Abstract
Improved prognostic stratification of patients with TNM stage II colorectal cancer (CRC) is desired, since 20–30% of high‐risk stage II patients may die within five years of diagnosis. This study was conducted to investigate REarranged during Transfection (RET) gene promoter CpG island methylation as a possible prognostic marker for TNM stage II CRC patients.
The utility of RET promoter CpG island methylation in tumors of stage II CRC patients as a prognostic biomarker for CRC related death was studied in three independent series (including 233, 231, and 294 TNM stage II patients, respectively) by using MSP and pyrosequencing. The prognostic value of RET promoter CpG island methylation was analyzed by using Cox regression analysis.
In the first series, analyzed by MSP, CRC stage II patients (n = 233) with RET methylated tumors had a significantly worse overall survival as compared to those with unmethylated tumors (HRmultivariable = 2.51, 95%‐CI: 1.42–4.43). Despite a significant prognostic effect of RET methylation in stage III patients of a second series, analyzed by MSP, the prognostic effect in stage II patients (n = 231) was not statistically significant (HRmultivariable = 1.16, 95%‐CI 0.71–1.92). The third series (n = 294), analyzed by pyrosequencing, confirmed a statistically significant association between RET methylation and poor overall survival in stage II patients (HRmultivariable = 1.91, 95%‐CI: 1.04–3.53). Our results show that RET promoter CpG island methylation, analyzed by two different techniques, is associated with a poor prognosis in stage II CRC in two independent series and a poor prognosis in stage III CRC in one series. RET methylation may serve as a useful and robust tool for clinical practice to identify high‐risk stage II CRC patients with a poor prognosis. This merits further investigation.
Keywords: REarranged during transfection (RET), DNA methylation, Methylation marker, Prognostic biomarker, Colon cancer, Colorectal cancer
Highlights
RET methylation predicts poor prognosis for TNM stage II colorectal cancer patients.
The poor prognosis in stage II patients is independent of MSI, CIMP and BRAF status.
RET methylation can be detected by MSP and pyrosequencing.
RET methylation can be detected in fresh‐frozen and FFPE tissue.
RET methylation may serve as a robust biomarker in clinical practice.
1. Introduction
Currently, the main instrument for providing information on prognosis and treatment protocols for colorectal cancer (CRC) patients is the tumor‐node‐metastasis (TNM) staging system (Graziano and Cascinu, 2003; Klump et al., 2004). TNM stage II patients (T3‐4, N0, M0) often do not receive adjuvant therapy, as no supporting evidence is available that this improves the outcome for these patients. Nevertheless, 20–30% of stage II CRC patients die within five years of diagnosis as a result of CRC (Fretwell et al., 2010). Current strategies to identify high‐risk stage II patients, who may benefit from adjuvant chemotherapy, include pathological characteristics, such as the number of examined lymph nodes (<12), tumor perforation, ‐differentiation, ‐budding, and vascular‐, lymphatic‐, or perineural invasion (Benson et al., 2004). However, a reproducible assessment of these variables is challenging (Benson et al., 2004; Betge and Langner, 2011; Compton and Greene, 2004; Lugli et al., 2012; Tsai et al., 2009).
The need for objective tools to identify high‐risk CRC patients is widely accepted. Over the last years, researchers have focused on identifying prognostic biomarkers. Nevertheless, data are still not fully conclusive and according to the ASCO guidelines, molecular markers, such as mutation of KRAS or BRAF and microsatellite instability (MSI) status, are not routinely recommended for prognostic characterization (Locker et al., 2006).
Hypermethylation of promoter CpG islands has been studied extensively as a contributor in CRC carcinogenesis and methylated genes have been suggested as biomarkers for early detection, prognosis or prediction of treatment outcomes for CRC patients (Draht et al., 2012; Kondo and Issa, 2004). So far, only a few genes have been studied as possible prognostic methylation markers for CRC and even less have been validated in larger cohorts or clinical trials (Chan et al., 2008; Krtolica et al., 2007; Mokarram et al., 2009; Tanaka et al., 2011; Umetani et al., 2004; Yi et al., 2011b).
In a genome‐wide analysis of epigenetically altered genes in breast cancer and CRC, the RET gene (REarranged during Transfection) was identified as methylated in CRC (11% methylation) (Chan et al., 2008). In addition, an association between decreased RET expression and poor prognosis in CRC has been described (Chan et al., 2008). These data led us to investigate RET as a possible prognostic methylation marker for CRC.
RET is located on chromosome 10q11.2 and encodes a transmembrane tyrosine kinase receptor (TKR) that has three isoforms, produced by alternative splicing (Runeberg‐Roos and Saarma, 2007). Four ligands, GDNF, Neurturin, Persephin and Artemin, can bind and activate RET, leading to activation of downstream signalling pathways, such as PI3K or MAPK (Arighi et al., 2005; Runeberg‐Roos and Saarma, 2007). The RET gene is highly expressed during embryogenesis and is involved in the development of the enteric nervous system and the kidney, as well as in spermatogenesis and regulation of the thyroid (Arighi et al., 2005; Runeberg‐Roos and Saarma, 2007; Sasselli et al., 2012). Gain‐of‐function mutations of RET are associated with papillary thyroid carcinoma (PTC) and medullary thyroid cancers (MTC) (Phay and Shah, 2010), whereas loss‐of‐function mutations are associated with Hirschsprung disease, which is characterized by a loss of enteric neurons (Phay and Shah, 2010; Plaza‐Menacho et al., 2006).
Recently, RET was identified as a possible tumor suppressor gene in CRC. The authors found that aberrant methylation of RET and mutational inactivation of RET promote CRC formation (Luo et al., 2012). Although RET is proven to be methylated in CRC (Chan et al., 2008; Mokarram et al., 2009; Yi et al., 2011a) and a relationship between RET expression and prognosis in CRC has been identified (Chan et al., 2008), the influence of RET methylation on prognosis in CRC is unclear.
The aim of the current study was to examine RET promoter CpG island methylation as a possible prognostic marker for early stage CRC, employing two different methylation analysis techniques in three independent series consisting of 758 stage II colorectal cancer patients.
2. Methods
2.1. Cell culture
Two human CRC cell lines (HCT116, HT‐29) (ATCC, Manassas, VA, USA) were maintained in DMEM (Invitrogen, Breda, the Netherlands) enriched with 10% FBS (HyClone, Etten‐Leur, the Netherlands).
2.2. Study series and design
RET promoter methylation was initially investigated by bisulfite sequencing and methylation‐specific PCR (MSP) of two fresh‐frozen, primary CRC tissue samples and matched normal colon mucosa tissue samples collected from the archive of the Department of Pathology, Maastricht University Medical Center. RET promoter CpG island methylation as a prognostic marker was investigated in three independent well‐characterized tissue series by two different methylation analysis techniques (Table 1). Archival material was used in compliance with the institutional ethical regulations for use of patient material and national guidelines.
Table 1.
Summary of characteristics and analyses corresponding to series 1, 2 and 3.
| Series 1 | Series 2 | Series 3 |
|---|---|---|
| • All TNM stages (incl. 233 TNM stage II CRC patients) | • TNM stage II & III (incl. 231 TNM stage II CRC patients) | • 294 TNM stage II colon adenocarcinoma patients |
| • Sample collection during 1989–1994 | • Sample collection during 1996–2005 | • Sample collection during 1998–2005 |
| • DNA isolated from FFPE tissue | • DNA isolated from FFPE tissue | • DNA isolated from fresh‐frozen tissue |
| • Methylation detected by MSP | • Methylation detected by MSP | • Methylation detected by Pyrosequencing |
| • Follow‐up data available for more than 10 years | • Follow‐up data available for up to 10 years | • Follow‐up data available for up to 5 years |
| • Data available for OS and DS analysis | • Data available for OS analysis | • Data available for OS analysis |
2.2.1. Series 1
We used formalin‐fixed, paraffin‐embedded (FFPE) tumor tissue samples from a tissue collection of the Netherlands Cohort Study on diet and cancer (NLCS). This cohort study started in 1986 with 120,852 healthy men and women between 55 and 69 years old. Incident cancer cases were identified by computerized linkage with the Netherlands Cancer Registry and PALGA, a nationwide network and registry of histopathology and cytopathology (Van den Brandt et al., 1990b). The NLCS design has been described in more detail elsewhere (van den Brandt et al., 1990a). Briefly, from 1989 until 1994, with the exclusion of the first 2.3‐year of follow‐up, 925 incident CRC cases (ICD‐O: 153.0–154.1) were identified. Of the initial 925 cases, 815 could be linked to a PALGA report. FFPE tumor tissue was collected from 54 pathology registries throughout the Netherlands after approval of the Ethical Review Board of Maastricht University Medical Center. We were able to collect blocks with sufficient carcinoma tissue of 773 CRC cases (Brink et al., 2003). Information on tumor localization, staging, differentiation grade, and incidence date was available through the Netherlands Cancer Registry. Vital status was retrieved from the Central Bureau of Genealogy and the municipal population registries and could be obtained for 716 cases until May 2005. Causes of death for all 716 cases were retrieved through linkage with Statistics Netherlands. CRC related deaths were defined as deaths as a result of a carcinoma in the colon, rectosigmoid, rectum, gastro‐intestinal tract (non‐specific) or liver metastases. In case of gastro‐intestinal (non‐specific) or liver metastases, we used the information from the cancer registry and PALGA to eliminate the possibility of another primary cancer as the cause of death. According to Dutch guidelines, adjuvant treatment was not specifically recommended at the time patients were diagnosed with CRC in the NLCS study. Consequently, the proportion of patients that received adjuvant treatment was low. Within stage I and II cases, only 9% received adjuvant chemotherapy, for the advanced‐stage patients this was 31% for stage III and 19% for stage IV.
We assessed RET as a possible prognostic marker in 716 TNM stage I‐IV CRC patients using MSP. This series consisted of a comparable number of men (n = 395) and women (n = 321) with a mean age of 68 years. TNM stage II was diagnosed in 233 (32.5%) of all patients.
2.2.2. Series 2
The second series consisted of a total of 870 patients that underwent surgical resection for CRC at the Kennemer Gasthuis hospital in Haarlem, the Netherlands, between 1996 and 2005 (Belt et al., 2012). Of these, 554 were classified as TNM stage II (T3‐4, N0, M0) or III (T1‐4, N1‐2, M0) according to the TNM staging system (Hermanek et al., 1987). Patients with positive resection margins and those who were lost to follow‐up or died within three months after surgery were excluded. Patient data were collected from clinical reports and included sex, date of birth, date of surgery, date of death, location of the primary tumor, tumor differentiation grade and adjuvant therapy. Adjuvant 5‐FU‐based chemotherapy was administered to 11.0% of stage II patients and 37.7% of stage III patients. Collection, storage and use of tissue and patient data were performed in agreement with the ‘Code for Proper Secondary Use of Human Tissue in The Netherlands’. The methylation status of FFPE material of a total of 425 TNM stage II and III patients was assessed by MSP and statistical analysis could be done for a total of 393 patients. TNM stage II tumor material was available for 117 male patients and 114 female patients with a mean age of 71,8 years at diagnosis.
2.2.3. Series 3
As a third series, we used resected tumor tissue provided by the Ferdinand Cabanne Biological Resources Centre (Dijon, France). Fresh frozen colon adenocarcinoma tissue from patients diagnosed with colon cancer, who underwent surgical resection between January 1998 and December 2005, was provided by three pathology laboratories covering the area: Dijon University Hospital, Comprehensive Cancer Centre, and a private Center. Five fragments, considered as surgical waste in accordance with French ethical laws (L.1211‐3 to L.1211‐9), were obtained from each tumor and were immediately frozen in liquid nitrogen (Barault et al., 2008). From the digestive cancer Registry of Burgundy, data were provided on patients' characteristics, cancer site, stage at diagnosis, treatment, recurrence, and date of death. TNM stage II (Sobin and Fleming, 1997) colon adenocarcinoma tissue was available from 294 patients (172 men and 122 women; mean age = 71.9 years) and was used for RET methylation analysis by pyrosequencing. 13.6% of 294 patients received adjuvant chemotherapy.
2.3. DNA isolation
Genomic DNA of cell lines was extracted by using the Puregene DNA isolation kit (Gentra Systems, MN, USA) according to the manufacturer's protocol. DNA from FFPE tissue of series 1 was extracted using the Puregene DNA isolation kit (Gentra Systems), as previously described (Brink et al., 2003). DNA from FFPE tissue of series 2 was isolated by using the QIAamp microkit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. In order to isolate DNA from fresh‐frozen tissue, the Nucleospin®96 tissue kit (Macherey–Nagel, Düren, Germany) was used as previously described (Tournier et al., 2012).
2.4. Determination of microsatellite instability (MSI), CpG island methylator phenotype (CIMP) and BRAF mutation status
MSI, CIMP or BRAF mutation status of series 1 and series 3 has already been investigated and was known from previous studies (Barault et al., 2008; Simons et al., 2013). This was also the case for MSI status in series 2 (Belt et al., 2010). However, for the current study, CIMP in series 2 was investigated by MSP with the marker panel proposed by Weisenberger et al. (2006). CRC samples were defined as CIMP positive when ≥3/5 analyzed markers (CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1) were methylated. BRAF mutation status was investigated by sequencing.
2.5. Sodium bisulfite conversion, MSP and bisulfite sequencing
Sodium bisulfite modification of genomic DNA was performed using the EZ DNA methylation kit (ZYMO Research Corporation, Orange, CA, USA; in case of series 1 & 2) or the EpiTect Bisulfite kit (Qiagen, Hilden, Germany; in case of series 3) according to the manufacturer's instructions. Bisulfite sequencing was conducted as previously described (Melotte et al., 2009). Nested MSP was performed as described in detail elsewhere (Derks et al., 2004; Herman et al., 1996). Primer pairs were designed near the putative transcriptional start site (TSS; Figure 1a). The primer sequences of RET are provided in Supplementary Table 1.
Figure 1.
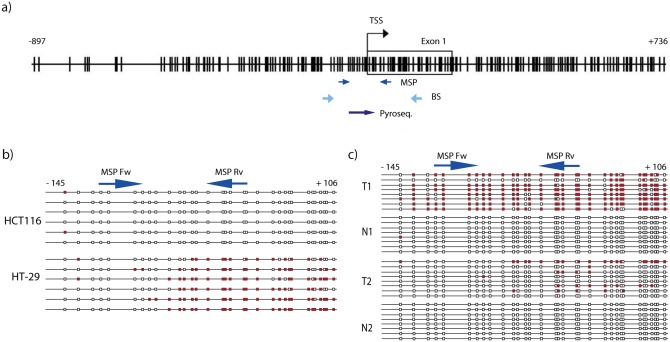
RET promoter structure, promoter CpG island methylation in CRC cell lines and primary tissue. a) Schematic representation of the RET promoter region. Vertical lines represent the location of CpG dinucleotides. Location of MSP primers, bisulfite sequencing primers (BS) and pyrosequencing region (pyroseq.), are indicated by arrows (TSS = transcription start site). b) Schematic representation of bisulfite sequencing results of two human CRC cell lines. Bisulfite sequencing of HCT116 and HT‐29 was performed to investigate the methylation pattern of our region of interest. Six different bacterial clones were sequenced. Each row represents an individual cloned allele that was sequenced following bisulfite conversion. Each box indicates a CpG dinucleotide (red box = methylated CpG site; white box = unmethylated CpG site). c) Bisulfite sequencing of primary CRC (T) and matched normal colon (N) tissue of two patients. At least eight different bacterial clones were sequenced. Each row represents an individual cloned allele that was sequenced. Each column represents a CpG dinucleotide (red box = methylated; white box = unmethylated). Arrows indicate the location of MSP primers (MSP Fw = forward primer; MSP Rv = reverse primer). In order to investigate the RET promoter region by MSP, primers were designed 115 base pairs (bp) around the TSS (−98 and +17 bp relative to the TSS). MSP confirmed the results obtained by bisulfite sequencing.
2.6. Pyrosequencing assay
Pyrosequencing primers were designed within the MSP region (Figure 1a). Pyrosequencing was performed on series 3 using the PyroMark Q24 kit (Qiagen, Hilden, Germany). Templates for pyrosequencing were obtained by amplifying bisulfite‐treated DNA. PCR reactions were carried out in a 25 μL final volume comprising 12.5 μL of PyroMark Master Mix, 2.5 μL of Coral Load buffer, 0.5 μL of forward and biotinylated reverse primers (0.2 μM final concentration), 8 μL of RNase‐free water and 1 μL of bisulfite‐treated DNA (20 ng). The biotinylated PCR products were processed as previously described (Tournier et al., 2012). Results were analyzed using PyroMark Q24 2.0.6 Software. To ensure successful bisulfite conversion, an internal conversion control that corresponded to the position of a non‐CG cytosine was present in the dispensation sequence. Pyrosequencing can measure the methylation level of single CpG sites in a promoter. For data analysis, we averaged the percentage of methylation of the first 6 CpG sites of the analyzed region. To determine a cut‐off value for RET methylation, we compared 30 adenocarcinoma samples (subset from series 3) with both pyrosequencing and MSP (supplementary figure S1). Consequently, we defined a mean methylation level of 20% or higher, as methylation‐positive.
2.7. Data analysis
Disease‐specific (DS) survival was defined as time from cancer diagnosis until CRC‐related death or end of follow‐up. Overall survival (OS) was defined as time from cancer diagnosis until death of all causes. DS and OS analyses were performed on series 1, whereas for series 2 and 3, only OS analyses were possible. Follow‐up data of series 1 and 2 were available for at least 10 years, whereas for series 3, follow‐up data were available for up to five years after diagnosis (Table 1). Analyses were performed for three and five years, since results from series 1 and 2 indicated higher prognostic effects between three and five years. Kaplan–Meier curves and log‐rank tests were used to estimate the influence of methylation on overall survival (OS) and DS. Hazard Ratios (HR) and corresponding 95% confidence intervals (CI) were assessed by use of Cox proportional hazard model adjusted for potential confounders. Factors were considered possible confounders if they were known prognostic factors for CRC and influenced the crude HR. Possible confounders that were included in the model were age at diagnosis (continuous), sex, tumor differentiation grade (well, moderate, poor and undifferentiated) and location (proximal, distal, rectosigmoid and rectum). The proportional hazard assumption was tested using the Schoenfeld residuals and the log(‐log) hazards plots. All analyses were done with the statistical package STATA 11.0.
3. Results
3.1. RET promoter CpG island methylation
Our region of interest is located in a dense CpG island located in the promoter region of RET and is frequently methylated in colon cancer tissue, but not in normal colon tissue (Figure 1). MSP primers were designed based on methylation data of human CRC cell lines and primary tissue, obtained by bisulfite sequencing (Figure 1b and c).
3.2. RET promoter CpG island methylation in TNM stage II CRC and colon cancer patients
The utility of RET promoter CpG island methylation in tumors of stage II CRC patients as a prognostic biomarker for CRC related death was studied in three independent series. The clinical and pathological characteristics of TNM stage II CRC patients stratified for RET promoter methylation, are summarized in Table 2. No statistically significant differences in tumor sub‐location, tumor differentiation grade and sex were found between methylated and unmethylated cases in series 1 and 2. Series 3 did not include rectal cancer cases and significantly more methylated tumors were found in the proximal colon (p = 0.006).
Table 2.
Clinicopathologic characteristics of TNM stage II CRC patients within the three study cohorts.
| RET (TNM stage II) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Series 1 | Series 2 | Series 3 | ||||||||
| U | M | p‐value | U | M | p‐value | U | M | p‐value | ||
| N (%) | 195 (83.7) | 38 (16.3) | 172 (74.46) | 59 (25.5) | 257 (87.41) | 37 (12.59) | ||||
| Sublocation, N (%) | Proximal | 71 (36.8) | 21 (55.3) | 72 (41.86) | 32 (54.24) | 110 (42.80) | 26 (70.27) | |||
| Distal | 65 (33.7) | 9 (23.7) | 76 (44.19) | 21 (35.59) | 97 (37.74) | 6 (16.22) | ||||
| Rectosigmoid | 27 (14.0) | 3 (7.9) | 24 (13.95) | 6 (10.17) | 0.253 | 50 (19.46) | 5 (13.51) | 0.006 | ||
| Rectum | 30 (15.5) | 5 (13.2) | 0.192 | |||||||
| Differentiation grade | Well | 15 (8.4) | 1 (2.9) | 14 (8.14) | 4 (6.78) | * | * | |||
| Moderate | 139 (78.1) | 30 (88.2) | 135 (78.49) | 50 (84.75) | * | * | ||||
| Poor | 23 (12.9) | 3 (8.8) | 23 (13.37) | 5 (8.47) | 0.552 | * | * | |||
| Undifferentiated | 1 (0.6) | – | 0.554 | * | * | |||||
| Sex, N (%) | Male | 111 (56.9) | 22 (57.9) | 0.912 | 91 (51.91) | 26 (44.07) | 0.241 | 155 (60.31) | 17 (45.94) | 0.097 |
| Age at diagnosis, mean (SD) | 68.2 (4.5) | 68.7 (4.7) | 0.506 | 71.4(12.31) | 73.1 (9.92) | 0.345 | 71.68 (11.98) | 73.7 (9.79) | 0.164 | |
* Data not available.
3.2.1. Series 1
First, we assessed RET as a possible prognostic marker in series 1 (consisting of 716 TNM stage I‐IV CRC patients) using MSP. Seventeen percent of all patients and 16.3% of stage II patients showed RET methylation.
Kaplan–Meier analyses showed a statistically significant association between RET promoter CpG island methylation and disease‐specific survival (DS) in the total series (p = 0.002; Figure 2a), and this was also seen in age and sex adjusted Cox regression analysis (HRage/sex 1.55; 95%‐CI: 1.17–2.02). However, a multivariable analysis adjusted for age, sex, tumor location and tumor differentiation grade, showed no significant association between RET methylation and DS (HRmultivariable 1.36; 95%‐CI 0.98–1.87). As microsatellite instability (MSI) is an independent prognostic factor for CRC, we excluded MSI cases to investigate whether a prognostic value of RET methylation was restricted to microsatellite stable (MSS) tumors (n = 567). In MSS cases a significant difference was observed between cases with and without RET methylation (Figure 2b, p = 0.016; HRage/sex 1.49; 95%‐CI 1.08–20.4). However, this effect was not significant in the multivariable analysis (HRmultivariable 1.21; 95%‐CI: 0.85–1.73). Overall survival (OS) analysis results did not alter our conclusions (data not shown).
Figure 2.
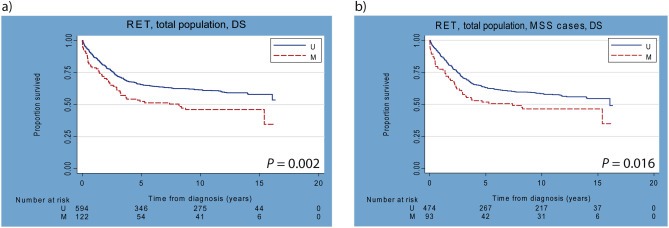
Kaplan–Meier curves of total series 1 with and without MSI cases. a) Disease‐specific (DS) survival analysis of total series 1 (n = 716; TNM stage I–IV). b) DS survival analysis of total series 1, excluding MSI cases (MSS only: n = 567).
Figure 3a shows the Kaplan–Meier curves for stage II CRC patients for OS analysis at five‐year follow‐up, indicating that patients with RET methylation have a poorer prognosis than the unmethylated cases (p = 0.002), independent of age, sex, sub‐location or differentiation grade of the tumor (HRage/sex 2.28; 95%‐CI: 1.35–3.87; HRmultivariable 2.51; 95%‐CI: 1.42–4.43). DS analysis showed similar results (Table 3 and Figure S2; p = 0.008; HRage/sex 2.31; 95%‐CI 1.25–4.28; HRmultivariable 2.49; 95%‐CI: 1.31–4.74). Results at three‐year follow‐up did not differ from results at five‐year follow‐up (OS: HRage/sex 2.20; 95%‐CI: 1.16–4.17; HRmultivariable 2.46; 95%‐CI: 1.26–4.81; DS: HRage/sex 2.28; 95%‐CI: 1.14–4.60; HRmultivariable 2.56; 95%‐CI: 1.23–5.32). Exclusion of MSI, CIMP‐positive and BRAF mutant cases did not alter our conclusion (data not shown). Also, exclusion of patients, who received adjuvant treatment, did not alter our conclusion (Table 3). For other TNM stages, no statistically significant associations were observed (data not shown). Overall, these data suggested that the association between RET methylation and poor prognosis is limited to stage II CRC patients. This hints at the suitability of RET as a possible prognostic marker. No significant difference in survival was found between stage II cases with different tumor locations. However, a survival analysis for colon versus rectal cancer subgroups was not possible, due to small number of stage II rectal cancer cases in series 1.
Figure 3.
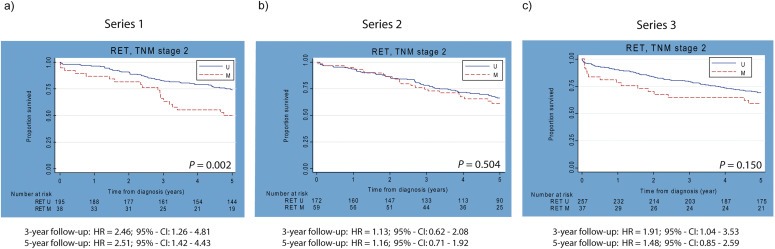
Kaplan–Meier curves for TNM stage II patients of three independent series. a) Kaplan–Meier curves for OS analysis of series 1 (n = 233) at five years of follow‐up. b) Kaplan–Meier curve for OS analysis of series 2 (n = 231) at five years of follow‐up. c) Kaplan–Meier curve for OS analysis of series 3 (n = 294) at five years of follow‐up. Corresponding p‐values are depicted in each Kaplan–Meier curve. Multivariable adjusted hazard ratios (HRs) for three‐ and five‐year follow up are indicated below Kaplan–Meier curves. U = unmethylated, M = methylated.
Table 3.
Multivariable adjusted hazard ratios (HR) and corresponding 95% Confidence Intervals (CI) for RET methylation in TNM stage II patients of three independent series.
| Series 1 | Series 2 | Series 3 | |
|---|---|---|---|
| Overall survival | |||
| 3‐year follow‐up | 2.46 (1.26–4.81) | 1.13 (0.62–2.08) | 1.91 (1.04–3.53) |
| 2.60 (1.18–5.69)a | 1.06 (0.57–1.99)a | 2.23 (1.17–4.24)a | |
| 5‐year follow‐up | 2.51 (1.42–4.43) | 1.16 (0.71–1.92) | 1.48 (0.85–2.59) |
| 2.69 (1.42–5.09)a | 1.06 (0.63–1.79)a | 1.69 (0.95–3.02)a | |
| Disease‐specific survival | |||
| 3‐year follow‐up | 2.56 (1.23–5.32) | n/a | n/a |
| 3.28 (1.44–7.48)a | |||
| 5‐year follow‐up | 2.49 (1.31–4.74) | ||
| 3.13 (1.54‐6.35)a | |||
HRs for untreated patients only.
3.2.2. Series 2
To validate our findings, we investigated RET as a possible prognostic methylation marker in a second independent series, consisting of 393 CRC TNM stage II and III patients, using the same MSP primers. RET promoter methylation was observed in 59 (25.5%) of 231 TNM stage II patients and 37 (22.8%) of 162 TNM stage III patients. Figure 3b displays the Kaplan–Meier curve for OS analysis of stage II patients for five‐year follow‐up, indicating no statistically significant difference between methylated and unmethylated cases (p = 0.504). In contrast to series 1, an association between RET methylation and poor overall survival in series 2 was statistically not significant (HRage/sex 1.20, 95%‐CI: 0.73–1.97; HRmultivariable 1.16, 95%‐CI: 0.71–1.92). Exclusion of other molecular alterations that might be associated with survival (MSI present in 21.1%, CIMP present in 45.0%, BRAF mutations present in 18.5%) did not alter our conclusion (data not shown). Survival analyses for three years follow‐up showed similar results for stage II cases (HRage/sex 1.20; 95%‐CI: 0.66–2.20; HRmultivariable 1.13; 95%‐CI: 0.62–2.08). Analyses excluding patients that received adjuvant therapy also showed no survival influence of RET methylation (3‐year follow‐up: HRmultivariable 1.06; 95%‐CI: 0.57–1.99; 5‐year follow‐up: HRmultivariable 1.06; 95%‐CI: 0.63–1.79; Table 3).
Strikingly, analyses for TNM stage III patients showed a strong prognostic effect of RET methylation for five years of follow‐up (Kaplan–Meier p = 0.0008; HRage/sex 2.08; 95%‐CI: 1.29–3.37; HRmultivariable 2.04; 95%‐CI: 1.23–3.37) and for three years of follow‐up (HRage/sex 1.86; 95%‐CI: 1.07–3.26; HRmultivariable 1.75; 95%‐CI: 0.97–3.17).
3.2.3. Series 3
To further elucidate the potential of RET methylation as a possible biomarker, we investigated RET promoter CpG island methylation using quantitative pyrosequencing method in a third independent series, consisting of 294 fresh‐frozen colon adenocarcinoma tissue samples from stage II patients only. RET was methylated in 12.6% of all patients. Figure 3c depicts the Kaplan–Meier curve for five‐year follow‐up. Although a trend was observed, a significant association between RET methylation and poor prognosis after five years of follow‐up, was absent and this was also seen by Cox proportional hazard analysis (Table 3; HRage/sex 1.55; 95%‐CI: 0.88–2.75; HRmultivariable 1.48; 95%‐CI: 0.85–2.59). However, at three years after diagnosis, a significant association between RET methylation and poor overall survival was observed (Table 3). Cox proportional hazard analysis adjusted for age and sex showed a significant association between methylated RET and poor prognosis in stage II patients (HRage/sex 1.87, 95%‐CI: 1.00–3.48). Multivariable survival analyses showed a significantly poorer prognosis (HRmultivariable 1.91; 95%‐CI: 1.04–3.53). Analyses excluding patients that received adjuvant therapy showed poorer overall survival for RET methylated cases (3‐year follow‐up: HRmultivariable 2.23; 95%‐CI: 1.17–4.24; 5‐year follow up: HRmultivariable 1.69; 95%‐CI: 0.95–3.02, Table 3). Analysis of CIMP negative cases only, was not possible in this population. Exclusion of BRAF mutant cases did not affect the prognostic effect of RET methylation (data not shown). Thus, RET promoter CpG island methylation detected by pyrosequencing, was associated with poorer overall survival after three years of follow‐up.
4. Discussion
In the last decade many studies have been conducted to seek for molecular biomarkers to identify high‐risk stage II CRC patients. Recently developed gene expression profiling (GEP) assays, such as the ColoPrint (Agendia BV, Amsterdam, The Netherlands) (Salazar et al., 2011), Oncotype DX® Colon Cancer Assay (Genomic Health, Los Angeles, USA) (Clark‐Langone et al., 2010) and OncoDefender™–CRC (Everist Genomics, Ann Arbor, USA) (Lenehan et al., 2012), are commercially available molecular marker assays to predict disease prognosis for high‐risk stage II CRC patients, but up till now they are not conventionally used in clinical practice. So far, the level of evidence for the OncotypeDX Colon Cancer assay and the OncoDefender‐CRC assay is still low, whereas the Coloprint assay has been validated in two independent studies and a prospective trial is ongoing. However, limitations of these GEP assays are that the tests cannot be used in‐house and an elaborately processing of material is required, which can take 10–12 working days of generating results and costs per test range between USD 3000 and 4000. In addition, cost effectiveness analyses for these tests have yet to be performed.
Considering these limitations and as it remains to be observed how these assays will be implemented in daily clinical practice, the search for novel prognostic biomarkers for stage II CRC patients is still essential. Here we demonstrate for the first time, that RET promoter CpG island methylation is associated with a poor five‐year and three‐year overall survival in two independent series (n = 233, n = 294). The prognostic effect of RET methylation was also present even after excluding known molecular markers, such as MSI, CIMP and BRAF mutation. Strikingly, in series 2, RET methylation was associated with survival in stage III patients. Remarkable is that RET methylation was more abundant in series 2 (25.5%) as compared to series 1 (16.3%) and series 3 (12.6%). Alongside, it is also notably that CIMP positive and BRAF mutated cases were highly prevalent in series 2. Series 2 has been collected retrospectively from a restricted geographical location, whereas series 1 and series 3 have been prospectively collected nation‐wide or from a wider geographical area, respectively. This might indicate a positive selection towards a specific type of patient in series 2.
We also showed that RET promoter CpG island methylation is a robust marker, associated with a poor prognosis in stage II CRCs and colon cancers, measured by two different techniques (Table 1). MSP is a qualitative technique and is commonly used to detect DNA methylation in biomarker studies (Derks et al., 2004; Herman et al., 1996). Pyrosequencing is a quantitative method and has single nucleotide resolution. In the current study, the prognostic potential of RET was detected in DNA obtained from tissue preserved by two different methods, the FFPE‐ and the fresh‐frozen tissue preservation. In the last years, both, the preservation method and the method to detect DNA methylation have been subject of discussion (Claus et al., 2012; Tournier et al., 2012). However, our study shows that RET methylation has prognostic potential for stage II CRC patients regardless of preservation method and detection method. In addition, the prognostic potential of RET was found in two independent series, analyzed in two different laboratories. The suitability of both, techniques and tissue types for methylation analysis, could be advantageous for implementation into daily clinical practice, as compared to above described GEP assays. Many biomarker studies report, that previous findings cannot be reproduced using different techniques or different tissues. Here, we describe a robust methylation marker suitable for both FFPE and fresh‐frozen tissue.
Current strategies to identify high‐risk CRC patients suggest that more than 12 lymph nodes should be investigated. It has been found that a high lymph node yield is associated with a favorable prognosis in stage II patients. However, high lymph node yields cannot always be accomplished in clinical practice. This also holds true for one of our series, as sample collection started 30 years ago. An association between RET promoter methylation and the lymph node yield, as well as other pathologic prognostic factors has still to be investigated.
Understanding the biological role of a biomarker and its role in tumorigenesis could aid in interpreting study results, for instance in case of contradictory results, and to make treatment decisions. So far, the biological role of RET methylation in CRC tumorigenesis is not completely understood. Luo et al. recently showed that RET has tumor‐suppressive functions in CRC, and is silenced by hypermethylation of the RET promoter (Luo et al., 2012). However, their region of interest was located about 410 bp more upstream from our region. RET encodes a tyrosine kinase, involved in the enteric nervous system development (Sasselli et al., 2012). As investigated by Luo et al., RET, when activated, leads to the induction of apoptosis in CRC. RET belongs to the family of dependence receptors, having the ability to trigger apoptosis if expressed in the absence of ligands, thereby controlling tumor progression (Cabrera et al., 2011). Although the function of dependence receptors is not completely understood yet, it is hypothesized that tumor development is inhibited by inducing apoptosis of tumor cells growing outside the region of ligand availability, thereby avoiding metastasis. Most dependence receptors indeed seem to act as tumor suppressors and it is thought that loss of the dependence receptor is necessary for metastasis (Diaz‐Rodriguez et al., 2012; Goldschneider and Mehlen, 2010; Mehlen and Puisieux, 2006).
In various studies, it has been reported that overexpression of the RET protein and the RET receptor ligand GDNF, leads to perineural invasion in pancreatic cancer (Cavel et al., 2012; Zeng et al., 2008). However, the role of RET during perineural invasion in CRC needs still to be elucidated. Most likely, a tumor suppressive function of RET is blocked by promoter CpG island methylation, leading to downregulation of RET mRNA expression and RET protein expression. Taken together, our findings and those of others suggest that RET, and in particular RET methylation, plays an important role in CRC progression, explaining the prognostic role for high‐risk stage II patients.
Here, we described RET promoter methylation as a possible prognostic marker for stage II CRC patients in two independent series, using qualitative MSP and quantitative pyrosequencing, reproduced in two different laboratories. Despite RET methylation was also associated with a poorer prognosis in stage III patients in one series, RET seems to be a promising, robust and affordable prognostic marker for CRC patients being at high risk of disease recurrence. To increase the level of evidence of RET as a prognostic marker, our results need to be validated in an observational prospective study. The future of optimal CRC staging and disease management might cover a combination of clinicopathologic prognostic factors and prognostic biomarkers, such as RET promoter methylation.
Conflict of interest statement
The authors declare to have no conflicts of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary Figure 1 Defining the methylation cut‐off for pyrosequencing. Pyrosequencing results of 30 adenocarcinoma samples from series 3 were compared by MSP. Pyrosequencing results were divided into three groups: clearly methylated (>five CG sites showed methylation of 15% or higher), clearly unmethylated (no CG site showed methylation above 10%) and intermediate or no clear methylation status (outlying methylation of single CGs above 15%). Clearly methylated samples with an average methylation above 20% by pyrosequencing showed also methylation by MSP, whereas clearly unmethylated samples by pyrosequencing were also unmethylated by MSP. Samples with intermediate/no clear methylation status, were unmethylated by MSP. Accordingly, a pyrosequencing cut‐off was set to an average methylation of 20% or higher. Each row represents results for one colon adenocarcinoma sample. Squares represent the location of CG sites.
Supplementary Figure 2 Kaplan–Meier curve for disease specific (DS) survival in stage II patients from series 1. Kaplan–Meier curve for the association of RET promoter CpG island methylation and DS at 5‐year follow up. U = unmethylated, M = methylated.
Acknowledgements
This work was performed within the framework of CTMM, the Center for Translational Molecular Medicine project DeCoDe (grant 03O‐101).
We would like to pay tribute to Prof. Françoise Piard, who enthusiastically initiated the collaboration between the Pathology Departments of Dijon and Maastricht.
We thank Dr. Hanneke Niessen for her advice with the manuscript and Kim van Straeten and Kathleen Daenen, for their excellent technical support with the tissue collection and MSP experiments.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.01.011
Draht Muriel X.G., Smits Kim M., Tournier Benjamin, Jooste Valerie, Chapusot Caroline, Carvalho Beatriz, Cleven Arjen H.G., Derks Sarah, Wouters Kim A.D., Belt Eric J.T., Stockmann Hein B.A.C., Bril Herman, Weijenberg Matty P., van den Brandt Piet A., de Bruïne Adriaan P., Herman James G., Meijer Gerrit A., Piard Françoise, Melotte Veerle and van Engeland Manon, (2014), Promoter CpG island methylation of RET predicts poor prognosis in stage II colorectal cancer patients, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.01.011.
References
- Arighi, E. , Borrello, M. , Sariola, H. , 2005. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev.. 16, 441–467. [DOI] [PubMed] [Google Scholar]
- Barault, L. , Charon-Barra, C. , Jooste, V. , de la Vega, M.F. , Martin, L. , Roignot, P. , Rat, P. , Bouvier, A.M. , Laurent-Puig, P. , Faivre, J. , Chapusot, C. , Piard, F. , 2008. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res.. 68, 8541–8546. [DOI] [PubMed] [Google Scholar]
- Belt, E.J. , te Velde, E.A. , Krijgsman, O. , Brosens, R.P. , Tijssen, M. , van Essen, H.F. , Stockmann, H.B. , Bril, H. , Carvalho, B. , Ylstra, B. , Bonjer, H.J. , Meijer, G.A. , 2012. High lymph node yield is related to microsatellite instability in colon cancer. Ann. Surg. Oncol.. 19, 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belt, E.J. , van Stijn, M.F. , Bril, H. , de Lange-de Klerk, E.S. , Meijer, G.A. , Meijer, S. , Stockmann, H.B. , 2010. Lymph node negative colorectal cancers with isolated tumor deposits should be classified and treated as stage III. Ann. Surg. Oncol.. 17, 3203–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, A.B. , Schrag, D. , Somerfield, M.R. , Cohen, A.M. , Figueredo, A.T. , Flynn, P.J. , Krzyzanowska, M.K. , Maroun, J. , McAllister, P. , Van Cutsem, E. , Brouwers, M. , Charette, M. , Haller, D.G. , 2004. American society of clinical oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J. Clin. Oncol.. 22, 3408–3419. [DOI] [PubMed] [Google Scholar]
- Betge, J. , Langner, C. , 2011. Vascular invasion, perineural invasion, and tumour budding: predictors of outcome in colorectal cancer. Acta Gastro-enterologica Belgica. 74, 516–529. [PubMed] [Google Scholar]
- Brink, M. , de Goeij, A.F. , Weijenberg, M.P. , Roemen, G.M. , Lentjes, M.H. , Pachen, M.M. , Smits, K.M. , de Bruine, A.P. , Goldbohm, R.A. , van den Brandt, P.A. , 2003. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 24, 703–710. [DOI] [PubMed] [Google Scholar]
- Cabrera, J.R. , Bouzas-Rodriguez, J. , Tauszig-Delamasure, S. , Mehlen, P. , 2011. RET modulates cell adhesion via its cleavage by caspase in sympathetic neurons. J. Biol. Chem.. 286, 14628–14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavel, O. , Shomron, O. , Shabtay, A. , Vital, J. , Trejo-Leider, L. , Weizman, N. , Krelin, Y. , Fong, Y. , Wong, R.J. , Amit, M. , Gil, Z. , 2012. Endoneurial macrophages induce perineural invasion of pancreatic cancer cells by secretion of GDNF and activation of RET tyrosine kinase receptor. Cancer Res.. 72, 5733–5743. [DOI] [PubMed] [Google Scholar]
- Chan, T.A. , Glockner, S. , Yi, J.M. , Chen, W. , Van Neste, L. , Cope, L. , Herman, J.G. , Velculescu, V. , Schuebel, K.E. , Ahuja, N. , Baylin, S.B. , 2008. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med.. 5, e114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Langone, K.M. , Sangli, C. , Krishnakumar, J. , Watson, D. , 2010. Translating tumor biology into personalized treatment planning: analytical performance characteristics of the Oncotype DX Colon Cancer Assay. BMC Cancer. 10, 691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus, R. , Wilop, S. , Hielscher, T. , Sonnet, M. , Dahl, E. , Galm, O. , Jost, E. , Plass, C. , 2012. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics. 7, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, C.C. , Greene, F.L. , 2004. The staging of colorectal cancer: 2004 and beyond. CA Cancer J. Clin.. 54, 295–308. [DOI] [PubMed] [Google Scholar]
- Derks, S. , Lentjes, M.H. , Hellebrekers, D.M. , de Bruine, A.P. , Herman, J.G. , van Engeland, M. , 2004. Methylation-specific PCR unraveled. Cell Oncol.. 26, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodriguez, E. , GarcÃa-Lavandeira, M. , Perez-Romero, S. , Senra, A. , Cañibano, C. , Palmero, I. , Borrello, M. , Dieguez, C. , Alvarez, C. , 2012. Direct promoter induction of p19Arf by Pit-1 explains the dependence receptor RET/Pit-1/p53-induced apoptosis in the pituitary somatotroph cells. Oncogene. 31, 2824–2835. [DOI] [PubMed] [Google Scholar]
- Draht, M.X. , Riedl, R.R. , Niessen, H. , Carvalho, B. , Meijer, G.A. , Herman, J.G. , van Engeland, M. , Melotte, V. , Smits, K.M. , 2012. Promoter CpG island methylation markers in colorectal cancer: the road ahead. Epigenomics. 4, 179–194. [DOI] [PubMed] [Google Scholar]
- Fretwell, V.L. , Ang, C.W. , Tweedle, E.M. , Rooney, P.S. , 2010. The impact of lymph node yield on Duke's B and C colorectal cancer survival. Colorectal Dis.. 12, 995–1000. [DOI] [PubMed] [Google Scholar]
- Goldschneider, D. , Mehlen, P. , 2010. Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene. 29, 1865–1882. [DOI] [PubMed] [Google Scholar]
- Graziano, F. , Cascinu, S. , 2003. Prognostic molecular markers for planning adjuvant chemotherapy trials in Dukes' B colorectal cancer patients: how much evidence is enough?. Ann. Oncol.. 14, 1026–1038. [DOI] [PubMed] [Google Scholar]
- Herman, J.G. , Graff, J.R. , Myohanen, S. , Nelkin, B.D. , Baylin, S.B. , 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U S A. 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanek, P. , Scheibe, O. , Spiessl, B. , Wagner, G. , 1987. TNM classification of malignant tumors: a new 1987 edition. Der Pathologe. 8, 137 [PubMed] [Google Scholar]
- Klump, B. , Nehls, O. , Okech, T. , Hsieh, C.J. , Gaco, V. , Gittinger, F.S. , Sarbia, M. , Borchard, F. , Greschniok, A. , Gruenagel, H.H. , Porschen, R. , Gregor, M. , 2004. Molecular lesions in colorectal cancer: impact on prognosis? Original data and review of the literature. Int. J. Colorectal Dis.. 19, 23–42. [DOI] [PubMed] [Google Scholar]
- Kondo, Y. , Issa, J.P. , 2004. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev.. 23, 29–39. [DOI] [PubMed] [Google Scholar]
- Krtolica, K. , Krajnovic, M. , Usaj-Knezevic, S. , Babic, D. , Jovanovic, D. , Dimitrijevic, B. , 2007. Comethylation of p16 and MGMT genes in colorectal carcinoma: correlation with clinicopathological features and prognostic value. World J. Gastroenterol. WJG. 13, 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenehan, P.F. , Boardman, L.A. , Riegert-Johnson, D. , De Petris, G. , Fry, D.W. , Ohrnberger, J. , Heyman, E.R. , Gerard, B. , Almal, A.A. , Worzel, W.P. , 2012. Generation and external validation of a tumor-derived 5-gene prognostic signature for recurrence of lymph node-negative, invasive colorectal carcinoma. Cancer. 118, 5234–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker, G.Y. , Hamilton, S. , Harris, J. , Jessup, J.M. , Kemeny, N. , Macdonald, J.S. , Somerfield, M.R. , Hayes, D.F. , Bast, R.C. , Asco, 2006. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol.. 24, 5313–5327. [DOI] [PubMed] [Google Scholar]
- Lugli, A. , Karamitopoulou, E. , Zlobec, I. , 2012. Tumour budding: a promising parameter in colorectal cancer. Br. J. Cancer. 106, 1713–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Tsuchiya, K.D. , Il Park, D. , Fausel, R. , Kanngurn, S. , Welcsh, P. , Dzieciatkowski, S. , Wang, J. , Grady, W.M. , 2012. RET is a potential tumor suppressor gene in colorectal cancer. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen, P. , Puisieux, A. , 2006. Metastasis: a question of life or death. Nat. Rev. Cancer. 6, 449–458. [DOI] [PubMed] [Google Scholar]
- Melotte, V. , Lentjes, M.H. , van den Bosch, S.M. , Hellebrekers, D.M. , de Hoon, J.P. , Wouters, K.A. , Daenen, K.L. , Partouns-Hendriks, I.E. , Stessels, F. , Louwagie, J. , Smits, K.M. , Weijenberg, M.P. , Sanduleanu, S. , Khalid-de Bakker, C.A. , Oort, F.A. , Meijer, G.A. , Jonkers, D.M. , Herman, J.G. , de Bruine, A.P. , van Engeland, M. , 2009. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J. Natl. Cancer Inst.. 101, 916–927. [DOI] [PubMed] [Google Scholar]
- Mokarram, P. , Kumar, K. , Brim, H. , Naghibalhossaini, F. , Saberi-firoozi, M. , Nouraie, M. , Green, R. , Lee, E. , Smoot, D.T. , Ashktorab, H. , 2009. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 4, e7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phay, J.E. , Shah, M.H. , 2010. Targeting RET receptor tyrosine kinase activation in cancer. Clin. Cancer Res.. 16, 5936–5941. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho, I. , Burzynski, G.M. , de Groot, J.W. , Eggen, B.J. , Hofstra, R.M. , 2006. Current concepts in RET-related genetics, signaling and therapeutics. Trends Genet.. 22, 627–636. [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos, P. , Saarma, M. , 2007. Neurotrophic factor receptor RET: structure, cell biology, and inherited diseases. Ann. Med.. 39, 572–580. [DOI] [PubMed] [Google Scholar]
- Salazar, R. , Roepman, P. , Capella, G. , Moreno, V. , Simon, I. , Dreezen, C. , Lopez-Doriga, A. , Santos, C. , Marijnen, C. , Westerga, J. , Bruin, S. , Kerr, D. , Kuppen, P. , van de Velde, C. , Morreau, H. , Van Velthuysen, L. , Glas, A.M. , Van't Veer, L.J. , Tollenaar, R. , 2011. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J. Clin. Oncol.. 29, 17–24. [DOI] [PubMed] [Google Scholar]
- Sasselli, V. , Pachnis, V. , Burns, A. , 2012. The enteric nervous system. Dev. Biol.. 366, 64–73. [DOI] [PubMed] [Google Scholar]
- Simons, C.C. , Hughes, L.A. , Smits, K.M. , Khalid-de Bakker, C.A. , de Bruine, A.P. , Carvalho, B. , Meijer, G.A. , Schouten, L.J. , van den Brandt, P.A. , Weijenberg, M.P. , van Engeland, M. , 2013. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann. Oncol.. 24, 2048–2056. [DOI] [PubMed] [Google Scholar]
- Sobin, L.H. , Fleming, I.D. , 1997. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 80, 1803–1804. [DOI] [PubMed] [Google Scholar]
- Tanaka, M. , Chang, P. , Li, Y. , Li, D. , Overman, M. , Maru, D.M. , Sethi, S. , Phillips, J. , Bland, G.L. , Abbruzzese, J.L. , Eng, C. , 2011. Association of CHFR promoter methylation with disease recurrence in locally advanced colon cancer. Clin. Cancer Res.. 17, 4531–4540. [DOI] [PubMed] [Google Scholar]
- Tournier, B. , Chapusot, C. , Courcet, E. , Martin, L. , Lepage, C. , Faivre, J. , Piard, F. , 2012. Why do results conflict regarding the prognostic value of the methylation status in colon cancers? The role of the preservation method. BMC Cancer. 12, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.L. , Chu, K.S. , Huang, Y.H. , Su, Y.C. , Wu, J.Y. , Kuo, C.H. , Chen, C.W. , Wang, J.Y. , 2009. Predictive factors of early relapse in UICC stage I-III colorectal cancer patients after curative resection. J. Surg. Oncol.. 100, 736–743. [DOI] [PubMed] [Google Scholar]
- Umetani, N. , Takeuchi, H. , Fujimoto, A. , Shinozaki, M. , Bilchik, A.J. , Hoon, D.S. , 2004. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin. Cancer Res.. 10, 7475–7483. [DOI] [PubMed] [Google Scholar]
- van den Brandt, P.A. , Goldbohm, R.A. , van 't Veer, P. , Volovics, A. , Hermus, R.J. , Sturmans, F. , 1990. A large-scale prospective cohort study on diet and cancer in The Netherlands. J. Clin. Epidemiol.. 43, 285–295. [DOI] [PubMed] [Google Scholar]
- Van den Brandt, P.A. , Schouten, L.J. , Goldbohm, R.A. , Dorant, E. , Hunen, P.M. , 1990. Development of a record linkage protocol for use in the Dutch cancer registry for epidemiological research. Int. J. Epidemiol.. 19, 553–558. [DOI] [PubMed] [Google Scholar]
- Weisenberger, D.J. , Siegmund, K.D. , Campan, M. , Young, J. , Long, T.I. , Faasse, M.A. , Kang, G.H. , Widschwendter, M. , Weener, D. , Buchanan, D. , Koh, H. , Simms, L. , Barker, M. , Leggett, B. , Levine, J. , Kim, M. , French, A.J. , Thibodeau, S.N. , Jass, J. , Haile, R. , Laird, P.W. , 2006. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet.. 38, 787–793. [DOI] [PubMed] [Google Scholar]
- Yi, J. , Dhir, M. , Van Neste, L. , Downing, S. , Jeschke, J. , GlÃckner, S. , de Freitas Calmon, M. , Hooker, C. , Funes, J. , Boshoff, C. , Smits, K. , van Engeland, M. , Weijenberg, M. , Iacobuzio-Donahue, C. , Herman, J. , Schuebel, K. , Baylin, S. , Ahuja, N. , 2011. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin. Cancer Res. An Official J. Am. Assoc. Cancer Res.. 17, 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, J.M. , Dhir, M. , Van Neste, L. , Downing, S.R. , Jeschke, J. , Glockner, S.C. , de Freitas Calmon, M. , Hooker, C.M. , Funes, J.M. , Boshoff, C. , Smits, K.M. , van Engeland, M. , Weijenberg, M.P. , Iacobuzio-Donahue, C.A. , Herman, J.G. , Schuebel, K.E. , Baylin, S.B. , Ahuja, N. , 2011. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin. Cancer Res.. 17, 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Q. , Cheng, Y. , Zhu, Q. , Yu, Z. , Wu, X. , Huang, K. , Zhou, M. , Han, S. , Zhang, Q. , 2008. The relationship between overexpression of glial cell-derived neurotrophic factor and its RET receptor with progression and prognosis of human pancreatic cancer. J. Int. Med. Res.. 36, 656–664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary Figure 1 Defining the methylation cut‐off for pyrosequencing. Pyrosequencing results of 30 adenocarcinoma samples from series 3 were compared by MSP. Pyrosequencing results were divided into three groups: clearly methylated (>five CG sites showed methylation of 15% or higher), clearly unmethylated (no CG site showed methylation above 10%) and intermediate or no clear methylation status (outlying methylation of single CGs above 15%). Clearly methylated samples with an average methylation above 20% by pyrosequencing showed also methylation by MSP, whereas clearly unmethylated samples by pyrosequencing were also unmethylated by MSP. Samples with intermediate/no clear methylation status, were unmethylated by MSP. Accordingly, a pyrosequencing cut‐off was set to an average methylation of 20% or higher. Each row represents results for one colon adenocarcinoma sample. Squares represent the location of CG sites.
Supplementary Figure 2 Kaplan–Meier curve for disease specific (DS) survival in stage II patients from series 1. Kaplan–Meier curve for the association of RET promoter CpG island methylation and DS at 5‐year follow up. U = unmethylated, M = methylated.


