Abstract
Breast cancers (BC) carry a complex set of gene mutations that can influence their gene expression and clinical behavior. We aimed to identify genes driven by the TP53 mutation status and assess their clinical relevance in estrogen receptor (ER)‐positive and ER‐negative BC, and their potential as targets for patients with TP53 mutated tumors. Separate ROC analyses of each gene expression according to TP53 mutation status were performed. The prognostic value of genes with the highest AUC were assessed in a large dataset of untreated, and neoadjuvant chemotherapy treated patients. The mitotic checkpoint gene MPS1 was the most significant gene correlated with TP53 status, and the most significant prognostic marker in all ER‐positive BC datasets. MPS1 retained its prognostic value independently from the type of treatment administered. The biological functions of MPS1 were investigated in different BC cell lines. We also assessed the effects of a potent small molecule inhibitor of MPS1, SP600125, alone and in combination with chemotherapy. Consistent with the gene expression profiling and siRNA assays, the inhibition of MPS1 by SP600125 led to a reduction in cell viability and a significant increase in cell death, selectively in TP53‐mutated BC cells. Furthermore, the chemical inhibition of MPS1 sensitized BC cells to conventional chemotherapy, particularly taxanes. Our results collectively demonstrate that TP53‐correlated kinase MPS1, is a potential therapeutic target in BC patients with TP53 mutated tumors, and that SP600125 warrant further development in future clinical trials.
Keywords: Breast cancer subtypes, TP53 mutation status, Tumor relapse, MPS1 protein kinase, SP600125, Chemotherapy
Highlights
TP53 status is associated with two sets of genes in ER+ and ER− breast tumors.
Genes associated with TP53 status were correlated to RFS in ER+ patients.
MPS1 was the most significant gene associated with TP53 status and poor prognosis.
MPS1 inhibition affected cell viability and apoptosis in TP53 mutated cells.
Targeting MPS1 by SP600125 sensitizes TP53 mutated cells to chemotherapy.
1. Introduction
Breast cancer (BC) is a heterogeneous disease comprising different molecular subtypes characterized by different types of genetic alterations (Cancer Genome Atlas Network, 2012; Santarpia et al., 2012; Sotiriou and Pusztai, 2009). Activation of distinct gene and signaling pathways are associated with prognosis and different responses to therapies (Iwamoto et al., 2011; Santarpia et al., 2013). The tumor suppressor TP53 is the most frequently altered gene in human cancer, including BC. The importance of TP53 as a tumor suppressor in BC is highlighted also by the occurrence of TP53 mutations in the majority of HER2‐positive, and basal‐like BC subtypes, and in an important percentage of luminal A and B subtypes (Cancer Genome Atlas Network, 2012). TP53 mutations in BC have been found associated with earlier onset, increased aggressiveness of tumors, aneuploidy, lack of response to endocrine therapies, and in general with poor prognosis (Olivier et al., 2006). Few studies have analyzed a limited number of BC samples and identified prognostic gene signatures correlated with TP53 status (Bonnefoi et al., 2011; Coutant et al., 2011; Miller et al., 2005; Troester et al., 2006). TP53 is an important regulator of several cellular signaling pathways and the disruption of TP53 functions has been associated with the impairment of DNA repair, cell cycle arrest and apoptosis, which ultimately leads to genomic instability and cancer progression (Turner et al., 2013; Walerych et al., 2012).
Restoring endogenous TP53 function holds a lot of promise, although it is still a challenging task due to the complex signaling and multiple cellular functions of TP53. However, considering the relevant role of TP53 signaling pathway in BC, a potential targeting approach could be through the modulation of TP53‐regulated downstream signals. This would be particularly relevant to the tailoring of individual therapies, especially when the treatment agents affect TP53‐dependent biological responses.
In this study, we integrated a large cohort of BC patients, constructed a complete database to identify a more robust gene signature of target genes associated with TP53 mutation status (Gyorffy et al., 2009). Several BC datasets for gene expression data containing TP53 mutation status and clinical informations of patients with long‐term follow‐up were simultaneously analyzed. We combined different levels of data to identify important genes potentially regulated by mutant TP53 and having clinical relevance. Therefore, we assessed the role of the most relevant gene that arose from our analysis, the human monopolar spindle 1 MPS1, in TP53‐proficient and ‐deficient BC cell lines. Finally, we used a potent inhibitor of MPS1, SP600125, alone and in combination with chemotherapy, which resulted in reduced proliferation and a significant cell death of BC cells. Moreover, inhibition of MPS1 caused an important chemosensitization of cancer cells.
2. Materials and methods
2.1. Datasets
We structured the collection of public datasets according to the “Preferred Reporting Items for Systematic Reviews and Meta‐Analyses” (PRISMA) guidelines (Moher et al., 2009). The complete workflow of the performed analyses is summarized in Figure 1. We used a similar approach as previously reported (Coutant et al., 2011) and in a very large cohort of BC patients. To assess the prognostic value of the mutant TP53‐related genes, five independent BC datasets containing TP53 mutation status, gene expression and clinical data with follow‐up were used as the discovery cohort (estrogen receptor (ER)‐positive = 511, and ER‐negative = 184) (Supplementary Table 1) (Bertheau et al., 2007; Desmedt et al., 2011; Iwamoto et al., 2011; Miller et al., 2005; Reme et al., 2013).
Figure 1.
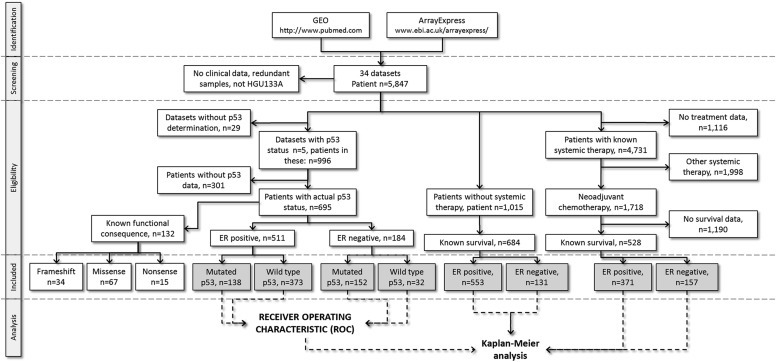
General workflow of the study and data processing.
To evaluate the association between mutant TP53‐related gene expressions and tumor relapse in BC patients, we tested the expression of selected genes in patients who did not receive any treatment by analyzing three different lymph node negative BC datasets (Mainz GSE11121, Wang GSE2034, and TRANSBIG GSE7390) for a total of 684 patients (Supplementary Table 1). We also selected the most significant gene that arose from our in silico analysis, a specific kinase gene, MPS1 and assessed its prognostic role in BC patients stratified for ER (after correction for HER2) who received different chemotherapy regimens. Specifically, we analyzed six independent cohorts of patients (n = 528) who received neoadjuvant chemotherapy (Supplementary Table 1) (Desmedt et al., 2009; Hatzis et al., 2011; Karn et al., 2011).
2.2. Array processing
All arrays were downloaded and normalized in the R environment using the affy Bioconductor package as previously described (Gyorffy and Schafer, 2009). For genes measured by several probe sets, the most reliable probe set was selected using JetSet (Li et al., 2011). Only probe sets reaching a MAS5 expression value of 1000 in at least one of the samples were used in the statistical computations. Additionally, all microarray files were compared using the ranked expression of all genes to spot microarrays re‐published in different studies. The ER status was identified as previously described (Gyorffy et al., 2012).
2.3. Statistical analysis
The ROC analysis was performed in the R statistical environment (http://www.r‐project.org) using the ROC Bioconductor library (http://www.bioconductor.org). A Bonferroni correction was applied to account for multiple testing. The statistical significance was set at p < 0.001.
The Kaplan–Meier survival analysis was performed as previously described (Gyorffy et al., 2010). The Kaplan–Meier survival plot and Cox regression models were computed in WinStat 2013 (Robert K. Fitch Software, Germany). The functional annotation and pathway analysis was performed using the NCBI DAVID server (Huang et al., 2009, 2009) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000).
Correlative studies between the most significant mutant TP53‐correlated genes and proliferation (MKI67 probe [212021_s_at] expression) were performed on two independent BC datasets (Miller et al., 2005; Iwamoto et al., 2011) by Pearson correlation analysis.
2.4. Cell culture and treatments
The human BC cell lines MCF7, MDA‐MB‐231 and T47D were obtained from American Type Cell Collection (ATCC). MCF7 and MDA‐MB‐231 cells were cultured in DMEM medium, and T47D cells were cultured in RMPI‐1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. For the experiments, the MCF7, MDA‐MB‐231 and T47D cells were grown in phenol red‐free culture medium under serum‐free conditions. 2H‐Dibenzo[cd,g]indazol‐6‐one (SP600125) was selected as a MPS1 inhibitor from Vichem's Chemical Validation Library (Bennett Brydon L. et al., US PAt. App. Publ., 20040072888, 15 Apr. 2004). SP600125 was synthesized in a one‐step cyclization of 1‐chloro‐anthraquinone with hydrazine hydrate, as previously described (Kim and Wiemer, 2004). SP600125 was solubilized in DMSO and was used at a final concentration of 10 μM; paclitaxel and doxorubicin were used at concentration of 1 μM and 4 μM, respectively. The appropriate amount of DMSO was employed for the negative control condition.
2.5. Western blotting
The cells were harvested and lysed in a RIPA buffer containing protease inhibitors (Sigma–Aldrich). The total protein (30 μg) was denatured, separated by 4%–20% SDS‐PAGE (BioRad) and transferred to Immuno‐Blot™ polyvinylidene difluoride (PVDF) membranes (BioRad). After blocking the membranes in 5% non‐fat milk powder dissolved in phosphate‐buffered saline (PBS), they were incubated overnight in 0.5% non‐fat milk powder at 4 °C with primary anti‐MPS1 antibody (Millipore). Afterwards, the membranes were incubated for one hour with an HRP‐conjugated anti‐mouse IgG (Millipore). After washing, a chemiluminescent substrate (Millipore) was added to the membranes, which were then exposed in the Chemidoc XRS station (Biorad). To control for protein loading, membranes were stripped and reprobed with an anti‐β actin antibody (Life Technologies). Band intensities were analyzed and calculated using the Quantity One 4.6 software (BioRad).
2.6. Cell viability and proliferation assays
Viable cells were identified using the 3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide assay (MTT; Sigma). Briefly, cells were plated (3 × 103 cells/well) on 96‐well plates in growth medium supplemented with 10% serum, and SP600125 or DMSO (for control cells) was added 24 h later. Seventy‐two hours after the SP600125 treatment, the MTT reagent (5 mg/ml in PBS) was added to each well, followed by incubation for 4 h at 37 °C. The MTT crystals in each well were solubilized in DMSO. The absorbance was read at 560 nm with an iMark plate reader (BioRad). All the treatments were performed in triplicate, and cell viability was expressed as a percentage of the untreated controls (mean ± S.D.). For proliferation assay, serum starved BC cell lines (approximately 80% density) were treated with SP600125 or DMSO. Cell proliferation was assessed using a commercial kit (cell proliferation enzyme‐linked immunosorbent assay, 5‐bromo‐2′‐deoxyuridine (BrdU); Roche Applied Science) according to the manufacturer's instructions. Briefly, after 48 h of incubation with SP600125 or DMSO, the cells were labeled with BrdU for 3 h at 37 °C. The cells were fixed and incubated with peroxidase‐conjugated anti‐BrdU antibody. Then, the peroxidase substrate 3,3′,5,5′‐tetramethylbenzidine was added, and BrdU incorporation was quantitated by the difference in the absorbance at 370 and 492 nm.
2.7. Assessment of apoptosis by Annexin V/propidium iodide double‐staining assay
Cells were incubated for 48 h with SP600125 and then collected, washed with PBS, and resuspended in Annexin V binding buffer (1 × 106 cells/ml final concentration). The amount of apoptotic cells were identified by double supravital staining with recombinant fluorescein isothiocyanate‐conjugated Annexin V antibody (Dako) and propidium iodide. Flow cytometric analysis was performed immediately after the supravital staining. The data acquisition and analysis were performed in a FACSCalibur flow cytometer using CellQuest software (BD Biosciences).
2.8. siRNA transfections
MPS1 was targeted with two distinct siRNAs (Dharmacon Inc.) that were delivered into MCF7, MDA‐MB‐231 and T47D BC cells by oligofectamine (Invitrogen). After 48 h, the effect of silencing on protein expression levels was determined by immunoblotting. We optimized the transfection conditions for each cell lines, as previously described, with minimum modifications (Bianchini et al., 2010). Briefly, we used three different types of negative controls, including cells grown in regular OptiMEM medium (Invitrogen), cells grown in the presence of transfection reagents only, and cells transfected with control siRNA, including four different constructs and four different siRNA constructs (Dharmacon Inc.). Different positive controls (40 nmol/L final concentration) were used to define the optimal transfection efficiency conditions. Gene silencing was performed in three replicates in 96‐well plates using predetermined optimal transfection conditions. The cells were seeded at a density that yielded 70–80% confluence in the control wells at 96 h. Each plate included wells with target siRNAs (40 nmol/L per well in a 50 μL total volume) and negative and positive siRNA controls; the plates were incubated at 37 °C for 96 h.
2.9. Immunoprecipitation/kinase assay
The immunoprecipitation/kinase assay was performed as previously described (Huang et al., 2009, 2009). Briefly, MCF7, MDA‐MB‐231 and T47D cells were lysed, and MPS1 was immunoprecipitated from the cell lysates with anti‐MPS1 antibody (Acris Antibodies GmbH) and washed three times in CSK Buffer (10 mM piperazine‐N,N‐bis(2‐ethanesulfonic acid) (pH 7.0), 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.1% NP‐40) to which 0.7 M LiCl was added for the second wash. After two washes in kinase buffer, the kinase reaction was carried out using His‐TP53 as substrate. The reaction was stopped by the addition of a 0.5 volume of protein sample buffer, and the proteins were resolved by SDS‐PAGE. The phosphorylation of TP53 was visualized by Western blotting using an anti‐phospho‐TP53 antibody (Cell Signaling).
3. Results
3.1. Identification of genes correlated with TP53 status
Five independent BC datasets containing informations on TP53 mutation status, gene expression and clinical data were used as the discovery cohort (Supplementary Table 1). We independently assessed ER‐positive (n = 511) and ER‐negative (n = 184) patients to identify genes associated with mutant TP53 using ROC analysis. We identified 4172 and 794 mutant TP53‐associated genes in ER‐positive and ER‐negative groups, respectively (p < 0.001; Supplementary Table 2A and B). The ROC results for the top ranking genes for ER‐positive and ER‐negative BC patients are listed in Table 1. The top‐ranked gene in ER‐positive tumors, the MPS1 kinase, was the most significant one associated with TP53 status (AUC = 0.814, p < 1E‐16). MPS1 remained significantly associated with TP53 status even when the individual datasets were analyzed separately (GSE3494: AUC = 0.852, p < 1E‐16; GSE22093: AUC = 0.78, p = 1.45E‐06; E‐MTAB‐43: AUC = 0.82, p = 9.0E‐04; E‐MTAB‐365: AUC = 0.77, p = 7.42E‐14). To evaluate the impact of the most significant genes associated with TP53 status we used the DAVID server. We identified specific and significantly enriched canonical signaling pathways potentially activated by mutations of TP53 (p < 0.01) (Supplementary Table 3). Functional annotation clustering for all the significantly mutant TP53‐associated genes in ER‐positive tumors revealed “Cell cycle” (p = 9.2E‐6), “DNA replication” (p = 1.0E‐3), “Mismatch repair” (p = 1.9E‐3), “TP53 signaling pathway” (p = 2.8E‐3) and “Nucleotide excision repair” (p = 1.3E‐2). The most relevant enriched pathways among the mutant TP53‐correlated genes in the ER‐negative patients were “Mismatch repair” (p = 1.1E‐6), “DNA replication” (p = 1.2E‐6), “Cell cycle” (p = 1.4E‐6), “Pathways in cancer” (p = 6.0E‐3), “Nucleotide excision repair” (p = 7.0E‐3) and “ECM‐receptor interaction” (p = 1.1E‐2). Of note, cell cycle was the category hit by most of the genes; therefore, we used the KEGG cell cycle pathway map to visualize the genes with altered expression in TP53 mutant cancers (Supplementary Figure 1). TP53 mutations were more frequent in ER‐negative tumors (Chi‐square test p = 0.0001).
Table 1.
Receiver Operating Characteristic analysis in ER‐positive and ER‐negative breast cancer patients. *p < 0.001. MPS1 was the gene with the highest AUC value in ER‐positive tumors.
| Affymetrix ID | Gene symbol | AUC | p‐value |
|---|---|---|---|
| ER‐positive | |||
| 204822_at | MPS1 | 0.814 | <1E‐16* |
| 202870_s_at | CDC20 | 0.804 | <1E‐16* |
| 210052_s_at | TPX2 | 0.803 | <1E‐16* |
| 204962_s_at | CENPA | 0.802 | <1E‐16* |
| 202705_at | CCNB2 | 0.790 | <1E‐16* |
| 201710_at | MYBL2 | 0.789 | <1E‐16* |
| 208079_s_at | AURKA | 0.785 | <1E‐16* |
| 203438_at | STC2 | 0.782 | <1E‐16* |
| 209773_s_at | RRM2 | 0.781 | <1E‐16* |
| ER‐negative | |||
| 201774_s_at | NCAPD2 | 0.779 | 1.29E‐12* |
| 209710_at | GATA2 | 0.759 | 2.01E‐10* |
| 201286_at | SDC1 | 0.729 | 4.96E‐09* |
| 203233_at | IL4R | 0.765 | 1.15E‐08* |
| 203758_at | CTSO | 0.759 | 1.28E‐08* |
| 209421_at | MSH2 | 0.733 | 5.90E‐08* |
| 219076_s_at | PXMP2 | 0.739 | 6.48E‐08* |
| 208763_s_at | TSC22D3 | 0.741 | 8.01E‐08* |
| 211623_s_at | FBL | 0.759 | 9.26E‐08* |
3.2. Survival analysis in untreated breast cancer patients and patients treated with systemic chemotherapy
To evaluate the prognostic value of the genes associated with TP53 status we downloaded the expression data for 684 lymph node negative untreated patients including 553 ER‐positive and 132 ER‐negative patients of three independent datasets (Supplementary Table 1). In the ER‐positive patients, low MPS1 expression was associated with longer relapse‐free survival (HR = 1.89, p = 1.85E‐05, Supplementary Figure 2). In the untreated BC series, after correction for HER2 (Cox multivariate regression model), MPS1 was still significant prognostic (HR = 1.87, p = 3.99E‐05). MPS1 retained its prognostic significance even after stratification for lymph node status (lymph node positive patients: HR = 2.4, p = 4.9E‐04; lymph node negative patients: HR = 11.9, p = 0.0022).
We also assessed the association of these classes of genes with clinical outcome in patients treated with systemic therapy. Survival data available for 528 BC patients who received neoadjuvant chemotherapy (Supplementary Table 1) were used for further validation by employing Kaplan–Meier analysis (Figure 1). None of these validation samples was included in the discovery cohort. The Kaplan–Meier plots for three of the top genes in each cohort are depicted in Figure 2. We performed Cox regression analysis in ER‐positive and ER‐negative BC patients on the genes with the highest AUC values (Table 2). We found that the gene with the highest AUC value in ER‐positive tumors, MPS1, was also strongly associated with patient relapse‐free survival (HR = 2.5, p = 4.0E‐05; Table 2). Analysis in ER‐negative tumors did not reveal any gene correlated with TP53 status significantly associated with risk of relapse (Table 2 and Figure 2). In addition, to demonstrate the robustness of the median approach used in our study we randomly divided the neoadjuvant treated ER‐positive cohort into two cohorts of equal size. The median expression of MPS1 in the training cohort was used in the Cox regression analysis for both the training and the testing cohorts; the gene retained its significant association with relapse‐free survival in both cohorts (Supplementary Figure 3). Pearson correlation analysis demonstrated that almost all of the top TP53‐associated genes were correlated with proliferation (Supplementary Table 4).
Figure 2.
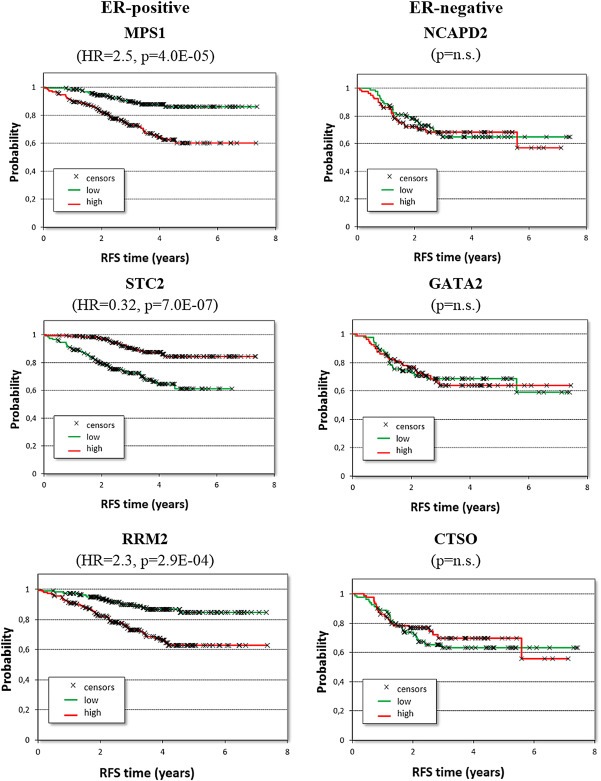
Survival analysis in neoadjuvant chemotherapy‐treated breast cancer patients for three of the top ranking genes identified using ROC analysis. (n.s. = not significant; RFS = relapse‐free survival).
Table 2.
Cox regression analyses in ER‐positive and ER‐negative breast cancer patients performed on the genes ranked by the highest AUC values. *p < 0.001. MPS1 was the most significant gene associated with poor outcome in ER‐positive tumors.
| Affymetrix ID | Gene symbol | HR | p‐value |
|---|---|---|---|
| ER‐positive | |||
| 204822_at | MPS1 | 2.5 | 4.0E‐05* |
| 202870_s_at | CDC20 | 2.1 | 1.2E‐03 |
| 210052_s_at | TPX2 | 1.47 | 0.08 |
| 204962_s_at | CENPA | 2.1 | 1.3E‐03 |
| 202705_at | CCNB2 | 2.2 | 3.7E‐04* |
| 201710_at | MYBL2 | 1.31 | 0.22 |
| 208079_s_at | AURKA | 2.2 | 3.9E‐04* |
| 203438_at | STC2 | 0.32 | 7.0E‐07* |
| 209773_s_at | RRM2 | 2.3 | 2.9E‐04* |
| ER‐negative | |||
| 201774_s_at | NCAPD2 | 1.14 | 0.56 |
| 209710_at | GATA2 | 1.30 | 0.23 |
| 201286_at | SDC1 | 1.02 | 0.94 |
| 203233_at | IL4R | 1.20 | 0.41 |
| 203758_at | CTSO | 0.73 | 0.16 |
| 209421_at | MSH2 | 0.82 | 0.36 |
| 219076_s_at | PXMP2 | 0.86 | 0.5 |
| 208763_s_at | TSC22D3 | 0.84 | 0.43 |
| 211623_s_at | FBL | 0.88 | 0.54 |
3.3. Loss of MPS1 results in decreased survival and induction of apoptosis in breast cancer cells
We analyzed a large BC cell line dataset (http://www.ebi.ac.uk/arrayexpress/, accession No. E‐TABM‐157) (Neve et al., 2006) and verified that MPS1 was expressed, though at different levels, in all BC cell lines (data not shown). For in vitro analysis, we selected one TP53 wild‐type cell line (MCF7, luminal) and two TP53‐mutated BC cell lines (T47D, luminal and MDA‐MB‐231, basal‐like). To determine the basal MPS1 activity levels and the effect of the MPS1 inhibitor SP600125 on BC cells, in vitro kinase assays were employed using MPS1 immunoprecipitated from MCF7, T47D and MDA‐MB‐231 cells. The phosphorylation of TP53, a downstream target of MPS1, at threonine‐18 was assessed by Western blotting. As shown in Figure 3, MPS1 activity was detected in all BC cell lines and was found to be consistently elevated in T47D cells. Furthermore, SP600125 at 10 μM significantly reduced the MPS1 activity in all BC cells (Figure 3), confirming MPS1 as a specific target of SP600125.
Figure 3.
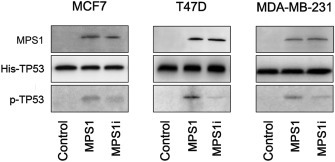
MPS1 activity and the effect of SP600125 (MPS1i) in breast cancer cell lines. Immunoprecipitated endogenous MPS1 phosphorylated TP53 on Thr18. Immunoprecipitation/kinase assays were performed using MPS1 immunoprecipitated from MCF7, T47D and MDA‐MB‐231 cells. Normal rabbit immunoglobulin G was used as control. Phosphorylation of the substrate (His‐TP53) was detected by Western blotting using an anti‐phospho‐Thr18‐TP53 antibody.
Reduction in MPS1 levels has been found to decrease the viability of several human cancer cell lines (Janssen et al., 2009). To explore the role of MPS1 in BC cells, we examined the effect of MPS1 gene silencing in MCF7, MDA‐MB‐231 and T47D cell lines. Treatment with MPS1‐siRNA resulted in a reduced expression of the MPS1 protein in all of the cell lines tested (Figure 4A), whereas control cells and non‐targeting siRNA transfected cells retained normal MPS1 protein expression (Figure 4A), confirming the MPS1 silencing efficiency of the MPS1‐siRNA. The cell viability and cell proliferation were then assessed by the MTT assay (Figure 4B) and BrdU incorporation (Figure 4C), respectively. We found that MPS1 silencing had no effect on the proliferative potential of MCF7 and a slight effect (p < 0.05) on cell viability and proliferation in both of the TP53‐mutant cell lines. This result indicates that the TP53‐mutated BC cells and TP53 wild‐type MCF7 cells have differential requirements for MPS1, potentially corresponding to their TP53 functional status.
Figure 4.
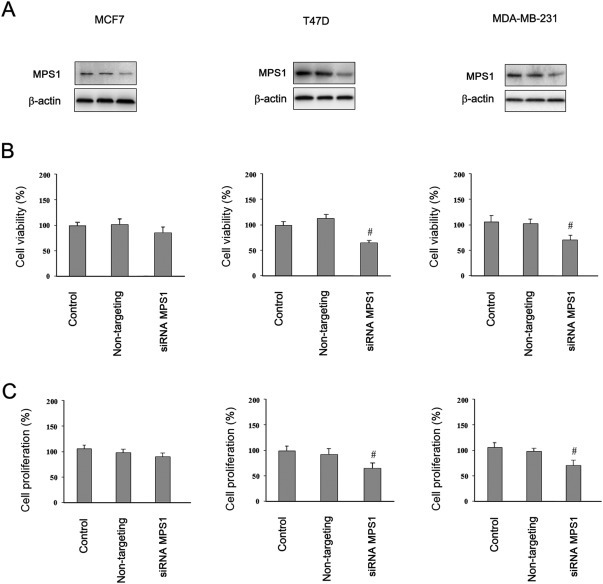
Viability and proliferation of breast cancer cells after the reduction of MPS1 expression levels with siRNA. (A) Western blot analysis showing protein levels of parental control (transfection reagents only), non‐targeting construct‐transfected, and MPS1 siRNA‐transfected after 48 h. Cell viability (B) and cell proliferation (C) after silencing of MPS1 were determined by MTT assay and BrdU incorporation, respectively. Error bars represent the S.D. of triplicate measurements. #p < 0.05.
Similar results were obtained with the MPS1 inhibitor SP600125 (Figure 5A). Inhibition of MPS1 was not sufficient to consistently reduce the viability of MCF7 cells and showed only marginal effects (p < 0.05) on the viability of the TP53‐mutated BC cell lines (Figure 5A). In the presence of the MPS1 inhibitor, the TP53 wild‐type MCF7 cells did not manifest any increased staining for Annexin V (Figure 5B). Conversely, both TP53‐mutated BC cell lines showed a markedly increased rate of apoptosis (Figure 5B), consistent with the concept that a reduction in MPS1 causes cellular apoptosis rather than simply a reduction in cell growth rates. Furthermore, MPS1 inhibition was more efficient (p < 0.01) in increasing apoptosis in ER‐positive T47D cells compared with ER‐negative MDA‐MB‐231 cells (p < 0.05). These results, in line with the in silico analyses, establish that MPS1 inhibition preferentially impaired TP53‐mutated cells, and that MPS1 is a potential therapeutic target for TP53‐mutated ER‐positive breast cancers.
Figure 5.
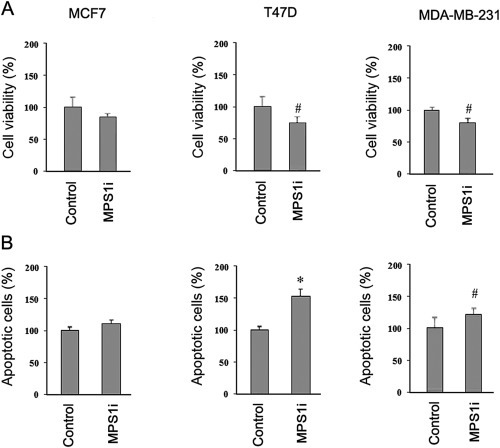
Effects of SP600125 (MSP1i) on viability and apoptosis in breast cancer cell lines. MCF7, T47D and MDA‐MB‐231 cells were treated with 10 μM SP600125 or DMSO alone (control) for 72 h. (A) Cell viability was analyzed by MTT assay. (B) Apoptotic cells were detected by propidium iodide and Annexin V staining. Histogram comparing the percent of apoptotic cells in each of the different treatments are shown. Error bars represent the S.D. of triplicate measurements. #p < 0.05, *p < 0.01.
To further assess the potential therapeutic value of MPS1 inhibition in combined therapy, we analyzed the effect of SP600125 in combination with conventional chemotherapeutic drugs in different BC cell lines (Figure 6). SP600125 alone consistently reduced TP53‐mutated cells survival (p < 0.05), but had no significant effects on MCF7 survival (Figure 6). Of note, MPS1 inhibition had a synergistic effect on cell survival in mutant TP53 cells either in combination with doxorubicin (p < 0.01), or with paclitaxel (p < 0.001). Although the combination with taxanes and anthracyclines caused a decrease of cell survival also in MCF7 (p < 0.05), the observed synergistic effect was negligible with respect to TP53‐mutated cells, suggesting that this type of cells is sensitive at less extent to the inhibition of MPS1 (Figure 6).
Figure 6.
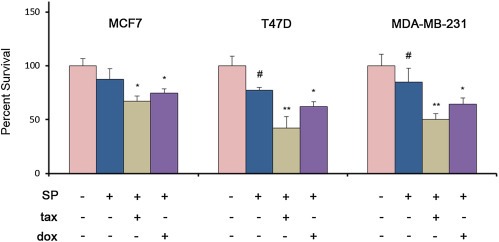
Effects of SP600125 (SP) in combination with taxane (tax) or anthracycline (dox) on breast cancer cells survival assessed by MTT assay. MCF7, T47D and MDA‐MB‐231 cells were treated with 10 μM SP alone or in combination with paclitaxel (1 μM) or doxorubicin (4 μM). Error bars indicate the S.D. (n = 3). #p < 0.05, *p < 0.01, **p < 0.001.
4. Discussion
TP53 is an important prognostic indicator in BC, and dysfunctions of this gene have been demonstrated to affect different gene patterns. Moreover, BC with TP53 mutations tend to have fewer mutations in other driver cancer genes, suggesting a crucial role of the TP53 signaling pathway in these cancer cells (Cancer Genome Atlas Network, 2012).
In this study, we assembled several different BC datasets to assess the effect of TP53 status on the potential regulation of specific genes, to evaluate their prognostic value, and identify genes to target in TP53 mutated ER‐positive and ER‐negative BC subtypes. We established two distinct sets of genes associated with TP53 status in ER‐positive and ER‐negative BCs and demonstrated that single genes were able to predict TP53 dysfunction in both subtypes. This suggests a potential intersection in the activation of transcriptional programs regulated by TP53 in both cancer groups. However, genes correlated with mutant TP53 were associated with relapse‐free survival only in ER‐positive tumors, suggesting that the expression of some genes in TP53 mutated tumors may be dependent on ER status. The prognostic genes associated with TP53 status and identified in the discovery dataset were confirmed in a large cohort of lymph‐node negative untreated BC patients.
The prognostic value of the two sets of genes was confirmed in ER‐positive and ER‐negative BC patients treated with neoadjuvant chemotherapies, including taxane‐anthracycline based regimens. The genes associated with TP53 status were confirmed to be prognostic only in ER‐positive tumors and independently from the type of chemotherapy received. The same analysis performed for TP53‐genes on the ER‐negative group had no consistent prognostic value. Some of the genes associated with TP53 status are also linked to cell cycle and proliferation, making the latter a potential confounding variable in the identification of prognostic biomarker. However, important studies have demonstrated that despite TP53 mutations are directly linked to high tumor proliferation rates, both factors remain independently associated with BC prognosis (Allred et al., 1993; Yerushalmi et al., 2010). In our results, as expected, we found that numerous top‐ranked genes linked to TP53 status were also significantly associated with proliferation and tumor relapse. To make our approach reliable and robust, from our data we selected MPS1 (also named TTK), the gene with the highest AUC value linked with TP53 status, and most significantly associated with poor prognosis. MPS1 was previously included in different prognostic proliferation‐based signatures (Sotiriou et al., 2006; Bianchini et al., 2010), and in a 16‐kinase signature that was able to further distinguish two subgroups of luminal A tumors with different survival (Finetti et al., 2008). In spite of the strong link between MPS1 and proliferation, this kinase has also recently been established as part of a gene module dependent on TP53 status, which further stratified luminal tumors, independently of proliferation. These observations shed light on the biological consequences of specific defective cell cycle checkpoints upon regulation of TP53‐modulated kinase genes, such as MPS1, in this subgroup of luminal tumors, which are most likely characterized by higher genomic instability (Fredlund et al., 2012). These findings indirectly support the correctness of the approach and gene selection adopted in our study, and suggest that this poor prognosis luminal subgroup could be enriched for distinct genomically unstable TP53‐mutated tumors.
These observations also suggest that TP53 mutations may activate specific transcriptional checkpoint programs that in turn could destabilize the interactions between TP53 and ER thus explaining the resistance of specific ER‐positive tumors to apoptosis (Bailey et al., 2012). Accordingly, in our study, the top‐scored genes associated with TP53 status in ER‐positive tumors were enriched for functions related to mitotic spindle assembly and checkpoint regulation (MPS1, CDC20, TPX2, CCNB2, and AURKA). Our data are also in agreement with a recent study demonstrating that a subset of poor prognostic luminal tumors was linked to TP53‐regulated programs, and genomic instability (Ciriello et al., 2013).
TP53 is capable to regulate a number of checkpoint and mitotic kinases; few of these have been recently investigated as potential targets for therapies (Ha and Breuer, 2012). The MPS1 gene encodes for a dual serine/threonine kinase, which is involved in the mitotic spindle assembly checkpoint (SAC) (Jones et al., 2005; Weiss and Winey, 1996), chromosome stability (Leng et al., 2006), DNA damage checkpoints (Wei et al., 2005) and the TP53‐dependent mitotic checkpoint (Huang et al., 2009c). In this study, the prognostic value of MPS1 in BC was confirmed in both ER‐positive untreated patients, and in ER‐positive patients treated with neoadjuvant chemotherapy. MPS1 was prognostic after stratification for lymph node status, and as TP53 status, independently from the type of therapy administrated (Bonnefoi et al., 2011). Our in vitro results paralleled and supported the in silico data, suggesting that MPS1 is a potential TP53‐regulated kinase to target in BC. Accordingly, in our study, the inhibition of MPS1 by siRNA and a chemical inhibitor, strengthened the important role played by this kinase in a subset of TP53‐mutated ER‐positive tumors characterized by poor prognosis. We demonstrated that all BC cell lines expressed MPS1 and that the loss of MPS1 resulted in decreased cell viability and proliferation. The TP53‐mutated (MDA‐MB‐231 and T47D) and TP53 wild‐type (MCF7) BC cells had differential requirements for MPS1, potentially corresponding to their different TP53 functional status. The potent biological activity of SP600125, a small pan‐kinase mitotic inhibitor, demonstrated that the inhibition of MPS1 resulted in decreased cell viability and induction of apoptosis in BC cell lines with dysfunctions of TP53. MPS1, regulates checkpoint activity that is selectively required to sustain the proliferation of aneuploid tumors, and it is associated with chromosomal instability in different cancer types (Carter et al., 2006; Daniel et al., 2011). Our data suggest that the checkpoint inactivation leading to aneuploidy may serve as a mechanism by which MPS1 inhibition causes the loss of cell viability and the induction of apoptosis in mutated BC cells. In line with our findings, MPS1 has recently been suggested as a potential target for tumors characterized by high genomic instability, such as melanomas mutated for BRAF‐V600E (Liu et al., 2013), basal‐like BC (Maire et al., 2013), and glioblastomas (Tannous et al., 2013).
In this study, we confirmed a significant high activity of SP600125 in TP53 mutated BC cell lines, which are known to be characterized by high genomic instability and aneuploidy. Additional studies using these classes of anti‐checkpoint/mitotic inhibitors in these types of cancer cells support the consistency of our findings (Colombo et al., 2010).
Our data also proved that SP600125 added to cytotoxic drugs has an important effect on cell survival, enhancing the sensitivity of BC cells to chemotherapy, particularly to anti‐tubulin drugs. Comparable results were reported on recent different studies, which showed that MPS1 depletion was highly toxic in cancer cell lines if added in combination with chemotherapy (Janssen et al., 2009; Jemaà et al., 2013). TP53‐incompetent cells can override mitotic checkpoints becoming aneuploid, and contributing to chromosomal instability and resistance to therapy (Weaver and Cleveland, 2005; Cheok et al., 2011; Murray et al., 2012).
Consequently, it will be important to select compounds that preferentially kill TP53‐incompetent cells (Brown et al., 2009). Accordingly, SP600125 could selectively target TP53‐mutated proliferating mitotic cells in tumors, enhancing cell sensitivity to current chemotherapeutic drugs. Moreover, SP600125 may exert these cytotoxic effects through the concurrent inhibition of few serine/threonine kinases; although, the inhibition of MPS1 appears to be co‐responsible for the preferential effects of SP600125 on the cell cycle of TP53 −/− cells (Jemaà et al., 2012; Brown et al., 2009). Accordingly, we have demonstrated that SP600125 kills TP53‐defective cells more efficiently than their TP53‐proficient counterparts in vitro. Our data also suggest that mitotic associated checkpoint drugs including, SP600125 (or other drugs with “similar” activity, such as, NMS‐P715, MPI‐0479605, AZ3146 etc.), could be suitable candidates for further development toward clinical studies in genomically unstable breast tumors.
In conclusion, altogether our data demonstrate that TP53 status is associated with important key mitiotic‐checkpoint kinases, such as MPS1, involved in tumor relapse in a specific subset of ER‐positive tumors. MPS1 is predominantly expressed and activated by the loss of function mutations of TP53 in human breast tumors, and the inhibition of this marker efficiently increases the apoptosis of TP53 defective BC cell lines, particularly those expressing the estrogen receptor. Finally, the concurrent inhibition of important kinases, including MPS1, by SP600125 could have a synergistic cellular proapoptotic effect, which may be exploited for the therapeutic advantage of increasing cancer cell toxicities.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary Figure 1 Genes with significantly altered expression in TP53 mutant cancers mapped to the KEGG cell cycle pathway. The genes with altered expression in ER‐positive breast cancers are marked in red; the genes with altered expression in ER‐negative cancers only are in blue. The gene MPS1 (TTK) is marked by a wide black box. TP53 status affects several cyclin dependent kinase gene functions.
Supplementary Figure 2 Correlation between MPS1 mRNA levels and survival in untreated ER‐positive and ER‐negative breast cancer patients.
Supplementary Figure 3 Survival analysis in neoadjuvant treated ER‐positive breast cancer patients stratified for MPS1 expression. The 371 ER‐positive patients in the neoadjuvant dataset were divided into a training cohort (n = 211) and a testing cohort (n = 160) to perform survival analysis.
Acknowledgements
This study was supported by the OTKA PD 83154 Grant (to B.G.), the Predict project (Grant no. 259303 of the EU Health.2010.2.4.1.‐8 call to B.G.), Associazione Italiana per la Ricerca sul Cancro (AIRC Grant‐6251 to L.S.).
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.12.018
Győrffy Balázs, Bottai Giulia, Lehmann-Che Jacqueline, Kéri György, Őrfi László, Iwamoto Takayuki, Desmedt Christine, Bianchini Giampaolo, Turner Nicholas C., de Thè Hugues, André Fabrice, Sotiriou Christos, Hortobagyi Gabriel N., Di Leo Angelo, Pusztai Lajos and Santarpia Libero, (2014), TP53 mutation‐correlated genes predict the risk of tumor relapse and identify MPS1 as a potential therapeutic kinase in TP53‐mutated breast cancers, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.12.018.
References
- Allred, D.C. , Clark, G.M. , Elledge, R. , Fuqua, S.A. , Brown, R.W. , Chamness, G.C. , Osborne, C.K. , McGuire, W.L. , 1993. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J. Natl. Cancer Inst.. 85, 200–206. [DOI] [PubMed] [Google Scholar]
- Bailey, S.T. , Shin, H. , Westerling, T. , Liu, X.S. , Brown, M. , 2012. Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc. Natl. Acad. Sci. U S A. 109, 18060–18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertheau, P. , Turpin, E. , Rickman, D.S. , Espié, M. , de Reyniès, A. , Feugeas, J.P. , Plassa, L.F. , Soliman, H. , Varna, M. , de Roquancourt, A. , Lehmann-Che, J. , Beuzard, Y. , Marty, M. , Misset, J.L. , Janin, A. , de Thé, H. , 2007. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS. Med.. 4, e90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini, G. , Iwamoto, T. , Qi, Y. , Coutant, C. , Shiang, C.Y. , Wang, B. , Santarpia, L. , Valero, V. , Hortobagyi, G.N. , Symmans, W.F. , Gianni, L. , Pusztai, L. , 2010. Prognostic and therapeutic implications of distinct kinase expression patterns in different subtypes of breast cancer. Cancer Res.. 70, 8852–8862. [DOI] [PubMed] [Google Scholar]
- Bonnefoi, H. , Piccart, M. , Bogaerts, J. , Mauriac, L. , Fumoleau, P. , Brain, E. , Petit, T. , Rouanet, P. , Jassem, J. , Blot, E. , Zaman, K. , Cufer, T. , Lortholary, A. , Lidbrink, E. , André, S. , Litière, S. , Lago, L.D. , Becette, V. , Cameron, D.A. , Bergh, J. , Iggo, R. , EORTC 10994/BIG 1-00 Study Investigators2011. TP53 status for prediction of sensitivity to taxane versus non-taxane neoadjuvant chemotherapy in breast cancer (EORTC 10994/BIG 1-00): a randomised phase 3 trial. Lancet Oncol.. 12, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.J. , Lain, S. , Verma, C.S. , Fersht, A.R. , Lane, D.P. , 2009. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer. 9, 862–873. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network, 2012. Comprehensive molecular portraits of human breast tumors. Nature. 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, S.L. , Eklund, A.C. , Kohane, I.S. , Harris, L.N. , Szallasi, Z. , 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet.. 38, 104–1048. [DOI] [PubMed] [Google Scholar]
- Cheok, C.F. , Verma, C.S. , Baselga, J. , Lane, D.P. , 2011. Translating p53 into the clinic. Nat. Rev. Clin. Oncol.. 8, 25–37. [DOI] [PubMed] [Google Scholar]
- Ciriello, G. , Sinha, R. , Hoadley, K.A. , Jacobsen, A.S. , Reva, B. , Perou, C.M. , Sander, C. , Schultz, N. , 2013. The molecular diversity of Luminal A breast tumors. Breast Cancer Res. Treat. 141, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, R. , Caldarelli, M. , Mennecozzi, M. , Giorgini, M.L. , Sola, F. , Cappella, P. , Perrera, C. , Depaolini, S.R. , Rusconi, L. , Cucchi, U. , Avanzi, N. , Bertrand, J.A. , Bossi, R.T. , Pesenti, E. , Galvani, A. , Isacchi, A. , Colotta, F. , Donati, D. , Moll, J. , 2010. Targeting the mitotic checkpoint for cancer therapy with NMS-P715, an inhibitor of MPS1 kinase. Cancer Res.. 70, 10255–10264. [DOI] [PubMed] [Google Scholar]
- Coutant, C. , Rouzier, R. , Qi, Y. , Lehmann-Che, J. , Bianchini, G. , Iwamoto, T. , Hortobagyi, G.N. , Symmans, W.F. , Uzan, S. , Andre, F. , de Thé, H. , Pusztai, L. , 2011. Distinct p53 gene signatures are needed to predict prognosis and response to chemotherapy in ER-positive and ER-negative breast cancers. Clin. Cancer Res.. 17, 2591–2601. [DOI] [PubMed] [Google Scholar]
- Daniel, J. , Coulter, J. , Woo, J.H. , Wilsbach, K. , Gabrielson, E. , 2011. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc. Natl. Acad. Sci. U S A. 108, 5384–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmedt, C. , Giobbie-Hurder, A. , Neven, P. , Paridaens, R. , Christiaens, M.R. , Smeets, A. , Lallemand, F. , Haibe-Kains, B. , Viale, G. , Gelber, R.D. , Piccart, M. , Sotiriou, C. , 2009. The gene expression grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC. Med. Genomics. 2, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmedt, C. , Di Leo, A. , de Azambuja, E. , Larsimont, D. , Haibe-Kains, B. , Selleslags, J. , Delaloge, S. , Duhem, C. , Kains, J.P. , Carly, B. , Maerevoet, M. , Vindevoghel, A. , Rouas, G. , Lallemand, F. , Durbecq, V. , Cardoso, F. , Salgado, R. , Rovere, R. , Bontempi, G. , Michiels, S. , Buyse, M. , Nogaret, J.M. , Qi, Y. , Symmans, F. , Pusztai, L. , D'Hondt, V. , Piccart-Gebhart, M. , Sotiriou, C. , 2011. Multifactorial approach to predicting resistance to anthracyclines. J. Clin. Oncol.. 29, 1578–1586. [DOI] [PubMed] [Google Scholar]
- Finetti, P. , Cervera, N. , Charafe-Jauffret, E. , Chabannon, C. , Charpin, C. , Chaffanet, M. , Jacquemier, J. , Viens, P. , Birnbaum, D. , Bertucci, F. , 2008. Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis. Cancer Res.. 68, 767–776. [DOI] [PubMed] [Google Scholar]
- Fredlund, E. , Staaf, J. , Rantala, J.K. , Kallioniemi, O. , Borg, A. , Ringnér, M. , 2012. The gene expression landscape of breast cancer is shaped by tumor protein p53 status and epithelial-mesenchymal transition. Breast Cancer Res.. 14, R113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy, B. , Benke, Z. , Lánczky, A. , Balázs, B. , Szállási, Z. , Timár, J. , Schäfer, R. , 2012. Recurrence online: an online analysis tool to determine breast cancer recurrence and hormone receptor status using microarray data. Breast Cancer Res. Treat.. 132, 1025–1034. [DOI] [PubMed] [Google Scholar]
- Gyorffy, B. , Lanczky, A. , Eklund, A.C. , Denkert, C. , Budczies, J. , Li, Q. , Szallasi, Z. , 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat.. 123, 725–731. [DOI] [PubMed] [Google Scholar]
- Gyorffy, B. , Molnar, B. , Lage, H. , Szallasi, Z. , Eklund, A.C. , 2009. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS. One. 4, e5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy, B. , Schafer, R. , 2009. Meta-analysis of gene expression profiles related to relapse-free survival in 1,079 breast cancer patients. Breast Cancer Res. Treat.. 118, 433–441. [DOI] [PubMed] [Google Scholar]
- Ha, G.H. , Breuer, E.K. , 2012. Mitotic kinases and p53 signaling. Biochem. Res. Int.. 2012, 195903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis, C. , Pusztai, L. , Valero, V. , Booser, D.J. , Esserman, L. , Lluch, A. , Vidaurre, T. , Holmes, F. , Souchon, E. , Wang, H. , Martin, M. , Cotrina, J. , Gomez, H. , Hubbard, R. , Chacón, J.I. , Ferrer-Lozano, J. , Dyer, R. , Buxton, M. , Gong, Y. , Wu, Y. , Ibrahim, N. , Andreopoulou, E. , Ueno, N.T. , Hunt, K. , Yang, W. , Nazario, A. , DeMichele, A. , O'Shaughnessy, J. , Hortobagyi, G.N. , Symmans, W.F. , 2011. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 305, 1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.W. , Sherman, B.T. , Lempicki, R.A. , 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang, D.W. , Sherman, B.T. , Lempicki, R.A. , 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res.. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.F. , Chang, M.D. , Shieh, S.Y. , 2009. TTK/hMps1 mediates the p53-dependent postmitotic checkpoint by phosphorylating p53 at Thr18. Mol. Cell. Biol.. 29, 2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto, T. , Bianchini, G. , Booser, D. , Qi, Y. , Coutant, C. , Shiang, C.Y. , Santarpia, L. , Matsuoka, J. , Hortobagyi, G.N. , Symmans, W.F. , Holmes, F.A. , O'Shaughnessy, J. , Hellerstedt, B. , Pippen, J. , Andre, F. , Simon, R. , Pusztai, L. , 2011. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J. Natl. Cancer Inst.. 103, 264–272. [DOI] [PubMed] [Google Scholar]
- Janssen, A. , Kops, G.J. , Medema, R.H. , 2009. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. U S A. 106, 19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemaà, M. , Galluzzi, L. , Kepp, O. , Senovilla, L. , Brands, M. , Boemer, U. , Koppitz, M. , Lienau, P. , Prechtl, S. , Schulze, V. , Siemeister, G. , Wengner, A.M. , Mumberg, D. , Ziegelbauer, K. , Abrieu, A. , Castedo, M. , Vitale, I. , Kroemer, G. , 2013. Characterization of novel MPS1 inhibitors with preclinical anticancer activity. Cell. Death Differ.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemaà, M. , Vitale, I. , Kepp, O. , Berardinelli, F. , Galluzzi, L. , Senovilla, L. , Mariño, G. , Malik, S.A. , Rello-Varona, S. , Lissa, D. , Antoccia, A. , Tailler, M. , Schlemmer, F. , Harper, F. , Pierron, G. , Castedo, M. , Kroemer, G. , 2012. Selective killing of p53-deficient cancer cells by SP600125. EMBO. Mol. Med.. 4, 500–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H. , Huneycutt, B.J. , Pearson, C.G. , Zhang, C. , Morgan, G. , Shokat, K. , Bloom, K. , Winey, M. , 2005. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol.. 15, 160–165. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Goto, S. , 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn, T. , Pusztai, L. , Holtrich, U. , Iwamoto, T. , Shiang, C.Y. , Schmidt, M. , Müller, V. , Solbach, C. , Gaetje, R. , Hanker, L. , Ahr, A. , Liedtke, C. , Ruckhäberle, E. , Kaufmann, M. , Rody, A. , 2011. Homogeneous datasets of triple negative breast cancers enable the identification of novel prognostic and predictive signatures. PLoS. One. 6, e28403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. , Wiemer, D.F. , 2004. EDC-mediated condensations of 1-chloro-5-hydrazino-9,10-anthracenedione, 1-hydrazino-9,10-anthracenedione, and the corresponding anthrapyrazoles. Tetrahedron Lett.. 45, 4977–4980. [Google Scholar]
- Leng, M. , Chan, D.W. , Luo, H. , Zhu, C. , Qin, J. , Wang, Y. , 2006. MPS1-dependent mitotic BLM phosphorylation is important for chromosome stability. Proc. Natl. Acad. Sci. U S A. 103, 11485–11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Birkbak, N.J. , Gyorffy, B. , Szallasi, Z. , Eklund, A.C. , 2011. Jetset: selecting the optimal microarray probe set to represent a gene. BMC. Bioinfo.. 12, 474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Cheng, X. , Zhang, Y. , Li, S. , Cui, H. , Zhang, L. , Shi, R. , Zhao, Z. , He, C. , Wang, C. , Zhao, H. , Zhang, C. , Fisk, H.A. , Guadagno, T.M. , Cui, Y. , 2013. Phosphorylation of Mps1 by BRAFV600E prevents Mps1 degradation and contributes to chromosome instability in melanoma. Oncogene. 32, 713–723. [DOI] [PubMed] [Google Scholar]
- Maire, V. , Baldeyron, C. , Richardson, M. , Tesson, B. , Vincent-Salomon, A. , Gravier, E. , Marty-Prouvost, B. , De Koning, L. , Rigaill, G. , Dumont, A. , Gentien, D. , Barillot, E. , Roman-Roman, S. , Depil, S. , Cruzalegui, F. , Pierré, A. , Tucker, G.C. , Dubois, T. , 2013. TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS. One. 8, e63712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, L.D. , Smeds, J. , George, J. , Vega, V.B. , Vergara, L. , Ploner, A. , Pawitan, Y. , Hall, P. , Klaar, S. , Liu, E.T. , Bergh, J. , 2005. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U S A. 102, 13550–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher, D. , Liberati, A. , Tetzlaff, J. , Altman, D.G. , 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. Open. Med.. 3, e123–130. [PMC free article] [PubMed] [Google Scholar]
- Murray, S. , Briasoulis, E. , Linardou, H. , Bafaloukos, D. , Papadimitriou, C. , 2012. Taxane resistance in breast cancer: mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev.. 38, 890–903. [DOI] [PubMed] [Google Scholar]
- Neve, R.M. , Chin, K. , Fridlyand, J. , Yeh, J. , Baehner, F.L. , Fevr, T. , Clark, L. , Bayani, N. , Coppe, J.P. , Tong, F. , Speed, T. , Spellman, P.T. , DeVries, S. , Lapuk, A. , Wang, N.J. , Kuo, W.L. , Stilwell, J.L. , Pinkel, D. , Albertson, D.G. , Waldman, F.M. , McCormick, F. , Dickson, R.B. , Johnson, M.D. , Lippman, M. , Ethier, S. , Gazdar, A. , Gray, J.W. , 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell.. 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier, M. , Langerød, A. , Carrieri, P. , Bergh, J. , Klaar, S. , Eyfjord, J. , Theillet, C. , Rodriguez, C. , Lidereau, R. , Bièche, I. , Varley, J. , Bignon, Y. , Uhrhammer, N. , Winqvist, R. , Jukkola-Vuorinen, A. , Niederacher, D. , Kato, S. , Ishioka, C. , Hainaut, P. , Børresen-Dale, A.L. , 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res.. 12, 1157–1167. [DOI] [PubMed] [Google Scholar]
- Reme, T. , Hose, D. , Theillet, C. , Klein, B. , 2013. Modeling risk stratification in human cancer. Bioinformatics. 29, 1149–1157. [DOI] [PubMed] [Google Scholar]
- Santarpia, L. , Iwamoto, T. , Di Leo, A. , Hayashi, N. , Bottai, G. , Stampfer, M. , Andre, F. , Turner, N.C. , Symmans, W.F. , Hortobagyi, G.N. , Pusztai, L. , Bianchini, G. , 2013. DNA repair gene patterns as prognostic and predictive factors in molecular breast cancer subtypes.. Oncologist. 18, 1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia, L. , Qi, Y. , Stemke-Hale, K. , Wang, B. , Young, E.J. , Booser, D.J. , Holmes, F.A. , O'Shaughnessy, J. , Hellerstedt, B. , Pippen, J. , Vidaurre, T. , Gomez, H. , Valero, V. , Hortobagyi, G.N. , Symmans, W.F. , Bottai, G. , Di Leo, A. , Gonzalez-Angulo, A.M. , Pusztai, L. , 2012. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res. Treat.. 13, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou, C. , Pusztai, L. , 2009. Gene-expression signatures in breast cancer. N. Engl. J. Med.. 360, 790–800. [DOI] [PubMed] [Google Scholar]
- Sotiriou, C. , Wirapati, P. , Loi, S. , Harris, A. , Fox, S. , Smeds, J. , Nordgren, H. , Farmer, P. , Praz, V. , Haibe-Kains, B. , Desmedt, C. , Larsimont, D. , Cardoso, F. , Peterse, H. , Nuyten, D. , Buyse, M. , Van de Vijver, M.J. , Bergh, J. , Piccart, M. , Delorenzi, M. , 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst.. 98, 262–272. [DOI] [PubMed] [Google Scholar]
- Tannous, B.A. , Kerami, M. , Van der Stoop, P.M. , Kwiatkowski, N. , Wang, J. , Zhou, W. , Kessler, A.F. , Lewandrowski, G. , Hiddingh, L. , Sol, N. , Lagerweij, T. , Wedekind, L. , Niers, J.M. , Barazas, M. , Nilsson, R.J. , Geerts, D. , De Witt Hamer, P.C. , Hagemann, C. , Vandertop, W.P. , Van Tellingen, O. , Noske, D.P. , Gray, N.S. , Würdinger, T. , 2013. Effects of the selective MPS1 inhibitor MPS1-IN-3 on glioblastoma sensitivity to antimitotic drugs. J. Natl. Cancer Inst.. 105, 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troester, M.A. , Herschkowitz, J.I. , Oh, D.S. , He, X. , Hoadley, K.A. , Barbier, C.S. , Perou, C.M. , 2006. Gene expression patterns associated with p53 status in breast cancer. BMC. Cancer. 6, 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, N. , Moretti, E. , Siclari, O. , Migliaccio, I. , Santarpia, L. , D'Incalci, M. , Piccolo, S. , Veronesi, A. , Zambelli, A. , Del Sal, G. , Di Leo, A. , 2013. Targeting triple negative breast cancer: is p53 the answer?. Cancer Treat. Rev.. 39, 541–550. [DOI] [PubMed] [Google Scholar]
- Walerych, D. , Napoli, M. , Collavin, L. , Del Sal, G. , 2012. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 33, 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, B.A. , Cleveland, D.W. , 2005. Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell.. 8, 7–12. [DOI] [PubMed] [Google Scholar]
- Wei, J.H. , Chou, Y.F. , Ou, Y.H. , Yeh, Y.H. , Tyan, S.W. , Sun, T.P. , Shen, C.Y. , Shieh, S.Y. , 2005. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J. Biol. Chem.. 280, 7748–7757. [DOI] [PubMed] [Google Scholar]
- Weiss, E. , Winey, M. , 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell. Biol.. 132, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi, R. , Woods, R. , Ravdin, P.M. , Hayes, M.M. , Gelmon, K.A. , 2010. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol.. 11, 174–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary Figure 1 Genes with significantly altered expression in TP53 mutant cancers mapped to the KEGG cell cycle pathway. The genes with altered expression in ER‐positive breast cancers are marked in red; the genes with altered expression in ER‐negative cancers only are in blue. The gene MPS1 (TTK) is marked by a wide black box. TP53 status affects several cyclin dependent kinase gene functions.
Supplementary Figure 2 Correlation between MPS1 mRNA levels and survival in untreated ER‐positive and ER‐negative breast cancer patients.
Supplementary Figure 3 Survival analysis in neoadjuvant treated ER‐positive breast cancer patients stratified for MPS1 expression. The 371 ER‐positive patients in the neoadjuvant dataset were divided into a training cohort (n = 211) and a testing cohort (n = 160) to perform survival analysis.


