Abstract
Nonviral gene therapy represents a realistic option for clinical application in cancer treatment. This preclinical study demonstrates the advantage of using the small‐size MIDGE® DNA vector for improved transgene expression and therapeutic application. This is caused by significant increase in transcription efficiency, but not by increased intracellular vector copy numbers or gene transfer efficiency. We used the MIDGE‐hTNF‐alpha vector for high‐level expression of hTNF‐alpha in vitro and in vivo for a combined gene therapy and vindesine treatment in human melanoma models. The MIDGE vector mediated high‐level hTNF‐alpha expression leads to sensitization of melanoma cells towards vindesine. The increased efficacy of this combination is mediated by remarkable acceleration and increase of initiator caspase 8 and 9 and effector caspase 3 and 7 activation. In the therapeutic approach, the nonviral intratumoral in vivo jet‐injection gene transfer of MIDGE‐hTNF‐alpha in combination with vindesine causes melanoma growth inhibition in association with increased apoptosis in A375 cell line or patient derived human melanoma xenotransplant (PDX) models. This study represents a proof‐of‐concept for an anticipated phase I clinical gene therapy trial, in which the MIDGE‐hTNF‐alpha vector will be used for efficient combined chemo‐ and nonviral gene therapy of malignant melanoma.
Keywords: Cancer, Gene therapy, Melanoma, Nonviral gene transfer, TNF‐alpha
Highlights
The minimalistic MIDGE‐vector improves transcription compared to plasmid vectors.
This vector achieved high‐level hTNF‐alpha expression for melanoma sensitization.
This sensitization is mediated by accelerated and increased caspase activation.
Nonviral in vivo MIDGE‐vector gene transfer leads to efficient TNF‐expression.
Combined gene‐ and chemotherapy improves melanoma growth inhibition in vivo.
1. Introduction
The worldwide incidence of malignant melanoma is increasing, with the number of cases doubling in the past 20 years (Jemal et al., 2011). Early diagnosis of melanoma is associated with a high cure rate. However, once distant metastases/stage IV disease is documented, the median survival rate is 6–8 months and the 5‐year survival rate is less than 5%. Although surgery can provide benefit in some patients with stage IV disease, for most patients presence of multiple metastatic sites and/or comorbidities limits the applicability of this approach (Jaques et al., 1989). Up to 5–8% of melanoma patients develop in‐transit metastasis of their extremities, which are locally treated by combination of high dose cytostatic drug and hTNFα in isolated limb perfusion (ILP) procedures (Deroose et al., 2012).
Cytokines are long known as sensitizing agents for tumor treatment. Numerous studies demonstrated that combination of TNFα, IFN‐γ or IL‐2 with cytostatic drugs leads to improved therapeutic efficacies, particularly for treatment of melanoma (Kedar et al., 1992; Lasek et al., 1996; Mouawad et al., 2010; Regenass et al., 1987). However, systemic in vivo application of cytokines at concentrations, which generate such synergy, is often limited by severe side effects. To circumvent these problems, the ILP was developed to achieve higher local hTNFα concentration compared to systemic applications (Eggermont et al., 1996; Eggermont et al., 1998). This concept was successfully used in particular for treatment of in‐transit metastasis of melanoma patients, in which local high dose hTNFα is combined with melphalan chemotherapy (Deroose et al., 2011b; de Wilt et al., 2000). Numerous clinical studies suggest, that this high dose hTNFα is effective for chemosensitization of melanoma leading to improved local control or eradication of melanoma lesions (Deroose et al., 2011a). Based on this, the idea of local hTNFα gene transfer has emerged. This was of particular attractiveness, since gene therapy can generate hTNFα concentrations locally, which are sufficient for antitumoral effects. In this regard, the TNFerade gene therapy clinical trials, using adenoviral gene transfer for hTNFα expression in different tumor entities including melanoma demonstrated, that local expression of hTNFα has sensitizing effects for e.g. radiotherapy (Senzer et al., 2004). In addition it also seems to exert distal effects through interruption of metastatic pathways and influence on immune surveillance (Atkins, 2006). Apart from the use of adenoviral based hTNFα gene transfer, nonviral alternatives are of great interest. During the last decade significant improvements were made for nonviral vector systems, which led to the development of small‐size vectors, such as minicircle or MIDGE (Mayrhofer et al., 2009; Schakowski et al., 2007). These vectors are reduced to the essential expression cassette by omitting all or almost all unnecessary bacterial backbone, as well as CpG sequences. One such small‐size vector platform is MIDGE (minimalistic immunogenically defined gene expression), consisting of end‐sealed double stranded linear DNA, essentially reduced to the promoter and transgene unit, which is important for successful and safe gene therapy (Schakowski et al., 2007).
Here we use this MIDGE system for efficient expression of reporter genes and of hTNFα. The study demonstrates the superiority of MIDGE‐mediated transgene expression over plasmid driven gene expression and provides insight into the molecular mechanisms of this improved performance. Most importantly, we show the usefulness of the MIDGE vector for efficient in vitro and in vivo hTNFα expression leading to synergistic effects in combination with chemotherapeutic drugs in melanoma. We show, that hTNFα gene transfer rapidly triggers apoptosis in the melanoma if combined with vindesine chemotherapy. This study provides important data on the use of a small‐size vector for high‐level hTNFα expression in a combination approach, which holds promise for the conceivable clinical application in local gene therapy of malignant melanoma.
2. Materials and methods
2.1. Cell lines
The human melanoma cell lines A375 (ATCC CRL‐1619), MeWo (ATCC HTB‐65), were kept in DMEM + 10% FCS. The human melanoma cell lines SK‐MEL‐5 (ATCC HTB‐70) and SK‐MEL‐28 (ATCC HTB‐72) and the human colon carcinoma cell lines SW480 (ATCC CCL‐228), HCT116 (ATCC CCL‐247) were kept in RPMI + 10% FCS. All cell lines were cultured without antibiotics in a humidified incubator at 37 °C, 5% CO2. Identity of all lines was confirmed by STR DNA typing (DSMZ, Braunschweig, Germany).
2.2. Vectors
The plasmid‐based luciferase (Luc) pCMV‐Luc and green fluorescence protein (GFP) pCMV‐GFP encoding vectors were obtained from PlasmidFactory (Bielefeld, Germany). All other vectors (pMok‐plasmids and MIDGE vectors) were provided by MOLOGEN (Berlin, Germany). The generation of MIDGE synthesis was already described elsewhere (Schakowski et al., 2001). All vector preparations were free of endotoxin. The MIDGE‐vectors represent almost CpG‐free DNA and are unlikely to induce endogenous TNFα expression (data not shown) (Takai and Jones, 2002, 2003).
2.3. In vitro lipofection and electroporation
Transfection was done using Metafectene (Biontex Laboratories, Martinsried, Germany). For transfection 1 × 105 cells/ml were used. Metafectene (2.2 μl) and vector DNA (1 μg of pCMV‐Luc, all other vectors were used equimolar in relation to pCMV‐Luc, mixed with an empty vector to 1 μg total DNA) were solved in DMEM.
Electroporation was performed in 4 mm cuvettes using the square wave protocol of a Gene Pulser Xcell (Bio‐Rad Laboratories, Munich, Germany). 5 × 106 cells/ml were treated with one square wave pulse at 200 V/20 ms.
2.4. Luciferase assay
For measurement of luciferase activity the Steady‐Glo Luciferase Assay System (Promega, Madison, WI) was used according to manufacturer's recommendations. The assay was performed in 24 well plates using 5 × 104 cells transfected or electroporated as described. For measurement of luciferase activity the Tecan spectra Fluor plus (Tecan, Männedorf, Switzerland) was used.
2.5. DNA isolation
To isolate DNA from the nucleic cell fraction, cells were treated after gene transfer with DNase I and cell nuclei from 1 × 105 cells were isolated using the NE‐PER Nuclear and Cytoplasmatic Extraction Kit (Pierce/Fisher Scientific, Pittsburgh, PA) following manufactures recommendations. DNA from cell nuclei was isolated using the NucleoSpin Tissue XS Kit (Macherey & Nagel, Düren, Germany) without modifications. DNA from animal tissues and tumor samples was isolated using the NucleoSpin Tissue Kit (Macherey & Nagel).
2.6. RNA isolation
Total cellular RNA was isolated using the Trizol (Life Technologies, Carlsbad, CA) protocol following manufacturer's recommendations including DNaseI digestion. Isolated RNA was solved in nuclease‐free water and stored at −80 °C.
2.7. Reverse transcription and real‐time PCR (qPCR)
Reverse transcription (RT) was performed using 1 × PCR buffer II, 5 mM MgCl2, 1 mM dNTPs, 20 U RNase inhibitor, 50 U MuLV reverse transcriptase and 2.5 μM random hexamers from Applied Biosystems (Life Technologies) with 50 ng of total RNA for 5 min at 25 °C followed by 45 min at 42 °C.
Quantitative real‐time PCR was performed using the LightCycler480 (Roche Diagnostics) in 96 well format with a 10 μl reaction volume. Vector DNA and cDNA samples from RT‐reactions generated in in vitro experiments (luciferase gene transfer) were quantified using the GoTaq Master Mix (Promega, Madison, WI) using the primers luc fw 5′‐gggctcactgagactacatc‐3′ and luc rev 5′‐gtagccatccatccttgtc‐3′.
To quantify vector DNA and cDNA from RT‐reactions from in vivo experiments of hTNFα gene transfer the LightCycler FastStart DNA Master HybProbe Kit (Roche Applied Science, Mannheim, Germany) with the primer sequences hTNF‐α fw 5′‐ctctggcccaggcagtcaga‐3′, hTNF‐α rev 5′‐tcggcaaagtcgagatagtc‐3′ and probe sequences hTNFα FL 5′‐gcattggcccggcggttc‐3′ and hTNFα LC 5′‐ccactggagctgcccctcagct‐3′ was used. All primer and probes were synthesized by Tib Molbiol.
For quantification of hTNFα expression, the mRNA was normalized to G6PDH (Roche) levels. PCR conditions were 95 °C 2 min (GoTaq) 10 min (HybProbe), 95 °C 10s, 61 °C (Luc) 62 °C (TNF‐α) 20 s, 72 °C 10 s for 45 cycles. Prior to quantification of vector DNA, a restriction digest using Fast Digest EcoRI (Fermentas, St. Leon‐Rot, Deutschland) for pCMV‐Luc and Fast Digest KpnI/SacI (Fermentas) for pMok and MIDGE‐based vectors was performed to release the transgenes for detection.
2.8. hTNFα expression analysis by ELISA
Human TNFα of cell culture supernatants and tumor lysates was quantified using the human TNFα ELISA Kit (Invitrogen, Carlsbad, CA, USA) according to manufacturer's recommendations. Absorbance was measured at 492 nm using the Tecan spectra Fluor plus (Tecan). The in vivo hTNFα values were normalized to total protein content of tumor lysates.
2.9. DNA‐labeling and analysis of intracellular vector accumulation
The vector DNA was labeled with Label IT Nucleic Acid Labelling Kit, Cy3 (Mirus Bio, Madison, WI) according to manufacturer's instructions and used for gene transfer as described. After DNase I digestions of extracellular DNA cells were fixed and stained using DAPI (Life Technologies, Grand Island, NY) and Phalloidin‐AlexaFluor488 (Life Technologies). Images were taken with an Axio Observer Z1 microscope using AxioVision 4.7 (Carl Zeiss, Göttingen, Germany).
For FACS analysis of vector DNA distribution, cells were harvested at indicated time points after gene transfer and treated with DNase I. The LSRFortessa (Becton & Dickinson Biosciences, Franklin Lakes, NJ) was used for detection of Cy3‐labeled intracellular vector DNA. Data analysis was done with FlowJo 7.6 (Tree Star Inc., Ashland, OR).
2.10. Analysis of transfection efficiencies by FACS
To analyse the fraction of GFP positive cells, cells were collected after equimolar gene transfer at indicated time points. For quantitative analysis FACSCalibur (Becton & Dickinson) was used. The data were analysed by using CellQuest Pro 5.2 (BD) and FlowJo (Tree Star Inc. Ashland, OR, USA).
2.11. Cytotoxicity assay
For the MTT cytotoxicity assay, 1 × 104 non‐transfected and hTNFα gene transfected cells were seeded into 96‐well plates. After 24 h cells were treated with media with or without vindesine and incubated for 72 h. MTT (3‐(4,5‐dimethylthiazyol‐2yl)‐2,5‐diphenyltetrazolium bromide (Sigma, 5 mg/mL) was added and absorbance was measured in triplicates at 560 nm in a micro plate reader (Tecan SpectraFluor Plus, Thermo Fisher Scientific, Waltham, MA). Values are expressed as fold change of respective untreated controls.
2.12. Caspase 3, 7 and caspase 8, 9 assays
The activity of caspases 3, 7 (0–72 h after vindesine treatment), caspase 8 and 9 (0–24 h after vindesine treatment) was assessed using the respective Caspase‐Glo Assay kits (Promega, Madison, WI) following manufacturers recommendations. Luciferase readings were performed in a Tecan SpectraFluor Plus micro plate reader (Thermo Fisher Scientific). Values are expressed as fold change of respective untreated controls.
2.13. Lactate dehydrogenase (LDH) release assay
After gene transfer supernatants of transfected cells and control cells were collected at indicated times (3–72 h) and LDH‐release was determined using the Cytotoxicity detection kit (Roche Diagnostics) as recommended by the manufacturer. Absorbance readings at 492 nm were performed in a Tecan SpectraFluor Plus micro plate reader (Thermo Fisher Scientific). Values are expressed as fold change of a lysate from 1 × 104 untreated cells, indicating the maximum LDH release.
2.14. In vivo melanoma models and in vivo gene transfer
To establish tumors, 1 × 107 A375 cells or tumor pieces of 3 × 3 mm of a patient derived melanoma Mel9663_A were injected subcutaneously into the left flank of NMRI nu/nu mice. When tumors reached a volume of approximately 5 × 5 mm, intratumoral jet‐injection was performed as described earlier (Walther et al., 2002; Walther et al., 2008). Animals were sacrificed at indicated time points after gene transfer by cervical dislocation. Samples were snap frozen in liquid nitrogen.
In the therapeutic combination experiments using hTNFα gene transfer and vindesine the animals (n = 5 per group) were treated in consecutive weeks. Vindesine was applied by tail vein injection at a dose of 1 mg vindesine/kg body weight 24 h after gene transfer. Tumor volumes were calculated by measuring length and width using callipers, by the formula 0.5 × length × width2. The animal handling was performed according to the German Animal Protection law and with approval from the local responsible authorities.
2.15. Immunohistochemistry
For intratumoral hTNFα distribution analysis snap‐frozen tumors were cryosectioned (6 μm, microtome cryostat CM1900, Leica microsystems) and air dried (1 h room temperature). Following 1% H2O2 incubation, permeabilization (0.5% Triton X‐100, 2.5% BSA in PBS) and blocking (3.5% casein, 1.5% BSA in PBS) slides were incubated over night with primary goat anti‐hTNFα antibody (15 μg/μl, R&D, Wiesbaden, Germany) at 4 °C. For detection slides were incubated for 1 h with HRP‐labeled rabbit anti‐goat secondary antibody (Abcam, Cambridge, UK) followed by DAB HRP‐substrate (DAKO, Hamburg, Germany). Counterstaining was performed using hemalum. Images were taken using a light microscope (Axioplan 2, Zeiss, Göttingen, Germany).
2.16. TUNEL assay
For in situ apoptosis detection a TUNEL assay (TumorTACS, Trevigen, Gaithersburg, MD) was used following manufacturers recommendations. Frozen tumor samples were cryosectioned and air dried over night at room temperature. After rehydration using 100%, 95% and 70% ethanol samples were fixed with 3.7% formaldehyde. Following permeabilization with cytonin the TdT labeling reaction was performed. Images were taken using a light microscope (Axioplan 2, Zeiss).
2.17. Statistical analyses
The indicated statistical tests, e.g. 1‐way ANOVA, 2‐way ANOVA and calculation of IC50 values were performed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) and Systat 12 (Systat Software, Inc., Chicago, IL, USA). The values are presented as mean ± S.E.
3. Results
3.1. Analyses of MIDGE vector performance in vitro
To analyze the performance of the MIDGE vector in comparison to its conventional parental plasmid, equimolar gene transfer experiments were performed. The analysis at the level of transgene expression revealed the highest increase in reporter activity after MIDGE‐based gene transfer (Figure 1A, Supplementary Information Table S1). After equimolar lipofection this increase (MIDGE‐Luc vs. pCMV‐Luc) was in the range of up to 12‐fold (12 h after gene transfer), depending on the cell line assessed (Supplementary Information Table S1). After equimolar electroporation we found an increase of luciferase expression of more than 1500‐fold in A375 melanoma cells 48 h after gene transfer (Supplementary Information Table S1). Next, we asked why the transgene expression is increased by the MIDGE vector and analyzed this in more detail in A375 melanoma cells. Analysis of gene transfer efficiency (Figure 1B) after equimolar gene transfer of GFP‐encoding pCMV‐GFP and pMok‐GFP plasmid and MIDGE‐GFP vector revealed only a small, less than 3‐fold, but significant (p ≤ 0.01) increase in GFP‐positive cells after MIDGE‐GFP lipofection or electroporation.
Figure 1.
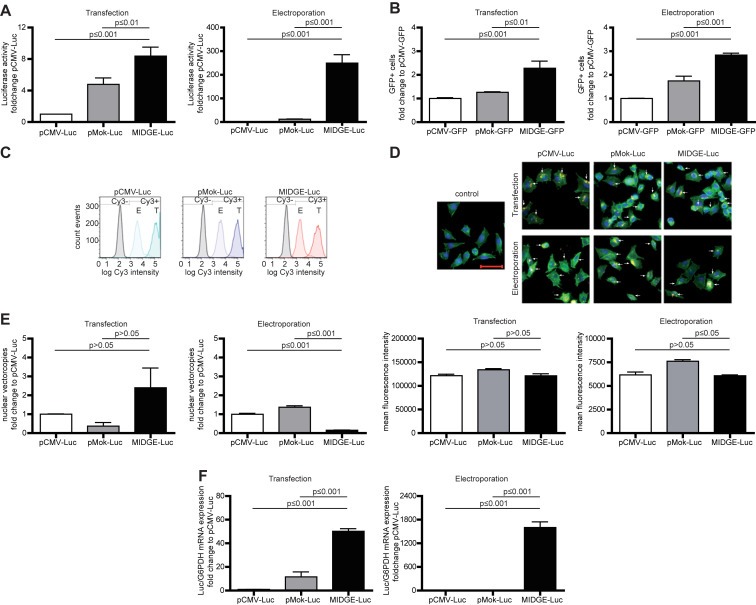
Luciferase reporter gene expression analyses in A375 human melanoma cells, in which the MIDGE vector leads to an increased reporter gene expression in vitro. (A) Overall reporter gene expression as analyzed by luciferase activity 24 h after equimolar gene transfer shows the highest luciferase activity when the MIDGE‐Luc is used. Expression data for all other cell lines are shown in Supplementary Information Table S1. (B) The small and optimized vectors increase the gene transfer efficiency 24 h after equimolar gene transfer at a low level only. (C) FACS analysis after equimolar gene transfer of Cy3‐labeled vector‐DNA shows no change in vector distribution and uptake. Transfection leads to a more than 10 times increased vector‐DNA uptake compared to electroporation. (D) Fluorescence microscopic analysis of cellular vector‐DNA uptake and distribution confirmed the FACS and qPCR analysis (C and D). Scale bar represents 50 μm. (E) Detailed analysis of intracellular vector‐DNA distribution shows only minor changes in nuclear uptake. Transfection leads to a higher nuclear amount in vector‐DNA compared to electroporation. (F) Real‐time RT‐PCR analysis of transgene expression reveals a strong increase of transgene transcription, particularly after transfer of the small MIDGE‐Luc vector. The data sets are in mean ± SEM. One way ANOVA followed by Bonferroni's Multiple Comparison Test was used for statistical analysis.
Next we analyzed the cellular vector uptake by using Cy3‐labeled vector DNA for gene transfer. The FACS analysis and fluorescence microscopy show a comparable load of the cells for all three vectors (Figure 1C and D). In addition to this, nuclear vector accumulation was only slightly altered after lipofection or electroporation transfer of the MIDGE‐Luc vector, as shown by qPCR analysis of isolated cell nuclei (Figure 1E). Since vector transfer efficiency, intracellular distribution and accumulation are insufficient to explain the observed increase in transgene expression, we analyzed luciferase expression at mRNA level (Figure 1F). For this we show up to 50‐ and 1500‐fold increase in the overall transgene transcription after lipofection as well as electroporation transfer of the MIDGE‐Luc vector compared to the plasmid vectors. This suggests that the minimal‐size MIDGE vector permits an increased transgene expression via enhanced transcription. We therefore used the MIDGE system for gene therapeutic applications in a sensitization strategy of human melanoma.
3.2. In vitro efficiency of MIDGE‐hTNFα gene transfer
Since the MIDGE system showed best performance for different human cell lines, we used this vector for the expression of the therapeutic transgene hTNFα. First we evaluated, if the MIDGE‐hTNFα vector is still superior compared to its parental plasmid pMok‐hTNFα. Again, the small‐size MIDGE vector shows superiority in terms of transgene expression after equimolar gene transfer. For A375 melanoma cells the maximum increase after transfection was more than 4‐fold (22.2 ng/ml vs. 93.5 ng TNFα/mL) and after electroporation up to 77‐fold (0.19 ng/ml vs. 14.9 ng TNFα/mL) compared to pMok‐hTNFα mediated expression levels (Figure 2A; Supplementary Information Table S2).
Figure 2.
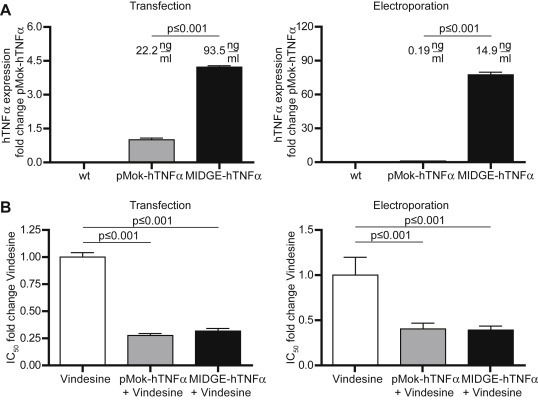
Analysis of the MIDGE‐hTNFα vector performance compared to its parental plasmid pMok‐hTNFα for hTNFα expression in vitro (A) The MIDGE‐hTNFα vector leads to an increase in hTNFα release after equimolar gene transfer. Although electroporation leads to a much lower vector transfer compared to transfection based gene transfer (Figure 1), the MIDGE vector produces comparable amounts of hTNFα underlining its improved performance. (B) The hTNFα gene transfer leads to a synergistic effect in combination with vindesine. The IC50 of vindesine is decreased to about 25%. The changes in IC50 for all other cell lines are given in Supplementary Information Table S3. The fold changes are in mean ± SEM. One way ANOVA followed by Bonferroni's Multiple Comparison Test was used for statistical analysis.
3.3. MIDGE‐hTNFα gene transfer for in vitro combination treatment of melanoma
One major focus of this study was to evaluate the potential of high‐level hTNFα expression by minimal‐size MIDGE vector in combination with the chemotherapeutic drug vindesine to increase treatment efficacy. As shown for A375 cells the combination of hTNFα gene transfer with vindesine leads to a decrease in the IC50 values (Figure 2B). In fact, this sensitizing effect was also shown for all other cell lines after transfection or electroporation, but was independent of the use of either MIDGE‐hTNFα or pMok‐hTNFα due to the very high level cytokine expression by both vectors (Supplementary Information Table S3).
To analyze the mechanism behind increased cell death we determined time‐dependent lactate dehydrogenase (LDH) release and caspase activation in A375 cells (Figure 3A–G). The LDH assay did not reveal an LDH release in cells treated with hTNFα gene transfer and vindesine at any time (Figure 3A). By contrast, we observed the acceleration of activation of initiator caspases 8 and 9, as well as the effector caspases 3 and 7, which were activated 12 h–24 h earlier, than by vindesine alone treatment (Figure 3B–G). More importantly, the activation of the initiator caspases 8 and 9 was stronger (up to 4‐fold for caspase 8, up to 5‐fold for caspase 9) than by the treatment with vindesine alone. We therefore conclude that in the combination treatment the cells rather enter apoptosis than necrosis leading to cell death.
Figure 3.
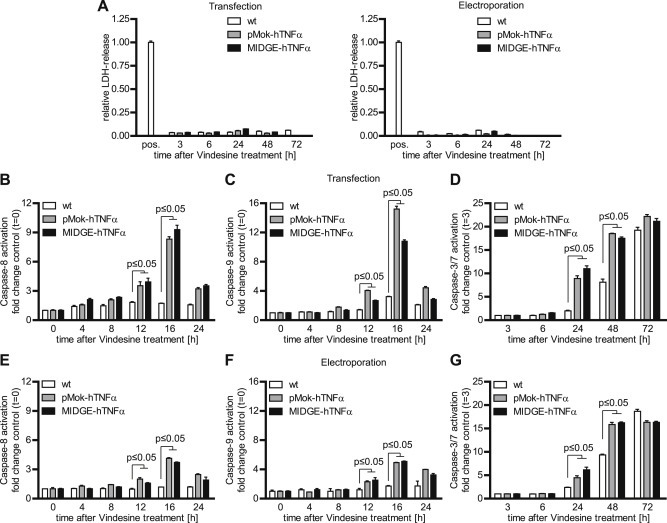
Mechanism of synergism in combined hTNFα and vindesine treatment in A375 melanoma cells (A) The combined treatment of hTNFα gene transfer and vindesine did not lead to LDH release, indicating the absence of necrotic processes. (B–G) The combined treatment with hTNFα gene transfer followed by vindesine leads to earlier and stronger activation of the initiator caspases 8 and 9 as well as effector caspases 3 and 7, indicating activation of apoptotic rather than necrotic signaling. The fold changes are in mean ± SEM. One way ANOVA followed by Bonferroni's Multiple Comparison Test was used for statistical analysis.
3.4. In vivo performance of the MIDGE‐hTNFα vector
We analyzed the overall gene transfer efficiency and safety properties of the MIDGE vector in the A375 human melanoma in vivo model xenografted in NMRI nu/nu mice. After equimolar jet‐injection of the parental plasmid and the MIDGE‐hTNFα vector we found an up to 30‐fold increase in transcription and up to 15‐fold elevated intratumoral hTNFα protein level after intratumoral jet‐injection of the MIDGE‐hTNFα vector (Figure 4A–B). As safety parameters we analyzed intratumoral time and dose dependent hTNFα expression and the clearance kinetics. We determined a time and vector dose dependent transgene expression (Figure 4C). This was confirmed by IHC staining of jet‐injected tumor cryosections. With increasing vector doses we found increased intratumoral hTNFα, which declined within 14 days (Figures 4C, 4E). Three days after gene transfer vector levels dropped below quantification limit of the qPCR. The intratumoral vector clearance was completed within 7–14 days (Figure 4D). A preclinical toxicology study at the laboratory of pharmacology and toxicology (LPT, Hamburg, Germany) proved, that the application even of the highest MIDGE‐hTNFα vector dose of 150 μg does not lead to any adverse effects (Supplementary Information Table S4).
Figure 4.
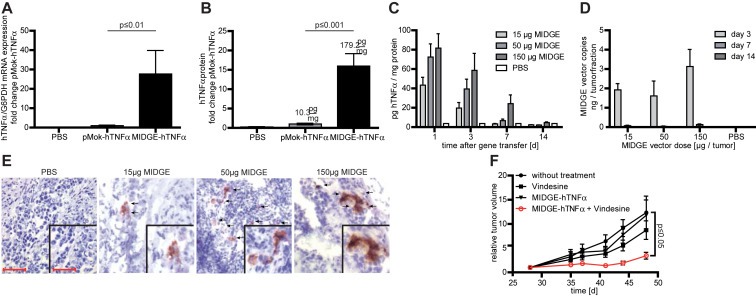
In vivo efficiency and safety analysis of nonviral gene transfer in the A375 melanoma xenograft model. (A) The intratumoral in vivo jet‐injection of the hTNFα encoding MIDGE‐hTNFα vector leads to increased transgene expression at the mRNA level compared to the parental plasmid. (B) The intratumoral in vivo jet‐injection of the hTNFα encoding MIDGE‐hTNFα vector leads to high‐level cytokine production. (C) Detailed ELISA analysis of hTNFα protein expression over time. Within 14 days hTNFαprotein expression declines even at the highest vector dose. (D) Intratumoral vector clearance was monitored over time with qPCR. Clearance is completed within 14 days. (E) Immunohistochemistry staining of A375 melanoma xenografts after treatment with PBS and three different doses of MIDGE‐hTNFα vector 24 h after gene transfer. Scale bar represents 50 μm and 25 μm (inset). (F) The in vivo combination of intratumoral jet‐injection MIDGE‐hTNFα gene transfer and vindesine leads to a significantly reduced tumor growth. The values are in mean ± SEM. One way ANOVA followed by Bonferroni's Multiple Comparison Test was used for statistical analysis. For the combination experiment the Dunnett's Multiple Comparison Test vs. untreated group was used.
Figure 5.
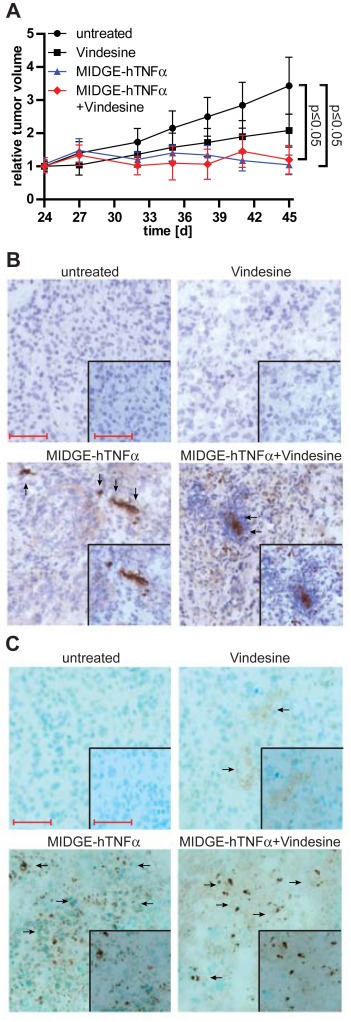
In vivo efficiency in a patient‐derived (PDX) melanoma xenograft model. (A) The application of the small‐size MIDGE‐hTNFα vector in PDX melanoma shows significantly reduced tumor growth in vivo. (B) Immunohistochemistry staining for hTNFα detection in the treated and non‐treated melanoma PDX. (C) Intratumoral detection of apoptosis in the treated and non‐treated melanoma PDX using TUNEL‐assay. The relative tumor volumes are in mean ± SEM. One way ANOVA followed by Dunnett's Multiple Comparison Test vs. untreated group was used for statistical analysis. Scale bar represents 100 μm and 50 μm (inset).
3.5. In vivo MIDGE‐hTNFα and vindesine combination therapy for melanoma
To analyze the combination of gene transfer and chemotherapy we used NMRI nu/nu mice xenografted s.c. with A375 human melanoma cells. Comparing untreated animals with the different treatment groups we found a significant decrease in tumor growth only after MIDGE‐hTNFα gene transfer in combination with vindesine (Figure 4F).
In a second in vivo model of Mel9663_A patient derived melanoma both the MIDGE‐hTNFα only and the combination (vindesine + MIDGE‐hTNFα gene transfer) treated group showed significant tumor growth inhibition (p ≤ 0.05) compared to the control or vindesine only treated animals (Figure 5A). The sensitivity of this particular melanoma model towards hTNFα only supports the potential of MIDGE hTNFα gene therapy. In this model strong TUNEL staining is observed in the MIDGE‐hTNFα only transfected tumors and even stronger staining is seen in the tumors treated with the combination (Figure 5). This TUNEL positivity is particularly associated with those tumor areas, which show TNFα expression and release in the respective immunohistochemistry (Figure 5B). This confirms the in vitro results, that the combined gene transfer and vindesine treatment enhances apoptosis in the melanoma, which results in efficient tumor growth inhibition.
4. Discussion
Nonviral gene therapy has developed to a clinically applicable option to treat cancer. For this access to safe and effective vectors is required. To address these issues for nonviral vectors the plasmid‐like minicircle and the linear double stranded MIDGE vector were developed (Boretti et al., 2000; Darquet et al., 1997; Choi et al., 2007). In both vectors resistance and bacterial backbone sequences are omitted, reducing the vector DNA to nearly the essential expression cassette. Compared to plasmids the small‐size MIDGE vector represents a linear double‐stranded DNA molecule with end‐sealing loops with respective transgenes driven by the cytomegalo virus (CMV) promoter (Supplementary Figure S1). This vector is significantly reduced in size and harbors almost exclusively the expression cassette.
We compared the effectiveness of the small‐size MIDGE vector to plasmid vectors regarding transgene expression in different human melanoma and colon carcinoma cell lines. For all cell lines we found an increase in the reporter gene activity after equimolar gene transfer. This is in line with comparable studies, which employed minimalistic vectors (Bigger et al., 2001; Chabot et al., 2013; Chen et al., 2003; Darquet et al., 1997; Kobelt et al., 2013; Schakowski et al., 2007, 2001). The fact, that small‐size backbone‐free vectors lead to improved overall gene expression efficiency is well accepted, but the mechanism is still poorly understood.
One mechanism, which might be responsible for elevated transgene expression is the increase in intracellular vector copies. Several studies have shown, that use of the small‐size vectors lead to increased vector uptake (Chabot et al., 2012; Schakowski et al., 2007). In our study however this does not explain the improved transgene expression of the cell lines analyzed. Especially for electroporation the fold change of luciferase activity is very much higher than the change in vector copy numbers, if any. Other related mechanisms, e.g. cellular association, intracellular trafficking and nuclear uptake have only limited impact (Chabot et al., 2012).
Furthermore, in analytical terms, the quantification of partially degraded small DNA vector molecules (e.g. minicircle or MIDGE) after gene transfer can be misinterpreted by qPCR, if intact small closed circular or linear (MIDGE) molecules are used for the generation of the standard curves. Intact molecules are less efficiently used as PCR template, possibly due to rapid reassociation during the annealing step. This leads to the detection of less DNA compared to relaxed DNA molecules. The discussed possible increases in vector copy number are in that range (Hou et al., 2010). In this study we used opened vector molecules for standard preparation and linearized the isolated vector molecules, which results in more reliable quantification of intracellular vector copies.
Decreased silencing could explain the optimized expression when using backbone free vectors (Chen et al., 2004, 2008). Chen et al. demonstrated that expression cassettes covalently bound to bacterial backbones are target for rapid silencing, mostly independent of the backbone or promoter used. Removal of the backbone or application of the backbone as extra DNA resulted in significantly increased transgene expression. This might also explain our observation that relatively unchanged numbers of transgene expressing cells with constant small‐size vector copies express more transgene at mRNA‐ and protein level.
It is well accepted that hTNFα is sensitizing melanoma towards chemotherapeutic drugs like melphalan, as applied in the isolated limb perfusion therapies (Deroose et al., 2012). We showed that the MIDGE vector mediated high‐level hTNFα expression is superior compared to its parental plasmid. The in vitro toxicity of hTNFα alone is very limited in various cancer cell lines (Belizario et al., 1993; Xu et al., 2006). Since vindesine is an approved second line therapy drug for the treatment of melanoma, we evaluated if the combination of hTNFα expression with this drug shows an improved therapeutic effect. The in vitro expression of hTNFα in combination with vindesine exerted increased cytotoxicity compared to both treatments alone. This is in line with other studies showing a synergistic effect of hTNFα in combination with chemotherapeutic drugs in different in vitro models (Lejeune et al., 1998; Mocellin et al., 2005; Stein and Walther, 1998; Walther et al., 1995). However, the cytokine expression levels achieved with either the parental plasmid or MIDGE are at such level, at which hTNFα is not a limiting factor for improved cytotoxicity of this combination in vitro.
To reveal the mechanism behind the increased cell death, we analyzed the downstream events in the death signaling. The mechanism behind the synergistic hTNFα effect is not fully understood, however molecular properties of the cells are of crucial importance which drive cells into cell death via apoptotic or necrotic pathways or alternatively trigger proliferation and survival through pathways such as NF‐kB‐signaling (Aggarwal, 2003).
In this study the expression of hTNFα in combination with vindesine led to an increase in cell death via apoptotic rather than necrotic processes. In this context, we observed the accelerated and increased activation of the initiator as well as the effector caspases in the melanoma cells, whereas LDH‐release was largely unaffected. This suggests that the high‐level hTNFα expression in combination with vindesine activates apoptotic signaling. One molecular mechanism for this phenomenon might be cell cycle inhibition by vindesine. During cell cycle progression proteins like c‐FLIP and others (e.g. NF‐κB dependent survival and anti‐apoptotic genes) are down regulated due to decreased protein expression. The c‐FLIP acts as an inhibitor of caspase‐8 activation leading to apoptosis resistance. Low levels of c‐FLIP allow hTNFα dependent caspase‐8 activation (extrinsic pathway). NF‐κB target genes act as survival and anti‐apoptotic factors, but need to be synthesized following NF‐κB activation. The missing NF‐κB signaling due to lack of protein expression was shown to permit apoptotic signaling (Guiet et al., 2002; Jin et al., 2008). We used various cell lines of two entities differing in their molecular properties. Independently all cell lines showed a decrease in the IC50 towards vindesine when hTNFα was present, indicating that the combination of hTNFα expression and vindesine seems to be a rather general feature.
In our in vivo experiments the hTNFα expression level was further elevated by the MIDGE vector (Schakowski et al., 2007). This amount of produced hTNFα was shown to be sufficient for chemosensitization in earlier studies (Walther et al., 2007).
To assess safety aspects, vector distribution and clearance were analyzed by qPCR. We found typical rapid clearance kinetics at the DNA‐, RNA‐ und protein level (Davis et al., 1996; Sheets et al., 2006; Walther et al., 2006, 2008; Wolff and Budker, 2005). In line with earlier clinical studies, we did not find vector molecules in blood samples and in the organs three days after gene transfer (Walther et al., 2008). Detailed preclinical analyses have shown, that there is a limited low level systemic dispersion via the blood stream to distant organs within 24 h–48 h after gene transfer (Galling et al., 2012). This might be attributed to the tissue damage caused by the high pressure used for jet‐injection and cancer related leaky vasculature (Baban and Seymour, 1998). The pressure is important to efficiently transfect DNA into the tissue overcoming the intratumoral hydrostatic pressure and preventing seepage (Walther et al., 2006). The applied jet‐injection leads to tolerable tissue damage in correlation with efficient gene transfer (Walther et al., 2008, 2002). However, the observed systemic vector biodistribution did not lead to gene expression at distant sites. Within three days systemic vector biodistribution was cleared. Even at high vector doses intratumoral vector clearance was completed within 7–14 days. Although more than 90% of the applied vector molecules are lost from the jet‐injected tumors within the first few days, efficient gene expression is maintained. To meet requirements for clinical application of this hTNFα encoding MIDGE vector, a preclinical toxicology study was performed, confirming the safe applicability of the MIDGE vector without vector related toxicities of adverse side effects. We furthermore evaluated in vitro and in vivo, if MIDGE vector DNA could induce endogenous human or murine TNFα expression. We did not observe such an effect, so that endogenous TNFα did not add to the effect of improved cytotoxicity (data not shown).
Systemic therapy with hTNFα is not possible due to its dose limiting toxicities (Hohenberger et al., 2003). Local applications (ILP, local gene transfer) have proven to be successful in patients (Barbour et al., 2009; Deroose et al., 2012, 2011b). In the experimental setup using human A375 melanoma cell xenografts we observed only very limited hTNFα based antitumoral activity and moderate vindesine effects. By contrast combined treatment showed pronounced antitumoral effect. Similarly, such improved combination effect was achieved in a number of clinical trials using local application of high‐dose recombinant hTNFα protein in combination with melphalan in isolated limb perfusion (ILP) particularly for melanoma and sarcoma patients (Deroose et al., 2012). Potentially, hTNFα might also contribute to improved intratumoral drug accumulation, which we did not analyze in this study (Baluk et al., 2009; Menon et al., 2006).
The patient‐derived xenograft (PDX) melanoma model allows a more clinically relevant evaluation of this combined treatment. Interestingly, hTNFα alone exerted a considerable effect on tumor growth, which was not further increased in combination with vindesine. This might be attributed to the composition of the PDX model, built not only of a homogenous cell mass of one type representing a rather heterogeneous patient tumor, but still reflects the therapeutic potential of this gene therapeutic approach.
Our preclinical data show, that MIDGE based local gene transfer of hTNFα in combination with vindesine is effective for local control of melanoma lesions. This is mainly achieved by activation of apoptosis signaling. These preclinical data provide the basis for clinical evaluation of this combination therapy.
Funding
This work was kindly supported by the H.W. & J. Hector foundation, grant M48.2 (to WW and PMS) and by EMS Medical Systems SA, Nyon, Switzerland. The authors declare no conflict of interests.
Supporting information
The following is the supplementary data related to this article:
Figure A1 Schematic representation of the different vectors used.
Table A1. Comparative in vitro luciferase gene transfer in different melanoma and colon carcinoma cell lines.
Table A2. Comparative in vitro hTNFα gene transfer in different melanoma and colon carcinoma cell lines.
Table A3. Vindesine IC50 in different melanoma and colon carcinoma cell lines after hTNFα gene transfer.
Table A4. Summary of acute toxicity signs after jet‐injection of MIDGE‐hTNFα vector solution, determined in a GLP toxicicty study.
Acknowledgments
We would like to thank the MDC Core Facilities (FACS, Microscopy) for giving support. We also thank our lab colleagues for technical support and critical discussions during the preparation of the manuscript.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.12.019.
Kobelt Dennis, Aumann Jutta, Schmidt Manuel, Wittig Burghardt, Fichtner Iduna, Behrens Diana, Lemm Margit, Freundt Greta, Schlag Peter M. and Walther Wolfgang, (2014), Preclinical study on combined chemo‐ and nonviral gene therapy for sensitization of melanoma using a human TNF‐alpha expressing MIDGE DNA vector, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.12.019.
The work was done at the Max‐Delbrück‐Center for Molecular Medicine, Berlin, Germany.
Contributor Information
Dennis Kobelt, Email: dennis.kobelt@mdc-berlin.de.
Wolfgang Walther, Email: wowalt@mdc-berlin.de.
References
- Aggarwal, B.B. , 2003. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol.. 3, 745–756. [DOI] [PubMed] [Google Scholar]
- Atkins, M.B. , 2006. Cytokine-based therapy and biochemotherapy for advanced melanoma. Clin. Cancer Res.. 12, 2353s–2358s. [DOI] [PubMed] [Google Scholar]
- Baban, D.F. , Seymour, L.W. , 1998. Control of tumour vascular permeability. Adv. Drug Deliv. Rev.. 34, 109–119. [DOI] [PubMed] [Google Scholar]
- Baluk, P. , Yao, L.C. , Feng, J. , Romano, T. , Jung, S.S. , Schreiter, J.L. , Yan, L. , Shealy, D.J. , McDonald, D.M. , 2009. TNF-alpha drives remodeling of blood vessels and lymphatics in sustained airway inflammation in mice. J. Clin. Invest.. 119, 2954–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour, A.P. , Thomas, J. , Suffolk, J. , Beller, E. , Smithers, B.M. , 2009. Isolated limb infusion for malignant melanoma: predictors of response and outcome. Ann. Surg. Oncol.. 16, 3463–3472. [DOI] [PubMed] [Google Scholar]
- Belizario, J.E. , Tilly, J.L. , Sherwood, S.W. , 1993. Caffeine potentiates the lethality of tumour necrosis factor in cancer cells. Br. J. Cancer. 67, 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger, B.W. , Tolmachov, O. , Collombet, J.M. , Fragkos, M. , Palaszewski, I. , Coutelle, C. , 2001. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem.. 276, 23018–23027. [DOI] [PubMed] [Google Scholar]
- Boretti, F.S. , Leutenegger, C.M. , Mislin, C. , Hofmann-Lehmann, R. , Konig, S. , Schroff, M. , Junghans, C. , Fehr, D. , Huettner, S.W. , Habel, A. , Flynn, J.N. , Aubert, A. , Pedersen, N.C. , Wittig, B. , Lutz, H. , 2000. Protection against FIV challenge infection by genetic vaccination using minimalistic DNA constructs for FIV env gene and feline IL-12 expression. AIDS. 14, 1749–1757. [DOI] [PubMed] [Google Scholar]
- Chabot, S. , Orio, J. , Schmeer, M. , Schleef, M. , Golzio, M. , Teissie, J. , 2013. Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery. Gene Ther.. 20, 62–68. [DOI] [PubMed] [Google Scholar]
- Chen, Z.Y. , He, C.Y. , Ehrhardt, A. , Kay, M.A. , 2003. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol. Ther.. 8, 495–500. [DOI] [PubMed] [Google Scholar]
- Chen, Z.Y. , He, C.Y. , Meuse, L. , Kay, M.A. , 2004. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther.. 11, 856–864. [DOI] [PubMed] [Google Scholar]
- Chen, Z.Y. , Riu, E. , He, C.Y. , Xu, H. , Kay, M.A. , 2008. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol. Ther.. 16, 548–556. [DOI] [PubMed] [Google Scholar]
- Choi, Y. , Jeon, Y.H. , Kang, J.H. , Chung, J.K. , Schmidt, M. , Kim, A.C. , 2007. MIDGE/hNIS vaccination generates antigen-associated CD8+IFN-gamma+ T cells and enhances protective antitumor immunity. Int J cancer. 120, 1942–1950. [DOI] [PubMed] [Google Scholar]
- Darquet, A.M. , Cameron, B. , Wils, P. , Scherman, D. , Crouzet, J. , 1997. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther.. 4, 1341–1349. [DOI] [PubMed] [Google Scholar]
- Davis, H.L. , McCluskie, M.J. , Gerin, J.L. , Purcell, R.H. , 1996. DNA vaccine for hepatitis B: evidence for immunogenicity in chimpanzees and comparison with other vaccines. Proc. Natl. Acad. Sci. U.S.A.. 93, 7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilt, J.H. , Soma, G. , ten Hagen, T.L. , Kanou, J. , Takagi, K. , Nooijen, P.T. , Seynhaevel, A.L. , Eggermont, A.M. , 2000. Synergistic antitumour effect of TNF-SAM2 with melphalan and doxorubicin in isolated limb perfusion in rats. Anticancer Res.. 20, 3491–3496. [PubMed] [Google Scholar]
- Deroose, J.P. , Eggermont, A.M. , van Geel, A.N. , de Wilt, J.H. , Burger, J.W. , Verhoef, C. , 2012. 20 years experience of TNF-based isolated limb perfusion for in-transit melanoma metastases: TNF dose matters. Ann. Surg. Oncol.. 19, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroose, J.P. , Eggermont, A.M. , van Geel, A.N. , Verhoef, C. , 2011. Isolated limb perfusion for melanoma in-transit metastases: developments in recent years and the role of tumor necrosis factor alpha. Curr. Opin. Oncol.. 23, 183–188. [DOI] [PubMed] [Google Scholar]
- Deroose, J.P. , Grunhagen, D.J. , van Geel, A.N. , de Wilt, J.H. , Eggermont, A.M. , Verhoef, C. , 2011. Long-term outcome of isolated limb perfusion with tumour necrosis factor-alpha for patients with melanoma in-transit metastases. Br. J. Surg.. 98, 1573–1580. [DOI] [PubMed] [Google Scholar]
- Eggermont, A.M. , 1998. TNF alpha in isolated perfusion systems: success in the limb, developments for the liver credits, debits and future perspectives. Anticancer Res.. 18, 3899–3905. [PubMed] [Google Scholar]
- Eggermont, A.M. , Schraffordt Koops, H. , Lienard, D. , Kroon, B.B. , van Geel, A.N. , Hoekstra, H.J. , Lejeune, F.J. , 1996. Isolated limb perfusion with high-dose tumor necrosis factor-alpha in combination with interferon-gamma and melphalan for nonresectable extremity soft tissue sarcomas: a multicenter trial. J. Clin. Oncol.. 14, 2653–2665. [DOI] [PubMed] [Google Scholar]
- Galling, N. , Kobelt, D. , Aumann, J. , Schmidt, M. , Wittig, B. , Schlag, P.M. , Walther, W. , 2012. Intratumoral dispersion, retention, systemic biodistribution, and clearance of a small-size tumor necrosis factor-alpha-expressing MIDGE vector after nonviral in vivo jet-injection gene transfer. Hum. Gene Ther. Part B. Methods. 23, 264–270. [DOI] [PubMed] [Google Scholar]
- Guiet, C. , Silvestri, E. , De Smaele, E. , Franzoso, G. , Vito, P. , 2002. c-FLIP efficiently rescues TRAF-2-/- cells from TNF-induced apoptosis. Cell Death Differ.. 9, 138–144. [DOI] [PubMed] [Google Scholar]
- Hohenberger, P. , Latz, E. , Kettelhack, C. , Rezaei, A.H. , Schumann, R. , Schlag, P.M. , 2003. Pentoxifyllin attenuates the systemic inflammatory response induced during isolated limb perfusion with recombinant human tumor necrosis factor-alpha and melphalan. Ann. Surg. Oncol.. 10, 562–568. [DOI] [PubMed] [Google Scholar]
- Hou, Y. , Zhang, H. , Miranda, L. , Lin, S. , 2010. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PloS One. 5, e9545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaques, D.P. , Coit, D.G. , Brennan, M.F. , 1989. Major amputation for advanced malignant melanoma. Surg. Gynecol. Obstet.. 169, 1–6. [PubMed] [Google Scholar]
- Jemal, A. , Bray, F. , Center, M.M. , Ferlay, J. , Ward, E. , Forman, D. , 2011. Global cancer statistics. CA Cancer J. Clinicians. 61, 69–90. [DOI] [PubMed] [Google Scholar]
- Jin, S. , Ray, R.M. , Johnson, L.R. , 2008. TNF-alpha/cycloheximide-induced apoptosis in intestinal epithelial cells requires Rac1-regulated reactive oxygen species. Am. J. Physiol. Gastrointest. Liver Physiol.. 294, G928–G937. [DOI] [PubMed] [Google Scholar]
- Kedar, E. , Rutkowski, Y. , Leshem, B. , 1992. Chemo-immunotherapy of murine solid tumors: enhanced therapeutic effects by interleukin-2 combined with interferon alpha and the role of specific T cells. Cancer Immunol. Immunother. CII. 35, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobelt, D. , Schleef, M. , Schmeer, M. , Aumann, J. , Schlag, P.M. , Walther, W. , 2013. Performance of high Quality minicircle DNA for in vitro and in vivo gene transfer. Mol. Biotechnol.. 53, 80–89. [DOI] [PubMed] [Google Scholar]
- Lasek, W. , Wankowicz, A. , Kuc, K. , Feleszko, W. , Giermasz, A. , Jakobisiak, M. , 1996. Augmentation of antitumor efficacy by the combination of actinomycin D with tumor necrosis factor-alpha and interferon-gamma on a melanoma model in mice. Oncology. 53, 31–37. [DOI] [PubMed] [Google Scholar]
- Lejeune, F.J. , Ruegg, C. , Lienard, D. , 1998. Clinical applications of TNF-alpha in cancer. Curr. Opin. Immunol.. 10, 573–580. [DOI] [PubMed] [Google Scholar]
- Mayrhofer, P. , Schleef, M. , Jechlinger, W. , 2009. Use of minicircle plasmids for gene therapy. Methods Mol. Biol.. 542, 87–104. [DOI] [PubMed] [Google Scholar]
- Menon, C. , Ghartey, A. , Canter, R. , Feldman, M. , Fraker, D.L. , 2006. Tumor necrosis factor-alpha damages tumor blood vessel integrity by targeting VE-cadherin. Ann. Surg.. 244, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocellin, S. , Rossi, C.R. , Pilati, P. , Nitti, D. , 2005. Tumor necrosis factor, cancer and anticancer therapy. Cytokine Growth Factor Rev.. 16, 35–53. [DOI] [PubMed] [Google Scholar]
- Mouawad, R. , Sebert, M. , Michels, J. , Bloch, J. , Spano, J.P. , Khayat, D. , 2010. Treatment for metastatic malignant melanoma: old drugs and new strategies. Crit. Rev. Oncol./Hematol.. 74, 27–39. [DOI] [PubMed] [Google Scholar]
- Regenass, U. , Muller, M. , Curschellas, E. , Matter, A. , 1987. Anti-tumor effects of tumor necrosis factor in combination with chemotherapeutic agents. Int. J. Cancer.. 39, 266–273. [DOI] [PubMed] [Google Scholar]
- Schakowski, F. , Gorschluter, M. , Buttgereit, P. , Marten, A. , Lilienfeld-Toal, M.V. , Junghans, C. , Schroff, M. , Konig-Merediz, S.A. , Ziske, C. , Strehl, J. , Sauerbruch, T. , Wittig, B. , Schmidt-Wolf, I.G. , 2007. Minimal size MIDGE vectors improve transgene expression in vivo. In Vivo. 21, 17–23. [PubMed] [Google Scholar]
- Schakowski, F. , Gorschluter, M. , Junghans, C. , Schroff, M. , Buttgereit, P. , Ziske, C. , Schottker, B. , Konig-Merediz, S.A. , Sauerbruch, T. , Wittig, B. , Schmidt-Wolf, I.G. , 2001. A novel minimal-size vector (MIDGE) improves transgene expression in colon carcinoma cells and avoids transfection of undesired DNA. Mol. Ther.. 3, 793–800. [DOI] [PubMed] [Google Scholar]
- Senzer, N. , Mani, S. , Rosemurgy, A. , Nemunaitis, J. , Cunningham, C. , Guha, C. , Bayol, N. , Gillen, M. , Chu, K. , Rasmussen, C. , Rasmussen, H. , Kufe, D. , Weichselbaum, R. , Hanna, N. , 2004. TNFerade biologic, an adenovector with a radiation-inducible promoter, carrying the human tumor necrosis factor alpha gene: a phase I study in patients with solid tumors. J. Clin. Oncol.. 22, 592–601. [DOI] [PubMed] [Google Scholar]
- Sheets, R.L. , Stein, J. , Manetz, T.S. , Duffy, C. , Nason, M. , Andrews, C. , Kong, W.P. , Nabel, G.J. , Gomez, P.L. , 2006. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol. Sci.. 91, 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, U. , Walther, W. , 1998. Cytokine-mediated reversal of multidrug resistance. Cytotechnology. 27, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, D. , Jones, P.A. , 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U.S.A.. 99, 3740–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, D. , Jones, P.A. , 2003. The CpG island searcher: a new WWW resource. In Silico Biol.. 3, 235–240. [PubMed] [Google Scholar]
- Walther, W. , Arlt, F. , Fichtner, I. , Aumann, J. , Stein, U. , Schlag, P.M. , 2007. Heat-inducible in vivo gene therapy of colon carcinoma by human mdr1 promoter-regulated tumor necrosis factor-alpha expression. Mol. Cancer Therapeut.. 6, 236–243. [DOI] [PubMed] [Google Scholar]
- Walther, W. , Minow, T. , Martin, R. , Fichtner, I. , Schlag, P.M. , Stein, U. , 2006. Uptake, biodistribution, and time course of naked plasmid DNA trafficking after intratumoral in vivo jet injection. Hum. Gene Ther.. 17, 611–624. [DOI] [PubMed] [Google Scholar]
- Walther, W. , Siegel, R. , Kobelt, D. , Knosel, T. , Dietel, M. , Bembenek, A. , Aumann, J. , Schleef, M. , Baier, R. , Stein, U. , Schlag, P.M. , 2008. Novel jet-injection technology for nonviral intratumoral gene transfer in patients with melanoma and breast cancer. Clin. Cancer Res.. 14, 7545–7553. [DOI] [PubMed] [Google Scholar]
- Walther, W. , Stein, U. , Fichtner, I. , Voss, C. , Schmidt, T. , Schleef, M. , Nellessen, T. , Schlag, P.M. , 2002. Intratumoral low-volume jet-injection for efficient nonviral gene transfer. Mol. Biotechnol.. 21, 105–115. [DOI] [PubMed] [Google Scholar]
- Walther, W. , Stein, U. , Pfeil, D. , 1995. Gene transfer of human TNF alpha into glioblastoma cells permits modulation of mdr1 expression and potentiation of chemosensitivity. Int. J. Cancer.. 61, 832–839. [DOI] [PubMed] [Google Scholar]
- Wolff, J.A. , Budker, V. , 2005. The mechanism of naked DNA uptake and expression. Adv. Genet.. 54, 3–20. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Zhou, J.Y. , Wu, G.S. , 2006. Tumor necrosis factor-related apoptosis-inducing ligand is required for tumor necrosis factor alpha-mediated sensitization of human breast cancer cells to chemotherapy. Cancer Res.. 66, 10092–10099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Figure A1 Schematic representation of the different vectors used.
Table A1. Comparative in vitro luciferase gene transfer in different melanoma and colon carcinoma cell lines.
Table A2. Comparative in vitro hTNFα gene transfer in different melanoma and colon carcinoma cell lines.
Table A3. Vindesine IC50 in different melanoma and colon carcinoma cell lines after hTNFα gene transfer.
Table A4. Summary of acute toxicity signs after jet‐injection of MIDGE‐hTNFα vector solution, determined in a GLP toxicicty study.


