Abstract
Chronic inflammation is a major risk factor for the development and metastatic progression of cancer. We have previously reported that the chemopreventive polyphenol Curcumin inhibits the expression of the proinflammatory cytokines CXCL1 and ‐2 leading to diminished formation of breast and prostate cancer metastases. In the present study, we have analyzed the effects of Curcumin on miRNA expression and its correlation to the anti‐tumorigenic properties of this natural occurring polyphenol.
Using microarray miRNA expression analyses, we show here that Curcumin modulates the expression of a series of miRNAs, including miR181b, in metastatic breast cancer cells. Interestingly, we found that miR181b down‐modulates CXCL1 and ‐2 through a direct binding to their 3′‐UTR. Overexpression or inhibition of miR181b in metastatic breast cancer cells has a significant impact on CXCL1 and ‐2 and is required for the effect of Curcumin on these two cytokines. miR181b also mediates the effects of Curcumin on inhibition of proliferation and invasion as well as induction of apoptosis. Importantly, over‐expression of miR181b in metastatic breast cancer cells inhibits metastasis formation in vivo in immunodeficient mice. Finally, we demonstrated that Curcumin up‐regulates miR181b and down‐regulates CXCL1 and ‐2 in cells isolated from several primary human breast cancers.
Taken together, these data show that Curcumin provides a simple bridge to bring metastamir modulation into the clinic, placing it in a primary and tertiary preventive, as well as a therapeutic, setting.
Keywords: Metastases prevention, Curcumin, Breast cancer, Inflammatory cytokines, microRNAs
Highlights
We show that Curcumin modulates expression of miR181b.
We show that miR181b overexpression inhibits breast cancer cell growth and invasion.
We show that miR181b modulates CXCL1 and ‐2, resulting in reduction of metastases.
Our data allow the introduction of metastamirs into the clinic.
Our data provide evidence for the use of curcumin for cancer prevention.
1. Introduction
Breast cancer is the most common cancer in women worldwide and the second leading cause of cancer‐related mortality in women. It is estimated that about 20% of all patients with invasive breast cancer develop metastatic disease, which is essentially incurable (Alvarez). In these cases the aims of therapy include palliation of symptoms, delay of disease progression, and prolongation of overall survival time without negatively impacting quality of life (Irvin et al.). To develop successful anti‐dissemination approaches, also in the context of cancer chemoprevention, we need to understand the molecular mechanisms of how tumors metastasize. Over the last decade several genes that regulate metastasis or act as molecular markers of metastatic disease have been discovered. These include metastasis genes (Nguyen et al., 2009) and metastasis repressor genes (Shoushtari et al., 2011) as well as multi‐gene or multi‐protein “signatures” often developed using microarray approaches (Pfeffer, 2013). In addition, a newly discovered class of small (19–25 nucleotides) non‐coding RNAs, the microRNAs (miRs or miRNAs) have been linked to several human diseases, in particular cancer (Kong et al., 2012; Lujambio and Lowe, 2012). miRNAs post‐transcriptionally regulate protein expression: each miRNA can control the expression of several proteins through binding to the mRNA 3′ UTR, thus resulting in mRNA degradation or inhibition of mRNA translation (Krol et al.). miRNAs involved in cancer can act as either tumor promoters (“oncomirs”) or tumor suppressors (tumor suppressor miRs) (Kong et al., 2012; Lujambio and Lowe, 2012). Several miRNAs have been associated to metastasis (“metastamirs”) with both prometastatic and antimetastatic effects (Hurst et al., 2009; White et al., 2011).
Translation of the information concerning metastasis associated genes and gene expression regulators into the clinic has been difficult (Kong et al., 2012; Shoushtari et al., 2011) since tumor cell dissemination and the formation of metastases begins long before the primary tumor is diagnosed and surgically removed. Studies on metastasis‐associated genes have largely focused on the cancer cells themselves, rather than genes that modify the microenvironment. The regulation of the microenvironment and the key role this plays both in permitting tumor development (Bissell and Hines, 2011; de Visser et al., 2006; DeNardo et al., 2010), regulating progression (Albini et al., 2012; Bierie and Moses, 2006; Mantovani and Sica, 2010; Noonan et al., 2008), and even in preparing the “soil” for metastatic dissemination (Joyce and Pollard, 2009; Psaila and Lyden, 2009) is now coming to light.
One approach to reducing the morbidity and mortality of cancer is through prevention, in particular chemoprevention (Sporn). The concept of chemoprevention has been recently extended to the microenvironment (Albini and Sporn, 2007; Albini et al., 2012), potentially preventing tumor insurgence, progression and metastasis. The largest group of potential chemoprevention molecules are the phytochemicals and their derivatives (Noonan et al., 2011), which include molecules with known clinical chemoprevention potential such as aspirin and metformin (Albini et al., 2012). Major targets include inflammation, a key element in tumor insurgence (de Visser et al., 2006; DeNardo et al., 2010), progression (Mantovani and Sica, 2010) and metastasis (Joyce and Pollard, 2009; Psaila and Lyden, 2009), as well as angiogenesis (Albini and Sporn, 2007; Albini et al., 2012), a process driven by inflammation (Albini et al., 2005; Noonan et al., 2008).
One of the most promising phytochemicals is Curcumin, a compound with pleiotropic activities targeting both tumor cells and the tumor microenvironment (Gupta et al., 2010; Noonan et al., 2011; Reuter et al., 2011). We have recently demonstrated that Curcumin inhibits metastatic dissemination in breast and prostate cancer models (Bachmeier et al., 2007; Killian et al., 2012) and we have identified Curcumin‐induced modulation of the proinflammatory cytokines CXCL1 and ‐2 as being critical to this anti‐metastatic effect (Bachmeier et al., 2008). Curcumin interrupts the feed‐back loop between NFκB, the chemokines CXCL1 and ‐2 and the pro‐metastatic players whose transcription is normally induced by the two chemokines (Bachmeier et al., 2008; Killian et al., 2012). Curcumin also acts as a phytoestrogen in estrogen receptor α positive breast cancer cells (Bachmeier et al., 2010b). Recently, Curcumin has been found to modulate miRNA expression in retinoblastoma cells (Sreenivasan et al.) and other cancer cells leading to abrogation of tumor growth (Neelakandan et al.) yet nothing is known on the effect of Curcumin modulated miRNAs on the interaction of the tumor cell with the microenvironment and metastasis.
Using microarray miRNA expression analyses, we show here that Curcumin induces miR181b expression in breast cancer cells that down‐regulates the expression of the pro‐metastatic cytokines CXCL1 and ‐2 repressing the metastatic potential. Our data show that Curcumin provides a simple bridge to bring metastamir modulation into the clinic, placing it in a primary and tertiary preventive, as well as a therapeutic, setting.
2. Materials and methods
2.1. Cells and culture conditions
Human metastatic breast cancer cells MDA‐MB‐231 (ATCC) (Zhang et al., 1991) were selected because of their sustained CXCL1 and ‐2 expression that is not observed in other commonly used breast cancer cell lines. The cells were grown as previously described (Killian et al., 2012).
2.2. Isolation of cells from primary breast cancers
Breast cancer tissue samples were collected following surgical resection of tumors larger than 2 cm in diameter from patients of the Ospedale di Circolo, Fondazione Macchi, Varese‐Italy, following informed consent in an ethics committee approved protocol. Samples were stored in phosphate buffered saline (LONZA, Basel, Switzerland) with 1% Pen/Strep (Sigma–Aldrich, St. Louis, MO, USA) at 4 °C for less than 18 h prior to processing. The patient characteristics are given in Table 1. Samples were processed as previously described (Bruno et al.). Briefly red blood cell aggregates and excess of adipose tissue were removed, tissues were mechanically minced by scissors and enzymatically digested with a cocktail containing DNAse (100 μg/mL, Roche, Mannheim, Germany), Collagenase IV (1 mg/ml, Sigma–Aldrich) and hyaluronidase (200 U/mL, Sigma–Aldrich) in RPMI 1640 (LONZA) supplemented with Pen/Strep for 2 h at 37 °C. Tissue fragments were processed in a tissue disassociator (GentleMACS dissassociator, Miltenyi, Bologna, Italy) and the suspension was filtered twice through 50 μm pore size cell strainers (Becton Dickinson, BD – San Jose, California, USA). The total single cell suspension was washed twice by centrifugation in PBS at 500 × g for 5 min at room temperature to remove residual enzymes and debris. Cell viability was determined by trypan Blue (LONZA) staining and the vital primary breast cancer cells were seeded into six‐well plates in RPMI 1640 supplemented with FBS (Euroclone, UK), 1% Pen/Strep and allowed to attach at 37 °C, 5% CO2 for 24 h. Cells were treated with 25 μM Curcumin or carrier alone for 24 h. Supernatants were collected and immediately stored at −80 °C for CXCL1 analyses. Cells were lysed with Quiazol (Qiagen, Hilden, Germany) and stored at −80 °C for subsequent miRNA analyses. Each experiment was performed in triplicate.
Table 1.
Patient data.
| Patient | Age | Grade | Estrogen receptor positive | Progesterone receptor positive | Erb‐B2 status | p53 status | Ki67 status | TNM | Histotype |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 64 | 3 | 100% | 98% | neg | 3% | 28% | T2 N0 (1s) | Infiltrating ductal carcinoma, poorly differentiated, angioinvasive |
| 2 | 54 | 2 | 100% | 2% (weak) | neg | neg | 9% | T2 N3 | Infiltrating lobular carcinoma, discreetly differentiated, no signs of angioinvasion or neuroinvasion. |
| 3 | 73 | 2 | 100% | 70% | neg | 15% | 15% | T2 N0 (SN) | Infiltrating lobular carcinoma, discreetly differentiated, no signs of angioinvasion or neuroinvasion. |
2.3. Curcumin treatment of cells
Curcumin, 95% pure, was purchased from Fluka (Buchs, Switzerland) and dissolved in 0.5 M NaOH. For the use in cell culture a 2.5 mM solution in sterile PBS was prepared. Curcumin was applied at an end concentration of 25 μM for all assays. For controls the carrier (0.5 M NaOH) was diluted in PBS according to the Curcumin working solutions.
2.4. Total and low molecular weight (LMW) RNA isolation
Isolation of separate fractions of LMW RNA and total RNA (>200 nt) from MDA‐MB‐231 cells was performed using QIAzol and MiRNeasy kit (Qiagen, Hilden, Germany) following the manufacturers' protocol.
2.5. Microarray miRNA expression analyses
miRCURY LNA probes (miRBase 9.2.) were spotted onto Ultra GAPS II slides (Corning) by using the OmniGrid Accent spotter (GeneMachines) and crosslinked by UV. The probe sets included spike‐in control probes, negative capture probes and the capture probes that hybridize to small nuclear RNAs. All hybridizations were made using a reference standard for the control channel prepared from an equimolar pool of short RNAs, extracted from large scale cultures of 20 different cell lines of ecotdermal, mesenchymal and endodermal origin. For labeling of the miRNAs, we used the miRCURY™ LNA microRNA Array Power labeling kit (Exiqon) according to the recommendations of the manufacturer. Hybridization at 56 °C for 16 h in the HS‐400 Pro system (Tecan) was followed by washing and nitrogen drying according to the manufacturer's protocol (Exiqon). For image acquisition the Axon Genepix 4000B scanner (Molecular Devices, Sunnyvale, CA) and dedicated software was used. Quality control was performed using Axon and R/BioConductor (Gentleman et al., 2004) software. Only spotted arrays with good quality were used for normalization and data analysis. Normalization was performed on data from individual arrays (intra‐slide normalization) using an internal positive control, whereas normalization of different arrays (inter‐slide normalization) was based on data from a set of arrays using the quantitative standard reported. All analyses were performed in quadruplicate.
2.6. In silico target gene identification – three public miRNA databases were used to identify putative target genes
TargetScanHuman (http://www.targetscan.org/) Release 6.2
Microrna.org (http://www.microrna.org/microrna/home.do) Release August 2010
MicroCosm Targets (http://www.ebi.ac.uk/enright‐srv/microcosm/htdocs/targets(v5/) Release Version 5
2.7. Real‐time RT‐PCR
mRNA or miRNA was reverse transcribed according to standard protocols using oligo‐dT primers and the first strand synthesis kit (GE Healthcare) or the miScript Reverse Transcription Kit together with the miScript Primer Assay (both Qiagen, Hilden, Gemany) respectively. Expression analysis of mRNA was performed using the Light Cycler technology according to standard protocols. Human RPII and HPRT were used as endogenous controls. All primers were designed using primer3 software (Rozen and Skaletsky, 2000) (see primer list, Supplementary Table 1). Expression of mature miRNAs was determined using miRNA specific quantitative real‐time PCR using a specific miR181b primer and RNUB6_2 for normalization (MiScript Reverse Transcription Kit and Primer Assay, Qiagen, Hilden, Germany) by Light Cycler technology according to the recommendations of the manufacturer.
2.8. Luciferase assay
The human CXCL1 and CXCL2 3′UTRs (607 bp and 654 bp from the STOP codon respectively) were both PCR amplified from human genomic DNA by using the primers “A” and “B” or primers “E” and “F” respectively. These two primer pairs contain each KpnI and NheI recognition sites at the 5′ end. The DNA fragments corresponding to the CXCL1 or the CXCL2 3′UTRs were cloned downstream of the luciferase gene in the pGL3‐cont PLK + vector (Fontana et al., 2007). From these two wildtype constructs, the mutant derivatives were generated by inverse PCR with primers “C” and “D” or “G” and “H”, respectively. HeLa cells were cotransfected with 0.4 g of wildtype or mutant constructs and 50 ng of Renilla luciferase vector (pRL‐TK Promega), together with 160 nM 2′‐O‐Methyl oligonucleotides (anti‐181 and anti‐control; Dharmacon, Lafayette, CO), with Lipofectamine 2000 (Invitrogen). Cells were harvested and assayed with Dual Luciferase Assay (Promega) 48 or 72 h after transfection by using Microlite TLX1 (Dynatech Laboratoires, Chantilly, CA). Three independent experiments were done in triplicate. All sequences all listed in Supplementary Table 1.
2.9. Oligonucleotides and transfection experiments
MDA‐MB‐231 cells were transfected with mature miR181b (miRIDIAN microRNA mimics), a specific miR181b inhibitor (miRIDIAN microRNA hairpin inhibitor) or the respective scrambled negative controls (miRIDIAN microRNA mimic negative control and hairpin inhibitor negative control) obtained from Thermo Scientific (Dharmacon, Lafayette, USA). In silencing experiments 30 nM or 50 nM specific small interfering oligonucleotids against CXCL1 and CXCL2 were used (Silencer Pre‐designed siRNA, Ambion, USA).
2.10. Generation of a stable miR181b overexpressing MDA‐MB‐231 cell clone
Double stranded DNA fragments were obtained by annealing the oligonucleotides miR181b‐s and miR181b‐as (Supplementary Table 1, synthesized by TibMolBiol, Berlin, Germany). This 71 base pair DNA fragment was cloned by restriction site ligation into the NheI and KpnI sites of the pcDNA6.2‐GW/EmGFPmod‐miR vector (Fontana et al., 2008) resulting in a miR181b overexpression vector (MDA‐MB‐231miR181b+). 25.000/well MDA‐MB‐231 cells were seeded into a 6‐well plate and after 24 h transfected with 6 μg MDA‐MB‐231miR181b+ or the corresponding empty vector (MDA‐MB‐231MOCK) vector DNA respectively by lipofection (Lipofectamine 2000, Invitrogen). Stable clones were selected with 1 mg/ml blasticidin S (Invitrogen). After 6 week of selection with 8 μg/ml blasticidin cells with highest fluorescence were further selected by FACS. Stable clones were maintained in growth medium containing 6 μg/ml blasticidin.
2.11. Preparation of conditioned media
Cell culture supernatants were collected and centrifuged 15 min at 4000 × g.
2.12. Determination of protein concentration
Protein concentrations were determined by the BCA protein assay (Pierce, Oud‐Bejierland, Netherlands) with bovine serum albumin as standard.
2.13. Western Blots
Blots were performed as previously described by us in detail (Bachmeier et al., 2008). Conditioned media were analyzed using antibodies against CXCL1, CXCL2 (both Dianova, Germany), MMP‐1 and MMP‐3 (both Sigma, Germany). In order to verify that equal amounts of total protein of conditioned media (β‐actin staining not possible) were loaded to each lane of the gels, we visualized the protein bands blotted onto the nitrocellulose membranes (after the gel run) by Ponceau staining (see Figures 1B, 2B, and 3B, lanes indicated as “loading control”). Semiquantitative evaluation of the bands was performed by densitometric analysis with the ImageJ software provided by the NIH (http://rsb.info.nih.gov/ij/).
Figure 1.
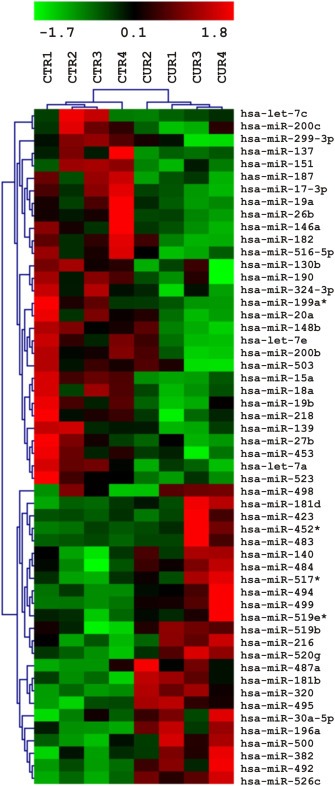
Curcumin modulates miRNA expression in human breast cancer cells. Microarray miRNA expression analysis. Four samples each of Curcumin treated (6 h) MDA‐MB‐231 cells and carrier‐treated controls were analyzed by hierarchical clustering applying euclidean distance and average linkage. Expression levels are indicated by a color code, green = expression below, red = expression above mean levels of expression, black = mean level.
Figure 2.
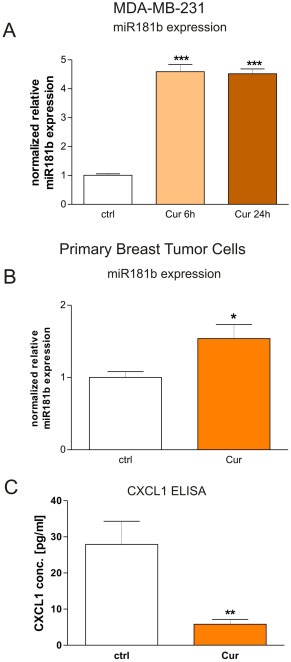
Curcumin modulates miR181b expression in human breast cancer cells. A: Quantitative RT‐PCR reveals that human metastatic breast cancer cells (MDA‐MB‐231) treated with Curcumin for 6 h (“Cur”) express four fold higher amounts of miRNA181b in respect to carrier‐treated cells (“ctrl”). ***P < 0.001, student's t‐test. B: miR181b expression was analyzed in breast cancer cells from human primary tumor samples, cultured in vitro and treated for 24 h with Curcumin with respect to carrier‐treated control cells from the same origin. For each case, we observed up‐regulation of miR181b after Curcumin treatment (data not shown) and by pooling the results from all patient data we obtained a mean miR181b up‐regulation rate of 50%, which was statistically significant (*p < 0.05; student's t‐test). Mean + SD from 3 different patients are shown. C: CXCL1 protein expression secreted from breast cancer cells isolated from primary human tumors that were treated with Curcumin in vitro was analyzed by ELISA. CXCL1 concentrations were statistically significantly down‐regulated about 3.5‐fold in Curcumin treated tumor cells (**p < 0.01; student's t‐test). Mean + SD from 3 different patients are shown.
Figure 3.
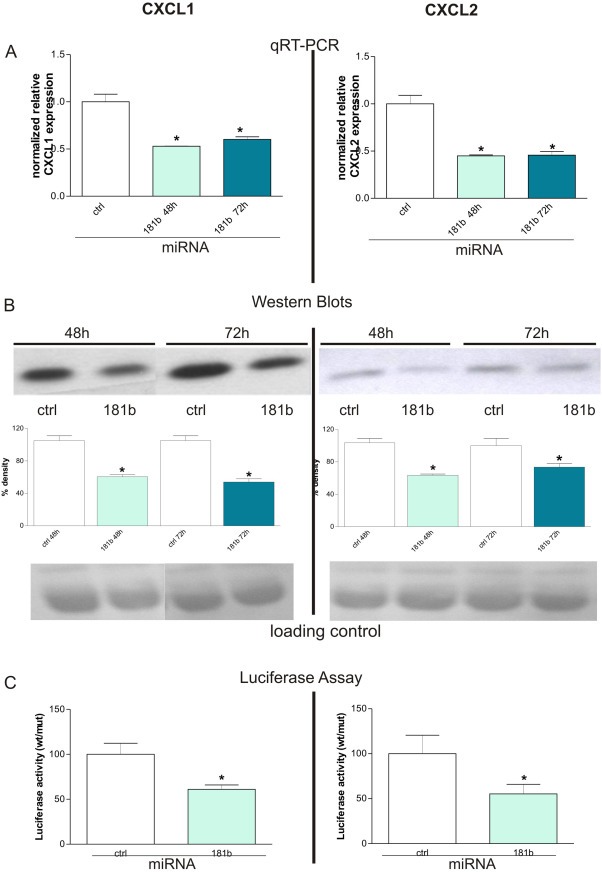
miR181b regulates CXCL1 expression through a direct binding to its 3′ UTR. A: Transient overexpression of miR181b in MDA‐MB‐231 breast cancer cells leads to a ∼50% down‐regulation of CXCL1 (left side) and CXCL2 (right side) transcripts as evidenced by qRT‐PCR (*P < 0.05, student's t‐test). Mean + SD from 3 independent experiments normalized to the respective controls are shown. B: On the corresponding protein level the same effect could be seen as indicated by Western Blot (upper panel) analysis 48 h and 72 h after transient introduction of miR181b into the tumor cells. Densitometric analysis of the bands (middle panel) reveals that CXCL1 (left side) and CXCL2 (right side) protein syntheses were statistically significantly (*P < 0.05, student's t‐test) impaired by about 50% 48 h after transfection. Ponceau staining was used as loading control to evaluate that equal amounts of proteins were applied (lower panel). Mean + SD from 3 independent experiments are shown. C: Luciferase assays demonstrated that regulation of CXCL1 and ‐2 occurs through direct binding of miR181b to their corresponding 3′UTRs. Co‐transfection of pGL3‐cont‐CXCL1 UTR‐wt or pGL3‐cont‐CXCL2 UTR‐wt together with miR‐181b, but not with a scrambled miR181b oligonucleotide, caused a decrease of the luciferase activity in He–La cells. Conversely, miR181b oligonucleotide did not inhibit luciferase activity of pGL3‐cont‐CXCL1 UTR‐mut or pGL3‐cont‐CXCL2 UTR‐mut. Mean + SD from 3 independent experiments are shown.
2.14. CXCL1 ELISA
CXCL1 was quantified in conditioned media of Curcumin treated and non‐treated cells isolated from primary breast cancers using the Quantikine Human CXCL1/GROα Immunoassay (R&D Systems, Minneapolis, USA) according to the recommendations of the manufacturer.
2.15. Apoptosis assay
Apoptotic cell death was determined by an enzyme‐linked immunoassay (Cell Death Detection ELISA PLUS, Roche) to detect fragmented DNA and histones. MDA‐MB‐231 cells were seeded on 24‐well plates 25 h after transfection with the miR181b mimic or the corresponding control oligos respectively in triplicates. After another 24 h cells were washed with PBS lysed and processed following the instructions of the manufacturer.
2.16. Proliferation assay
We used the BrdU Cell Proliferation ELISA (Roche, Germany) according to the recommendations of the manufacturer. Briefly, two thousand cells were seeded in a 96‐well plate and left overnight to attach. After several time periods, according to the experiment (e.g. 24 h, 48 h), BrdU was added and incubated for 2 h. Afterwards cells were fixed, stained and colorimetric analysis was performed with an ELISA plate reader (MPP 3408T, Mikrotek Laborsysteme GmbH, Germany).
2.17. Invasion assay
A fluorescent quantitative cell invasion assay in a 96‐well format (QCM™96‐Well Cell Invasion Assay, Chemicon, USA) was used to examine the ability of MDA‐MB‐231mock and MDA‐MB‐231miR1871b cells to penetrate the extracellular matrix (ECM). The assay was performed according to the recommendations of the manufacturer. Briefly, cells in a density of 1 × 106/ml were re‐suspended in serum‐free medium and added to the upper chambers while the lower chambers were filled with human serum (PAA, Germany) that served as a chemo‐attractant. Cells were then incubated for 6 h at 37 °C. After removal of cells on the upper surface of the membrane, cells on the lower surface of the membrane were lysed and stained with CyQuant DR Dye and fluorescence was read in a plate reader (safire2, Tecan, Switzerland). Data was expressed as the percentage of invasive cells as compared with the control. All experiments were performed in triplicates and results were expressed as mean ± SEM of three independent experiments.
2.18. Hematogenous metastases in immunodeficient mice
Animal studies and research protocols were reviewed and approved by the institutional ethics committee of the IRCCS San Martino‐IST and were conducted in accordance with the national current regulations and guidelines for the care and use of laboratory animals (D.L. 27/01/1992, n. 116). Five‐week‐old CD‐1 Foxn1nu female mice (Charles River Laboratories, Calco, Como, Italy) were maintained under specific pathogen‐free conditions and given sterile food and water ad libitum. Mice were anaesthetized and tumor cells inoculated intracardially. Health and survival rate was observed until their euthanasia. MDA‐MB‐231miR181b+ and MDA‐MB‐231MOCK were collected by trypsinization, washed and resuspended in PBS. 5 × 105 MDA‐MB‐231miR181b+ or MDA‐MB‐231MOCK cells were injected into the left cardiac ventricle of 15 mice. All mice in both groups were fed with standard diet (Mucedola, Italy). On day 35, mice were humanely sacrificed, and necropsied and analyzed for the formation of metastases as previously described by us in detail (Bachmeier et al., 2007; Killian et al., 2012).
2.19. Data analysis
Statistical significance was assessed by comparing mean (±SD) values, which were normalized to the control group with Student's t‐test for independent groups. One‐way ANOVA was used to test for statistical significance (p < 0.05), and when significance was determined, Bonferroni's Multiple Comparison Test was performed post hoc, as indicated in the figure legends. Statistical analysis was performed using the Prism software (GraphPad, San Diego, CA, USA).
2.20. Analysis of genes affecting BC progression
Association of the expression of miR181b regulated genes with disease free survival (DFS) of basal‐like human breast cancers was analyzed using the Breast Mark collection of breast cancer datasets and analysis tools (Madden et al.). The Hazard ratio was generated using Cox regression and a log rank test was used to assign significance.
3. Results
3.1. Curcumin modulates miRNA expression in human breast cancer cells
We performed Microarray analysis to evaluate if treatment of MDA‐MB‐231 human breast cancer cells with Curcumin leads to a modulation of miRNA expression. Of 351 miRNAs detected on the microarray, we found 58 miRNAs to be statistically significantly (*P < 0.05) differentially expressed in Curcumin treated cells as compared with controls (Figure 1). Among these 58 miRNAs, we selected by bioinformatics analysis those miRNAs with at least a 2.5‐fold increase or decrease in response to Curcumin treatment and found 10 miRNAs up‐regulated and 3 miRNAs down‐regulated (Supplementary Table 2).
By using different bioinformatic programs (see Material and Methods) we searched for putative target genes of the selected miRNAs. Interestingly, we found CXCL1 and CXCL2 as putative target genes of miR181b. We previously demonstrated that treatment of MDA‐MB‐231 cells with Curcumin leads to a down‐modulation of CXCL1 and CXCL2, which in turn mediates the anti‐metastatic effect of Curcumin (Bachmeier et al., 2008). Therefore, we hypothesized that Curcumin may regulate CXCL1 and CXCL2 expression through miR181b.
Subsequently we validated the microarray data by measuring levels of miR181b by qRT‐PCR. Consistently, Curcumin treatment (6 h) causes a four‐fold up‐modulation of miR181b in MDA‐MB‐231 cells (Figure 2A) corresponding very well with the microarray data.
We searched in depth for all possible targets of miR181 (see Supplementary Table 3 pages 1–3). In brief, since miR181a, b, c and d are identical in their seed regions, they all have the same target genes. We also performed a thorough data bank search to obtain a complete picture of all putative miRNAs binding to the 3′‐UTR of CXCL1 and ‐2 (see Supplementary Table 3 pages 4–6). Interpretation of our in silico data showed that the pro‐inflammatory cytokines CXCL1 and ‐2 are highly probable targets of miR181b and miR181d (see Supplementary Table 3, page 6).
In order to verify that Curcumin is able to regulate miR181b in primary human breast cancers, we isolated breast cancer cells from 3 human primary tumor samples, all of which were ER+ (Table 1). The tumor cells were treated for 24 h in vitro with Curcumin or carrier as controls, then analyzed for miR181b expression (Figure 2B). In each case, we observed up‐regulation of miR181b after Curcumin treatment that indicated a statistically significant miR181b up‐regulation of 50% (Figure 2B). This suggests that Curcumin regulation of miR181b is present in both ER‐ (the MDA‐MB‐231 model) and in ER+ (primary samples) breast cancers.
3.2. The metastasis‐related cytokine CXCL1 is down‐regulated by curcumin in primary breast cancer cells
We have previously published that curcumin regulates CXCL1 expression in breast cancer cell lines (Bachmeier et al., 2008), we therefore examined whether Curcumin was able to mediate CXCL1 protein down‐regulation in primary human tumor in vitro. ELISA results demonstrated that CXCL1 concentrations were statistically significantly down‐regulated about 3.5‐fold in Curcumin treated primary tumor cells with respect to controls (Figure 2C). These data obtained on the same samples show a potential relation between Curcumin effects on miR181b and CXCL1 modulation.
3.3. miR181b regulates CXCL1 and ‐2 expression through a direct binding to its 3′ UTR
We then hypothesized that CXCL1 and CXCL2 expression is regulated by miR181b. In order to test this hypothesis, we transiently transfected MDA‐MB‐231 cells with miR181b or a control miRNA. Transfection of MDA‐MB‐231 cells with miR181b oligos led to a down‐modulation of CXCL1 and ‐2 in breast cancer cells similar to that obtained by treatment of the cells with Curcumin, as published by us previously (Bachmeier et al., 2008). This effect is not observed in cells transfected with unspecific miRNAs. Furthermore qRT‐PCR and Western blot analysis showed very consistently that over‐expression of miR181b significantly down‐modulated expression of CXCL1 and CXCL2 about 50% at 48 and 72 h post‐transfection on the transcript (Figure 3A) as well as at the protein (Figure 3B) level.
To demonstrate that this regulation occurs through a direct binding of miR181b to the CXCL1 and CXCL2 3′ UTR, we cloned a portion of the CXCL1 or CXCL2 3′ UTR containing the miR181b putative binding sites into the pGL3‐Control vector downstream the luciferase gene, generating the pGL3‐cont‐CXCL1 UTR‐wt and pGL3‐cont‐CXCL2 UTR‐wt constructs. As a control, we cloned a region of CXCL1 or CXCL2 3′ UTR containing a mutated miR181b recognition site (pGL3‐cont‐CXCL1 UTR‐mut and pGL3‐cont‐CXCL2 UTR‐mut). Co‐transfection of pGL3‐cont‐CXCL1 UTR‐wt or pGL3‐cont‐CXCL2 UTR‐wt together with miR181b, but not with a scrambled miR181b oligonucleotide, caused a decrease of the luciferase activity in He–La cells (Figure 3C). Conversely, miR181b oligonucleotide did not inhibit luciferase activity of pGL3‐cont‐CXCL1 UTR‐mut or pGL3‐cont‐CXCL2 UTR‐mut, demonstrating that mutation of the miR181b binding site in the CXCL1 or CXCL2 3′ UTR abolished the ability of miR181b to regulate their expression (Figure 3C).
3.4. miR181b mediates Curcumin‐related down‐modulation of CXCL1 and CXCL2
In order to investigate the role of miR181b in the regulation of CXCL1 and CXCL2 expression by Curcumin, MDA‐MB‐231 cells were treated with Curcumin in the presence or absence of a miR181b inhibitor (miRIDIAN microRNA hairpin inhibitor, Dharmacon, Lafayette, USA (see also material and methods). Curcumin down‐modulates CXCL1 (Figure 4 left panel) and CXCL2 (Figure 4 right panel) at both transcript (Figure 4A) and protein (Figure 4B) levels. However, in the presence of the miR181b inhibitor this down‐modulation was abolished. Transfection of MDA‐MB‐231 cells with a hairpin inhibitor directed against mir181b induces CXCL1 (Figure 4 left panel) and CXCL2 (Figure 4 right panel) expression when compared to cells transfected with an unspecific scrambled hairpin inhibitor control. These results clearly indicate that Curcumin inhibits CXCL1 and CXCL2 through the induction of miR181b. Interestingly, in the presence of the miR181b inhibitor, Curcumin‐mediated down‐regulation of CXCL1 was completely restored (left panel) thus indicating that Curcumin regulates CXCL1 exclusively through miR181b. Conversely, CXCL2 expression was only partially restored in cells treated with Curcumin plus the miR181b inhibitor (right panel), suggesting that additional regulation mechanisms may exist. The data obtained from analysis on protein level (Western Blots) and transcript level (qRT‐PCR) are in strong agreement, supporting the hypothesis that miR181b mediates the down‐modulation of CXCL1 and ‐2 by Curcumin.
Figure 4.
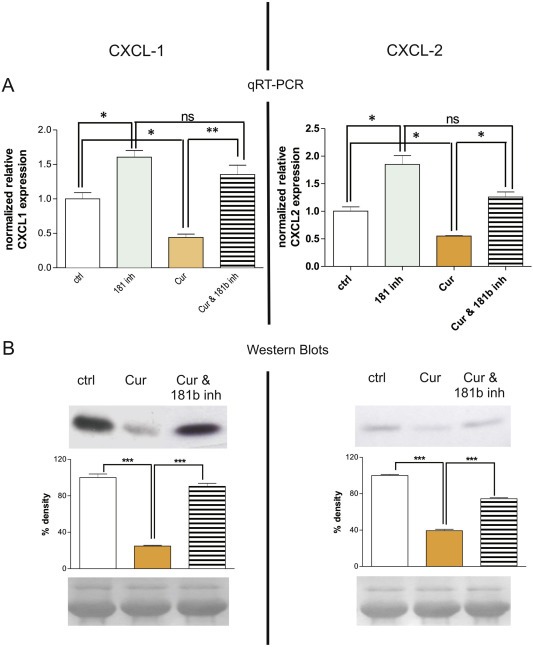
miR181b mediates Curcumin‐related down‐modulation of CXCL1 and CXCL2. A: qRT‐PCR results reveal that treatment of MDA‐MB‐231 cells with Curcumin for 24 h leads to inhibition of CXCL1 expression (left side, “Cur”) that can be reverted to original expression levels (“ctrl”) by concomitant application of a miR181b hairpin inhibitor (“Cur &181b inh”). Conversely, CXCL2 expression (right side) was only partially restored in cells treated with Curcumin plus the miR181b hairpin inhibitor, indicating that additional regulation mechanisms may exist. Inhibition of mir181b by specific small hairpin inhibitors leads to induction of CXCL1 and ‐2 expression compared to control cells transfected with scrambled unspecific hairpin inhibitors (*P < 0.05; **P < 0.001; ANOVA with Bonferroni's post‐test). Mean + SD from 3 independent experiments are shown. "ns”: not significant. B: The corresponding analysis on protein level (Western Blots, upper panel) confirm the data obtained from qRT‐PCR, strengthening the evidence that miR181b mediates down‐modulation of CXCL1 (left side) and CXCL2 (right side) by Curcumin. Densitometric analysis of the bands (middle panel) reveals that differences in expression levels of CXCL1 and ‐2 after Curcumin treatment were statistically significant (***P < 0.001; ANOVA with Bonferroni's post‐test). Loading controls (bottom panel) indicate that equal amounts of total protein were subjected to each lane of the gel. Mean + SD from 3 independent experiments are shown.
3.5. Curcumin inhibits proliferation via miR181b
We wished to know whether miR181b triggers the anti‐proliferative effect of Curcumin (Bachmeier et al., 2010a), and therefore modulated miR181b expression using small hairpin inhibitors directed against miR181b along with the corresponding controls in the presence of Curcumin, in double modulation experiments. Curcumin inhibited MDA‐MB‐231 cell proliferation (Figure 5A), however, Curcumin treatment together with the specific miR181b hairpin‐inhibitor abolished the anti‐proliferative effect, returning cell proliferation to control‐levels. The effect of miR181b expression on proliferation was highly statistically significant. Thus inhibition of miR181b abrogates the ability of Curcumin to inhibit proliferation in MDA‐MB‐231 cells, strongly indicating that miR181b is responsible for Curcumin's anti‐proliferative effects.
Figure 5.
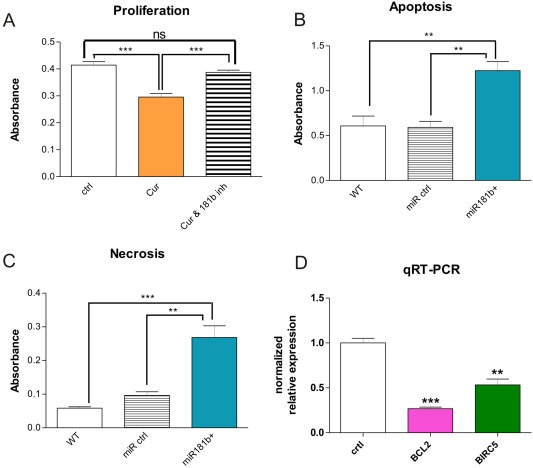
Involvement of miR181b in tumor cell proliferation, apoptosis and invasion. A: Curcumin inhibits proliferation via miR181b. Treatment of MDA‐MB‐231 breast cancer cells for 24 h with Curcumin (“Cur”) inhibits cell proliferation. However, treatment with Curcumin together with a specific miR181b hairpin‐inhibitor (“Cur & 181b inh”) abolishes the anti‐proliferative effect of Curcumin, returning cell proliferation to control‐levels. The effect of miR181b expression on proliferation was statistically highly significant as evidenced by student's t‐test (***P < 0.001). "ns”: not significant. B, C: miR181b overexpression induces apoptosis and necrosis in breast cancer cells. Functional apoptosis/necrosis assays revealed that miR181b over‐expression led to enhanced apoptosis (left panel) as well as necrosis (middle panel). 24 h after transfection with miR181b oligos apoptosis rate in MDA‐MB‐231 cells was doubled as compared to wildtype MDA‐MB‐231 cells or to MDA‐MB‐231 cells transfected with an appropriate control oligo (left panel). Likewise, necrosis rate was increased approximately 5 fold in miR181b over‐expressing MDA‐MB‐231 cells as compared to wildtype cells and over 2.5 fold as compared to MDA‐MB‐231 cells expressing an appropriate control oligo (**P < 0.01; ***P < 0.001; ANOVA with Bonferroni's post test). Mean + SD from 3 independent experiments are shown. D: Using quantitative RT‐PCR, expression of the apoptosis related factors BCL2 and survivin/BIRC5 in MDA‐MB‐231 over‐expressing miR181b showed a statistically significant down‐regulation achieved 72 h after transfection with a double stranded miR181b oligo as compared to the appropriate controls (right panel) (**P < 0.01 and ***P < 0.001; student's t‐test). Mean + SD (SEM?) from 3 independent experiments are shown.
3.6. miR181b induces apoptosis and necrosis in breast cancer cells
Since Curcumin induces tumor cell apoptosis (Bachmeier et al., 2010a), a function critical for suppression of tumor formation and metastasis, we examined if miR181b is likewise able to induce apoptosis. Functional apoptosis/necrosis assays revealed that miR181b over‐expression led to enhanced apoptosis (Figure 5B) as well as necrosis (Figure 4C) in a highly statistically significant manner. Over‐expression of miR181b for 24 h in MDA‐MB‐231 cells doubled the apoptosis rate as compared to wildtype MDA‐MB‐231 cells or to MDA‐MB‐231 cells transfected with an appropriate control oligo (Figure 5B). Likewise, necrosis rate was increased approximately 5 fold in miR181b over‐expressing MDA‐MB‐231 cells as compared to wildtype cells and over 2.5 fold as compared to MDA‐MB‐231 cells expressing an appropriate control oligo (Figure 5C).
Biostatistic analysis of apoptosis related miR181 target genes revealed that BCL2 was among the putative candidates (see Supplementary Table 3, page 4). Using qRT‐PCR, we found a statistically significant down‐regulation of the apoptosis related factors BCL2 and survivin/BIRC5 in MDA‐MB‐231 over‐expressing miR181b, 72 h after transfection with a double stranded miR181b oligo as compared to the appropriate controls (Figure 5D).
3.7. miR181b inhibits expression of MMPs in metastatic breast cancer cells
We evaluated the correlation between miR181b and the expression of extracellular matrix degrading proteases (MMPs – matrix metalloproteinases), as expression of MMPs is a prerequisite for tumor growth, invasion and metastasis. In an initial step we found that MMPs are present among the in silico targets of miR181 (Supplementary Table 3, page 5). Furthermore, our qRT‐PCR data revealed that MMP‐1 and ‐3 transcripts were down‐regulated by 93% and 75% respectively, in MDA‐MB‐231 cells stably transfected with a miR181b over‐expression vector when compared to cells stably transfected with the appropriate control vector (Figure 6A). Western blots (Figure 6B) of conditioned media from MDA‐MB‐231 cells stably over‐expressing miR181b have a lower MMP‐1 and ‐3 protein expression than appropriate control cells. Quantification of the bands by densitometry indicated that the differences in expression levels were approximately 15% and 34% for MMP‐1 and MMP‐3 (respectively) and were statistically significant.
Figure 6.
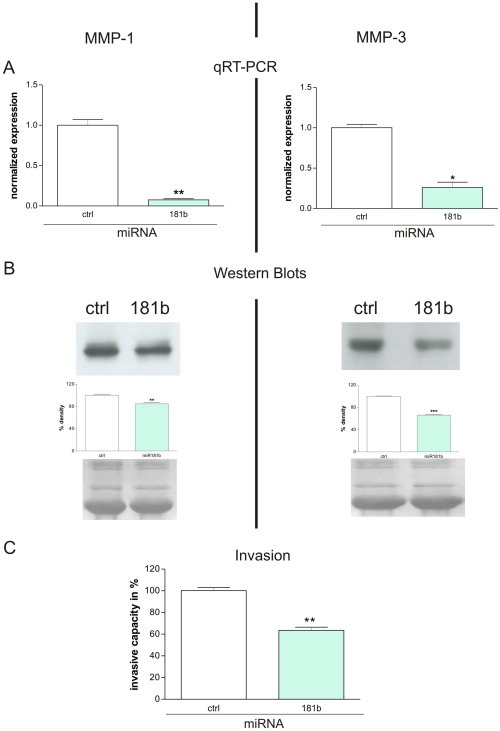
miR181b impairs the expression of MMP‐1 and MMP‐3. A: qRT‐PCR shows that MMP‐1 (left side) and MMP‐3 (right side) transcripts were downregulated 93% (**P = 0.0059; student's t‐test) and 75% (*P = 0.0109; student's t‐test) respectively in MDA‐MB‐231 cells stably transfected with a miR181b overexpression vector (lanes indicated with 181b) when compared to cells stably transfected with an appropriate control vector (lanes indicated with ctrl). Mean + SD from 3 independent experiments are shown. B: Western blots (upper panel) of conditioned media from MDA‐MB‐231 cells stably transfected with a miR181b overexpression vector (lanes indicated with 181b), reveal a downregulation of MMP‐1 and ‐3 protein when compared to cells stably transfected with an appropriate control vector (lanes indicated with ctrl). This effect was quantified by subsequent densitometry (middle panel) showing that the differences in expression levels were about 15% (**P = 0.0068; student's t‐test) for MMP‐1 and about 34% (***P < 0.0001; student's t‐test) for MMP‐3. Equal amounts of total protein were loaded to each lane of the gels and to verify this, we visualized the protein bands blotted onto the nitrocellulose membranes (after the gel run) by Ponceau staining (lower panel). Mean + SD from 3 independent experiments are shown. C: miR181b over‐expression impairs the invasive capacity of breast cancer cells. Invasion of MDA‐MB‐231 breast cancer cells stably over‐expressing miR181b through a reconstituted basement membrane (Matrigel) was significantly reduced (**P < 0.01; student's t‐test) by about 50% after an incubation period of 6 h. Mean + SD from 3 independent experiments are shown.
3.8. miR181b impairs the invasive capacity of breast cancer cells
Given the significant effect on MMP expression, we investigated the influence of miR181b expression on invasive capacity using the chemoinvasion assay (Albini et al.). As the invasion chambers are covered with a reconstituted basement membrane (Matrigel) active invasion as result of proteolytic digestion of the matrix is required for the tumor cells to pass the barrier. Invasion of MDA‐MB‐231 breast cancer cells stably over‐expressing miR181b was significantly reduced by about 50% after an incubation period of only 6 h (Figure 6C). This establishes a functional consequence of decreased MMP production.
3.9. Breast cancer cells over‐expressing miR181b have a lower capacity to metastasize in vivo
We previously reported that Curcumin prevents the formation of hematogenous breast cancer metastases in immunodeficient mice in a highly significant manner (Bachmeier et al., 2007). We therefore examined whether miR181b is responsible for the diminished formation of lung metastases by creating a stably miR181b over‐expressing cell clone from MDA‐MB‐231 cells (MDA‐MB‐231miR1871b) along with the corresponding MOCK control cell clone (MDA‐MB‐231mock) (see material and methods). As an in vivo model we used mice injected with either the miR181b over‐expressing or the MOCK cell clones and divided them into two study groups accordingly. At the end of the experimental time period, brains, humeri, femurae and vertebral columns of the mice were free of metastases. Tumor cells/cell aggregates were found in the intrapulmonary and the peripulmonary compartment, however since peripulmonary metastases are more likely to be derived from direct dissemination during the intercardiac injection, we limited our analysis to intrapulmonary metastases of hematogenous origin.
Breast cancer cells over‐expressing miR181b had a highly statistically significant reduction (Mann–Whitney test, p = 0.0070) in lung metastasis (Figure 7A). The average number of metastasis in mice belonging to the control group was approximately 15 per animal. Animals inoculated with MDA‐MB‐231miR181b cells developed an average of only 6 metastases per animal. Tumors of animals in both study groups were similar in dimension, morphology and histology. The tumor cell morphology showed characteristic atypia, significant expression of human p53 protein was observed only in human tumor cells. Vitality of the tumor cells was confirmed by a high number of proliferating Ki‐67 positive cells thus excluding tumor cell dormancy (Figure 7B). The application of antagomirs in this type of experimental setting of hematogenous metastasis was not possible, due to the undesired toxicity of antagomirs in systemic application (data not shown).
Figure 7.
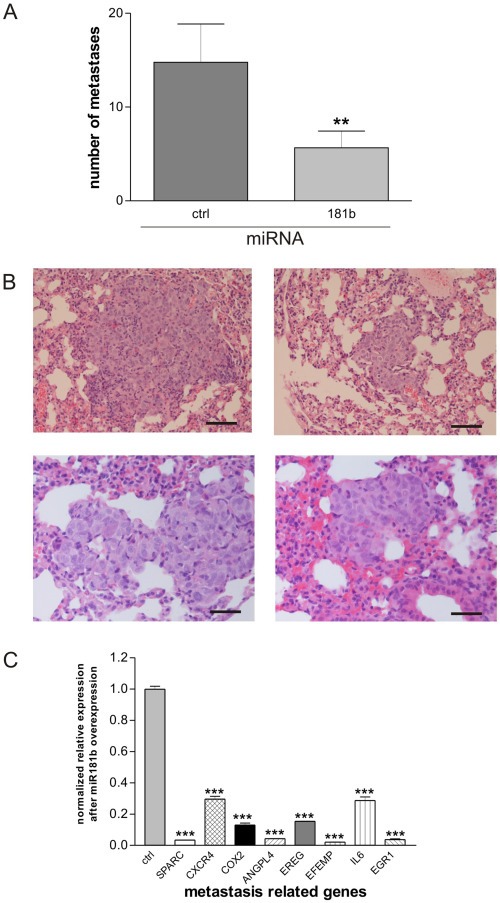
Breast cancer cells over‐expressing miR181b have a lower capacity to metastasize in vivo. Breast cancer cells over‐expressing miR181b had a statistically significant (Mann–Whitney test, p = 0.0070) reduction in their capacity to metastasize into the lung of the animals (A). While the average number of metastasis in mice belonging to the control group was approximately 15 per animal, animals carrying MDA‐MB‐231miR1871b cells developed an average of only 6 metastases per animal. The tumor cell morphology showed characteristic atypia, significant expression of human p53 protein was observed only in human tumor cells. Vitality of the tumor cells was confirmed by a high number of proliferating Ki‐67 positive cells thus excluding tumor cell dormancy (B). Bars in the right lower corners of all photos are equivalent to 50 μm. Expression analysis (qRT‐PCR) of a series of metastasis‐related genes in MDA‐MB‐231 cells transiently over‐expressing miR181b in comparison to corresponding MDA‐MB‐231 control cells showed that expression of the metastasis‐related genes SPARC, CXCR4, COX2, ANGPL4, EFEMP, IL‐6, and EGR1 to be statistically highly significantly down‐regulated (C) 72 h after transfection (***p < 0.001; student's t‐test). Mean + SD from 3 independent experiments are shown.
To identify the molecular players involved in the diminished metastatic capacity of breast cancer cells over‐expressing miR181b, we analyzed the expression of a series of metastasis‐related genes in MDA‐MB‐231 cells transiently over‐expressing miR181b through transfection with double stranded miRNA oligos in comparison to MDA‐MB‐231 cells transfected with the corresponding control oligos. We chose genes whose functional involvement has been shown through the analysis of highly metastatic tumor cells with pulmonary tropism (Minn et al., 2005). In this context we found expression of the metastasis‐related genes SPARC (osteonectin), CXCR4, PTGS2 (prostaglandin‐endoperoxide‐synthase 2, COX2), ANGPTL4 (angiopoetin‐like 4), EFEMP1 (EGF containing fibulin like extracellular matrix protein 1), IL‐6, and EGR1 to be statistically highly significantly down‐regulated 72 h after transfection (Figure 7C). These genes are comprised in the general metastasis signature developed by Ramaswamy et al. (2003) and SPARC, EFEMP1 and ANGPTL4 are also present in a signature of extracellular matrix genes that predict breast cancer metastasis (Albini et al., 2008). In addition, we analyzed the association of the expression of these genes with disease free survival in 424 cases of basal like human breast cancers (with 124 events). Low expression of SPARC (p = 0.002), PTGS2 (p = 0.013) and ANGPTL4 (p = 0.016) was significantly associated with disease free survival, IL6 (p = 0.054) showed a similar trend whereas EFEMP1, CXCR4, IL6 and EGR were not significantly associated with DFS of basal like breast cancer (see Supplementary Figure 1).
4. Discussion
In the metastatic process tumor cells are not the only participants; tumor‐associated cells like macrophages, vascular and lymphatic endothelial cells or fibroblasts that reside in the tumor microenvironment of the host are clearly involved (Peinado et al., 2008). Inflammation is considered as a major factor for tumor progression (Balkwill and Mantovani, 2001) and recruitment of host cells from the microenvironment mediated by chemokines and their receptors play a critical role in this context. The two pro‐inflammatory cytokines CXCL1 and ‐2 have recently been found to be linked to breast (Bachmeier et al., 2008; Minn et al., 2005) and prostate (Killian et al., 2012) cancer metastasis to the lung involving NFκB and CXCR2 (Bachmeier et al., 2008; Killian et al., 2012; Minn et al., 2005). Very recently CXCL1 and its cognate receptor CXCR2 have been shown to facilitate homing of mammary cancer cells (Halpern et al.) and CXCL1 has been found to be up‐regulated in lymphatic endothelial cells from lymph node metastases suggesting a role in lymphangiogenesis (Xu et al.). Chemokine production is common for but not limited to inflammatory breast cancer and characterizes a specific subset of triple negative breast cancer for which the MDA‐MB‐231 cells are a suitable model.
Curcumin is a plant‐derived compound that is particularly suited for prevention of tumor progression in inflammatory cancer because it is known to act on the central activator of inflammation, NFκB (Bachmeier et al., 2007, 2010, 2008, 2012). In previous studies we have shown that Curcumin prevents formation of breast and prostate metastasis by targeting CXCL1 and ‐2 (Bachmeier et al., 2008; Killian et al., 2012). In this study we focused on the involvement of small non‐coding RNAs (miRNAs) in the anti‐metastatic effect of Curcumin. We addressed this problem in particular as little is yet known about the effect of this natural polyphenol on miRNA expression. Curcumin has been shown to modulate the expression of a variety of miRNAs in cancer cells in vitro of different origin (Gandhy et al.; Sreenivasan et al.; Sun et al., 2008; Yang et al., 2010). This in turn has a functional impact on proliferation, migration and apoptosis of the tumor cells (Sreenivasan et al.; Subramaniam et al.). However, these reports do not address how Curcumin regulates miRNA expression in the context of metastasis.
Our microarray and bioinformatics analyses identified miR181b to be modulated by Curcumin and linked to CXCL1 and ‐2. We validated the causal link between miR181b and CXCL1 and ‐2 expression by promoter binding assays and knock‐in/knock‐out experiments in a cell model of human metastatic breast cancer and found that miR181b mediates Curcumin‐related down‐modulation of the inflammatory cytokines CXCL1 and CXCL2, which are both tightly related to metastases (Bachmeier et al., 2008; Killian et al., 2012).
Curcumin is a known inhibitor of NFκB activation that most likely acts through the inhibition of degradation of the Inhibitor of Kappa Light Polypeptide Gene Enhancer In B‐Cells, IκB. We have previously shown that Curcumin reduces NFκB activation and translocation in breast (Bachmeier et al., 2007, 2008) and prostate cancer cells (Killian et al., 2012) thereby affecting the expression of pro‐metastatic genes, among which CXCL1 and –2. The Curcumin mediated induction of miR181b described here has similar effects. Figure 4 shows that the addition of an inhibitor of miR181b to Curcumin treated MDA‐MB‐231 cells only partially restores the CXCL1 and ‐2 expression levels of untreated cells. This is particularly evident for CXCL2, and at least in this case, Curcumin shows effects that cannot entirely be explained by miR181b induction, most likely due to inhibition of NFκB. On the other hand, miR181b on its own has been described as an inhibitor of NFκB therefore the fact that Curcumin induces miR181b is expected to result in a strong inhibition of NFκB activation (Olarerin‐George et al.; Sun et al.).
Our results also demonstrate that miR181b inhibits expression of the matrix degrading enzymes MMPs (matrix metalloproteinases) which leads to reduced invasion. Given the presence of miR181b target sequences in the transcripts of MMP1 and ‐3 their downregulation in miR181b over‐expressing cells is likely to be direct. However, indirect, CXCL1 mediated effects might contribute through miR181b dependent disruption of the positive feedback loop between CXCL1 and NFκB (Bachmeier et al., 2008; Killian et al., 2012) that also affects MMP expression.
Additionally we show that the anti‐proliferative effect of Curcumin on breast cancer cells is mediated via miR181b and that apoptosis and necrosis – both processes dysregulated in metastatic tumor cells – are induced in metastatic breast cancer cells over‐expressing miR181b.
We found that miR181b over‐expression in metastatic breast cancer cells down‐regulates expression of genes belonging to the breast cancer lung metastases signature (Minn et al., 2005) and miR181b over‐expression in breast cancer cells inhibits metastases formation in vivo. So far there are only a few reports regarding the role of miR181 expression in tumor progression and it seems that miR181b has both oncomir and tumor suppressor functions depending on tumor type and context with inflammation (Bisso et al.; Seoudi et al.; Taylor et al.).
Finally we verified the clinical relevance of our results by showing identical responses to Curcumin treatment in tumor cells from primary breast cancers. We show here that Curcumin impacts on miRNA expression in primary tumors inducing miR181b expression, which translates into a down‐regulation of the pro‐inflammatory cytokine CXCL1 and subsequent loss of metastatic potential. The data presented here together with our previously published data (Bachmeier et al., 2007, 2008) can be easily translated into the clinic, in particular because Curcumin can be easily administered orally and dosages up to 8 g per day have been shown to be safe (Dhillon et al., 2008). For example breast cancer patients that are either at high risk for, in treatment for or in remission from breast cancer, could receive a combination of Curcumin prevention with standard therapeutic approaches in controlled clinical trials eventually stratifying patients for ABCA1 mediated resistance to Curcumin (Bachmeier et al., 2009).
Supporting information
The following are the supplementary data related to this article:
Supplementary Table 1 Primer and oligo sequences.
Supplementary Table 2 Altered Expression of miRNAs after Curcumin treatment. Microarray data of miRNA expression in Curcumin treated (6 h) and carrier treated (6 h) human MDA‐MB‐231 breast cancer cells was analyzed using MA plot with M as a parameter to express how many times each miR is regulated against the control and P as parameter for the statistical significance.
Supplementary Table 3 In silico analysis of miRNA targets.
Supplementary Figure 1 Kaplan Meier curves for the association of the expression of metastasis related and miR181b regulated genes in human breast cancers. 424 basal‐like human breast cancers with 124 events from the Breast Mark collection of breast cancer datasets were analyzed. Cases were assigned to low (red) and high (blue) expression classes based on the median level of expression. Log rank test p‐values are indicated.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.01.005.
Kronski Emanuel, Fiori Micol E., Barbieri Ottavia, Astigiano Simonetta, Mirisola Valentina, Killian Peter H., Bruno Antonino, Pagani Arianna, Rovera Francesca, Pfeffer Ulrich, Sommerhoff Christian P., Noonan Douglas M., Nerlich Andreas G., Fontana Laura and Bachmeier Beatrice E., (2014), miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down‐regulation of the inflammatory cytokines CXCL1 and ‐2, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.01.005.
References
- Albini, A., Indraccolo, S., Noonan, D.M., Pfeffer, U., Functional genomics of endothelial cells treated with anti-angiogenic or angiopreventive drugs. Clin. Exp. Metastasis 27, 419–439. [DOI] [PubMed]
- Albini, A. , Mirisola, V. , Pfeffer, U. , 2008. Metastasis signatures: genes regulating tumor-microenvironment interactions predict metastatic behavior. Cancer Metastasis Rev.. 27, 75–83. [DOI] [PubMed] [Google Scholar]
- Albini, A. , Sporn, M.B. , 2007. The tumour microenvironment as a target for chemoprevention. Nat.Rev. Cancer. 7, 139–147. [DOI] [PubMed] [Google Scholar]
- Albini, A. , Tosetti, F. , Benelli, R. , Noonan, D.M. , 2005. Tumor inflammatory angiogenesis and its chemoprevention. Cancer Res.. 65, 10637–10641. [DOI] [PubMed] [Google Scholar]
- Albini, A. , Tosetti, F. , Li, V.W. , Noonan, D.M. , Li, W.W. , 2012. Cancer prevention by targeting angiogenesis. Nat. Rev. Clin. Oncol.. 9, 498–509. [DOI] [PubMed] [Google Scholar]
- Alvarez, R.H., Present and future evolution of advanced breast cancer therapy. Breast Cancer Res. 12 Suppl 2, S1. [DOI] [PMC free article] [PubMed]
- Bachmeier, B. , Nerlich, A.G. , Iancu, C.M. , Cilli, M. , Schleicher, E. , Vene, R. , Dell'Eva, R. , Jochum, M. , Albini, A. , Pfeffer, U. , 2007. The chemopreventive polyphenol Curcumin prevents hematogenous breast cancer metastases in immunodeficient mice. Cell Physiol. Biochem. 19, 137–152. [DOI] [PubMed] [Google Scholar]
- Bachmeier, B.E. , Iancu, C.M. , Killian, P.H. , Kronski, E. , Mirisola, V. , Angelini, G. , Jochum, M. , Nerlich, A.G. , Pfeffer, U. , 2009. Overexpression of the ATP binding cassette gene ABCA1 determines resistance to Curcumin in M14 melanoma cells. Mol. Cancer. 8, 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmeier, B.E. , Killian, P. , Pfeffer, U. , Nerlich, A.G. , 2010. Novel aspects for the application of Curcumin in chemoprevention of various cancers. Front. Biosci.. 2, 697–717. [DOI] [PubMed] [Google Scholar]
- Bachmeier, B.E. , Mirisola, V. , Romeo, F. , Generoso, L. , Esposito, A. , Dell'eva, R. , Blengio, F. , Killian, P.H. , Albini, A. , Pfeffer, U. , 2010. Reference profile correlation reveals estrogen-like trancriptional activity of Curcumin. Cell Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol.. 26, 471–482. [DOI] [PubMed] [Google Scholar]
- Bachmeier, B.E. , Mohrenz, I.V. , Mirisola, V. , Schleicher, E. , Romeo, F. , Hohneke, C. , Jochum, M. , Nerlich, A.G. , Pfeffer, U. , 2008. Curcumin downregulates the inflammatory cytokines CXCL1 and -2 in breast cancer cells via NFkappaB. Carcinogenesis. 29, 779–789. [DOI] [PubMed] [Google Scholar]
- Balkwill, F. , Mantovani, A. , 2001. Inflammation and cancer: back to Virchow?. Lancet. 357, 539–545. [DOI] [PubMed] [Google Scholar]
- Bierie, B. , Moses, H.L. , 2006. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer. 6, 506–520. [DOI] [PubMed] [Google Scholar]
- Bissell, M.J. , Hines, W.C. , 2011. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med.. 17, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisso, A., Faleschini, M., Zampa, F., Capaci, V., De Santa, J., Santarpia, L., Piazza, S., Cappelletti, V., Daidone, M., Agami, R., Del Sal, G., Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle 12, 1679–1687. [DOI] [PMC free article] [PubMed]
- Bruno, A., Focaccetti, C., Pagani, A., Imperatori, A.S., Spagnoletti, M., Rotolo, N., Cantelmo, A.R., Franzi, F., Capella, C., Ferlazzo, G., Mortara, L., Albini, A., Noonan, D.M., The proangiogenic phenotype of natural killer cells in patients with non-small cell lung cancer. Neoplasia 15, 133–142. [DOI] [PMC free article] [PubMed]
- de Visser, K.E. , Eichten, A. , Coussens, L.M. , 2006. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer. 6, 24–37. [DOI] [PubMed] [Google Scholar]
- DeNardo, D.G. , Andreu, P. , Coussens, L.M. , 2010. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev.. 29, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon, N. , Aggarwal, B.B. , Newman, R.A. , Wolff, R.A. , Kunnumakkara, A.B. , Abbruzzese, J.L. , Ng, C.S. , Badmaev, V. , Kurzrock, R. , 2008. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin. Cancer Res.. 14, 4491–4499. [DOI] [PubMed] [Google Scholar]
- Fontana, L. , Fiori, M.E. , Albini, S. , Cifaldi, L. , Giovinazzi, S. , Forloni, M. , Boldrini, R. , Donfrancesco, A. , Federici, V. , Giacomini, P. , Peschle, C. , Fruci, D. , 2008. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 3, e2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana, L. , Pelosi, E. , Greco, P. , Racanicchi, S. , Testa, U. , Liuzzi, F. , Croce, C.M. , Brunetti, E. , Grignani, F. , Peschle, C. , 2007. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol.. 9, 775–787. [DOI] [PubMed] [Google Scholar]
- Gandhy, S.U., Kim, K., Larsen, L., Rosengren, R.J., Safe, S., Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (Sp) transcription factors by targeting microRNAs. BMC Cancer 12, 564. [DOI] [PMC free article] [PubMed]
- Gentleman, R.C. , Carey, V.J. , Bates, D.M. , Bolstad, B. , Dettling, M. , Dudoit, S. , Ellis, B. , Gautier, L. , Ge, Y. , Gentry, J. , Hornik, K. , Hothorn, T. , Huber, W. , Iacus, S. , Irizarry, R. , Leisch, F. , Li, C. , Maechler, M. , Rossini, A.J. , Sawitzki, G. , Smith, C. , Smyth, G. , Tierney, L. , Yang, J.Y. , Zhang, J. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol.. 5, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S.C. , Kim, J.H. , Prasad, S. , Aggarwal, B.B. , 2010. Regulation of survival, proliferation, invasion, angiogenesis, and metastasis of tumor cells through modulation of inflammatory pathways by nutraceuticals. Cancer Metastasis Rev.. 29, 405–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern, J.L., Kilbarger, A., Lynch, C.C., Mesenchymal stem cells promote mammary cancer cell migration in vitro via the CXCR2 receptor. Cancer Lett. 308, 91–99. [DOI] [PMC free article] [PubMed]
- Hurst, D.R. , Edmonds, M.D. , Welch, D.R. , 2009. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res.. 69, 7495–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin, W., Jr., Muss, H.B., Mayer, D.K., Symptom management in metastatic breast cancer. Oncologist 16, 1203–1214. [DOI] [PMC free article] [PubMed]
- Joyce, J.A. , Pollard, J.W. , 2009. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 9, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian, P.H. , Kronski, E. , Michalik, K.M. , Barbieri, O. , Astigiano, S. , Sommerhoff, C.P. , Pfeffer, U. , Nerlich, A.G. , Bachmeier, B.E. , 2012. Curcumin inhibits prostate cancer metastasis in vivo by targeting the inflammatory cytokines CXCL1 and -2. Carcinogenesis. 33, 2507–2519. [DOI] [PubMed] [Google Scholar]
- Kong, Y.W. , Ferland-McCollough, D. , Jackson, T.J. , Bushell, M. , 2012. microRNAs in cancer management. Lancet Oncol.. 13, e249–258. [DOI] [PubMed] [Google Scholar]
- Krol, J., Loedige, I., Filipowicz, W., The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. [DOI] [PubMed]
- Lujambio, A. , Lowe, S.W. , 2012. The microcosmos of cancer. Nature. 482, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, S.F., Clarke, C., Gaule, P., Aherne, S.T., O'Donovan, N., Clynes, M., Crown, J., Gallagher, W.M., BreastMark: an integrated approach to mining publicly available transcriptomic datasets relating to breast cancer Outcome. Breast Cancer Res. 15, R52. [DOI] [PMC free article] [PubMed]
- Mantovani, A. , Sica, A. , 2010. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol.. 22, 231–237. [DOI] [PubMed] [Google Scholar]
- Minn, A.J. , Gupta, G.P. , Siegel, P.M. , Bos, P.D. , Shu, W. , Giri, D.D. , Viale, A. , Olshen, A.B. , Gerald, W.L. , Massague, J. , 2005. Genes that mediate breast cancer metastasis to lung. Nature. 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelakandan, K., Babu, P., Nair, S., Emerging roles for modulation of microRNA signatures in cancer chemoprevention. Curr. Cancer Drug Targets 12, 716–740. [DOI] [PubMed]
- Nguyen, D.X. , Bos, P.D. , Massague, J. , 2009. Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer. 9, 274–284. [DOI] [PubMed] [Google Scholar]
- Noonan, D.M. , De Lerma Barbaro, A. , Vannini, N. , Mortara, L. , Albini, A. , 2008. Inflammation, inflammatory cells and angiogenesis: decisions and indecisions. Cancer Metastasis Rev.. 27, 31–40. [DOI] [PubMed] [Google Scholar]
- Noonan, D.M. , Sogno, I. , Albini, A. , 2011. Plants and plant-derived products as cancer chemopreventive agents. In Bagetta G., Cosentino M., Corasaniti M.T., Sakurada S.(Eds.), Herbal Medicines: Development and Validation of Plant-derived Medicines for Human Health. CRC Press Inc; 285–306. ISBN-10: 1439837686 ISBN-13: 9781439837689 [Google Scholar]
- Olarerin-George, A.O., Anton, L., Hwang, Y.C., Elovitz, M.A., Hogenesch, J.B., A functional genomics screen for microRNA regulators of NF-kappaB signaling. BMC Biol. 11, 19. [DOI] [PMC free article] [PubMed]
- Peinado, H. , Rafii, S. , Lyden, D. , 2008. Inflammation joins the "niche". Cancer Cell. 14, 347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, U. , 2013. Cancer Genomics: Molecular Classification, Prognosis and Response Prediction Springer Science and Business Media; Dordrecht: [Google Scholar]
- Psaila, B. , Lyden, D. , 2009. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer. 9, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy, S. , Ross, K.N. , Lander, E.S. , Golub, T.R. , 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet.. 33, 49–54. [DOI] [PubMed] [Google Scholar]
- Reuter, S. , Gupta, S.C. , Park, B. , Goel, A. , Aggarwal, B.B. , 2011. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr.. 6, 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. , Skaletsky, H. , 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol.. 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Seoudi, A.M., Lashine, Y.A., Abdelaziz, A.I., MicroRNA-181a – a tale of discrepancies. Expert Rev Mol Med 14, e5. [DOI] [PubMed]
- Shoushtari, A.N. , Szmulewitz, R.Z. , Rinker-Schaeffer, C.W. , 2011. Metastasis-suppressor genes in clinical practice: lost in translation?. Nat. Rev. Clin. Oncol.. 8, 333–342. [DOI] [PubMed] [Google Scholar]
- Sporn, M.B., Perspective: the big C – for Chemoprevention. Nature 471, S10–S11. [DOI] [PubMed]
- Sreenivasan, S., Thirumalai, K., Danda, R., Krishnakumar, S., Effect of curcumin on miRNA expression in human Y79 retinoblastoma cells. Curr. Eye Res. 37, 421–428. [DOI] [PubMed]
- Subramaniam, D., Ponnurangam, S., Ramamoorthy, P., Standing, D., Battafarano, R.J., Anant, S., Sharma, P., Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PLoS ONE 7, e30590. [DOI] [PMC free article] [PubMed]
- Sun, M. , Estrov, Z. , Ji, Y. , Coombes, K.R. , Harris, D.H. , Kurzrock, R. , 2008. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol. Cancer Ther.. 7, 464–473. [DOI] [PubMed] [Google Scholar]
- Sun, X., Icli, B., Wara, A.K., Belkin, N., He, S., Kobzik, L., Hunninghake, G.M., Vera, M.P., Blackwell, T.S., Baron, R.M., Feinberg, M.W., MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J. Clin. Invest 122, 1973–1990. [DOI] [PMC free article] [PubMed]
- Taylor, M.A., Sossey-Alaoui, K., Thompson, C.L., Danielpour, D., Schiemann, W.P., TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J. Clin. Invest 123, 150–163. [DOI] [PMC free article] [PubMed]
- White, N.M. , Fatoohi, E. , Metias, M. , Jung, K. , Stephan, C. , Yousef, G.M. , 2011. Metastamirs: a stepping stone towards improved cancer management. Nat. Rev. Clin. Oncol.. 8, 75–84. [DOI] [PubMed] [Google Scholar]
- Xu, J., Zhang, C., He, Y., Wu, H., Wang, Z., Song, W., Li, W., He, W., Cai, S., Zhan, W., Lymphatic endothelial cell-secreted CXCL1 stimulates lymphangiogenesis and metastasis of gastric cancer. Int. J. Cancer 130, 787–797. [DOI] [PubMed]
- Yang, J. , Cao, Y. , Sun, J. , Zhang, Y. , 2010 Dec. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med Oncol. 27, (4) 1114–1118. 10.1007/s12032-009-9344-3 Epub 2009 Nov 12 [DOI] [PubMed] [Google Scholar]
- Zhang, R.D. , Fidler, I.J. , Price, J.E. , 1991. Relative malignant potential of human breast carcinoma cell lines established from pleural effusions and a brain metastasis. Invasion Metastasis. 11, 204–215. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary Table 1 Primer and oligo sequences.
Supplementary Table 2 Altered Expression of miRNAs after Curcumin treatment. Microarray data of miRNA expression in Curcumin treated (6 h) and carrier treated (6 h) human MDA‐MB‐231 breast cancer cells was analyzed using MA plot with M as a parameter to express how many times each miR is regulated against the control and P as parameter for the statistical significance.
Supplementary Table 3 In silico analysis of miRNA targets.
Supplementary Figure 1 Kaplan Meier curves for the association of the expression of metastasis related and miR181b regulated genes in human breast cancers. 424 basal‐like human breast cancers with 124 events from the Breast Mark collection of breast cancer datasets were analyzed. Cases were assigned to low (red) and high (blue) expression classes based on the median level of expression. Log rank test p‐values are indicated.


