Abstract
High‐risk human papillomavirus (HPV) infection is the principal risk factor for the development of cervical cancer. The HPV E6 oncoprotein has the ability to target and interfere with several PSD‐95/DLG/ZO‐1 (PDZ) domain‐containing proteins that are involved in the control of cell polarity. This function can be significant for E6 oncogenic activity because a deficiency in cell polarisation is a marker of tumour progression. The establishment and control of polarity in epithelial cells depend on the correct asymmetrical distribution of proteins and lipids at the cell borders and on specialised cell junctions. In this report, we have investigated the effects of HPV E6 protein on the polarity machinery, with a focus on the PDZ partitioning defective 3 (Par3) protein, which is a key component of tight junctions (TJ) and the polarity network. We demonstrate that E6 is able to bind and induce the mislocalisation of Par3 protein in a PDZ‐dependent manner without significant reduction in Par3 protein levels. In addition, the high‐risk HPV‐18 E6 protein promotes a delay in TJ formation when analysed by calcium switch assays. Taken together, the data presented in this study contribute to our understanding of the molecular mechanism by which HPVs induce the loss of cell polarity, with potential implications for the development and progression of HPV‐associated tumours.
Keywords: HPV, E6 protein, PDZ, Cell polarity, Par3
Highlights
HPV‐18 E6 oncoprotein alters the Par‐3 cell localization in a PDZ‐binding dependent manner.
HPV‐18 E6 oncoprotein targets Par3 protein in vivo without inducing proteasome‐mediated degradation.
HPV‐18 E6 protein induces a redistribution of Par3 subcellular pools.
High risk HPV‐18 E6 protein interferes with tight junction restoration.
Abbreviations
- DLG1
human Disc large
- E6AP
E6 associated protein
- GFP
green fluorescent protein
- HA
Influenza Virus Hemagglutinin epitope
- HPV
Human Papillomavirus
- MAGUKs
membrane-associated guanylate kinase homologues
- Par
Partitioning defective
- PATJ
PALS1 associated tight junction protein
- PBM
PDZ-binding motif
- PDZ
PSD-95/DLG/ZO-1 domains
- RhPV
Rhesus papillomavirus
- Scrib
Scribble
- TJ
tight junction
- ZO-1
zonula occludens 1
- ZO-2
zonula occludens 2
1. Introduction
Persistent infection with high‐risk HPVs, such as HPV‐16 and HPV‐18, is the principal risk factor for the development of cervical cancer, which is the third most common cancer in women, with over 500,000 cases being reported globally each year (zur Hausen, 2002; Lowy and Schiller, 2012). In addition, HPV infection has been associated with the development of other malignancies, including: vulvar, vaginal, penile, anal, and oropharyngeal tumours (Lowy and Schiller, 2012).
Oncogenic HPV functions depend on the combined and complementary action of both HPV E6 and E7 oncoproteins, whose continuous expression is required for the maintenance of the transformed phenotype in carcinoma‐derived cell lines (Yoshinouchi et al., 2003; Jonson et al., 2008). High‐risk HPV E6 is a multifunctional protein that has the ability to bind and interfere with key cellular proteins, and these interactions are required for E6 to demonstrate full transforming activity. An interesting characteristic of HPV E6 oncoproteins is the presence, in the carboxy terminal region, of a conserved PDZ‐binding motif (PBM) that is able to recognise and bind PDZ interaction domains (Thomas et al., 2008; Pim et al., 2012). PDZ‐containing proteins are involved in diverse biological processes and many of them are scaffolding proteins that allow the assembly of multiprotein complexes at the cell membrane (Humbert et al., 2003, 2008). There is a growing list of PDZ proteins that are targeted by E6, including human Disc Large 1 (DLG1), Scribble (Scrib), PALS1‐associated tight junction protein (PATJ), and membrane‐associated guanylate kinase with inverted domain structure 1 (Gardiol et al., 1999; Nakagawa and Huibregtse, 2000; Storrs and Silverstein, 2007; Kranjec and Banks, 2011). Most of these proteins are involved in cell junction assembly, control of cell signalling and establishment of apicobasal polarity, and have been characterised as potential tumour suppressor proteins (Facciuto et al., 2012). Interestingly, the interaction of E6 with certain of these PDZ proteins can result in their degradation and/or mislocalisation, which has implications for polarity deregulation and HPV carcinogenesis (Thomas et al., 2008; Pim et al., 2012). Moreover, this activity is restricted to E6 derived from high‐risk HPV because the PBM is absent in low‐risk HPV‐derived E6 proteins (which are associated with benign lesions), highlighting the fact that disruption of cell polarity is potentially a key event in the progression toward malignancy (Banks et al., 2012).
Epithelial cell polarity is defined by the interplay of three protein complexes: the Scribble, the Crumbs, and the Par complexes, which determine the basolateral domain, apical domain, and apical‐lateral cell border, respectively (Assemat et al., 2008). The mammalian Par complex comprises the Par3, Par6, and atypical protein kinase C (aPKCs) proteins and is required for the establishment and maintenance of cell polarity (Goldstein and Macara, 2007). Among these components, Par3 is the central organiser for complex assembly; it contains three PDZ domains and is necessary for both TJ formation and spatial regulation of important signalling pathways (Feng et al., 2007; Goldstein and Macara, 2007; Pieczynski and Margolis, 2011). Par3 is a multi‐modular scaffold protein that interacts with diverse cell polarity regulators and these specific interactions ensure that Par3 is localized at specific membrane domains (Chen and Zhang, 2013).
Furthermore, recent findings emphasise the importance of Par3 in cancer development (Facciuto et al., 2012). Reduced Par3 expression, in association with tumour progression and poor prognosis, was observed in several human cancers, including primary oesophagus tumours, glioblastomas, breast carcinomas, and skin cancer (Zen et al., 2009). Recently, two studies have reported that Par3 protein is an important suppressor of tumourigenesis and metastasis, highlighting its significant role in human breast cancer progression (McCaffrey et al., 2012; Xue et al., 2012).
Additionally, it is important to note that Rhesus papillomavirus (RhPV), which causes anogenital malignancy in Rhesus Macaque monkeys, presents a PBM in the C‐ terminus of the E7 protein instead of E6, as it is for HPV. This motif confers PDZ‐binding activity and directs the interaction of RhPV E7 with Par3, suggesting that the targeting of cell polarity components is evolutionary conserved among PVs (Tomaic et al., 2008).
Considering i) that Par3 is critical for the establishment of TJs and apicobasal polarity and appears to be an oncosuppressor, ii) that HPV E6 is able to target and interfere with PDZ proteins involved in polarity machinery and, specifically, to members of the polarity protein complexes (e.g., Scrib, DLG1, and PATJ), and iii) that a finely tuned interplay among the different components of such complexes has been established, we initiated a series of studies to investigate the effect of HPV E6 on the Par polarity complex.
We show that the expression of high‐risk HPV E6 results in a dramatic change in Par3 cellular distribution in a PBM dependent manner. We observe that HPV‐18 E6 oncoprotein and Par3 interact in vivo and that this protein binding does not result in a significant reduction in Par3 protein level. Moreover, HPV E6 interferes with TJ formation in calcium switch assays. Overall, the data presented in this study contribute to the understanding of HPV E6 activities as they relate to interference of cell polarity during HPV‐mediated cell transformation.
2. Materials and methods
2.1. Cell culture and transfection
HEK293, HaCaT and HeLa cells were grown in Dulbecco's modified Eagle's medium DMEM (Gibco, NY, USA) supplemented with 10% fetal bovine serum (PAA Laboratories GmbH, Pasching, Austria). HEK293 and HaCaT cells were transfected using calcium phosphate precipitation (Matlashewski et al., 1987) or EcoTransfect reagent, respectively (OZ Biosciences, Marseille, France). To generate stable cell lines expressing HA‐E6 fusion proteins (Influenza Virus Hemagglutinin epitope [HA] tagged‐HPV E6 proteins), HaCaT cells were transfected with pcDNA3‐HA‐E6 and selected with G418 (Sigma Aldrich, Saint Louis, USA, 500 μg/ml). Single colonies were analysed for HA‐E6 expression by RT‐PCR and immunofluorescence (IF) analysis. Parallel transfections and selections were performed using an empty expression vector as a control. For 3D Matrigel culture, HaCaT cells were grown using Matrigel Basement Membrane Matrix (BD Biosciences, San Jose, USA). Briefly, cells were trypsinised and suspended in complete medium containing 2% Matrigel to a concentration of 1.2 × 105 cells/dish. Cell suspensions were seeded into 35 mm plastic tissue culture plates containing coverslips pre‐coated with Matrigel. The cells were then covered with complete medium and grown at 37 °C under 5% CO2 for 72 h (Debnath et al., 2003).
For the delivery of all siRNAs (Dharmacon, Thermo Fisher Scientific, Rockford, USA), the cells were seeded on six well dishes at a confluence of 1.2 × 105 and were transfected using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) with siRNA against either luciferase, HPV‐18 E6/E7 (5′‐CAUUUACCAGCCCGACGAG), HPV‐18 E6 (5′‐CUCUGUGUAUGGAGACACATT) or E6AP (relevant Dharmacon Smart Pools).
2.2. Plasmids
The respective HA‐E6 protein was cloned under the control of the CMV promoter into the pCDNA‐3 expression plasmid (Invitrogen, Grand Island, NY, USA) and its identity was confirmed by DNA sequencing. Specific point mutations were introduced into the PBM of the HPV‐18 E6 protein by PCR‐directed mutagenesis. Plasmids encoding myc tagged‐mouse Par3 was generously provided by Dr. Mathieu Coureuil (Université Paris Descartes, Faculté de Médecine) (Joberty et al., 2000; Coureuil et al., 2009).
2.3. Antibodies
Antibodies used were mouse monoclonal anti‐γ tubulin (T6557), mouse anti‐α tubulin (T6199) (Sigma Aldrich, Saint Louis, USA), mouse monoclonal anti‐HA (12CA5) (Roche, Mannheim, Germany), rabbit polyclonal anti‐Par3 (H70), rabbit polyclonal anti zonula occludens 1 (ZO‐1) (H‐300), mouse monoclonal anti‐p53 (DO‐1), mouse monoclonal anti‐α actinin (H‐2), mouse monoclonal anti‐vimentin (V‐9), mouse anti‐c Myc (9E10), mouse anti‐transferrin receptor (3B82A1) (Santa Cruz Biotechnology, California, USA), rabbit polyclonal anti‐Par3 (07‐330) (Millipore, Temecula CA, USA), mouse monoclonal anti‐p84 (5E10) (Abcam) and anti‐β‐galactosidase (β‐Gal) (Promega, Wisconsin, USA).
2.4. Western blotting
Western blot analyses were carried out as described previously (Gardiol et al., 1999). Briefly, cells were harvested in extraction buffer (250 mM NaCl, 0.1% NP40, 50 mM HEPES pH 7.0, 1 mM MgCl2) containing Halt Protease Inhibitor single use cocktail (Thermo Scientific Pierce, Rockford, USA). Equal amounts of proteins were separated by SDS‐PAGE and transferred to nitrocellulose. Specific protein levels were determined by immunoblot analysis using the appropriate primary antibodies, as indicated in the text. Blots were developed using the SuperSignal West Pico Chemiluminescent Substrate reagent (Thermo Scientific Pierce, Rockford, USA). When specified, cells were treated with the proteasome inhibitor N‐CBZ‐LEU‐LEU‐LEU‐AL (CBZ), 40 μM (Sigma Aldrich, Saint Louis, USA) 2 h prior to protein extraction (Gardiol et al., 1999). Subcellular fractionation was performed using the ProteoExtract Fractionation Kit (Calbiochem, Darmstadt, Germany) according to the manufacturer's instructions. Protein band intensities were quantitated using the Image J quantification program.
2.5. Immunofluorescence and microscopy
Cells were grown on glass coverslips and fixed using 2% formaldehyde in phosphate‐buffered saline for 20 min at room temperature and processed, as previously described (Massimi et al., 2003). Endogenous Par3, p53 and ZO‐1 proteins were visualised using anti‐Par3, anti‐p53 and anti‐ZO‐1, respectively. HA‐ E6 expression was visualised using anti‐HA antibody. Secondary antibodies used were Alexa 488‐conjugated goat anti‐rabbit IgG (green, Molecular Probes, Grand Island, NY, USA) and Cy3 conjugated anti‐mouse IgG (red, Chemicon International, Temecula, USA). Slides were analysed with the laser Confocal microscope Nikon C1 (CLSM, Japan). When appropriate, z‐axis reconstructions of HaCaT cells grown on Matrigel were generated.
2.6. Immunoprecipitation
For co‐immunoprecipitation assays, HEK293 cells co‐transfected with myc‐Par3 and HA‐E6 expression plasmids were lysed in RIPA buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 0.1% NP‐40, 1% NaDC, 1 mM EDTA, and 1 mM PMSF). The supernatants were subsequently incubated with anti‐HA antibody and with Protein A‐Sepharose beads CL‐4B (Sigma Aldrich, Saint Louis, USA) for the precipitation of HA‐E6 proteins or with anti‐myc resins (Sigma Aldrich, Saint Louis, USA) for the precipitation of Par3. Immunocomplexes were collected, washed extensively, eluted from the beads, analysed by SDS‐PAGE, and immunoblotted using anti‐Par3 or anti‐HA antibodies as indicated, for the detection of E6‐bound Par3 protein.
2.7. Calcium switch assays and TJ assembly
Highly confluent HaCaT or G418‐resistant HaCaT‐HAE6.18 cells grown on glass coverslips were incubated with serum‐free DMEM for 2 h. Afterwards, extracellular Ca2+ was chelated with 2 mM EGTA at 37 °C for 20 min. Cells were then washed and switched back to complete DMEM medium and either fixed immediately (Time 0) or at different time points for TJ restoration analysis by IF using anti‐ ZO‐1 as the primary antibody (Massimi et al., 2012). To quantify the average ZO‐1 expression on cell borders, five to seven fields of cells were randomly selected from at least three independent experiments.
2.8. Statistical analysis
The statistical significance of the data from the quantification of ZO‐1 expression at cell borders in calcium switch assays was obtained by the Mann Whitney test. A P value <0.05 was considered to be significant.
3. Results
3.1. High‐risk HPV‐18 E6 protein interferes with Par3 protein localisation in a PDZ‐binding dependent manner
Although a number of PDZ domain‐containing proteins have been identified as targets of HPV E6, scarce information exists on the potential interference of these viral proteins with the TJ Par polarity complex (Pim et al., 2012). Considering that Par3 seems to be the principal PDZ cellular target of E7 protein derived from RhPV (Tomaic et al., 2008), we initiated a series of studies to analyse the expression of Par3 protein, the key component of the Par complex, in the presence of HPV E6.
We focused on the high‐risk HPV‐18 E6 (E6.18) protein since HPV‐18 was demonstrated as the most aggressive mucosal high‐risk HPV type tested in cultures (Lace et al., 2009) and a number of reports indicate poorer prognosis and a more aggressive clinical behaviour for tumours containing HPV 18 DNA rather that HPV16 DNA (Walker et al., 1989; Zhang et al., 1995). First, we evaluated the effect of the silencing of E6.18 protein upon the expression of Par3 in HPV‐18 positive HeLa cells that constitutively express E6.18. The E6 ablation was verified by analysing by IF the expression of a well‐characterized target of the high‐risk E6 proteins, the p53 tumour suppressor (Scheffner et al., 1993). As can be seen in Figure 1A, the levels of p53 were strongly increased in the nucleus of most of the siRNA E6 and siRNA E6/E7 treated cells, demonstrating the efficient silencing of E6 expression, which is in agreement with previous reports (Scheffner et al., 1993; Kranjec and Banks, 2011). The recovery of p53 in the nucleus was stronger in the si18E6/E7 cells than in the siE6 cells, indicating that the ablation of E6 protein expression is more efficient using siRNA E6/E7 than siRNA E6, and this is in agreement with previous studies (Figure 1A). The expression and subcellular distribution of endogenous Par3 protein was also assessed by IF. As shown in Figure 1A, Par3 exhibits a diffuse pattern of staining in the cytoplasm and nucleus and is very weakly expressed at the cell borders of cells transfected with control siRNA luciferase (Figure 1A) or no transfected HeLa cells (Figure 1S). However, in those E6 and E6/E7‐silenced/p53‐positive cells Par3 exhibited some increased cytoplasmic staining but is significantly enriched at the cell junctions, with this being more striking in the E6/E7‐silenced cells. This Par3 localization at the cell junctions is in agreement with previously reported studies where Par3 is also found at cell junctions in other epithelial cell types (Joberty et al., 2000). This result suggests that Par3 is mislocalized in HPV positive cells in an E6‐dependent manner.
Figure 1.
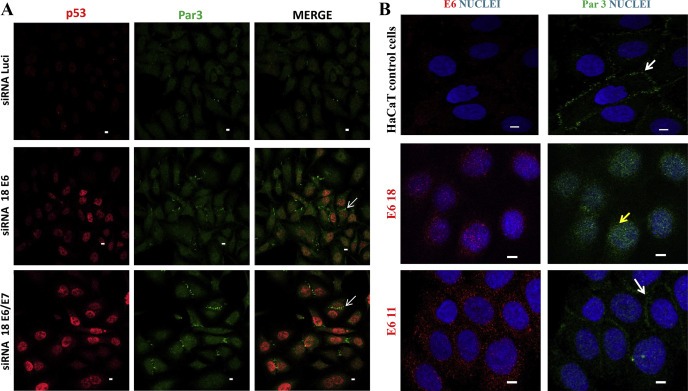
Expression of high‐risk HPV‐18 E6 oncoprotein is associated with the loss of normal Par3 localisation at cell junctions. A) Silencing of E6.18 restores Par3 expression at the cell–cell contacts in HeLa cells. HeLa cells were seeded on glass coverslips and were transfected with HPV‐18 E6/E7 siRNA, E6.18 siRNA or luciferase siRNA. Cells were grown for 48 h, fixed, incubated with anti‐Par3 and anti‐p53 antibodies, and counterstained with fluorescent secondary antibodies (Par3, green; p53, red). Confocal images were taken at wavelengths of 450 and 515 nm. The silencing of E6.18 was corroborated by the rescuing of p53 expression in the nucleus (white arrow for representative E6‐silenced cells). The images shown are representative of three independent experiments. B. IF staining for Par3 (green) and nuclear DAPI staining (blue) for HaCaT control cell line and HaCaT cell clones stably expressing HA‐E6 from low‐risk HPV‐11 or high‐risk HPV ‐18 (anti‐HA, red). White and yellow arrows indicate Par3 expression at cell borders or diffuse redistribution in high‐risk HPV E6‐expressing cells, respectively. Confocal images were taken at wavelengths of 450 and 515 nm. Scale bars: 5 μm.
In an attempt to confirm these findings, we generated HaCaT–derived clones stably expressing HA‐tagged E6.18 and for comparison the low risk HPV‐11 E6 (E6.11). We used immortalised HaCaT epithelial cells because they have been shown to be a good system for analysing TJ formation (Aono and Hirai, 2008).
Expression of the transgenes was confirmed by RT‐PCR (data not shown) and IF (Figure 1B). E6.18 protein was stably expressed and accumulated predominantly in the nucleus, whereas low‐risk HPV‐11 E6 could be found in both the nucleus and cytoplasm, as previously reported (Guccione et al., 2004; Mesplede et al., 2012). However, diffuse staining for E6.18 protein could be also observed in the cytoplasm of the stably transfected cells. Figure 1B shows that in HaCaT mock‐transfected cells, Par3 was predominantly localized at the cell borders in concordance with its normal localization and functions (Joberty et al., 2000). However, Par3 was diffusely redistributed to the cytoplasm with a marked reduction at cell junctions in E6.18‐expressing cells. The presence of E6.11 did not compromise the targeting of Par3 to cell–cell contacts when compared to control cells (Figure 1B). Even more striking, Par3 expression could be observed in the nucleus of 75% of the E6.18‐expressing cells (Figure 1B), in agreement with the data from experiments performed in HeLa cells and shown in Figure 1A.
Having shown that E6.18 oncoprotein can perturb Par3 distribution, we next performed experiments growing the cells on reconstituted basement membrane culture Matrigel. This system mimics the tissue architecture, resulting in a more relevant model in which to elucidate changes in the protein expression at cell junctions, generating well‐organized membrane domains with an apical‐basal polarity (Debnath et al., 2003; Chen and Zhang, 2013). We then performed Par3 IF analysis using cells grown on Matrigel for 72 h. As can be seen in Figure 2A, Par3 is expressed at cell junctions in HaCaT cells. The z‐axis reconstructions indicate that it is enriched at the apical domain of cell–cell contacts, most likely as a component of the TJ, as has been reported for other epithelial cell lines (Horikoshi et al., 2009). The same Par3 localisation was observed for E6.11‐expressing cells, whereas for E6.18‐derived clones, though some Par3 expression can be still observed at cell borders, Par3 has a clear redistribution to the cytoplasm. Interestingly, the Par3 localisation at the apical domain seemed to be lost in E6.18 clonal cells, where complete diffuse cytoplasmic and nuclear Par3 expression could be observed (Figure 2B). This observation can also be appreciated from the three dimensional reconstruction of the Z‐stack data, which is shown in Supplementary Figure 2 (Figure 2S).
Figure 2.
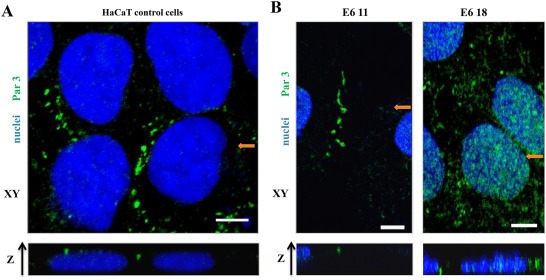
Expression of high‐risk HPV‐18 E6 promotes the loss of Par3 protein from the apical domain of cell junctions. IF staining for Par3 (green) and nuclear DAPI staining (blue). Image reconstruction along the z‐axis was performed from Z stacks of mock‐transfected HaCaT cells (A, control) or derivative clones stably expressing HA‐E6 proteins (B) that were cultured on Matrigel for 72 h. In each case, a representative section from the x‐y axis is shown. The lower panel presents an individual x‐z section along the position indicated in the upper image (orange arrow). Scale bars: 5 μm.
We next wanted to investigate if this effect is dependent on the ability of E6 oncoproteins to interact with PDZ domains. To do this, we generated an E6.18 mutant (E6.18 Mut) derivative where the E6 PBM sequence ETQV was mutated to EDQA because these amino acid changes were previously demonstrated to abolish PDZ interaction (Figure 3A) (Gardiol et al., 1999; Zhang et al., 2007). We transiently transfected HaCaT cells with either wild type or mutant E6.18, and endogenous Par3 distribution was analysed by IF. Interestingly, whilst the wild type E6.18 promotes Par3 mislocalisation from cell borders to the cytoplasm, the expression of E6.18 Mut did not induce any significant changes in the Par3 cellular distribution when compared to untransfected cells (Figure 3B). Taken together, the results presented above suggest that E6.18 oncoprotein has the ability to interfere with the proper distribution of Par3 in a PDZ‐binding dependent manner, with potential consequences for TJ organisation and polarity establishment.
Figure 3.
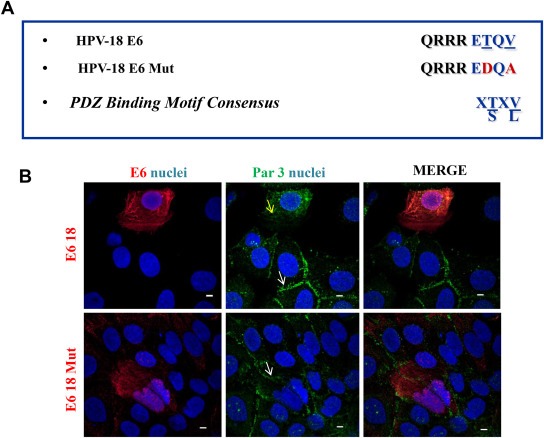
HPV‐18 E6 protein induces Par3 delocalisation in a PBM‐dependent manner. A. Sequence of the C terminal domain of the HPV E6 proteins used in this study. The consensus PBM of E6.18 protein is highlighted with blue letters. The amino acid substitutions introduced into E6.18 Mut are shown with respect to the wild type E6.18 sequence and are indicated with red letters. B. HaCaT cells transiently expressing HA‐E6.18 or HA‐E6.18 Mut were fixed and immunostained for Par3 (green) and HA‐E6 (red). Nuclear DAPI staining (blue) is also shown. White and yellow arrows indicate Par3 expression at cell borders in E6‐negative and E6.18 Mut, or diffuse redistribution in E6.18 transfected cells, respectively. The picture shown is representative of four independent experiments. Scale bar: 5 μm.
3.2. HPV‐18 E6 oncoprotein interacts with Par3 protein in vivo without inducing proteasome‐mediated degradation
We then performed experiments to determine if the effect of HPV E6 on Par3 distribution described above implies protein interactions between both proteins. Tomaic et al. had previously described a weak interaction of E6.18 with Par3 in vitro by GST‐pull down assays (Tomaic et al., 2008), so the interaction of E6.18 with Par3 in vivo was addressed by performing co‐immunoprecipitation assays. HEK293 cells were transiently co‐transfected with myc‐ tagged Par3‐expressing plasmid together with HA‐E6.11, HA‐E6.18 or HA‐E6.18 Mut expression constructs. As it can be appreciated, Par3 can be immunoprecipitated with anti‐HA antibody when co‐expressed with HA‐E6.18 but not with HA‐E6.11, which lacks the PBM (Figure 4 A, left panel). Mutating the PBM of E6.18 disrupted its association with Par3, indicating that this E6 region is involved in the interaction (Figure 4A). The same result was obtained when cell extracts were immunoprecipitated with anti‐myc antibody. In this case, HA‐E6.18 protein was identified in the myc tagged‐Par‐3 immunoprecipitates, but not with the PBM E6 mutant derivative (Figure 4B).
Figure 4.
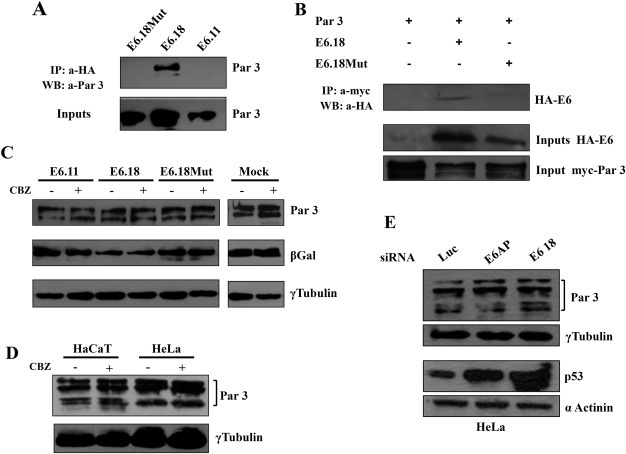
HPV‐18 E6 oncoprotein targets Par3 protein in vivo without inducing proteasome‐mediated degradation. A. High‐risk HPV‐18 E6 proteins in vivo interact with Par3. HEK293 cells were transfected with myc‐Par3 plasmid in the presence of HA‐E6.11, HA‐E6.18 and HA‐E6.18 Mut. Equal amounts of protein, extracted after 24 h, were either resolved directly (input) or immunoprecipitated with anti‐HA antibody followed by binding to Protein A‐Sepharose affinity beads. Following SDS‐PAGE, whole cell lysates (inputs, lower panel) and Protein A‐Sepharose bound complexes (upper panel) were probed with anti‐Par3 antibody. B. The same as panel A but cell extracts were analysed for myc‐Par3 and HA‐ E6 protein inputs or immunoprecipitated with anti‐myc resin. C. Expression of E6.18 oncoprotein does not affect Par3 protein levels. HEK293 cells were transfected with the Par3 expression plasmid in the absence (mock) or presence of E6.11, E6.18, or E6.18 Mut. After 24 h, cells were incubated for 2 h with or without CBZ proteasome inhibitor as indicated. Proteins were then extracted and equal amounts were separated by SDS‐PAGE. Protein levels were ascertained by western blotting analysis with anti‐Par3 (upper panel) or anti‐γ tubulin antibodies (as loading control, lower panel). The expression of β‐Gal was used as a control for transfection efficiency (middle panel). D. Par3 protein levels are not regulated by the proteasome pathway. HaCat and HeLa cells were incubated in the presence (+) or absence (−) of the proteasome inhibitor CBZ. The level of Par3 proteins were then ascertained by western blotting using anti‐Par3 antibody (upper panel). γ tubulin levels were determined as loading control, lower panel. E. The levels of Par3 protein do not change in HeLa cells upon E6.18 ablation. HeLa cells were transfected with siRNA directed against luciferase, E6AP or E6.18. After 48 h, cells were harvested and protein levels were assessed by western blotting using anti‐Par3 antibody, anti‐p53 (for control of E6.18 silencing) and anti‐γ tubulin or α‐actinin, for monitoring protein loading.
It is important to note that E6 was shown to be able to interact with and degrade several PDZ proteins belonging to the polarity complexes, including DLG1, Scrib and PATJ (Gardiol et al., 1999; Nakagawa and Huibregtse, 2000; Storrs and Silverstein, 2007). We observed some changes in Par3 abundance in the presence of E6 oncoproteins by IF but we wanted to assess this matter using a more quantitative in vivo degradation assay. HEK293 cells were co‐transfected with the Par3 expression plasmid plus empty vector, HA‐E6.11, or HA‐E6.18 or HA‐E6.18 Mut constructs. After 24 h, cells were treated with the proteasome inhibitor CBZ for 2 h prior to protein extraction. The cells were then harvested and Par3 protein expression was analysed by western blotting using anti‐Par3 antibodies. The results shown in Figure 4C (upper panel) indicate that the level of Par3 was not significantly affected by the presence of any of the E6 proteins. The addition of the proteasome inhibitor did not cause an increase in Par3 protein levels, suggesting that Par3 is not regulated to any substantial degree by the proteasome pathway. We corroborated the CBZ effect and the functional expression of the E6 oncoprotein by performing degradation assays for a well‐studied PDZ target of E6.18, DLG1 (Fig. S3) (Gardiol et al., 1999). Next we analysed the expression of endogenous Par3 protein using HaCat (HPV‐negative) or Hela (HPV‐18‐positive) epithelial cells, after incubation with CBZ. As it can bee seen in Figure 4D, and in agreement with the transfection experiments, the levels of Par3 protein did not change significantly in response to proteasome inhibitor treatment.
These results suggest that Par3 is not normally subject to proteasome degradation in epithelial cells in the presence or absence of HPV sequences.
In order to confirm the data presented above we used siRNA to block the expression of either E6 or the E6‐associated protein (E6AP) ubiquitin‐ligase in HeLa cells (Scheffner et al., 1993), considering that E6AP was shown to be involved in the degradation of several E6 cellular targets. Protein extracts from these cells were analysed by Western blotting. The E6 and E6AP ablation was verified by analysing the level of the p53 protein, a target of the E6‐E6AP complex (Scheffner et al., 1993; Kranjec and Banks, 2011). As can be seen in Figure 4E, the levels of p53 were increased in siRNA E6 and siRNA E6AP treated cells when compared with the control siRNA luciferase treated cells. Interestingly, Par3 levels did not change significantly after the silencing of E6 or E6‐AP indicating that E6 is not stimulating the proteasome‐mediated degradation of this particular E6.18 cellular partner.
Taken together, E6.18 can interact with Par3 through a PBM‐dependent mechanism in vivo without significantly affecting the levels of Par3 protein expression.
3.3. HPV‐18 E6 protein induces a redistribution of Par3 subcellular pools
Having found that E6.18 has the ability to alter Par3 cell localization without affecting significantly the total protein levels, we also compared the relative levels of expression of Par3 in different subcellular fractions in E6 positive and E6‐depleted HeLa cells. The cells were grown for 48 h and protein extracts were separated into cytosolic, membrane, nuclear and cytoskeletal fractions, and the pattern of endogenous Par3 expression was ascertained by western blotting. The results in Figure 5 show that Par3 levels are slightly reduced in the cytoplasmic fraction of E6 depleted cells in comparison to the siRNA luciferase treated cells. However, more interestingly, it can be observed an increase in Par3 levels in the membrane fraction together with a clear reduction in the nuclear fraction of the E6‐depleted, when comparing with control cells. Again, the major increase of p53 levels in the nucleus indicates the efficient ablation of E6 expression in the siRNA E6 treated HeLa cells. This experiment, together with the results presented above, indicates that E6.18 induces a subcellular redistribution of Par3 from the cell borders to the cytoplasm and nucleus with potential consequences over Par3 biological activities.
Figure 5.
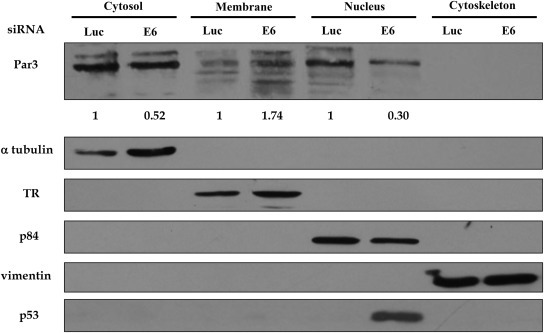
Subcellular fractionation shows that Par3 is redistributed in HPV‐positive cells in an E6‐dependent manner. HeLa cells were transfected with siRNA against either luciferase or E6.18. After 48 hs, cells were harvested and protein extracts were fractionated into cytosolic, membrane, nuclear and cytoskeletal fractions. The expression of Par3 and markers for the four subcellular fractions were assessed by western blotting. Expression of vimentin, p84, α‐tubulin, and transferrin receptor (TR) were used as control for the integrity of the cytoskeleton, nucleus, cytosolic and membrane fractions, respectively. The ablation of E6.18 was monitored by the expression of p53 in the nuclear fraction. The results shown are representative of three independent experiments. Numbers are folds of band intensity for Par3 in E6 depleted cells with respect to the luciferase siRNA control (considered as 1). The intensity of each band was normalized to the corresponding loading control in the different subcellular fraction.
3.4. High‐risk HPV‐18 E6 protein interferes with TJ restoration
The Par complex plays a critical role in both the establishment of cell asymmetry and TJ formation, and perturbation of its components results in alteration of TJ organisation (Aono and Hirai, 2008). Having demonstrated that HPV E6 oncoproteins can target Par3 protein, a key factor in TJs, we wanted to know how E6 could influence the formation of such intercellular junctions. Calcium switch assays were used to evaluate and compare the capacity of the cells to rescue TJ organisation and regulate apicobasal polarity. HaCaT control cells or stable E6.18‐expressing cells (Figure 1B) were cultured on coverslips overnight, serum depleted, and treated with EGTA to promote depolarisation and TJ disassembly (Figure 6A). After removal of the quelling agent and addition of complete medium, the TJs started to reorganise, and repolarisation was assayed over time by analysing ZO‐1 expression, which is a reliable marker of TJ assembly (Aono and Hirai, 2008). For control cells, TJ restoration started at 30 min and increased over time with strong ZO‐1 staining at cell borders after 60 min, indicating eventual polarisation (Figure 6B). However, the expression of high‐risk E6.18 significantly delayed repolarisation after the calcium switch when compared to control cells (Figure 6B). In contrast, ZO‐1 expression was comparable in the presence or absence of E6.18 6 h after the calcium switch, indicating that E6 may disturb polarity factors required for the initial steps in TJ formation.
Figure 6.
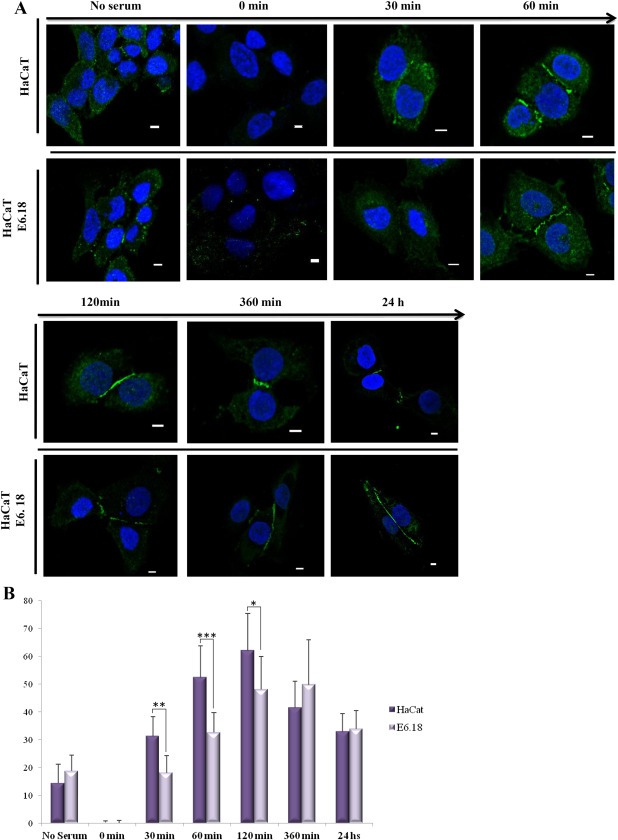
High‐risk HPV E6 compromises tight junction formation during calcium switch assays. A) HaCaT mock‐transfected cells or stable HA‐E6.18‐expressing cells were subjected to a calcium switch assay and stained for ZO‐1 (green) at various times after return to complete medium. Nuclear DAPI staining (blue) is also shown. Scale bar: 5 μm. B) ZO‐1 levels at cell junctions were normalised with respect to cytoplasmic expression using the MacBiophotonics ImageJ 1.43 m software (Wayne Rasband, National Institutes of Health, USA). Normalised quantification of ZO‐1 border localisation in control (dark bars) and HaCaT E6.18 (light bars) cells after re‐addition of calcium at the indicated time points is represented. Values are mean data ±standard deviation of randomly selected fields from imaging slides from at least three independent experiments. Asterisks denote significant difference among E6.18 and control cells (*p < 0.05, **p < 0.01, ***p < 0.001).
4. Discussion
In this study, we have provided further evidence of how high‐risk HPV E6 proteins can modulate the control of cell polarity. These studies are relevant for understanding HPV activities that lead to malignant progression because alterations in the maintenance of cell polarity are associated with a consequent loss of epithelial tissue architecture. High‐risk HPV E6 proteins were shown to interact with PDZ‐containing proteins through a PBM domain present at the C‐terminal region, which is conserved in all oncogenic mucosal HPV types. Several PDZ proteins belonging to the polarity control machinery have been shown to be targets of E6; however, the relevance of each E6‐PDZ interaction for viral replication and for the development of associated tumours is still unclear. Two members of the Par polarity complex, Par3 and Par6, belong to the PDZ protein family. This complex has traditionally been shown to be involved in different types of polarisation and signal transduction pathways; and several studies have shown that deregulation of Par complex activity is a key factor in the initiation of transformation (Aranda et al., 2008).
We demonstrated that the expression of high‐risk HPV‐18 derived E6 protein altered the proper localisation of Par3 at cell borders in a PDZ‐dependent manner (1, 2, 3).
Interestingly, depletion of E6.18 in HPV + HeLa cells restored the expression of Par3 at the cells contacts when compared with mock silenced cells (Figure 1A). More strikingly, some Par3 expression could be observed in the nucleus of E6‐expressing cells, both in HeLa and in stably transfected E6.18‐HaCat cells (Figures 1 and 2). Moreover, the subcellular fractionation experiment shown in Figure 5 demonstrated a clear redistribution of endogenous Par3 pools in the presence of E6.18 expression, with Par3 levels reduced at the membrane fraction together with an increase in the cytosolic and nuclear fractions. Par3 nuclear localisation in HeLa cells was previously reported by Fang et al. (2007). These researchers associated this nuclear distribution with a potential scaffolding role for Par3 in DNA‐dependent protein kinase activation during DNA repair following DNA damage (Fang et al., 2007). Sequence analysis of Par3 reveals the presence of potential nuclear localization signals, nevertheless whether Par3 can direct its own nuclear entry or uses an associated partner remains to be determined (Fang et al., 2007). Nonetheless, the contribution of E6 to Par3 redistribution, the biological significance, especially in the context of HPV‐positive cells, and the mechanism of Par3 translocation to the nucleus, remains to be elucidated and merit further investigation. Recently, it was reported that oncogenic HPV proteins trigger the mislocalization of the ZO‐2 TJ protein from the cell borders to the cytoplasm and nucleus in MDCK cells (Hernandez‐Monge et al., 2013); suggesting that delocalization of TJ proteins is an important mechanism for HPV‐mediated cell transformation and the development of cervical cancer.
More strikingly, the observations presented here are in agreement with our and other laboratories studies using cervical cancer tissues, where other PDZ polarity proteins, such as DLG1 and Scrib, instead of being localized at the epithelial cell–cell contacts are diffusely misdistributed into the cytoplasm (Cavatorta et al., 2004; Nakagawa et al., 2004).
We also demonstrated that the mislocalisation of Par3 in the presence of E6.18 was most likely due to the ability of these proteins to complex in vivo (Figure 4). However, at this stage we cannot rule out the possibility that E6.18 binds indirectly to Par3 through interaction via a common partner that allows the formation of multiprotein complexes, as long as both proteins have the ability to interact with several cell junction proteins.
Interestingly, no significant changes in Par3 levels were observed, which is in agreement with the notion that not all PDZ‐containing targets of E6 are equally susceptible to E6‐induced degradation in vivo (Figure 4). In fact, proteolytic degradation by E6 of its PDZ containing partners is highly specific, and interactions of E6 with some PDZ substrates can modulate their binding abilities or cell localization, thereby interfering with the PDZ protein function.
TJ formation is considered to be one of the critical steps in establishing cellular asymmetry in polarised cells, and defects in TJ formation can reflect defects in polarity establishment (Aono and Hirai, 2008). Par3 and other PDZ‐containing targets of E6 have been shown to be important components of TJs. Here, we performed classical calcium switch assays to evaluate the rescue of cell depolarisation in epithelial cells expressing E6.18 protein. The results shown in Figure 6 demonstrate that over a short period of time, E6‐expressing cells have an impaired ability to reform TJs with a delay in ZO‐1 accumulation at the cell borders, when compared with control cells. In contrast, longer after the calcium switch, ZO‐1 expression was comparable in the presence or absence of E6.18, indicating that E6 may disturb polarity factors required in the initial steps of TJ formation. It is tempting to speculate that the mislocalisation of Par3 in E6‐expressing cells contributes to this observation because it has recently been shown that the loss of Par3 impairs TJ maturation in early phases of junction formation (Iden et al., 2012).
During the last years a series of studies have reported the role for TJ proteins in cell proliferation, differentiation, transformation, and metastasis, connecting TJ factors to the carcinogenic process (Runkle and Mu, 2013). Though, due to the participation of the TJ in the control of cell proliferation and differentiation, the interference of E6 with TJ components, like Par3 protein, may be required for both virus replication and cell transformation. As localisation to the TJ in epithelia cells is a hallmark of Par3 activity, we could hypothesise that the expression of E6 is detrimental to the multiple functions of Par3, with negative consequences upon cell proliferation control and apico‐basal polarity.
5. Conclusions
In summary, we have shown that E6 is able to alter proper Par3 localisation. As Par3 protein expression has been shown to be important for tumour suppression, these findings significantly impact our understanding of malignant transformation in HPV‐infected cells. The change in Par3 localisation, primarily its absence from cell borders and loss of expression at the apical domain, can have a dramatic effect on the ability of Par3 to control the polarisation of epithelial cells. Par3 protein expression is frequently altered in a variety of human cancers (Facciuto et al., 2012) and has been associated with metastasis inhibition in breast cancer (Iden et al., 2012; McCaffrey et al., 2012). The targeting of Par3 by high‐risk HPV E6 proteins could have important consequences during the progression of HPV epithelial lesions; however, more in‐depth studies are needed to elucidate the HPV‐mediated mechanisms that interfere with the polarisation machinery and the signalling network during virus replication and tumour development.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Figure 1S Expression of Par3 protein in HeLa cells. IF staining for Par3 (green) and nuclear DAPI staining (blue) in HeLa (HPV‐18+) cell line. Scale bars: 5 μm. White arrow indicates Par3 nuclear expression.
Figure 2S Three‐dimensional reconstruction of z‐stack confocal optical slices, showing Par3 localization in E6‐expressing cells. IF staining for PAR3 (green) and nuclear DAPI staining (blue). Image reconstruction along the z‐axis was performed from Z stacks of derivative clones stably expressing HA‐E6.11 or HA‐E6.18 proteins that were cultured on Matrigel for 72 h. The three‐dimensional reconstruction of the z‐stack was performed using the NIS‐Elements Advanced Research 4.00.03 Program by Nikon.
Figure 3S E6.18 mediated degradation of DLG1. HEK293 cells were transfected with the HA‐tagged DLG1 expression plasmid (Gardiol et al., 1999) in the absence (mock) or presence of E6.18, or E6.18 Mut. After 24 h, cells were incubated for 2 h with or without CBZ proteasome inhibitor as indicated. Proteins were then extracted and equal amounts were separated by SDS‐PAGE. Protein levels were ascertained by western blotting analysis with anti‐HA (upper panel) or anti‐γ tubulin antibodies (as loading control, lower panel).
Acknowledgements
We gratefully acknowledge to Dolores Campos and Rodrigo Vena for excellent technical support and help with cell culture and confocal laser microscopy and image software, respectively. The authors wish to thank Dr. M. Coureuil for providing the Par3 expressing plasmid. This work was supported by a research grant from the Agencia de Promoción Científica y Tecnológica (Argentina, PICT 2008‐0421) to DG and the Associazione Italiana per la Ricerca sul Cancro to LB.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.01.002.
Facciuto Florencia, Bugnon Valdano Marina, Marziali Federico, Massimi Paola, Banks Lawrence, Cavatorta Ana Laura and Gardiol Daniela, (2014), Human papillomavirus (HPV)‐18 E6 oncoprotein interferes with the epithelial cell polarity Par3 protein, Molecular Oncology, 8, doi: 10.1016/j.molonc.2014.01.002.
References
- Aono, S. , Hirai, Y. , 2008. Phosphorylation of claudin-4 is required for tight junction formation in a human keratinocyte cell line. Exp. Cell. Res.. 314, 3326–3339. [DOI] [PubMed] [Google Scholar]
- Aranda, V. , Nolan, M.E. , Muthuswamy, S.K. , 2008. Par complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene. 27, 6878–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assemat, E. , Bazellieres, E. , Pallesi-Pocachard, E. , Le Bivic, A. , Massey-Harroche, D. , 2008. Polarity complex proteins. Biochim. Biophys. Acta. 1778, 614–630. [DOI] [PubMed] [Google Scholar]
- Banks, L. , Pim, D. , Thomas, M. , 2012. Human tumour viruses and the deregulation of cell polarity in cancer. Nat. Rev. Cancer. 12, 877–886. [DOI] [PubMed] [Google Scholar]
- Cavatorta, A.L. , Fumero, G. , Chouhy, D. , Aguirre, R. , Nocito, A.L. , Giri, A.A. , Banks, L. , Gardiol, D. , 2004. Differential expression of the human homologue of drosophila discs large oncosuppressor in histologic samples from human papillomavirus-associated lesions as a marker for progression to malignancy. Int. J. Cancer. 111, 373–380. [DOI] [PubMed] [Google Scholar]
- Coureuil, M. , Mikaty, G. , Miller, F. , Lecuyer, H. , Bernard, C. , Bourdoulous, S. , Dumenil, G. , Mege, R.M. , Weksler, B.B. , Romero, I.A. , Couraud, P.O. , Nassif, X. , 2009. Meningococcal type IV pili recruit the polarity complex to cross the brain endothelium. Science. 325, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Zhang, M. , 2013. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp. Cell Res.. 319, 1357–1364. [DOI] [PubMed] [Google Scholar]
- Debnath, J. , Muthuswamy, S.K. , Brugge, J.S. , 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30, 256–268. [DOI] [PubMed] [Google Scholar]
- Facciuto, F. , Cavatorta, A.L. , Valdano, M.B. , Marziali, F. , Gardiol, D. , 2012. Differential expression of PDZ domain-containing proteins in human diseases – challenging topics and novel issues. Febs. J.. 279, 3538–3548. [DOI] [PubMed] [Google Scholar]
- Fang, L. , Wang, Y. , Du, D. , Yang, G. , Tak Kwok, T. , Kai Kong, S. , Chen, B. , Chen, D.J. , Chen, Z. , 2007. Cell polarity protein Par3 complexes with DNA-PK via Ku70 and regulates DNA double-strand break repair. Cell Res.. 17, 100–116. [DOI] [PubMed] [Google Scholar]
- Feng, W. , Wu, H. , Chan, L.N. , Zhang, M. , 2007. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. Embo. J.. 26, 2786–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiol, D. , Kuhne, C. , Glaunsinger, B. , Lee, S.S. , Javier, R. , Banks, L. , 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 18, 5487–5496. [DOI] [PubMed] [Google Scholar]
- Goldstein, B. , Macara, I.G. , 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 13, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione, E. , Pim, D. , Banks, L. , 2004. HPV-18 E6*I modulates HPV-18 full-length E6 functions in a cell cycle dependent manner. Int. J. Cancer. 110, 928–933. [DOI] [PubMed] [Google Scholar]
- Hernandez-Monge, J. , Garay, E. , Raya-Sandino, A. , Vargas-Sierra, O. , Diaz-Chavez, J. , Popoca-Cuaya, M. , Lambert, P.F. , Gonzalez-Mariscal, L. , Gariglio, P. , 2013. Papillomavirus E6 oncoprotein up-regulates occludin and ZO-2 expression in ovariectomized mice epidermis. Exp. Cell Res.. 319, 2588–2603. [DOI] [PubMed] [Google Scholar]
- Horikoshi, Y. , Suzuki, A. , Yamanaka, T. , Sasaki, K. , Mizuno, K. , Sawada, H. , Yonemura, S. , Ohno, S. , 2009. Interaction between PAR-3 and the aPKC-PAR-6 complex is indispensable for apical domain development of epithelial cells. J. Cell Sci.. 122, 1595–1606. [DOI] [PubMed] [Google Scholar]
- Humbert, P. , Russell, S. , Richardson, H. , 2003. Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays. 25, 542–553. [DOI] [PubMed] [Google Scholar]
- Humbert, P.O. , Grzeschik, N.A. , Brumby, A.M. , Galea, R. , Elsum, I. , Richardson, H.E. , 2008. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 27, 6888–6907. [DOI] [PubMed] [Google Scholar]
- Iden, S. , van Riel, W.E. , Schafer, R. , Song, J.Y. , Hirose, T. , Ohno, S. , Collard, J.G. , 2012. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 22, 389–403. [DOI] [PubMed] [Google Scholar]
- Joberty, G. , Petersen, C. , Gao, L. , Macara, I.G. , 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol.. 2, 531–539. [DOI] [PubMed] [Google Scholar]
- Jonson, A.L. , Rogers, L.M. , Ramakrishnan, S. , Downs, L.S. , 2008. Gene silencing with siRNA targeting E6/E7 as a therapeutic intervention in a mouse model of cervical cancer. Gynecol. Oncol.. 111, 356–364. [DOI] [PubMed] [Google Scholar]
- Kranjec, C. , Banks, L. , 2011. A systematic analysis of human papillomavirus (HPV) E6 PDZ substrates identifies MAGI-1 as a major target of HPV type 16 (HPV-16) and HPV-18 whose loss accompanies disruption of tight junctions. J. Virol.. 85, 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lace, M.J. , Anson, J.R. , Klingelhutz, A.J. , Lee, J.H. , Bossler, A.D. , Haugen, T.H. , Turek, L.P. , 2009. Human papillomavirus (HPV) type 18 induces extended growth in primary human cervical, tonsillar, or foreskin keratinocytes more effectively than other high-risk mucosal HPVs. J. Virol.. 83, 11784–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy, D.R. , Schiller, J.T. , 2012. Reducing HPV-associated cancer globally. Cancer Prev. Res. (Phila). 5, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimi, P. , Gardiol, D. , Roberts, S. , Banks, L. , 2003. Redistribution of the discs large tumor suppressor protein during mitosis. Exp. Cell Res.. 290, 265–274. [DOI] [PubMed] [Google Scholar]
- Massimi, P. , Zori, P. , Roberts, S. , Banks, L. , 2012. Differential regulation of cell-cell contact, invasion and anoikis by hScrib and hDlg in keratinocytes. PLoS One. 7, e40279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlashewski, G. , Schneider, J. , Banks, L. , Jones, N. , Murray, A. , Crawford, L. , 1987. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. Embo. J.. 6, 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey, L.M. , Montalbano, J. , Mihai, C. , Macara, I.G. , 2012. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 22, 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesplede, T. , Gagnon, D. , Bergeron-Labrecque, F. , Azar, I. , Senechal, H. , Coutlee, F. , Archambault, J. , 2012. p53 degradation activity, expression, and subcellular localization of E6 proteins from 29 human papillomavirus genotypes. J. Virol.. 86, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, S. , Huibregtse, J.M. , 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell Biol.. 20, 8244–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, S. , Yano, T. , Nakagawa, K. , Takizawa, S. , Suzuki, Y. , Yasugi, T. , Huibregtse, J.M. , Taketani, Y. , 2004. Analysis of the expression and localisation of a LAP protein, human scribble, in the normal and neoplastic epithelium of uterine cervix. Br. J. Cancer. 90, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczynski, J. , Margolis, B. , 2011. Protein complexes that control renal epithelial polarity. Am. J. Physiol. Renal. Physiol.. 300, F589–F601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pim, D. , Bergant, M. , Boon, S.S. , Ganti, K. , Kranjec, C. , Massimi, P. , Subbaiah, V.K. , Thomas, M. , Tomaic, V. , Banks, L. , 2012. Human papillomaviruses and the specificity of PDZ domain targeting. Febs. J.. 279, 3530–3537. [DOI] [PubMed] [Google Scholar]
- Runkle, E.A. , Mu, D. , 2013. Tight junction proteins: from barrier to tumorigenesis. Cancer Lett.. 337, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner, M. , Huibregtse, J.M. , Vierstra, R.D. , Howley, P.M. , 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 75, 495–505. [DOI] [PubMed] [Google Scholar]
- Storrs, C.H. , Silverstein, S.J. , 2007. PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6. J. Virol.. 81, 4080–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M. , Narayan, N. , Pim, D. , Tomaic, V. , Massimi, P. , Nagasaka, K. , Kranjec, C. , Gammoh, N. , Banks, L. , 2008. Human papillomaviruses, cervical cancer and cell polarity. Oncogene. 27, 7018–7030. [DOI] [PubMed] [Google Scholar]
- Tomaic, V. , Gardiol, D. , Massimi, P. , Ozbun, M. , Myers, M. , Banks, L. , 2008. Human and primate tumour viruses use PDZ binding as an evolutionarily conserved mechanism of targeting cell polarity regulators. Oncogene. 28, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J. , Bloss, J.D. , Liao, S.Y. , Berman, M. , Bergen, S. , Wilczynski, S.P. , 1989. Human papillomavirus genotype as a prognostic indicator in carcinoma of the uterine cervix. Obstet. Gynecol.. 74, 781–785. [PubMed] [Google Scholar]
- Xue, B. , Krishnamurthy, K. , Allred, D.C. , Muthuswamy, S.K. , 2012. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat. Cell Biol.. 15, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinouchi, M. , Yamada, T. , Kizaki, M. , Fen, J. , Koseki, T. , Ikeda, Y. , Nishihara, T. , Yamato, K. , 2003. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by E6 siRNA. Mol. Ther.. 8, 762–768. [DOI] [PubMed] [Google Scholar]
- Zen, K. , Yasui, K. , Gen, Y. , Dohi, O. , Wakabayashi, N. , Mitsufuji, S. , Itoh, Y. , Zen, Y. , Nakanuma, Y. , Taniwaki, M. , Okanoue, T. , Yoshikawa, T. , 2009. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 28, 2910–2918. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Rose, B.R. , Thompson, C.H. , Jarrett, C. , Russell, P. , Houghton, R.S. , Cossart, Y.E. , 1995. Associations between oncogenic human papillomaviruses and local invasive patterns in cervical cancer. Gynecol. Oncol.. 57, 170–177. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Dasgupta, J. , Ma, R.Z. , Banks, L. , Thomas, M. , Chen, X.S. , 2007. Structures of a human papillomavirus (HPV) E6 polypeptide bound to MAGUK proteins: mechanisms of targeting tumor suppressors by a high-risk HPV oncoprotein. J. Virol.. 81, 3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen, H. , 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2, 342–350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Figure 1S Expression of Par3 protein in HeLa cells. IF staining for Par3 (green) and nuclear DAPI staining (blue) in HeLa (HPV‐18+) cell line. Scale bars: 5 μm. White arrow indicates Par3 nuclear expression.
Figure 2S Three‐dimensional reconstruction of z‐stack confocal optical slices, showing Par3 localization in E6‐expressing cells. IF staining for PAR3 (green) and nuclear DAPI staining (blue). Image reconstruction along the z‐axis was performed from Z stacks of derivative clones stably expressing HA‐E6.11 or HA‐E6.18 proteins that were cultured on Matrigel for 72 h. The three‐dimensional reconstruction of the z‐stack was performed using the NIS‐Elements Advanced Research 4.00.03 Program by Nikon.
Figure 3S E6.18 mediated degradation of DLG1. HEK293 cells were transfected with the HA‐tagged DLG1 expression plasmid (Gardiol et al., 1999) in the absence (mock) or presence of E6.18, or E6.18 Mut. After 24 h, cells were incubated for 2 h with or without CBZ proteasome inhibitor as indicated. Proteins were then extracted and equal amounts were separated by SDS‐PAGE. Protein levels were ascertained by western blotting analysis with anti‐HA (upper panel) or anti‐γ tubulin antibodies (as loading control, lower panel).


