townes, A. , Frederick, D.T. , Donahue, M.K. , Narayan, R. , Flaherty, K.T. , Wargo, J.A. , Root, D.E. , Garraway, L.A. , 2013 Dec 5. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Natur. 504, (7478) 138–142. http://dx.doi.org/10.1038/nature12688. Epub 2013 Nov 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott, U. , Downing, J.R. , Stratton, M.R. , 2011 Jan 27. Genomics and the continuum of cancer care. N. Engl. J. Med. 364, (4) 340–350. http://dx.doi.org/10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- Misale, S. , Yaeger, R. , Hobor, S. , Scala, E. , Janakiraman, M. , Liska, D. , Valtorta, E. , Schiavo, R. , Buscarino, M. , Siravegna, G. , Bencardino, K. , Cercek, A. , Chen, C.T. , Veronese, S. , Zanon, C. , Sartore‐Bianchi, A. , Gambacorta, M. , Gallicchio, M. , Vakiani, E. , Boscaro, V. , Medico, E. , Weiser, M. , Siena, S. , Di Nicolantonio, F. , Solit, D. , Bardelli, A. , 2012 Jun 28. Emergence of KRAS mutations and acquired resistance to anti‐EGFR therapy in colorectal cancer. Natur. 486, (7404) 532–536. http://dx.doi.org/10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza, M. , Dawson, S.J. , Tsui, D.W. , Gale, D. , Forshew, T. , Piskorz, A.M. , Parkinson, C. , Chin, S.F. , Kingsbury, Z. , Wong, A.S. , Marass, F. , Humphray, S. , Hadfield, J. , Bentley, D. , Chin, T.M. , Brenton, J.D. , Caldas, C. , Rosenfeld, N. , 2013 May 2. Non‐invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Natur. 497, (7447) 108–112. http://dx.doi.org/10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- Perez, C.A. , Velez, M. , Raez, L.E. , Santos, E.S. , 2014 May. Overcoming the resistance to Crizotinib in patients with Non‐Small Cell Lung Cancer harboring EML4/ALK translocation. Lung Cance. 84, (2) 110–115. http://dx.doi.org/10.1016/j.lungcan.2014.02.001. Epub 2014 Feb 8. [DOI] [PubMed] [Google Scholar]
- Poulikakos, P.I. , Zhang, C. , Bollag, G. , Shokat, K.M. , Rosen, N. , 2010 Mar 18. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild‐type BRAF. Natur. 464, (7287) 427–430. http://dx.doi.org/10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad, A. , Sun, C. , Huang, S. , Di Nicolantonio, F. , Salazar, R. , Zecchin, D. , Beijersbergen, R.L. , Bardelli, A. , Bernards, R. , 2012 Jan 26. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Natur. 483, (7387) 100–103. http://dx.doi.org/10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Reck, M. , Heigener, D.F. , Mok, T. , Soria, J.C. , Rabe, K.F. , 2013 Aug 24. Management of non‐small‐cell lung cancer: recent developments. Lancet 382, (9893) 709–719. http://dx.doi.org/10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- Rizos, H. , Menzies, A.M. , Pupo, G.M. , Carlino, M.S. , Fung, C. , Hyman, J. , Haydu, L.E. , Mijatov, B. , Becker, T.M. , Boyd, S.C. , Howle, J. , Saw, R. , Thompson, J.F. , Kefford, R.F. , Scolyer, R.A. , Long, G.V. , 2014 Apr 1. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin. Cancer Res. 20, (7) 1965–1977. http://dx.doi.org/10.1158/1078-0432.CCR-13-3122. Epub 2014 Jan 24 [DOI] [PubMed] [Google Scholar]
- Rubin, E.H. , Gilliland, D.G. , 2012 Feb 28. Drug development and clinical trials–the path to an approved cancer drug. Nat. Rev. Clin. Oncol. 9, (4) 215–222. http://dx.doi.org/10.1038/nrclinonc.2012.22. [DOI] [PubMed] [Google Scholar]
- Shalem, O. , Sanjana, N.E. , Hartenian, E. , Shi, X. , Scott, D.A. , Mikkelsen, T.S. , Heckl, D. , Ebert, B.L. , Root, D.E. , Doench, J.G. , Zhang, F. , 2014 Jan 3. Genome‐scale CRISPR‐Cas9 knockout screening in human cells. Scienc. 343, (6166) 84–87. http://dx.doi.org/10.1126/science.1247005 Epub 2013 Dec 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A.T. , Kim, D.W. , Mehra, R. , Tan, D.S. , Felip, E. , Chow, L.Q. , Camidge, D.R. , Vansteenkiste, J. , Sharma, S. , De Pas, T. , Riely, G.J. , Solomon, B.J. , Wolf, J. , Thomas, M. , Schuler, M. , Liu, G. , Santoro, A. , Lau, Y.Y. , Goldwasser, M. , Boral, A.L. , Engelman, J.A. , 2014 Mar 27. Ceritinib in ALK‐rearranged non‐small‐cell lung cancer. N. Engl. J. Med. 370, (13) 1189–1197. http://dx.doi.org/10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Hugo, W. , Kong, X. , Hong, A. , Koya, R.C. , Moriceau, G. , Chodon, T. , Guo, R. , Johnson, D.B. , Dahlman, K.B. , Kelley, M.C. , Kefford, R.F. , Chmielowski, B. , Glaspy, J.A. , Sosman, J.A. , van Baren, N. , Long, G.V. , Ribas, A. , Lo, R.S. , 2014 Jan. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 4, (1) 80–93. http://dx.doi.org/10.1158/2159-8290.CD-13-0642. Epub 2013 Nov 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, P.J. , Tarpey, P.S. , Davies, H. , Van Loo, P. , Greenman, C. , Wedge, D.C. , Nik‐Zainal, S. , Martin, S. , Varela, I. , Bignell, G.R. , Yates, L.R. , Papaemmanuil, E. , Beare, D. , Butler, A. , Cheverton, A. , Gamble, J. , Hinton, J. , Jia, M. , Jayakumar, A. , Jones, D. , Latimer, C. , Lau, K.W. , McLaren, S. , McBride, D.J. , Menzies, A. , Mudie, L. , Raine, K. , Rad, R. , Chapman, M.S. , Teague, J. , Easton, D. , Langerød, A. , Oslo Breast Cancer Consortium (OSBREAC), Lee, M.T. , Shen, C.Y. , Tee, B.T. , Huimin, B.W. , Broeks, A. , Vargas, A.C. , Turashvili, G. , Martens, J. , Fatima, A. , Miron, P. , Chin, S.F. , Thomas, G. , Boyault, S. , Mariani, O. , Lakhani, S.R. , van de Vijver, M. , van 't Veer, L. , Foekens, J. , Desmedt, C. , Sotiriou, C. , Tutt, A. , Caldas, C. , Reis‐Filho, J.S. , Aparicio, S.A. , Salomon, A.V. , Børresen‐Dale, A.L. , Richardson, A.L. , Campbell, P.J. , Futreal, P.A. , Stratton, M.R. , 2012 May 16. The landscape of cancer genes and mutational processes in breast cancer. Natur. 486, (7403) 400–404. http://dx.doi.org/10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton, M.R. , 2011 Mar 25. Exploring the genomes of cancer cells: progress and promise. Scienc. 331, (6024) 1553–1558. http://dx.doi.org/10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- Sullivan, R.J. , Flaherty, K.T. , 2013 Apr. Resistance to BRAF‐targeted therapy in melanoma. Eur. J. Cance. 49, (6) 1297–1304. http://dx.doi.org/10.1016/j.ejca.2012.11.019. Epub 2013 Jan 2 [DOI] [PubMed] [Google Scholar]
- Sun, C. , Hobor, S. , Bertotti, A. , Zecchin, D. , Huang, S. , Galimi, F. , Cottino, F. , Prahallad, A. , Grernrum, W. , Tzani, A. , Schlicker, A. , Wessels, L.F. , Smit, E.F. , Thunnissen, E. , Halonen, P. , Lieftink, C. , Beijersbergen, R.L. , Di Nicolantonio, F. , Bardelli, A. , Trusolino, L. , Bernards, R. , 2014 Apr 10. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Re. 7, (1) 86–93. http://dx.doi.org/10.1016/j.celrep.2014.02.045. Epub 2014 Mar 27 [DOI] [PubMed] [Google Scholar]
- Trunzer, K. , Pavlick, A.C. , Schuchter, L. , Gonzalez, R. , McArthur, G.A. , Hutson, T.E. , Moschos, S.J. , Flaherty, K.T. , Kim, K.B. , Weber, J.S. , Hersey, P. , Long, G.V. , Lawrence, D. , Ott, P.A. , Amaravadi, R.K. , Lewis, K.D. , Puzanov, I. , Lo, R.S. , Koehler, A. , Kockx, M. , Spleiss, O. , Schell‐Steven, A. , Gilbert, H.N. , Cockey, L. , Bollag, G. , Lee, R.J. , Joe, A.K. , Sosman, J.A. , Ribas, A. , 2013 May 10. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J. Clin. Oncol. 31, (14) 1767–1774. http://dx.doi.org/10.1200/JCO.2012.44.7888. Epub 2013 Apr 8 [DOI] [PubMed] [Google Scholar]
- Turajlic, S. , Furney, S.J. , Stamp, G. , Rana, S. , Ricken, G. , Oduko, Y. , Saturno, G. , Springer, C. , Hayes, A. , Gore, M. , Larkin, J. , Marais, R. , 2014 May. Whole‐genome sequencing reveals complex mechanisms of intrinsic resistance to BRAF inhibition. Ann. Oncol. 25, (5) 959–967. http://dx.doi.org/10.1093/annonc/mdu049. Epub 2014 Feb 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen, E.M. , Wagle, N. , Sucker, A. , Treacy, D.J. , Johannessen, C.M. , Goetz, E.M. , Place, C.S. , Taylor‐Weiner, A. , Whittaker, S. , Kryukov, G.V. , Hodis, E. , Rosenberg, M. , McKenna, A. , Cibulskis, K. , Farlow, D. , Zimmer, L. , Hillen, U. , Gutzmer, R. , Goldinger, S.M. , Ugurel, S. , Gogas, H.J. , Egberts, F. , Berking, C. , Trefzer, U. , Loquai, C. , Weide, B. , Hassel, J.C. , Gabriel, S.B. , Carter, S.L. , Getz, G. , Garraway, L.A. , Schadendorf, D. , Dermatologic Cooperative Oncology Group of Germany (DeCOG), 2014 Jan. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, (1) 94–109. http://dx.doi.org/10.1158/2159-8290.CD-13-0617. Epub 2013 Nov 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle, N. , Emery, C. , Berger, M.F. , Davis, M.J. , Sawyer, A. , Pochanard, P. , Kehoe, S.M. , Johannessen, C.M. , Macconaill, L.E. , Hahn, W.C. , Meyerson, M. , Garraway, L.A. , 2011 Aug 1. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, (22) 3085–3096. http://dx.doi.org/10.1200/JCO.2010.33.2312. Epub 2011 Mar 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, P.T. , Garnett, M.J. , Roe, S.M. , Lee, S. , Niculescu‐Duvaz, D. , Good, V.M. , Jones, C.M. , Marshall, C.J. , Springer, C.J. , Barford, D. , Marais, R. , Cancer Genome Project, 2004 Mar 19.. Mechanism of activation of the RAF‐ERK signaling pathway by oncogenic mutations of B-RAF. Cel. 116, (6) 855–867. [DOI] [PubMed] [Google Scholar]
- Whittaker, S.R. , Theurillat, J.P. , Van Allen, E. , Wagle, N. , Hsiao, J. , Cowley, G.S. , Schadendorf, D. , Root, D.E. , Garraway, L.A. , 2013 Mar. A genome‐scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 3, (3) 350–362. 10.1158/2159-8290.CD-12-0470 Epub 2013 Jan 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willyard, C. , 2013 Jun. 'Basket studies' will hold intricate data for cancer drug approvals. Nat. Med. 19, (6) 655 http://dx.doi.org/10.1038/nm0613-655. [DOI] [PubMed] [Google Scholar]
- Wilson, T.R. , Fridlyand, J. , Yan, Y. , Penuel, E. , Burton, L. , Chan, E. , Peng, J. , Lin, E. , Wang, Y. , Sosman, J. , Ribas, A. , Li, J. , Moffat, J. , Sutherlin, D.P. , Koeppen, H. , Merchant, M. , Neve, R. , Settleman, J. , 2012 Jul 26. Widespread potential for growth‐factor‐driven resistance to anticancer kinase inhibitors. Natur. 487, (7408) 505–509. http://dx.doi.org/10.1038/nature11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Wang, F. , Keats, J. , Zhu, X. , Ning, Y. , Wardwell, S.D. , Moran, L. , Mohemmad, Q.K. , Anjum, R. , Wang, Y. , Narasimhan, N.I. , Dalgarno, D. , Shakespeare, W.C. , Miret, J.J. , Clackson, T. , Rivera, V.M. , 2011 Dec. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem. Biol. Drug Des. 78, (6) 999–1005. http://dx.doi.org/10.1111/j.1747-0285.2011.01239.x Epub 2011 Oct 31 [DOI] [PMC free article] [PubMed] [Google Scholar]