Abstract
Short arm of chromosome 8 is a hot spot for chromosomal breaks, losses and amplifications in breast cancer. Although such genetic changes may have phenotypic consequences, the identity of candidate gene(s) remains to be clearly defined. Pol β gene is localized to chromosome 8p12‐p11 and encodes a key DNA base excision repair protein. Pol β may be a tumour suppressor and involved in breast cancer pathogenesis. We conducted the first and the largest study to comprehensively evaluate pol β in breast cancer. We investigated pol β gene copy number changes in two cohorts (n = 128 & n = 1952), pol β mRNA expression in two cohorts (n = 249 & n = 1952) and pol β protein expression in two cohorts (n = 1406 & n = 252). Artificial neural network analysis for pol β interacting genes was performed in 249 tumours. For mechanistic insights, pol β gene copy number changes, mRNA and protein levels were investigated together in 128 tumours and validated in 1952 tumours. Low pol β mRNA expression as well as low pol β protein expression was associated high grade, lymph node positivity, pleomorphism, triple negative, basal‐like phenotypes and poor survival (ps < 0.001). In oestrogen receptor (ER) positive sub‐group that received tamoxifen, low pol β protein remains associated with aggressive phenotype and poor survival (ps < 0.001). Artificial neural network analysis revealed ER as a top pol β interacting gene. Mechanistically, there was strong positive correlation between pol β gene copy number changes and pol β mRNA expression (p < 0.0000001) and between pol β mRNA and pol β protein expression (p < 0.0000001). This is the first study to provide evidence that pol β deficiency is linked to aggressive breast cancer and may have prognostic and predictive significance in patients.
Keywords: Pol β, Breast cancer, Prognostic factor, Predictive factor
Highlights
Pol β is a key base excision repair protein and may be involved in breast cancer.
Pol β gene, mRNA, and protein expression was investigated in large cohorts.
Pol β deficiency is associated with aggressive breast cancer and poor prognosis.
Pol β deficiency predicts response to endocrine therapy.
1. Introduction
Impaired DNA repair is a driving force for carcinogenesis. Base excision repair (BER) is required for the accurate removal of bases that have been damage by alkylation, oxidation or ring‐saturation (Dianov and Hubscher, 2013; Kim and Wilson, 2012; Wallace et al., 2012). DNA polymerase β (pol β) is a key factor in BER (Nicolay et al., 2012; Wallace et al., 2012; Yamtich and Sweasy, 2010). The pol β gene is localised to p11 region of chromosome 8, a hot spot for chromosomal breaks, losses and amplifications in breast cancer (Armes et al., 2004; Kerangueven et al., 1994; Pole et al., 2006; Sigbjornsdottir et al., 2000; Tagawa et al., 2003; Ugolini et al., 1999; Venter et al., 2005). The pol β gene encodes a 39 kDa protein with two functional domains; the N‐terminal domain is essential for the dRP lyase activity and the C‐terminal domain performs the nucleotidyl transferase activity during BER. Pol β interacts with several components of the BER machinery such as XRCC1, FEN1, PARP1, APE1 and ligase III to accomplish its biochemical functions (Nicolay et al., 2012; Wallace et al., 2012; Yamtich and Sweasy, 2010).
Pol β deficiency in mice is embryonically lethal (Gu et al., 1994) and embryonic fibroblasts derived from such mice are hypersensitive to alkylating agents (Poltoratsky et al., 2005; Sobol et al., 1996). Depletion of pol β expression by siRNA in human cancer cell lines is associated with sensitivity to chemotherapy (Albertella et al., 2005; Yang et al., 2010). On the other hand, pol β overexpression in pre‐clinical models is associated with relative resistance to DNA damaging agents (Canitrot et al., 1998; Nicolay et al., 2012). Germ line polymorphism of pol β gene (rs3136797) that encodes a pol β variant with a low catalytic activity has been recently shown to induce cellular transformation and may be associated with increased cancer susceptibility (Yamtich et al., 2012; Yamtich and Sweasy, 2010). About 30% of human tumours appear to express pol β variant proteins (such as K289M, I260M) which can induce cellular transformation in vitro, associated with aggressive mutator phenotype (Starcevic et al., 2004). Moreover, pol β mRNA expression may also be dysregulated in certain tumours. In a small study of 68 human tumours of diverse origin that also included nine breast tumours, pol β mRNA was found to be low in 20% of breast cancer samples (Albertella et al., 2005).
Our hypothesis is that in breast cancers with aberrations at chromosome 8p, alterations to pol β gene copy number and the consequent changes in pol β mRNA and/or protein levels could influence breast cancer pathogenesis and influence clinical outcomes in patients. In the current study, we have conducted a comprehensive evaluation of pol β by array comparative genomic hybridization (CGH), gene expression profiling and immunohistochemistry in large cohorts of breast cancer. We provide the first evidence that Pol β deficiency is associated with aggressive breast cancers. In ER positive breast cancers specifically, our data suggest that pol β could influence tamoxifen response and may allow therapy stratification.
2. Material and methods
2.1. Tumour samples
We have investigated multiple cohorts of breast tumours for the analyses described here. We have obtained appropriate consent and the studies were approved by the relevant research ethics boards.
2.1.1. Breast cancer cohorts
2.1.1.1. Discovery cohort
Pol β gene expression was investigated in Uppsala cohort that was originally composed of 315 women representing 65% of all breast cancers resected in Uppsala County, Sweden, from January 1, 1987, to December 31, 1989. Demographics are summarized in Supplementary Table S1 of supporting information and also described elsewhere (Chin et al., 2007). Tumour samples were microarray profiled on the Affymetrix U133A&B gene‐chips. Microarray analysis was carried out at the Genome institute of Singapore. All microarray data are accessible at NCBI‐GEO (http://www.ncbi.nlm.nih.gov/geo/) via series accession number (GSE4922). RNA preparation, microarray hybridization, and data processing were carried out essentially as previously described (Pawitan et al., 2005). All data were normalized using the global mean method (MAS5), and probe set signal intensities were natural log transformed and scaled by adjusting the mean signal to a target value of log 500. The expression intensity of the POLB probe 234907_x_at, located on the HG‐U133b gene chip was utilized for the analysis and to generate artificial neural network (ANN) model. Data were available for 249 patients.
2.1.1.1.1. Artificial neural network (ANN) model
A non‐linear, ANN modelling based, data mining approach was utilised for the Uppsala cohort to identify the best gene probes for sample classification. 47,293 probes were screened for each sample. The data mining algorithm comprised a three layer multilayer perception architecture modified with a feed forward back‐propagation algorithm and a sigmoidal transfer function, as previously described (Lancashire et al., 2010). The network momentum and learning rate were respectively set as 0.1 and 0.5. A parsimonious structure using two hidden nodes and three split Monte Carlo Cross validation were utilised to prevent over fitting. The output node was coded as 0 if a case was low Pol β expression (<the median) and 1 if high Pol β expression (>median). Inputs were ranked in ascending order based on their classification error. The top 100 predictive genes identified were then applied to an ANN based network inference algorithm as described in earlier studies (Lemetre et al., 2009). This model predicted a weighted link (direction and magnitude) between each of the top 100 gene probe markers associated with Pol β expression and every other marker in the top 100. The 100 strongest interactions based on the magnitude were then visualised as a map with Cyto‐scape (Smoot et al., 2011).
2.1.1.1.2. Ensemble classification and cross‐validation analysis
In a second bioinformatics analysis step in Uppsala cohort, we sought to obtain a robust ranking of genes that are differentially expressed between the mRNA Pol β + cases and the mRNA Pol β – and have high predictive power, by applying an ensemble sample classification method within a leave‐one‐out cross‐validation scheme. For this purpose, the 249 patient samples were first grouped into 249 different training/test set partitions, using 248 samples for the training sets and the remaining sample as the test set. For each of the 248 training sets differentially expressed genes were selected independently with the “Empirical Bayes moderated t‐statistic” (Smyth, 2004) and used to train a machine learning model, which was evaluated based on the left‐out sample (a procedure known as “external cross‐validation”). To classify the left‐out sample, the prediction results of four algorithms (Support Vector Machine, Random Forest, kNN and Prediction Analysis for Microarrays, with all parameters being optimised by using a grid search within a nested cross‐validation) (Tibshirani et al., 2002) were combined to a majority‐vote ensemble classifier as to compensate for the inevitable inherent biases and variances that exists amongst each of these machine learning algorithms. In order to rank the genes based on the cross‐validation results, their frequency of occurrence in the list of significantly differentially expressed genes (p‐value < 0.05) across different cross‐validation cycles was recorded, and genes received higher scores the more often they had been selected. All steps of the analysis were conducted using an in‐house web‐application for microarray analysis, available at www.arraymining.net.
2.1.1.2. METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) validation cohort
Pol β gene copy number changes as well as pol β mRNA gene expression was performed in METABRIC cohort which refers to a set of 1980 breast cancer samples with a minimum of 5 years of clinical follow up (Curtis et al., 2012). Patient demographics are summarized in Supplementary Table S2. ER positive and/or lymph‐node negative patients did not receive adjuvant chemotherapy. ER negative and/or lymph‐node positive patients received adjuvant chemotherapy. For this cohort, mRNA was hybridized to Illumina HT‐12 v3 platform (Bead Arrays) and the data were pre‐processed and normalized as described previously (Curtis et al., 2012). Genes copy number was assayed on the Affymetrix SNP 6.0 platform (data available through the European Genotype Archive (http://www.ebi.ac.uk/ega/page.php under accession Number: EGAS00000000082). Samples were classified into the intrinsic subtypes based on the PAM50 gene list. A description of the normalization, segmentation, and statistical analyses was previously described (Curtis et al., 2012). Real time PCR RT‐qPCR was performed on the ABI Prism 7900HT sequence detection system (Applied Biosystems) using SYBR1 Green reporter. All the samples were analysed as triplicates. The Chi‐square test was used for testing association between categorical variables and a multivariate Cox model was fitted to the data using as endpoint breast cancer specific death. Recursive partitioning (Hothorn et al., 2006) was used to identify a cut‐off in gene expression values such that the resulting subgroups have significantly different survival courses. Jonckheere's trend test was performed to evaluate correlation between pol β gene copy number and pol β m RNA expression in the METABRIC cohort.
2.1.1.3. Nottingham Tenovus Primary Breast Carcinoma Cohort (NTP‐BC)
Pol β protein expression was performed in a consecutive series of 1650 patients with primary invasive breast carcinomas who were diagnosed between 1986 and 1999 and entered into the Nottingham Tenovus Primary Breast Carcinoma series. All patients were treated uniformly in a single institution and have been investigated in a wide range of biomarker studies (Ellis et al., 1992; Elston and Ellis, 1991; Galea et al., 1992). Supplemental Table S3 summarizes patient demographics. Supplemental treatment data 1 summarizes various adjuvant treatments received by patients in this cohort.
2.1.1.4. Early primary ER negative (EP‐ER‐) BC cohort
We also evaluated Pol β protein expression in an independent series of 281 ER‐α negative invasive BCs diagnosed and managed at the Nottingham University Hospitals between 1999 and 2007. All patients were primarily treated with surgery, followed by radiotherapy and anthracycline chemotherapy. The characteristics of this cohort are summarised in Supplementary Table S4.
2.1.2. Mechanistic studies
2.1.2.1. Nottingham (NT) series
Pol β gene copy number changes, mRNA gene expression and protein expression were derived from the ‘NT‐series’ that was a set of 171 stage I and II primary operable invasive BC previously described by our group in several molecular profiling studies (Blenkiron et al., 2007; Chin et al., 2007). The raw and mode‐normalized data for gene copy number changes are available from National Centre for Biotechnology Information Gene Expression Omnibus (NCBI‐GEO)‐http://www.ncbi.nlm.nih.gov/geo/ under the series accession number GSE8757 and the expression data are available at the EBI website (http://www.ebi.ac.uk/miamexpress/) with the accession number E‐TABM‐576.
2.1.2.2. Pol β gene copy number changes
Copy number changes at Pol β locus (8p11.21 spanning from 42195973 to 42229321 with size of 33348 bases) were retrieved from oligonucleotide microarrays profiling previously described by our group (Chin et al., 2007). Briefly, DNA was extracted using the Promega DNA Wizard kit (Promega, UK) according to manufacturer's instructions. Labelled DNAs were hybridized to a customized 30,000 60‐mer oligo probes, for which 27,801 unique map positions were defined [Human Mar. 2006 assembly (hg18)]. The median interval between mapped elements was 39.4 kb, 75% of intervals were less than 104.2 kb and 95% were less than 402 kb.
2.1.2.3. Pol β mRNA expression
128 out of the 171 breast tumours were also profiled on Agilent gene expression arrays and have been previously described (Chin et al., 2007). Briefly, total RNA was extracted from a series of frozen breast cancers retrieved from Nottingham Hospitals NHS Trust Tumour Bank between 1986 and 1992. RNA integrity and DNA contamination were analysed using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Total RNA was biotin‐labelled using the Illumina TotalPrep RNA Amplification kit (Ambion, Austin, TX, USA) according to manufacturer's instructions. Biotin‐labelled cRNA (1.5 μg) was used for each hybridisation on Sentrix Human‐6 BeadChips (Illumina, San Diego, CA, USA) in accordance with the manufacturer's protocol. Illumina gene expression data containing 47,293 transcripts were analysed and summarised in the Illumina Bead Studio software. Analyses of the probe level data were done using the beadarray Bioconductor package.
2.1.2.4. Pol β protein expression and other biological biomarkers
The 128 cases which had both a‐CGH and mRNA data were then evaluated for Pol β protein expression by immunohistochemistry on tissue microarray (TMAs) as described below. Demographics of this cohort is summarised in Supplementary Table S5.
2.1.2.5. Integrated array CGH, mRNA gene expression and protein expression analysis
Gene‐dosage levels to gene expression were evaluated using Wilcoxon test to evaluate the significance of the association between copy number and aberrant expression. To determine whether mRNA expression levels correlated with protein levels, Pearson correlations were performed between Pol β mRNA expression log intensity values and Pol β protein expression (H‐score). Jonckheere's trend test was performed to evaluate correlation between pol β gene copy number and pol β m RNA expression in the METABRIC cohort.
2.2. Survival data
Survival data including survival time, disease‐free survival (DFS), and development of loco‐regional and distant metastases (DM) were maintained on a prospective basis. DFS was defined as the number of months from diagnosis to the occurrence of recurrence or DM relapse. BC specific survival (BCSS) was defined as the number of months from diagnosis to the occurrence of BC related‐death. Survival was censored if the patient was still alive, lost to follow‐up, or died from other causes.
The Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria were followed throughout this study.
2.3. Tissue microarrays (TMAs) and immunohistochemistry (IHC)
Tumours from NT‐BC, NTP‐BC and EP‐ER‐BC cohorts were arrayed in tissue microarrays (TMAs) constructed with 6 replicate 0.6 mm cores from the centre and periphery of the tumours for each patient. The TMAs were immunohistochemically profiled for Pol β and other biological antibodies (Supplementary Table S6). Immunohistochemical staining for Pol β was performed using the Thermo Scientific Shandon Sequenza chamber system (REF: 72110017), in combination with the Novolink Max Polymer Detection System (RE7280‐K: 1250 tests), and the Leica Bond Primary Antibody Diluent (AR9352), each used according to the manufacturer's instructions (Leica Microsystems). Pre‐treatment antigen retrieval was performed on the TMA sections using citrate buffer (pH 6.0), heated for 20 min at 95 °C in a microwave (Whirpool JT359 Jet Chef 1000W). TMA sections were then incubated for 60 min at room temperature with 1:200 anti‐Polβ rabbit polyclonal antibody (ab26343, Abcam, Cambridge, UK). Also, positive and negative (by omission of the primary antibody and IgG‐matched serum) controls were prepared for each set of samples. To validate the use of TMAs for immunophenotyping, full‐face sections of 40 cases were stained and protein expression levels of the different antibodies were compared. The concordance between TMAs and full‐face sections was excellent (k = 0.8). Positive and negative (by omission of the primary antibody and IgG‐matched serum) controls were included in each run. HER2, ER and PR expressions were assessed according to the new American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines. HER2 status was assessed using both IHC and fluorescence in situ hybridization (FISH).
2.3.1. Evaluation of pol β immunohistochemical staining
The tumour cores were evaluated by expert pathologists blinded to the clinico‐pathological characteristics of patients in two different settings. There was excellent intra and inter‐observer agreements (k > 0.8; Cohen's κ and multi‐rater κ tests, respectively). In this study, nuclei immunoreactivity was quantitatively evaluated with the Histoscore (H score). The absence (negative) of nuclear staining was given a score of 0 and the presence of nuclear stain was dependent on its intensity as 1, 2 or 3 equivalent to weak, moderate and strong nuclear stain respectively. The percentage staining of each core was defined as 0–100% and the H‐score was obtained by multiplying intensity of staining and percentage of staining. H‐score in a range of 0–300 was generated. The median H‐score of 100 was taken as the cut‐off and low pol β expression was classed as H‐score of ≤100 and >100 was classed as high for Pol β expression. Not all cores within the TMA were suitable for IHC analysis as some cores were missing or lacked tumour.
2.3.2. Breast cancer cell lines and Western blot analysis
MCF‐7, MDA‐MB‐231 and MDA‐MB‐436 breast cancer cell lines were purchased from ATCC and grown in RPMI medium supplemented with 10% FBS, 1% penicillin/streptomycin. Western blot analysis performed as described previously (Sultana et al., 2013). Primary antibodies used was anti‐Polβ rabbit polyclonal antibody (1:200 dilution, ab26343, Abcam, Cambridge, UK).
2.3.3. Statistical analysis
Data analysis was performed using SPSS (SPSS, version 17 Chicago, IL). Where appropriate, Pearson's Chi‐square, Fisher's exact, χ 2 for trend, Student's t and ANOVAs one way tests were performed using SPSS software (SPSS, version16 Chicago, IL). The Mann–Whitney test was used to compare the genomic copy number and Pol β protein expression. Pearson correlation coefficients assessed the correlation between array CGH and mRNA gene expression. Cumulative survival probabilities were estimated using the Kaplan–Meier method. Differences between survival rates were tested for significance using the log‐rank test. Multivariate analysis for survival was performed using the Cox hazard model. The proportional hazards assumption was tested using standard log–log plots. Each variable was assessed in univariate analysis as a continuous and categorical variable and the two models were compared using an appropriate likelihood ratio test. Hazard ratios (HR) and 95% confidence intervals (95% CI) were estimated for each variable. All tests were two‐sided with a 95% CI. P values for each test were adjusted with Benjamini and Hochberg multiple P‐value adjustment and an adjusted p value of <0.05 was considered significant.
3. Results
3.1. Low Pol β mRNA levels correlate to aggressive breast cancer
In the Uppsala cohort (n = 249), low pol β mRNA expression (29%) was associated with lymph node positive disease (p = 0.03) (Supplementary Table S7) and poor survival (p = 0.004) (Figure 1A). We then proceeded to investigate pol β mRNA expression in a large series of 1950 tumours comprising the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) cohort (Table 1). 10% (195/1950) of tumours had low Pol β mRNA expression in this cohort which was significantly associated with high histological grade (p = 0.001), pleomorphism (p = 0.04), glandular de‐differentiation (p = 0.006), lymph node positivity (0.039), HER2 overexpression (p < 0.0001), absence of ER(ER−) (p < 0.0001), presence of basal like phenotypes (p < 0.0001) and triple negative phenotypes (p < 0.0001). Low pol β mRNA mRNA expression was also found to be associated with previously described molecular phenotypes in breast cancer: PAM50.Her2 (p < 0.0001), PAM50.Basal (p < 0.0001) molecular phenotypes. However, PAM50.LumA (p < 0.0001) and PAM50.LumB (p = 0.0097) breast tumours where more likely in tumours that have high levels of pol β mRNA (p < 0.0001). Similarly, ER+/Her‐2‐/high proliferation (p = 0.001) and ER+/Her‐2‐/low proliferation tumours (p < 0.0001) were more likely in tumours that have high levels of pol β mRNA whereas low pol β mRNA levels was associated with ER‐/Her‐2‐ (p < 0.0001) and her‐2 positive tumours (p = 0.001). Low pol β mRNA expressing tumours had significantly worse breast cancer specific survival compared to tumours with high pol β mRNA levels (p < 0.0001) (Figure 1B).
Figure 1.
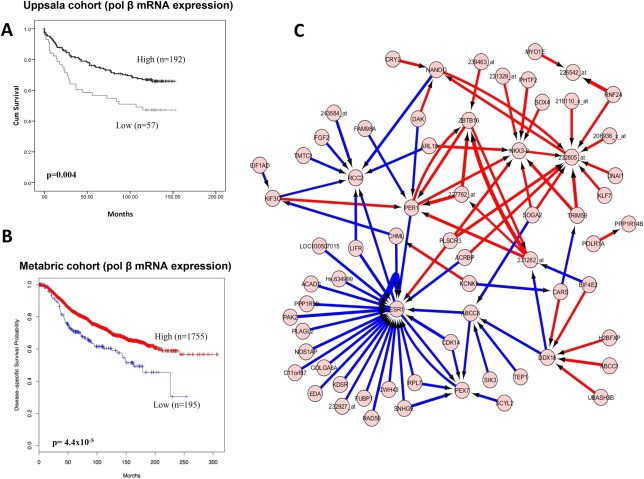
Pol β mRNA expression in breast cancer. A. Kaplan Meier curves showing breast cancer specific survival in the Uppsala cohort. B. Kaplan Meier curves showing breast cancer specific survival in the Metabric cohort. C. Artificial neural network analysis. Top pair‐wise interactions for gene probe markers associated with Pol β expression in 249 breast cancers is shown here. Each gene probe is represented by a node and the interaction weight between them as an edge, the width being defined by the magnitude of the weight. Interactions are directed from a source gene to a target gene as indicated by arrows. Red interactions indicate an excitatory interaction and blue indicates an inhibitory interaction. Highly linked genes represent hubs that are indicated to be highly influential or highly regulated in the Pol β system. See Supplementary data 2 for the biological functions of individual genes.
Table 1.
Pol β mRNA expression and breast cancer (METABRIC cohort, n = 1952).
| Variable | POLBmRNA expression | X2 p value | |
|---|---|---|---|
| Low N (%) | High N (%) | ||
| A) Pathological parameters | |||
| Lymph node stage | |||
| Negative | 90 (44.5%) | 922 (53%) | 0.023a |
| Positive | 112 (55.5%) | 822 (47%) | 0.039a |
| Grade b | |||
| G1 | 7 (3.6%) | 156 (9.2%) | 0.01a |
| G2 | 67 (34.2%) | 698 (41.5%) | 0.059 |
| G3 | 122 (62.2%) | 829 (49.3%) | 0.001a |
| Mitotic index | |||
| M1 (low; mitoses < 10) | 92 (54.5%) | 909 (62%) | 0.07 |
| M2 (medium; mitoses 10–18) | 41 (24.2%) | 337 (23%) | 0.852 |
| M3 (high; mitosis >18) | 36 (21.3%) | 220 (15%) | 0.05 |
| Pleomorphism | |||
| 1 (Small‐regular uniform) | 1 (0.6%) | 16 (1%) | 1.0 |
| 2 (Moderate variation) | 47 (27.8%) | 549 (37.4%) | 0.017a |
| 3 (Marked variation) | 121 (71.6%) | 902 (61.4%) | 0.044a |
| Tubule formation | |||
| 1 (>75% of definite tubule) | 5 (3%) | 52 (3.5%) | 0.84 |
| 2 (10%–75% definite tubule) | 35 (20.7%) | 319 (21.7%) | 0.77 |
| 3 (<10% definite tubule) | 129 (76.3%) | 1096 (74.7%) | 0.941 |
| Triple negative | |||
| (No) | 141 (69.1%) | 1498 (85.7%) | 1.8 × 10−9a |
| (Yes) | 63 (30.9%) | 250 (14.3%) | 1.8 × 10−9a |
| Basal like | |||
| (No) | 39 (19.1%) | 1653 (94.6%) | 7.6 × 10−13a |
| (Yes) | 165 (80.9%) | 95 (5.4%) | 7.6 × 10−13a |
| ER | |||
| (Negative) | 82 (41%) | 355 (20.6%) | 2.0 × 10−10a |
| (Positive) | 118 (59%) | 1367 (79.3%) | 1.9 × 10−10a |
| Genefu subtype | |||
| ER−/Her‐2 negative | 63 (35%) | 236 (15.0%) | 1.3 × 10−10a |
| ER+/Her‐2 negative/high proliferation | 45 (25%) | 584 (37.5%) | 0.001a |
| ER+/Her‐2 negative/low proliferation | 39 (19.5%) | 587 (37.5%) | 3.9 × 10−5a |
| Her‐2 positive | 33 (16.5%) | 156 (10%) | 0.001a |
| PAM50 subtype | |||
| PAM50.Her2 | 47 (26.4%) | 191 (12%) | 1.0 × 10−6a |
| PAM50.Basal | 57 (32.2%) | 265 (16.7%) | 5.2 × 10−6a |
| PAM50.LumA | 39 (21.9%) | 675 (42.7%) | 6.8 × 10−8a |
| PAM50.LumB | 35 (19.7%) | 449 (28.4%) | 0.0097 |
HER2: human epidermal growth factor receptor 2; ER: oestrogen receptor; PgR: progesterone receptor; CK: cytokeratin; Basal‐like: ER−, HER2 and positive expression of either CK5/6, CK14 or EGFR; Triple negative: ER−/PgR‐/HER2‐.
Statistically significant.
Grade as defined by NGS.
The data provides evidence that pol β mRNA expression may have prognostic significance in breast cancer.
3.2. Artificial neural network (ANN), ensemble classification and cross‐validation analysis reveals oesotrogen receptor (ESR1) as a novel pol β interaction gene
To investigate pol β interactions in breast cancer, a non‐linear, ANN modelling based, data mining approach was utilised to identify the gene probes most associated with changes in Pol β expression through a population of 249 breast tumours (Uppsala cohort). This was achieved by using the ANN modelling algorithm to predict the Pol β expression level.
The algorithm incorporated a constrained architecture and a 3 way Monte Carlo cross validation to prevent over fitting. Each of the 47,293 probes were utilised singly. The performance of each probe, and thus their association with Pol β was assessed based on the model performance. A rank order of probes was produced by ranking on the root mean squared error of the model for each probe (Figure 1C). This approach has been described in detail previously (Lancashire et al., 2010). A further ANN network inference algorithm based systems biology method (Lemetre et al., 2009) was run on the data for the top 100 probes to identify their interactions. This approach provides a magnitude and direction of interaction for each potential pair of probes. Out of 9900 possible interactions in the top 100 probes ((100 × 100) − 100) the 100 strongest are presented in Supplementary data 2 and shown in Figure 1C. Oestrogen receptor (ESR1) gene was identified as having the highest number of strongest connections, defining it as a highly influential factor in the pol β interactome. The majority of the interactions were negative suggesting the probes associated with pol β are generally driving ERS1 expression down. We therefore proceeded to investigate pol β protein expression in breast cancer with a specific focus on ER positive breast cancers.
3.3. Low Pol β protein levels correlate to aggressive ER + breast cancers
A total of 1406 tumours were suitable for pol β protein expression analysis. We first investigated specificity of the antibody used in the current study as well as pol β protein expression in a panel of breast cancer cell lines. Compared to MCF‐7 and MDA‐MB‐231 breast cancer cells, MDA‐MB‐436 had more than 80% reduction in pol β protein expression (Figure 2A1). In human tumours, 540/1406 (38.4%) of the tumours had low pol β protein expression whereas 866/1406 (61.6%) expressed high pol β protein expression (Figure 2A2 and A3) (Table 2). Low pol β protein expression is significantly associated with high tumour grade (p < 0.0001), high mitotic index (p < 0.001), pleomorphism (p < 0.0001), de‐differentiation (p < 0.0001), triple negative phenotype (p < 0.001), basal‐like phenotype (p < 0.0001), presence of cytokeratin (CK) 6 (p < 0.0001), CK14 (p = 0.002) and CK18 (p = 0.006), absence of hormone receptors including ER− (p < 0.001), progesterone receptor (PgR‐) (p < 0.001) and androgen receptor (AR‐) (p < 0.001). Low pol β protein expression is also significantly associated with low expression of other DNA repair proteins such as BRCA1 (p < 0.0001), XRCC1 (p < 0.001), SMUG1 (p = 0.01) and FEN1 (p < 0.0001). Low pol β protein expression was significantly associated with high proliferation index (high MIB1, p < 0.0001) and absence of anti‐apoptotic factor (Bcl‐2, p=<0.001). Low pol β was significantly associated with poor breast cancer specific survival (BCSS) (p < 0.0001) (Figure 2B1) and disease‐free survival (DFS) (p < 0.0001) (Supplementary Figure 1A). In multivariate cox regression analysis, low pol β protein expression was an independent predictor for BCSS (p = 0.022) (Supplementary Table S8).
Figure 2.
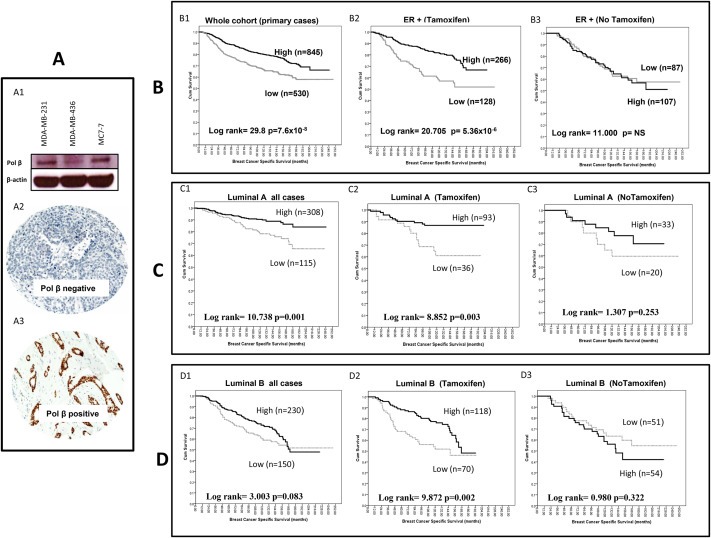
Pol β protein expression in breast cancer. A. Western blot showing pol β in breast cancer cell lines (A1). Microphotographs of Pol β protein expression in breast cancer tissue (magnification × 200) showing tumour with low pol β expression (A2) and high pol β expression (A3). B. Kaplan Meier curves showing breast cancer specific survival in whole cohort (B1), ER + treated with tamoxifen (B2), ER + no tamoxifen (B3). C. Kaplan Meier curves showing breast cancer specific survival in luminal A sub‐group whole cohort (C1), luminal A treated with tamoxifen (C2), luminal A no tamoxifen (C3). D. Kaplan Meier curves showing breast cancer specific survival in luminal B sub‐group whole cohort (D1), luminal A treated with tamoxifen (D2), luminal A no tamoxifen (D3).
Table 2.
Pol β protein expression and breast cancer (Nottingham Cohort, n = 1406).
| Variable | POL β protein expression total n = 1406 | P‐valve | |
|---|---|---|---|
| Low N (%) | High N (%) | ||
| A) Pathological parameters | |||
| Tumour size | |||
| <1 cm | 39 (7.2%) | 115 (13.4%) | 0.002 |
| >1–2 cm | 271 (50.4%) | 429 (49.9%) | |
| >2–5 cm | 210 (39.0%) | 297 (34.5%) | |
| >5 cm | 18 (3.3%) | 19 (2.2%) | |
| Tumour stage | |||
| 1 | 327 (60.7%) | 549 (63.7%) | 0.012 |
| 2 | 148 (27.5) | 251 (29.1%) | |
| 3 | 64 (11.9%) | 62 (7.2%) | |
| Tumour grade | |||
| G1 | 58 (10.8%) | 167 (19.4%) | 1.4 × 10−14 |
| G2 | 133 (24.7%) | 326 (37.9%) | |
| G3 | 347 (64.5%) | 367 (42.7%) | |
| Mitotic index | |||
| M1 (low; mitoses < 10) | 122 (22.9%) | 361 (42.1%) | <0.001 |
| M2 (medium; mitoses 10–18) | 84 (15.8%) | 180 (21.0%) | |
| M3 (high; mitosis >18) | 326 (61.3%) | 316 (36.9%) | |
| Pleomorphism | |||
| 1 (Small‐regular uniform) | 9 (1.7%) | 28 (3.3%) | 1.0 × 10−6 |
| 2 (Moderate variation) | 156 (29.4%) | 360 (42.1%) | |
| 3 (Marked variation) | 366 (68.9%) | 468 (54.7%) | |
| Tubule formation | |||
| 1 (>75% of definite tubule) | 15 (2.8%) | 63 (7.4%) | 1.0 × 10−6 |
| 2 (10%–75% definite tubule) | 161 (30.3%) | 291 (34.0%) | |
| 3 (<10% definite tubule) | 356 (66.9%) | 503 (58.7%) | |
| Tumour type | |||
| IDC‐NST | 307 (67.0%) | 402 (54.3%) | 1.4 × 10−5 |
| Tubular carcinoma | 69 (15.1%) | 182 (24.6%) | |
| Medullary carcinoma | 16 (3.5%) | 13 (1.8%) | |
| ILC | 32 (7.0%) | 74 (10.0%) | |
| Others | 34 (7.4%) | 70 (9.4%) | |
| Lymphovascular invasion | |||
| No | 335 (63.7%) | 591 (69.1) | 0.037 |
| Yes | 191 (36.3%) | 264 (30.9%) | |
| B) Aggressive phenotype | |||
| Her2 overexpression | |||
| No | 446 (85.1%) | 757 (90.1%) | 0.005 |
| Yes | 78 (14.9%) | 83 (9.9%) | |
| Triple negative phenotype | |||
| No | 381 (74.0%) | 729 (85.9%) | <0.001 |
| Yes | 134 (26.0%) | 120 (14.1%) | |
| Basal like phenotype | |||
| No | 412 (83.6%) | 753 (90.7%) | 1.1 × 10−4 |
| Yes | 81 (16.4%) | 77 (9.3%) | |
| Cytokeratin 6 (CK6) | |||
| Negative | 355 (79.1%) | 982 (84.4%) | 8.0 × 10−4 |
| Positive | 94 (20.9%) | 182 (15.6%) | |
| Cytokeratin 14 (CK14) | |||
| Negative | 372 (83.8%) | 642 (89.9%) | 0.002 |
| Positive | 72 (16.2%) | 72 (10.1%) | |
| Cytokeratin 18 (CK18) | |||
| Negative | 57 (13.8%) | 57 (8.5%) | 0.006 |
| Positive | 355 (86.2%) | 610 (91.5%) | |
| C) Hormone receptors | |||
| ER | |||
| Negative | 192 (36.9%) | 171 (20.0%) | <0.001 |
| Positive | 329 (63.1%) | 684 (80.0%) | |
| PgR | |||
| Negative | 265 (53.8%) | 284 (35.2%) | <0.001 |
| Positive | 228 (46.2%) | 523 (64.8%) | |
| AR | |||
| Negative | 197 (47.0%) | 201 (30.2%) | <0.001 |
| Positive | 222 (53.0%) | 465 (69.8%) | |
| D) DNA repair | |||
| ATM | |||
| Absent | 178 (54.6%) | 266 (52.8%) | 0.607 |
| Normal | 148 (45.4%) | 238 (47.2%) | |
| BRCA1 | |||
| Absent | 101 (27.4%) | 92 (15.4%) | 5.0 × 10−6 |
| Normal | 267 (72.6%) | 506 (84.6%) | |
| XRCC1 | |||
| Low | 102 (26.1%) | 55 (9.0%) | <0.001 |
| High | 289 (73.9%) | 554 (91.0%) | |
| FEN1 | |||
| Low | 288 (79.3%) | 388 (68.7%) | 3.6 × 10−4 |
| High | 75 (20.7%) | 252 (27.2%) | |
| SMUG1 | |||
| Low | 155 (43.5%) | 188 (34.9%) | 0.010 |
| High | 201 (56.5%) | 350 (65.1%) | |
| E) Cell cycle/apoptosis regulators | |||
| MIB1 | |||
| Low | 125 (28.0%) | 295 (40.9%) | 9 × 10−6 |
| High | 321 (72.0%) | 427 (59.1%) | |
| P53 | |||
| Low expression | 316 (73.7%) | 564 (82.3%) | 0.001 |
| High expression | 113 (26.3%) | 121 (17.7%) | |
| Bcl‐2 | |||
| Negative | 215 (45.6%) | 216 (28.3%) | <0.001 |
| Positive | 257 (54.4%) | 546 (71.7%) | |
| TOP2A | |||
| Low | 193 (51.3%) | 252 (42.1%) | 0.005 |
| Overexpression | 183 (48.7%) | 347 (57.9%) | |
Grade as defined by NGS; BRCA1: Breast cancer 1, early onset; HER2: human epidermal growth factor receptor 2; ER: oestrogen receptor; PgR: progesterone receptor; CK: cytokeratin; Basal‐like: ER−, HER2 and positive expression of either CK5/6, CK14 or EGFR; Triple negative: ER−/PgR‐/HER2‐.
3.3.1. Pol β and ER + breast cancer
We then proceeded to perform subgroup analysis in various sub‐types of breast cancers. A total of 1009 ER positive tumours were first investigated. 328/1009 (32.5%) of tumours had low pol β protein expression whereas 681/1009 (67.5%) of the tumours had high pol β expression (Table 3). Low DNA pol β expression was significantly associated with adverse pathological parameters including; high grade (p < 0.001), high mitotic index (p < 0.001), glandular de‐differentiation (p = 0.019) and pleomorphisim (p = 0.02). Low pol β was associated with low BRCA1 (p < 0.0001), low XRCC1 (p < 0.001), low FEN1 (p = 0.001) and low SMUG1 (p = 0.025) protein expression. Low pol β expression was also significantly associated with high proliferation index (MIB1, p = 0.005), mutant p53 (p = 0.016) and absence of anti‐apoptotic factor (Bcl‐2, p = 0.002). Low pol β was significantly associated with poor BCSS (p < 0.0001) (Supplementary Figure 1B) and DFS (p = 0.001) (Supplementary Figure 1C) in all ER + tumours that are high risk (NPI>3.4). Interestingly, in ER + tumours that received tamoxifen therapy, low pol β remains significantly associated with poor BCSS (p < 0.0001) (Figure 2B2) and DFS (Supplementary Figure 1D). On the other hand, ER + tumours that did not receive tamoxifen, there was no difference in BCSS between high and low pol β expressing tumours (Figure 2B3) as well as DFS (Supplementary Figure 1E).
Table 3.
Pol β protein expression and ER positive breast cancer (n = 1009).
| Variable | Pol β protein expression | P‐valve | |
|---|---|---|---|
| Low N (%) | High N (%) | ||
| A) Pathological parameters | |||
| Tumour size | |||
| <1 cm | 25 (7.6%) | 102 (15.0%) | 0.005 |
| >1–2 cm | 176 (53.7%) | 349 (51.2%) | |
| >2–5 cm | 117 (35.7%) | 219 (32.2%) | |
| >5 cm | 10 (3.0%) | 11 (1.6%) | |
| Tumour stage | |||
| 1 | 197 (59.9%) | 436 (63.8%) | 0.062 |
| 2 | 97 (29.5%) | 203 (29.7%) | |
| 3 | 35 (10.6%) | 44 (6.4%) | |
| Tumour grade | |||
| G1 | 53 (16.2%) | 158 (23.2%) | <0.001 |
| G2 | 112 (34.1%) | 304 (44.6%) | |
| G3 | 163 (49.7%) | 219 (32.2%) | |
| Mitotic index | |||
| M1 (low; mitoses < 10) | 109 (33.4%) | 341 (50.1%) | <0.001 |
| M2 (medium; mitoses 10–18) | 64 (19.6%) | 158 (23.2%) | |
| M3 (high; mitosis >18) | 153 (46.9%) | 181 (26.6%) | |
| Pleomorphism | |||
| 1 (Small‐regular uniform) | 8 (2.5%) | 26 (3.8%) | 0.020 |
| 2 (Moderate variation) | 138 (42.5%) | 342 (50.3%) | |
| 3 (Marked variation) | 179 (55.1%) | 312 (45.9%) | |
| Tubule formation | |||
| 1 (>75% of definite tubule) | 13 (4.0%) | 58 (8.5%) | 0.019 |
| 2 (10%–75% definite tubule) | 123 (37.7%) | 266 (39.1%) | |
| 3 (<10% definite tubule) | 190 (58.3%) | 356 (52.4%) | |
| Tumour type | |||
| IDC‐NST | 172 (59.1%) | 280 (47.5%) | 3.9 × 10−4 |
| Tubular carcinoma | 61 (21.0%) | 175 (29.7%) | |
| Medullary carcinoma | 4 (1.4%) | 0 (0.0%) | |
| ILC | 27 (9.3%) | 71 (12.0%) | |
| Others | 27 (9.3%) | 64 (10.8%) | |
| Lymphovascular invasion | |||
| No | 200 (61.9%) | 474 (70.0%) | 0.011 |
| Yes | 123 (38.1%) | 203 (30.0%) | |
| B) Aggressive phenotype | |||
| Her2 overexpression | |||
| No | 291 (90.7%) | 623 (93.8%) | 0.071 |
| Yes | 30 (9.3%) | 41 (6.2%) | |
| Cytokeratin 6 (CK6) | |||
| Negative | 261 (92.2%) | 546 (94.8%) | 0.138 |
| Positive | 22 (7.8%) | 30 (5.2%) | |
| Cytokeratin 14 (CK14) | |||
| Negative | 260 (92.5%) | 534 (93.5%) | 0.588 |
| Positive | 21 (7.5%) | 37 (6.5%) | |
| Cytokeratin 18 (CK18) | |||
| Negative | 16 (5.9%) | 12 (2.2%) | 0.007 |
| Positive | 253 (94.1%) | 524 (97.8%) | |
| C) Hormone receptors | |||
| PgR | |||
| Negative | 81 (26.6%) | 125 (19.3%) | 0.012 |
| Positive | 224 (73.4%) | 521 (80.7%) | |
| AR | |||
| Negative | 94 (34.6%) | 104 (19.4%) | 2.0 × 10−6 |
| Positive | 178 (65.4%) | 432 (80.6%) | |
| D) DNA repair | |||
| ATM | |||
| Absent | 104 (51.7%) | 194 (49.4%) | 0.583 |
| Normal | 97 (48.3%) | 199 (50.6%) | |
| BRCA1 | |||
| Absent | 58 (23.9%) | 52 (10.9%) | 5.0 × 10−6 |
| Normal | 185 (76.1%) | 424 (89.1%) | |
| XRCC1 | |||
| Low | 52 (21.6%) | 32 (6.6%) | <0.001 |
| High | 189 (78.4%) | 455 (93.4%) | |
| FEN1 | |||
| Low | 180 (79.3%) | 300 (66.5%) | 0.001 |
| High | 47 (20.7%) | 151 (33.5%) | |
| SMUG1 | |||
| Low | 84 (37.7%) | 121 (29.0%) | 0.025 |
| High | 139 (62.3%) | 296 (71.0%) | |
| E) Cell cycle/apoptosis regulators | |||
| MIB1 | |||
| Low | 103 (36.3%) | 266 (46.3%) | 0.005 |
| High | 181 (63.7%) | 308 (53.7%) | |
| P53 | |||
| Low expression | 227 (83.8%) | 493 (89.6%) | 0.016 |
| High expression | 44 (16.2%) | 57 (10.4%) | |
| Bcl‐2 | |||
| Negative | 80 (26.8%) | 109 (17.9%) | 0.002 |
| Positive | 218 (73.2%) | 499 (82.1%) | |
| TOP2A | |||
| Low | 125 (55.3%) | 203 (43.0%) | 0.002 |
| Overexpression | 101 (44.7%) | 269 (57.0%) | |
Grade as defined by NGS; BRCA1: Breast cancer 1, early onset; HER2: human epidermal growth factor receptor 2; ER: oestrogen receptor; PgR: progesterone receptor; CK: cytokeratin; Basal‐like: ER−, HER2 and positive expression of either CK5/6, CK14 or EGFR; Triple negative: ER−/PgR‐/HER2‐.
In Luminal A ER + breast cancers, low pol β expression remains associated with high mitotic index (p = 0.01), HER‐2 overexpression (p = 0.002), low BRCA1 (p = 0.019), XRCC1 (p < 0.0001) and FEN1 (p = 0.015) (Supplementary Table S9). In high risk tumours (NPI>3.4), low pol β was significantly associated with poor BCSS (p = 0.001) (Figure 2C1) and DFS (p = 0.006) (Supplementary Figure 2A). In luminal A tumours that received tamoxifen therapy, low pol β remains significantly associated with BCSS (p = 0.003) (Figure 2C2) and DFS (p = 0.004) (Supplementary Figure 2B). On the other hand, luminal A tumours that did not receive tamoxifen, there was no difference in BCSS between high and low pol β expressing tumours (Figure 2C3) and DFS (Supplementary Figure 2C).
In Luminal B ER + breast cancers, low pol β expression was associated with high grade (p = 0.006), high mitotic index (p = 0.008), HER‐2 overexpression (p = 0.002), CK6 (p = 0.009), low BRCA1 (p = 0.001), XRCC1 (p < 0.0001) and FEN1 (p = 0.038) (Supplementary Table S10). In luminal B tumours that received tamoxifen therapy, low pol β was significantly associated with poor BCSS (p = 0.002) (Figure 2D2) but not for DFS (p = 0.112) (Supplementary Figure 2E). In luminal B tumours that did not receive tamoxifen, there was no difference in BCSS between high and low pol β expressing tumours (Figure 2D3) and DFS (Supplementary Figure 2F).
Taken together the data provides evidence that low pol β protein expression may have clinicopathological, prognostic and predictive significance in ER positive breast cancers.
3.3.2. Pol β and ER− breast cancer
We then proceeded to investigate pol β protein expression in 363 ER‐ breast tumours. 192/363 (52.9%) of tumours had low pol β protein expression whereas 171/363 (47.1%) of the tumours had high pol β expression (Supplementary Table S11). There were no significant clinicopathological correlations although there were associations with ATM, XRCC1 and DNA‐PK expression (Supplementary Table S11). We then investigated pol β protein expression in another independent cohort of 347 ER‐ breast cancers. 229/347 (66%) of tumours had low pol β protein expression whereas 118/347 (34%) of the tumours had high pol β expression (Supplementary Table S12). There were no significant clinicopathological correlations although there were some associations with ATM, XRCC1, BRCA1 and Bcl‐2 expression. There were no significant associations between pol β protein expression and survival in ER− tumours in both cohorts (Supplementary Figure 3).
Taken together the data suggests that pol β expression has no clinical significance in ER negative breast cancers. This is in contrast to ER positive tumours where pol β deficiency may have poor prognostic as well as a predictive significance.
3.4. Mechanistic insights
3.4.1. Pol β gene copy number and pol β mRNA levels
Integrated array CGH and mRNA gene expression analysis was conducted in 125 breast tumours (Nottingham cohort) where pol β gene copy number and pol β mRNA expression data were all available. 29%, 8% and 11% of tumours showed gain, amplification and losses of pol β gene locus at chromosome 8p12‐p11 respectively. 52% of tumours were neutral for pol βgene copy number. There was a strong correlation between pol β gene copy number changes and pol β mRNA expression in tumours (adjusted p value = 0.00126).
In the METABRIC cohort, integrated array CGH and mRNA gene expression analysis was conducted in 1952 breast tumours. 16.2%, 3.6% and 5.0% showed gain, amplification and losses of pol β gene locus at chromosome 8p12‐p11 respectively. 75% of tumours were neutral for pol β gene copy number in the METABRIC cohort. A very strong correlation between gene copy number and mRNA expression was demonstrated in the METABRIC cohort (p = 5.6 × 10−93) (Figure 3A). Together the data provides evidence that low pol β mRNA may be due to loss of pol β gene copy number in a proportion of breast tumours.
Figure 3.
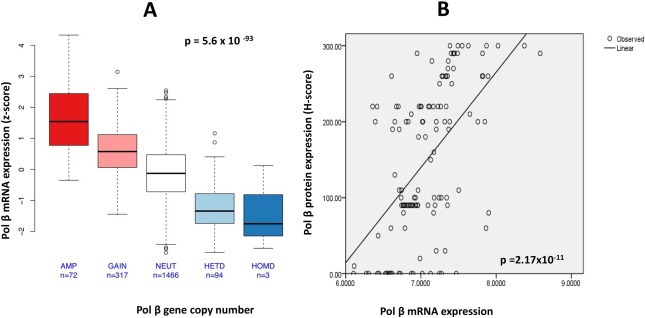
A. Correlation between Pol β gene copy number and Pol β mRNA expression in the Metabric cohort [AMP = amplification, NEUT = neutral, HETD = heterozygous deletion, HOMD = homozygous deletion]. B. Correlation between Pol β mRNA and Pol β protein expression in the Nottingham cohort.
3.4.2. Pol β mRNA and pol β protein levels
We compared pol β mRNA levels and protein expression in 125 breast tumours (Nottingham cohort). As shown in Figure 3B, there was positive correlation between pol β mRNA and pol β protein expression (r = 0.55, p = 2.17 × 10−11) in tumour. The data suggests that low pol β protein may be a consequence of low pol β mRNA in a proportion of breast tumours. However, it is also likely that there may be other factors influencing pol β protein such as post‐transcriptional and post‐translational mechanisms regulating pol β protein expression.
4. Discussion
Pol β is a key player in base excision repair and promotes genomic stability. Pol β gene is localised to chromosome 8p12‐p11 which is a hot spot for genetic changes in breast cancer (Armes et al., 2004; Kerangueven et al., 1994; Pole et al., 2006; Sigbjornsdottir et al., 2000; Tagawa et al., 2003; Ugolini et al., 1999; Venter et al., 2005). In the current study we have demonstrated for the first time that pol β gene deletion at chromosome 8 may be associated with low pol β mRNA levels as well low pol β protein levels in a proportion of breast cancers. Low pol β mRNA was significantly linked to aggressive features such as high grade, high mitotic index, pleomorphism, triple negative, PAM50. Her2, PAM50. Basal, PAM50. Luminal A and PAM50. Luminal B molecular phenotypes. At the protein level, similarly, low pol β expression was associated with aggressive clinical phenotype. Interestingly, low pol β protein also associated with other DNA repair factors such as absent BRCA1, low XRCC1, low FEN1 and low SMUG1 protein expression. The data provides evidence that pol β loss may be associated with genomic instability in breast tumours. In sub‐group analysis in ER positive tumour we also observed consistent association with aggressive clinicopathological features. In ER negative tumours, however, no significant associations were evident. Together the data provides evidence for a novel role for pol β in ER positive breast tumours. This hypothesis is further supported by artificial neural network analysis of gene expression in 249 breast tumours, where oestrogen receptor was found to be a top pol β interacting gene. In ER positive tumours we also found that pol β expression predicts resistance to endocrine therapy as evidenced by poor survival in patients whose tumours that had low pol β expression and received tamoxifen therapy. The poor predictive significance was seen in luminal A as well as luminal B tumours implying that pol β is a novel predictive biomarker and is likely related to the role of pol β in cell proliferation. Taken together, our data provides evidence that pol β may be involved in breast cancer pathogenesis.
Previous studies have shown that about 30% of human solid tumours may harbour pol β variants that have sub‐optimal DNA repair capacity, promote genomic instability and confer a mutator phenotype that is associated with aggressive clinical behaviour (Starcevic et al., 2004; Yamtich and Sweasy, 2010). Whether such pol β variants exist in breast cancer is unknown. Nevertheless, the data presented in our study suggests that deletion at the chromosome 8p12‐p11 locus is a mechanism for pol β deficiency which is associated with an aggressive phenotype in breast cancer. The link between pol β and ER was surprising. However, emerging evidence does suggest a potential association between oestrogen and base excision repair (BER). For example, oestrogen induced oxidative stress may play a key role in oestrogen driven carcinogenesis (Bhat et al., 2003). Oxidative stress is a major source of oxidative base damage which is a strong stimulus for activation of BER (Amouroux et al., 2010; Cabelof et al., 2002; Unnikrishnan et al., 2011). In preclinical models, pol β is strongly induced in response to oxidative stress. Accumulation of DNA damage has been demonstrated in cells with reduced pol β levels (Cabelof et al., 2002). We therefore speculate that in oestrogen/oestrogen receptor driven breast cancers with impaired BER, the consequent genomic instability and accelerated accumulation of mutations may drive an aggressive cancerous phenotype. However, confirmation of such a pol β related mechanism would need detailed pre‐clinical studies which is an area of on‐going investigation in our laboratory. The association with endocrine therapy resistance and low pol β levels in ER positive tumours implies that pol β may be a promising predictive biomarker. Although the mechanism for resistance is unclear, the high proliferation rate consistently observed in pol β deficient tumours in our study may account for the endocrine resistance seen in patients.
We have recently investigated XRCC1, a key BER factor, in breast cancer. Loss of XRCC1 was associated with high grade, loss of hormone receptors, triple negative, basal like phenotypes and poor survival. In ER + cohort that received adjuvant endocrine therapy, low XRCC1 remained associated with poor survival (Sultana et al., 2013). The pol β expression study presented here as well as the XRCC1 data in breast cancer supports the hypothesis that BER pathway may not only operate as a tumour suppressor (Sweasy et al., 2006) but BER down‐regulation may also result in an aggressive phenotype in breast cancer. Moreover, in XRCC1 deficient cells we also demonstrated a novel synthetic lethality application using inhibitors of DNA double strand break repair such as those targeting ATM, DNA‐Pk and ATR (Sultana et al., 2013). Taken together, our data suggests that identification of BER deficiency in breast cancers could be a new approach for a personalized treatment strategy.
Conflict of interest
The authors disclose no potential conflicts of interest.
Authors' contributions
TA.F., S.M. provided intellectual input, conceptual framework, designed the study and helped writing the manuscript. T.A.F, P.M., D.A., G.B., and C.P. collected protein expression, gene expression data and performed data analysis. R.R, C.C. performed polb gene expression analysis in the metabric cohort. N. A performed cell based experiments. S.C. and I.O.E., performed data analysis and helped writing the manuscript. All authors reviewed and approved the final version of the manuscript.
Supporting information
The following are the supplementary material related to this article:
Supplementary Figure S1 Kaplan Meier curves showing disease free survival in whole cohort (A), breast cancer specific survival in ER+, all cases [NPI > 3.4] (B), disease free survival in ER+, all cases [NPI > 3.4] (C), disease free survival in ER + cases that received tamoxifen (D), and disease free survival in ER + cases that did not receive tamoxifen (D).
Supplementary Figure S2 Kaplan Meier curves showing disease free survival in luminal A cases (A), disease free survival in luminal A cases that received tamoxifen (B), disease free survival in luminal A cases that did not receive tamoxifen (C), disease free survival in luminal B cases (D), disease free survival in luminal B cases that received tamoxifen (E) and disease free survival in luminal A cases that did not receive tamoxifen (D).
Supplementary Figure S3 Kaplan Meier curves showing breast cancer specific survival in ER− cohort (primary series) that received no chemotherapy (A), breast cancer specific survival in ER− cohort (primary series) that received chemotherapy (B), breast cancer specific survival in a separate ER− cohort that received no chemotherapy (C), breast cancer specific survival in a separate ER− cohort that received chemotherapy (D).
Supplementary data
Supplementary data
Supplementary material 1.
1.1.
Supplementary material related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.01.001.
Abdel-Fatah Tarek M.A., Russell Roslin, Agarwal Devika, Moseley Paul, Abayomi Michael Ayotunde, Perry Christina, Albarakati Nada, Ball Graham, Chan Stephen, Caldas Carlos, Ellis Ian O. and Madhusudan Srinivasan, (2014), DNA polymerase β deficiency is linked to aggressive breast cancer: A comprehensive analysis of gene copy number, mRNA and protein expression in multiple cohorts, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.01.001.
References
- Albertella, M.R. , Lau, A. , O'Connor, M.J. , 2005. The overexpression of specialized DNA polymerases in cancer. DNA Repair. 4, 583–593. [DOI] [PubMed] [Google Scholar]
- Amouroux, R. , Campalans, A. , Epe, B. , Radicella, J.P. , 2010. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res.. 38, 2878–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armes, J.E. , Hammet, F. , de Silva, M. , Ciciulla, J. , Ramus, S.J. , Soo, W.K. , Mahoney, A. , Yarovaya, N. , Henderson, M.A. , Gish, K. , Hutchins, A.M. , Price, G.R. , Venter, D.J. , 2004. Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene. 23, 5697–5702. [DOI] [PubMed] [Google Scholar]
- Bhat, H.K. , Calaf, G. , Hei, T.K. , Loya, T. , Vadgama, J.V. , 2003. Critical role of oxidative stress in estrogen-induced carcinogenesis. Proc. Natl. Acad. Sci. U.S.A.. 100, 3913–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron, C. , Goldstein, L.D. , Thorne, N.P. , Spiteri, I. , Chin, S.F. , Dunning, M.J. , Barbosa-Morais, N.L. , Teschendorff, A.E. , Green, A.R. , Ellis, I.O. , Tavare, S. , Caldas, C. , Miska, E.A. , 2007. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol.. 8, R214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof, D.C. , Raffoul, J.J. , Yanamadala, S. , Guo, Z. , Heydari, A.R. , 2002. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 23, 1419–1425. [DOI] [PubMed] [Google Scholar]
- Canitrot, Y. , Cazaux, C. , Frechet, M. , Bouayadi, K. , Lesca, C. , Salles, B. , Hoffmann, J.S. , 1998. Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. U.S.A.. 95, 12586–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, S.F. , Teschendorff, A.E. , Marioni, J.C. , Wang, Y. , Barbosa-Morais, N.L. , Thorne, N.P. , Costa, J.L. , Pinder, S.E. , van de Wiel, M.A. , Green, A.R. , Ellis, I.O. , Porter, P.L. , Tavare, S. , Brenton, J.D. , Ylstra, B. , Caldas, C. , 2007. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol.. 8, R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, C. , Shah, S.P. , Chin, S.F. , Turashvili, G. , Rueda, O.M. , Dunning, M.J. , Speed, D. , Lynch, A.G. , Samarajiwa, S. , Yuan, Y. , Graf, S. , Ha, G. , Haffari, G. , Bashashati, A. , Russell, R. , McKinney, S. , Langerod, A. , Green, A. , Provenzano, E. , Wishart, G. , Pinder, S. , Watson, P. , Markowetz, F. , Murphy, L. , Ellis, I. , Purushotham, A. , Borresen-Dale, A.L. , Brenton, J.D. , Tavare, S. , Caldas, C. , Aparicio, S. , 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov, G.L. , Hubscher, U. , 2013. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res.. 41, 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, I.O. , Galea, M. , Broughton, N. , Locker, A. , Blamey, R.W. , Elston, C.W. , 1992. Pathological prognostic factors in breast cancer. II. Histological type. Relationship with survival in a large study with long-term follow-up. Histopathology. 20, 479–489. [DOI] [PubMed] [Google Scholar]
- Elston, C.W. , Ellis, I.O. , 1991. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 19, 403–410. [DOI] [PubMed] [Google Scholar]
- Galea, M.H. , Blamey, R.W. , Elston, C.E. , Ellis, I.O. , 1992. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res. Treat.. 22, 207–219. [DOI] [PubMed] [Google Scholar]
- Gu, H. , Marth, J.D. , Orban, P.C. , Mossmann, H. , Rajewsky, K. , 1994. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science (New York, N.Y. 265, 103–106. [DOI] [PubMed] [Google Scholar]
- Hothorn, T. , Hornik, K. , Zeileis, A. , 2006. Unbiased recursive partitioning: a conditional inference framework. J. Comput. Graph Stat.. 15, 651–674. [Google Scholar]
- Kerangueven, F. , Adelaide, J. , Dib, A. , Longy, M. , Lidereau, R. , Pebusque, M. , Jacquemier, J. , Birnbaum, D. , 1994. Allelic loss at chromosome 8p in human breast-cancer. Oncol. Rep.. 1, 393–395. [DOI] [PubMed] [Google Scholar]
- Kim, Y.J. , Wilson, D.M. , 2012. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol.. 5, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancashire, L.J. , Powe, D.G. , Reis-Filho, J.S. , Rakha, E. , Lemetre, C. , Weigelt, B. , Abdel-Fatah, T.M. , Green, A.R. , Mukta, R. , Blamey, R. , Paish, E.C. , Rees, R.C. , Ellis, I.O. , Ball, G.R. , 2010. A validated gene expression profile for detecting clinical outcome in breast cancer using artificial neural networks. Breast Cancer Res. Treat.. 120, 83–93. [DOI] [PubMed] [Google Scholar]
- Lemetre, C. , 2009. Artificial neural network based algorithm for biomolecular interaction modeling. Bio-inspired systems: computational and ambient intellegence. Lecture Notes Comput Sci.. 5517, 877–885. [Google Scholar]
- Nicolay, N.H. , Helleday, T. , Sharma, R.A. , 2012. Biological relevance of DNA polymerase beta and translesion synthesis polymerases to cancer and its treatment. Curr. Mol. Pharmacol.. 5, 54–67. [PubMed] [Google Scholar]
- Pawitan, Y. , Bjohle, J. , Amler, L. , Borg, A.L. , Egyhazi, S. , Hall, P. , Han, X. , Holmberg, L. , Huang, F. , Klaar, S. , Liu, E.T. , Miller, L. , Nordgren, H. , Ploner, A. , Sandelin, K. , Shaw, P.M. , Smeds, J. , Skoog, L. , Wedren, S. , Bergh, J. , 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res.. 7, R953–R964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pole, J.C. , Courtay-Cahen, C. , Garcia, M.J. , Blood, K.A. , Cooke, S.L. , Alsop, A.E. , Tse, D.M. , Caldas, C. , Edwards, P.A. , 2006. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene. 25, 5693–5706. [DOI] [PubMed] [Google Scholar]
- Poltoratsky, V. , Horton, J.K. , Prasad, R. , Wilson, S.H. , 2005. REV1 mediated mutagenesis in base excision repair deficient mouse fibroblast. DNA Repair. 4, 1182–1188. [DOI] [PubMed] [Google Scholar]
- Sigbjornsdottir, B.I. , Ragnarsson, G. , Agnarsson, B.A. , Huiping, C. , Barkardottir, R.B. , Egilsson, V. , Ingvarsson, S. , 2000. Chromosome 8p alterations in sporadic and BRCA2 999del5 linked breast cancer. J. Med. Genet.. 37, 342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot, M.E. , Ono, K. , Ruscheinski, J. , Wang, P.L. , Ideker, T. , 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics (Oxford, England). 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. , 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol.. 3, Article3 [DOI] [PubMed] [Google Scholar]
- Sobol, R.W. , Horton, J.K. , Kuhn, R. , Gu, H. , Singhal, R.K. , Prasad, R. , Rajewsky, K. , Wilson, S.H. , 1996. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 379, 183–186. [DOI] [PubMed] [Google Scholar]
- Starcevic, D. , Dalal, S. , Sweasy, J.B. , 2004. Is there a link between DNA polymerase beta and cancer?. Cell Cycle (Georgetown, Tex). 3, 998–1001. [PubMed] [Google Scholar]
- Sultana, R. , Abdel-Fatah, T. , Abbotts, R. , Hawkes, C. , Albarakati, N. , Seedhouse, C. , Ball, G. , Chan, S. , Rakha, E.A. , Ellis, I.O. , Madhusudan, S. , 2013. Targeting XRCC1 deficiency in breast cancer for personalized therapy. Cancer Res.. 73, 1621–1634. [DOI] [PubMed] [Google Scholar]
- Sweasy, J.B. , Lang, T. , DiMaio, D. , 2006. Is base excision repair a tumor suppressor mechanism?. Cell Cycle. 5, 250–259. [DOI] [PubMed] [Google Scholar]
- Tagawa, Y. , Yasutake, T. , Ikuta, Y. , Oka, T. , Terada, R. , 2003. Chromosome 8 numerical aberrations in stage II invasive ductal carcinoma: correlation with patient outcome and poor prognosis. Med. Oncol. (Northwood, London, England). 20, 127–136. [DOI] [PubMed] [Google Scholar]
- Tibshirani, R. , Hastie, T. , Narasimhan, B. , Chu, G. , 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U.S.A.. 99, 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini, F. , Adelaide, J. , Charafe-Jauffret, E. , Nguyen, C. , Jacquemier, J. , Jordan, B. , Birnbaum, D. , Pebusque, M.J. , 1999. Differential expression assay of chromosome arm 8p genes identifies Frizzled-related (FRP1/FRZB) and Fibroblast Growth Factor Receptor 1 (FGFR1) as candidate breast cancer genes. Oncogene. 18, 1903–1910. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan, A. , Prychitko, T.M. , Patel, H.V. , Chowdhury, M.E. , Pilling, A.B. , Ventrella-Lucente, L.F. , Papakonstantinou, E.V. , Cabelof, D.C. , Heydari, A.R. , 2011. Folate deficiency regulates expression of DNA polymerase beta in response to oxidative stress. Free Radic. Biol. Med.. 50, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, D.J. , Ramus, S.J. , Hammet, F.M. , de Silva, M. , Hutchins, A.M. , Petrovic, V. , Price, G. , Armes, J.E. , 2005. Complex CGH alterations on chromosome arm 8p at candidate tumor suppressor gene loci in breast cancer cell lines. Cancer Genet. Cytogenet.. 160, 134–140. [DOI] [PubMed] [Google Scholar]
- Wallace, S.S. , Murphy, D.L. , Sweasy, J.B. , 2012. Base excision repair and cancer. Cancer Lett.. 327, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamtich, J. , Nemec, A.A. , Keh, A. , Sweasy, J.B. , 2012. A germline polymorphism of DNA polymerase beta induces genomic instability and cellular transformation. PLoS Genet.. 8, e1003052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamtich, J. , Sweasy, J.B. , 2012. DNA polymerase family X: function, structure, and cellular roles. Biochim. Biophys. Acta. 1804, 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Parsons, J. , Nicolay, N.H. , Caporali, S. , Harrington, C.F. , Singh, R. , Finch, D. , D'Atri, S. , Farmer, P.B. , Johnston, P.G. , McKenna, W.G. , Dianov, G. , Sharma, R.A. , 2010. Cells deficient in the base excision repair protein, DNA polymerase beta, are hypersensitive to oxaliplatin chemotherapy. Oncogene. 29, 463–468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary material related to this article:
Supplementary Figure S1 Kaplan Meier curves showing disease free survival in whole cohort (A), breast cancer specific survival in ER+, all cases [NPI > 3.4] (B), disease free survival in ER+, all cases [NPI > 3.4] (C), disease free survival in ER + cases that received tamoxifen (D), and disease free survival in ER + cases that did not receive tamoxifen (D).
Supplementary Figure S2 Kaplan Meier curves showing disease free survival in luminal A cases (A), disease free survival in luminal A cases that received tamoxifen (B), disease free survival in luminal A cases that did not receive tamoxifen (C), disease free survival in luminal B cases (D), disease free survival in luminal B cases that received tamoxifen (E) and disease free survival in luminal A cases that did not receive tamoxifen (D).
Supplementary Figure S3 Kaplan Meier curves showing breast cancer specific survival in ER− cohort (primary series) that received no chemotherapy (A), breast cancer specific survival in ER− cohort (primary series) that received chemotherapy (B), breast cancer specific survival in a separate ER− cohort that received no chemotherapy (C), breast cancer specific survival in a separate ER− cohort that received chemotherapy (D).
Supplementary data
Supplementary data


