Abstract
Adjuvant cisplatin‐based chemotherapy only marginally improves survival in patients with completely resected non‐small‐cell lung cancer (NSCLC). We have evaluated the predictive value of mutations in TP53, encoding the tumour suppressor p53, in the International Adjuvant Lung Cancer Trial (IALT), a randomized trial of adjuvant cisplatin‐based chemotherapy against observation. TP53 (exons 4–8) was sequenced in 524 archived specimens of IALT patients with a median follow‐up of 7.5 years. Predictive analyses were based on Cox models adjusted for clinical and pathological variables. P‐values ≤0.01 were considered as significant. Mutations were detected in 221 patients (42%) and had no predictive value for the effect of chemotherapy (interaction between TP53 and treatment: p = 0.17 for Overall Survival (OS); p = 0.06 for Disease‐Free Interval, (DFS)). However, among patients with mutations, outcome appeared worse in treatment compared to observation arms (HR for OS = 1.36 (95% CI [0.97–1.31), p = 0.08; DFS = 1.40 (95% CI [1.01–1.95]), p = 0.04). When grouping mutations into classes according to predicted effects on protein structure, the tendency towards worse outcomes was restricted to “structure” mutations affecting residues of the hydrophobic core that are not located at the p53 protein‐DNA interface (HR for death in this class vs wild‐type T53 = 1.66; 95% CI [1.10–2.52], p = 0.02). Overall, TP53 mutations are not significant predictors of outcome in this trial of cisplatin‐based chemotherapy, although a specific class of structural mutations may be associated with a tendency towards worse outcomes upon treatment.
Keywords: TP53, Mutations, NSCLC, Chemotherapy, Cisplatin, Randomized trial
Highlights
Chemotherapy only marginally improves survival in non‐small‐cell lung cancer.
In a large randomized trial, TP53 mutation does not predict survival.
Patients with mutations have worse survival in treatment than observation.
A class of structural mutations may carry a detrimental effect upon treatment.
1. Introduction
Non‐small cell lung cancer (NSCLC) includes essentially squamous cell carcinoma, adenocarcinoma and large cell carcinoma, which collectively represent the leading cause of cancer death worldwide (Bray et al., 2013). Early‐stage NSCLC patients who undergo complete tumor resection develop distant metastases in 50%–70% of the cases, resulting in 5‐year survival rates of about 40% (Farjah et al., 2009; Goldstraw et al., 2011). The International Adjuvant Lung Cancer Trial (IALT) was designed to evaluate the effect of cisplatin‐based adjuvant chemotherapy against observation on survival after complete resection of NSCLC. This trial demonstrated an absolute benefit of 4.1% on 5‐year overall survival among 1867 randomized patients (Arriagada et al., 2004; Dunant et al., 2005). In addition to IALT, two other randomized trials (JBR.10, ANITA) showed a modest but significant benefit of cisplatin‐based chemotherapy in patients with stage IB‐III NSCLC after complete resection (Douillard et al., 2006; Winton et al., 2005), while a third study, using carboplatin‐paclitaxel (CALGB9633), did not confirm this effect (Strauss et al., 2008). Overall a pooled analysis of the data of five randomized studies including IALT and these 3 trials concluded to the benefit of chemotherapy (Pignon et al., 2008). However, an updated follow‐up of IALT has shown that the benefit of chemotherapy on overall survival did not exist anymore beyond 5‐years of follow‐up, essentially due to increased mortality from non‐lung cancer causes in the treatment group (Arriagada et al., 2010a). Therefore, there is an urgent need for appropriate markers to identify patients who are most likely to benefit from adjuvant therapy before its administration.
The IALT Biological Working Group (IALT‐Bio) was established to identify potential biomarkers predicting treatment outcomes using available archival specimens from a subset of IALT patients. It was shown in IALT‐Bio that cases with low levels of immunohistological expression of excision repair cross‐complementation group 1 protein (ERCC1), as well as low levels of p27kip1 expression, appear benefit from adjuvant chemotherapy (Bepler et al., 2011; Filipits et al., 2007; Olaussen et al., 2006). Recently, low immunohistochemical expression of a panel of DNA repair proteins (XPF, BRCA1, ERCC1, MSH2, p53, PARP1, and ATM) was shown to have a borderline predictive effect on 5‐years outcomes, restricted to the squamous cell carcinoma subgroup (Pierceall et al., 2012). However, so far none of these potential biomarkers have been independently validated in other randomized trials.
TP53, the most frequently mutated gene in human cancer, encodes the nuclear phosphoprotein, p53, which exerts multiple suppressive effects by mediating growth arrest, apoptosis, senescence and differentiation, and by controlling several aspects of bioenergetics metabolism (Goldstein et al., 2011; Hollstein and Hainaut, 2010). TP53 mutations occur in about 50% of stage II–III NSCLC (Scoccianti et al., 2012). Most mutations are missense and occur in the DNA‐binding domain (residues 102–296), turning the p53 protein into a transcriptionally inactive form that often accumulates in the nucleus of cancer cells (Petitjean et al., 2007). Accumulated mutant protein may exert dominant effects by inhibiting the protein encoded by the remaining wild‐type allele or by suspected gain‐of‐function properties (Oren and Rotter, 2010; Solomon et al., 2011). Such properties have been identified in experimental cell and animal systems but their relevance and clinical impact in human cancer has not been demonstrated (Goldstein et al., 2011).
In the JBR.10 trial, a study of TP53 mutations and p53 protein expression in 397 patients has shown that, whereas mutation had no prognostic or predictive value, p53 protein overexpression was a significant prognostic marker of shortened survival, as well as a significant predictive marker for greater benefit from adjuvant chemotherapy (Tsao et al., 2007). In the present study we have used IALT‐Bio specimens to investigate whether TP53 mutations alone or in conjunction with p53 protein expression may have a prognostic role or provide predictive indications on the benefit from cisplatin‐based adjuvant chemotherapy in completely resected NSCLC with a median survival of 7.5 years.
2. Methods
2.1. Patients and study design
All patients had participated in the IALT, which compared adjuvant cisplatin‐based chemotherapy with observation among patients with non‐small cell lung cancer. Inclusion criteria and the results of the IALT have been reported previously (Arriagada et al., 2004, 2010, 2005). IALT‐Bio was subsequently designed by a steering committee to examine whether tumor markers could be used to predict a survival benefit from chemotherapy. The study was conducted according to a protocol, which defined a number of blocks to collect in order to guarantee a sufficient power and detailed a plan for statistical analysis. Formalin‐fixed, paraffin‐embedded tumor samples were collected from patients recruited at IALT participating centers that had enrolled more than 10 patients. Twenty‐eight centers in 14 countries contributed at total of 864 specimens. Approval was obtained from the local institutional review boards, according to the legal regulations in each participating country. All tumors were centrally reviewed at the Department of Pathology, Centre Hospitalier Universitaire Albert Michallon, Grenoble, France, according to the 2004 histopathological classification of the World Health Organization (WHO) (Brambilla et al., 2005).
2.2. TP53 mutation analysis
Mutations were analysed using genomic DNA isolated from paraffin‐embedded archived tissue sections. DNA was extracted by standard QIAamp DNA extraction Kit (QIAGEN, Hilden, Germany). TP53 mutations were detected by direct sequencing with BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) using primers and conditions described in the IARC TP53 standard sequencing protocol (http://p53.iarc.fr/ProtocolsAndTools.aspx). Each PCR product was bidirectionally sequenced and analyzed using a 16‐capillary automated sequencer (ABI PRISM® 3100 Genetic Analyzer, Applied Biosystems). All sequence variations were confirmed by running and second, independent PCR and sequencing analysis of the same sample. Only samples with confirmed results and with sequence data including the entire coding sequence of exons 4–8 and flanking splice sites were taken into account in the final analysis. In the IARC TP53 database, these exons comprise over 90% of the mutations detected in tumors that were sequence for the entire coding sequence. Variations were compared to previously described polymorphisms and mutations and mutation patterns were analysed the using on‐line sequence comparison tools available at http://p53.iarc.fr/TP53GeneVariations.aspx. Variations were considered as mutations if (1) not previously annotated as polymorphism in IARC TP53 database, (2) predicting a change in protein sequence. Samples with silent mutations were classified as “wild‐type”.
TP53 mutations in exons 4 to 8 were further separated in classes according to their position, nature, and suspected effect on protein structure and activity as described previously (Olivier et al., 2006). The following classes were defined: (1) Non‐missense mutations, including any mutation other than missense such as nonsense mutations (stop codons), deletions and insertions (in‐frame or producing a frameshift), and substitutions at splice donor or acceptor sites; (2) Missense DNA‐binding Motif (DBM) mutations, resulting in a single amino acid change in structural elements of the wild‐type protein that make direct contact with target DNA; these elements include the L3 loops (codons 237–250) involved in contacts in the minor groove of the DNA double helix, and the Loop‐Sheet‐Helix (LSH) motif (codons 119–135 and codons 272–287) involved in DNA contacts in the major groove. This category includes mutations at residues directly binding to DNA backbone or bases (residues at positions 119, 120, 121, 239, 241, 243, 247, 248, 273, 275, 276, 27, 280, and 283); (3) Missense non‐DBM mutations, resulting in a single amino‐acid change in structural elements that are not in direct contact with DNA but support the DNA binding surface, including beta‐strands and short intervening loops that define the hydrophobic core of the DNA‐biding domain of the protein. This classification has been previously validated against methods analysing amino‐acid substitutions such as Align‐GVGD and SIFT (Mathe et al., 2006a).
2.3. Immunohistochemistry for p53
Immunostaining for p53 was performed on serial sections from the same paraffin block from which DNA was extracted for TP53 mutation analysis. Indirect immunostaining was performed with the anti‐p53 monoclonal antibody DO‐7 (1/75, DAKO, Glostrup, Denmark) using the Ventana automated immunostainer (Ventana Corp., Tucson, AZ, USA) with specified procedures and reagents (Pierceall et al., 2012; Scoccianti et al., 2012). Scoring was performed by multiplying the percentage of positively stained nuclei (0–100%) by intensity of staining 0–3 evaluated on a scale of 0–3 (0: absent; 1: <10%; 2: 10–50%; 3: 50–100%), leading to a composite multiplicative global score of 0–300. A tumor was considered as positive for p53 when the composite global score was greater than 100 (e.g. tumors with a staining intensity of 2 in at least 51% of tumor cells or with a staining intensity of 3 in at least 34% of tumor cells).
2.4. Statistical analyses
Analyses were performed with the long‐term survival data (7.5 years mean follow‐up) of The International Adjuvant Lung Cancer Trial (IALT) (Arriagada et al., 2010b). The distribution of baseline data according to TP53 mutation status was compared using logistic analyses stratified by centre in the univariate and multivariate setting. To test for differences in a given characteristic between patients with or without TP53 mutation, conditional logistic regression on an aggregate center variable was used both for univariate and multivariate analyses. The prognostic values of TP53 mutation status and chemotherapy for survival were studied using the Cox model. Similar to the original IALT analysis, the Cox model included each factor used in the stratified randomization (center, tumor stage, type of surgery (pneumonectomy, lobectomy, segmentectomy), clinical and histological prognostic factors, age (<55 yrs, 55–64 yrs, >64 yrs), gender, WHO performance status, nodal status, lymphoid cell infiltration (not intense, intense) and the revised histopathological type (adenocarcinoma, squamous cell carcinoma, other NSCLC)). All other factors that were statistically related to TP53 mutation status in a multivariate logistic model (P < 0.05) were added to the survival Cox model. The predictive value of TP53 was studied by testing the interaction between TP53 status and the attributed treatment (chemotherapy or observation) in the same Cox model. For prognostic analysis, the Hazard Ratio (HR) of death (Overall Survival, OS) or Disease‐Free Survival event (DFS) was estimated for each mutation class relative to the wild‐type class. For predictive analysis, the HR of OS or DFS in chemotherapy relative to observation arm was estimated in each mutation class and in the wild‐type class. Heterogeneity tests of the HRs were performed to assess the prognostic (or predictive) role. All P values were two‐sided. According to IALT‐Bio statistical analysis plan, only P values below 0.01 were considered statistically significant in order to limit the risk of false positive results. Survival rates were estimated using the Kaplan–Meier method (p values indicated besides Kaplan–Meier curves were adjusted p values corresponding to the Cox analysis). All analyses and survival curves were performed using SAS software version 9.1 (SAS Institute Inc. Cary NC, USA).
3. Results
3.1. Patients and tumor characteristics in relation with TP53 mutation status
Overall, the 28 centers with 10 patients or more in IALT that agreed to participate in IALT‐Bio had included a total of 1045 of the 1867 patients of the original IALT trial. Archival FFPE materials were available for 867 of them (83%). These 867 patients and the remaining 178 had similar base‐line characteristics and overall rates of survival. The amount and quality of material were adequate for serial sectioning in 824 of the 867 specimens. Among these blocks, 783 contained tumor material corresponding to non‐small‐cell lung cancer and were included in the study. Of these samples, 53 did not yield DNA in sufficient amounts or quality to generate PCR products. A further 181 samples could not be entirely sequenced for exons 4 to 8 of TP53. In addition, variations found at the first analysis could not be confirmed by a second, independent analysis, in 25 cases. Thus, TP53 mutations were compiled in a total of 524 patients (Supplementary Figure 1). Of these tumors, 221 (42%) were classified as containing mutant TP53, including 18 patients whose tumors revealed multiple mutations (16 with two mutations, one with three and one with four mutations). When classified according to type, position and predicted effect on the p53 protein, the 221 mutant specimens were further sub‐divided into 49 cases with Class 1 (non‐missense) mutations, 92 with Class 2 (missense‐DBM) mutations and 80 with Class 3 (missense‐non‐DBM mutations) mutations (Figure 1). The pattern of base changes was typical of TP53 mutations in lung cancers of smokers, in agreement with the hypothesis that the vast majority of patients recruited in IALT were current or former smokers (Supplementary Figure 2).
Figure 1.
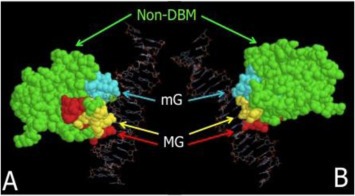
Classes of missense mutations according to their position in the structure of the DNA‐binding domain of p53. Two views of the tertiary structure of p53 of the p53 core‐domain (residues 96–292) in contact with target DNA are shown, structure B being rotated 180° as compared to structure A. Residues belonging to the DNA‐Binding‐Motifs (DBM) are colored in cyan (Loop 3, residues 237–250, biding in the DNA minor groove (mG)), yellow (loop/helix 272–287) and red (loop/sheet 119–135), both binding in the DNA major groove (MG). Residues belonging to non‐DNA –Binding‐Motifs (non‐DBM) are colored in green. Model created using RasMol v2.6 using PDB structure 1TUP (Protein Brookhaven Database, http://www.rcsb.org/pdb/home/home.do), modified to remove p53 monomers A and C.
The characteristics of the study population based on TP53 status are described in Supplementary Table 1. In univariate analysis, TP53 mutations (all classes) appeared to be significantly associated with age (more common in age group 55–64 years), Stage (more common in Stage III), T of TNM (more common in T3 tumors), histology (less common in ADC) and quality of tissue section as evaluated after Haematoxylin‐Eosin staining (less common in sections of average quality). Other variables (including gender, N of TNM, vascular, pleural or lymphatic invasion, type of surgery, intensity of lymphoid cell infiltration, WHO performance status) were not significantly associated with mutation status. When all significant variables were introduced in a logistic model stratified by center, the variables age, gender, TNM stage, T of TNM and quality after final H&E were retained to explain TP53 status.
3.2. Prognostic and predictive analysis
TP53 mutation had no prognostic effect on overall survival (p = 0.50) or disease‐free survival (p = 0.71) of IALT‐Bio patients for a median follow‐up of 7.5 years (Supplementary Table 2). In the subset of 524 IALT‐Bio patients evaluated for TP53 mutations, there was no effect of chemotherapy (HR = 1.13 (0.91–1.42), p = 0.27). The predictive analysis showed no significant difference in the effect of chemotherapy between patients with TP53 mutation and wild‐type patients (test for interaction between TP53 status and treatment: p = 0.17 on OS; p = 0.06 on DFS) (Table 1A and B, Figure 2, Supplementary Figure 2). However, among patients with mutant TP53, there was a non‐significant tendency for worse outcomes in the treatment group than in the control group (HR for death (OS) in therapy versus observation group: 1.36; 95% CI [0.97–1.91], p = 0.08; HR for relapse or death (DFS) in therapy versus observation group: 1.40; 95% CI [1.01–1.95], p = 0.04). Such a tendency was not seen in patients with wild‐type TP53. Table 2 shows the OS rates at 5 years and 8 years in the treatment and observation groups according to TP53 mutation status. Of note, at 8 years, the OS rate of treated patients with mutant TP53 was 24%, 95% CI [16%–33%], compared to 40%, 95% CI [29%–50%] in observation patients with mutant TP53. There was no significant difference in the predictive effect of TP53 mutation on OS in patients with adenocarcinoma when compared to non‐adenocarcinoma histologies (p = 0.88) (Table 3). The tendency for worse outcomes in patients with TP53 mutations was detected in both histological subgroups.
Table 1.
Predictive analysis of interaction between TP53 and treatment on Overall Survival (A: OS) and Disease‐Free Interval (B: DFS).
| A | Treatment arm | Observation arm | HR, treatment vs observation | [95% CI] | p‐value | |||
|---|---|---|---|---|---|---|---|---|
| OS | Deaths | Patients | Deaths | Patients | ||||
| Wild Type (WT) | 89 | 144 | 88 | 159 | 0.99 | 0.73–1.34 | 0.93 | |
| Mutant (MT) | 87 | 120 | 60 | 101 | 1.36 | 0.97–1.91 | 0.08 | |
| HR, MT vs WT | 1.27 | 0.92 | Test for interaction TP53*Treatment | 0.17 | ||||
| [95% CI] | (0.93–1.73) | (0.65–1.29) | ||||||
| p‐value | 0.14 | 0.63 | ||||||
| B | Treatment arm | Observation arm | HR, Treatment vs Observation | [95% CI] | p‐value | |||
| DFS | Events | Patients | Events | Patients | ||||
| Wild Type (WT) | 92 | 144 | 98 | 159 | 0.93 | (0.69–1.24) | 0.60 | |
| Mutant (MT) | 89 | 120 | 63 | 101 | 1.40 | (1.01–1.95) | 0.04 | |
| HR, MT vs WT | 1.29 | 0.85 | Test for interaction TP53*Treatment | 0.06 | ||||
| [95% CI] | (0.95–1.75) | (0.61–1.18) | ||||||
| p‐value | 0.11 | 0.33 | ||||||
Figure 2.
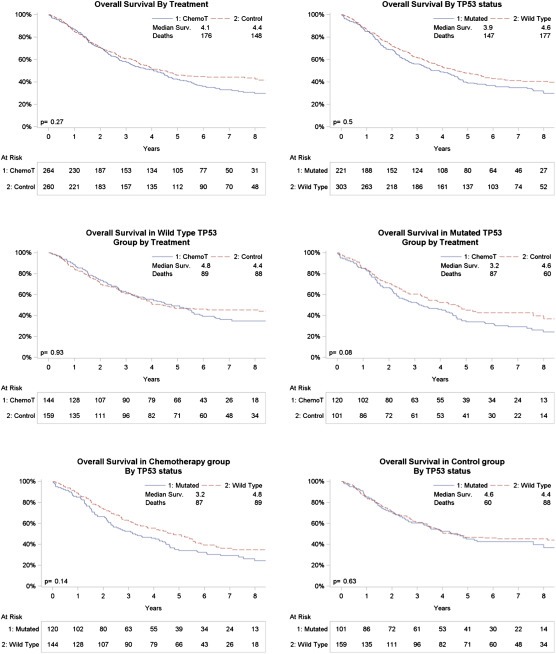
Overall Survival curves of the 524 IALT‐Bio patients with known TP53 mutation status Kaplan–Meier Overall Survival curves according treatment (top left), TP53 status (top right) are shown. Middle graphs: OS according to treatment in patients with wild‐type (middle left) or mutant (middle right) status. Bottom graphs: OS according to TP53 mutation status in chemotherapy (bottom left) or observation (bottom right) arms. P‐values each graph are: adjusted p‐value from Cox model.
Table 2.
5‐year and 8‐year Overall Survival (OS) rates according to treatment and TP53 mutation status.
| Wild type T53 | Treatment arm | Observation arm | Both arms | |||
|---|---|---|---|---|---|---|
| Rate (a) | [95% CI] | Rate (a) | [95% CI] | Rate (a) | [95% CI] | |
| 5 year survival | 49% | 41 %–57% | 47% | 39 %–54% | 48% | 42 %–53% |
| 8 year survival | 35% | 26 %–43% | 45% | 37 %–53% | 41% | 35%–46% |
| Median survival (years) | 4.8 | 4.4 | 3.2 | |||
| Mutant TP53 | ||||||
| 5 year survival | 34% | 26 %–43% | 45% | 35 %–55% | 39% | 33 %–46% |
| 8 year survival | 24% | 16 %–33% | 40% | 29 %–50% | 31% | 24 %–38% |
| Median survival (years) | 2.6 | 3.5 | 3.1 | |||
| Total | ||||||
| 5 year survival | 42% | 36 %–48% | 41% | 40 %–52% | ||
| 8 year survival | 30% | 24 %–36% | 36% | 37 %–50% | ||
| Median survival (years) | 3.1 | 3.3 | ||||
(a) Estimated with Kaplan–Meier method.
Table 3.
Predictive effect of TP53 mutation status on overall survival (OS) according to histology.
| Non‐Adenocarcinoma (n = 349) | Adenocarcinoma (n = 175) | Treatment effect by mutation status | ||||
|---|---|---|---|---|---|---|
| Treatment am | Observation arm | Treatment arm | Observation arm | HR, [95% CI] (a) | p‐value | |
| Wild‐Type TP53 | ||||||
| nb deaths/total | 53/90 | 53/96 | 36/54 | 35/63 | 1.00, [0.74–1.35] | p = 1.00 |
| HR, [95% CI] (a) | 0.99, [0.67–1.45] (b) | 1.00, [0.62–1.62] (b) | ||||
| p‐value | p = 0.95 | p = 1.00 | ||||
| Mutant TP53 | ||||||
| nb deaths/total | 66/91 | 44/72 | 21/29 | 16/29 | 1.36, [0.97–1.91] | p = 0.07 |
| HR, [95% CI] (a) | 1.33, [0.90–1.96] (b) | 1.46, [0.74–2.88] (b) | ||||
| p‐value | p = 0.15 | p = 0.28 | ||||
| Treatment effect by histology | ||||||
| HR, [95% CI] (a) | 1.15 [0.87–1.51] | 1.13 [0.76–1.68] | ||||
| p‐value | p = 0.32 | p = 0.55 | ||||
(a): HR for death, treatment vs Observation, [95% CI].
(b): test for equality of interaction of treatment by TP53 status in the 2 histology subgroups : p = 0.88.
3.3. Predictive effects according to classes of mutations
Mutations within exons 4 to 8 were classified in different groups according to the predicted effect of the mutation on the secondary or tertiary structure of the protein (Figure 1). Table 4 shows the prognostic and predictive analysis of this classification on overall survival (OS). When considering each mutation class in comparison with wild‐type status, a borderline significant effect was seen for class 3 (Missense‐nonDBM) in relation with chemotherapy (HR for death in this class as compared with class 1 (wild‐type T53; HR = 1.66; 95% CI [1.10–2.52], p = 0.02). Several other groupings of mutations were tested according to functional properties of mutant p53 protein, as well as structural or conservation properties of mutant residues (Mathe et al., 2006, 2006). None of these classification detected significant difference on predictive or prognosis effect.
Table 4.
Effects on Overall Survival (OS) according to classes of mutations. A: prognostic effect by treatment arm; B: treatment effect by mutation class.
| A class | Mutation status | Treatment arm (a) | Observation arm (a) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | [95% CI] | p | HR | [95% CI] | p | ||||
| 1 | Wild‐Type (WT) | REF = 1 | REF = 1 | Test for interaction TP53*treatment | p = 0.30 | ||||
| 2 | Mutant (MT), Non‐Missense | 1.27 | 0.78–2.07 | 0.33 | 1.15 | 0.62–2.15 | 0.65 | ||
| 3 | Mutant (MT), Missense‐nonDBM | 1.66 | 1.10–2.52 | 0.02 | 0.90 | 0.55–1.48 | 0.67 | ||
| 4 | Mutant (MT), Missense‐DBM | 1.00 | 0.65–1.53 | 1.00 | 0.87 | 0.55–1.36 | 0.53 | ||
| Heterogeneity test | p = 0.09 | p = 0.84 | |||||||
| B class | Mutation status | Treatment arm (nb/total) | Observation arm (nb/total) | Treatment effect by class (b) | Treatment effect, HR, [95% CI] (a) p‐value | ||||
| HR | [95% CI] | p‐value | |||||||
| 1 | Wild‐Type (WT) | 89/144 | 88/159 | 0.99 | 0.73–1.33 | p = 0.93 | 1.13 [0.91–1.42] p = 0.27 | ||
| 2 | Mutant (MT), Non‐Missense | 22/30 | 12/19 | 1.09 | 0.52–2.26 | p = 0.82 | |||
| 3 | Mutant (MT), Missense‐nonDBM | 35/43 | 21/37 | 1.82 | 1.05–3.18 | p = 0.03 | |||
| 4 | Mutant (MT), Missense‐DBM | 30/47 | 27/45 | 1.14 | 0.66–1.94 | p = 0.64 | |||
(a): HR [95% CI] for death in class vs wild‐type TP53.
(b): HR, [95% CI] for death in treatment arm vs observation arm.
3.4. Combination between TP53 mutation status and p53 protein expression
Of the 783 available specimens, 4 could not be evaluated for p53 immunohistological staining. Both immunohistological staining and mutation status was available for 522 patients (Table 5). Mutation and positivity for p53 protein were strongly associated (Pearson' Chi square: p < 0.001). The distribution of mutation scores according to wild type or mutant TP53 mutation status (all mutations taken as a single group) is shown in Supplementary Figure 3. Table 5 shows the HR for OS of treatment vs observation in 4 the groups defined according to TP53 mutation and p53 protein expression status. The test for equality of the four HR was not significant (p = 0.28). Furthermore, the 3 HR corresponding to groups with either mutant TP53 and/or p53 immuno‐positivity were not different (p = 0.97), while they were jointly borderline different from the wild‐type TP53/p53 immuno‐negative group (p = 0.06).
Table 5.
Predictive effect of TP53 mutation status on overall survival (OS) according to p53 protein expression level.
| p53‐negative | p53‐positive | Treatment effect by mutation status | ||||
|---|---|---|---|---|---|---|
| Treatment am | Observation arm | Treatment arm | Observation arm | HR, [95% CI] (a) | p‐value | |
| Wild‐Type TP53 | ||||||
| nb deaths/total | 57/91 | 61/101 | 31/52 | 28/57 | 1.00, [0.74–1.35] | p = 0.97 |
| HR, [95% CI] (a) | 0.85, [0.58–1.23] (b) | 1.28, [0.75–2.18] (b) | ||||
| p‐value | p = 0.38 | p = 0.37 | ||||
| Mutant TP53 | ||||||
| nb deaths/total | 34/46 | 28/42 | 53/74 | 32/59 | 1.37, [0.97–1.91] | p = 0.07 |
| HR, [95% CI] (a) | 1.35, [0.80–2.26] (b) | 1.40, [0.89–2.19] (b) | ||||
| p‐value | p = 0.26 | p = 0.14 | ||||
| Treatment effect by p53 expression status | ||||||
| HR, [95% CI] (a) | 0.99 [0.74–1.34] | 1.38 [0.98–1.94] | ||||
| p‐value | p = 0.95 | p = 0.07 | ||||
(a): HR for death, treatment vs Observation, [95% CI].
(b): Test of the equality of the four HR: p = 0.28.
4. Discussion
TP53 mutations are one of the most extensively investigated somatic mutations in Non‐Small Cell Lung cancer. However, their predictive or prognostic significance remain unclear (Tsao et al., 2007). Meta‐analysis of p53 protein expression has suggested that it may be a weak prognosis marker (Steels et al., 2001). However, studies on TP53 mutations have shown inconsistent results. A recent analysis of TP53 mutation status in a large prospective cohort, EUELC (European Early Lung Cancer cohort) showed no prognostic effect of TP53 mutations in NSCLC (Scoccianti et al., 2012). In the present study, we have used the set‐up of IALT‐Bio, a biomarker study nested in a phase III randomized trial of cisplatin‐based adjuvant therapy against observation (Arriagada et al., 2004), to assess the prognostic and predictive significance of TP53 mutations in a large group of NSCLC patients. This is, to our knowledge, the largest structured study to date on the predictive significance of TP53 mutations in NSCLC.
Of a total of 783 biological samples available in IALT‐Bio, 524 were completely analysed for mutations in exons 4 to 8 and a total of 221 (42%) were confirmed by at least two independent analyses to contain mutations predicted to alter p53 protein sequence (19 samples containing silent mutations were classified as “wild‐type”). These exons (and their flanking splice sites) were retained in the final analysis because data for other exons were incomplete. Based on data compiled in the IARC TP53 database, mutations in exons 4–8 (residues 33–306) represent over 90% all mutations reported in NSCLC samples that have been fully sequenced for the entire TP53 coding sequence. Mutations were significantly associated (p ≤ 0.01) with age (p trend = 0.02), stage (p trend ≤ 0.001), T but not N of TNM (p trend < 0.001), histology (p = 0.01) and quality of the tissue section as evaluated after Haematoxylin‐Eosin staining (p < 0.001). Of the 54 specimens scored as “average for quality of section, only 10 (19%) had mutant TP53, as compared to 211/470 (45%) in specimens scored as “good quality”, suggesting a bias against mutation detection in specimens judged as of “average” quality. The TP53 mutation pattern of IALT‐Bio patients was compatible with the one of smokers in the IARC TP53 mutation database, with in particular 30% of G:C to T:A transversions, compared to 27.4% in corresponding NSCLC histologies in the IARC database, and with mutational G:C to T:A “hotspots” at codons 157, 158, 248 and 273. These mutations have been experimentally described as resulting from adduct formation by metabolites of polycyclic aromatic hydrocarbons from tobacco smoke (Hainaut et al., 2001; Pfeifer et al., 2002).
The prognostic and predictive value of TP53 mutations was analysed using a Cox model including all co‐variables used in the stratified randomization plus all prognostic variables and all variables that were statistically related to TP53 mutation status at p < 0.05 in a multivariate logistic model. Only p values below 0.01 were considered as statistically significant in order to limit the risk of false positive results. Using this stringent statistical approach, TP53 mutations had no significant prognostic effect and were not predictive of the outcome of chemotherapy with a 7.5‐year median follow‐up (test for interaction between TP53 status and treatment: p = 0.17 for OS and p = 0.06 for DFS). Of note, there was no significant effect of chemotherapy in patients with wild‐type TP53, although this observation should be interpreted with caution due to the limited power of this study to identify small effects. Within this analysis, however, a tendency for worse outcomes emerged for patients with TP53 mutations in the chemotherapy group (OS, HR: 1.36; 95% CI [0.97–1.91], p = 0.08; DFS,HR: 1.40; 95% CI [1.01–1.91], p = 0.04). This observation suggests that TP53 mutation may have a detrimental effect on treatment outcome. This is also reflected in the overall survival rates at 8‐years showing a 16% absolute decrease to 24% (95% CI [16%–33%]) in patients with TP53 mutation who received therapy, compared to 40% (95% CI [29%–50%]) in patients with TP53 mutation in the observation group.
TP53 mutations are diverse in their types, location in coding sequence and predicted effects on the p53 protein. Whereas non‐missense mutations (including nonsense, splice site and frameshift mutation) primarily disrupt p53 protein synthesis (predicting a p53‐null phenotype), missense mutations may be classified in subgroups according to structure/function effects. Computational approaches have been developed to score the structural impact of mutations and predict their effects on p53 DNA‐biding and transactivation capacity (Mathe et al., 2006, 2006). In practice, however, the simplest and most effective method to compare mutations is to group them into simple classes according to their location in the DNA binding domain, as previously performed for evaluating the prognostic effect of TP53 mutations in breast cancer (Olivier et al., 2006). This classification separates mutations into a class of non‐missense mutations (frameshift, splice site, stop codons) that are thought to prevent wild‐type p53 protein expression, and two classes of missense mutations according to whether they interfere with p53 DNA binding capacity either directly by affecting residues within the protein DNA binding surface (missense‐DBM mutations) or indirectly, by altering the architecture of the hydrophobic domain of the protein (missense‐nonDBM mutations). In breast cancer, tumors with missense‐nonDBM mutation class were found to have a significantly better prognosis than missense‐DBM or non‐missense mutations but their predictive significance has not been evaluated (Olivier et al., 2006). In the present lung cancer study, the same mutation classes had no prognostic effect and they did not predict treatment outcomes. However, when compared to wild‐type TP53, missense‐nonDBM mutations had a tendency to be associated with a worse prognosis in the treatment arm, whereas they was no effect in the observation arm (HR for treatment effect for missense‐nonDBM mutations = 1.82, 95% CI [1.05–3.18]), p = 0.03). Other mutation classes did not show any tendency to differ from wild‐type TP53, whether in treatment or observation group. This observation would be consistent with a negative effect on treatment response in tumors that contain structurally altered forms of p53 due to mutations affecting the folding of the hydrophobic core of the protein. Thus, an interesting hypothesis is that these mutations may exert a type of dominant, gain‐of‐function effect that interferes with response to cisplatin‐base chemotherapy. Recent studies on radiation responses in Squamous Carcinoma of the Head and Neck (HNSCC) support that responses to therapy may vary according to the structural and functional impact of mutations (Sano et al., 2011; Skinner et al., 2012). Using a series of cell lines derived from primary HNSCC, Skinner et al. (2012) reported that cell lines with TP53 mutations considered as “disruptive” were associated with radiation resistance in vitro and that this class of mutation was predictive of loco‐regional recurrence in patients (Skinner et al., 2012). Further studies are needed to determine whether and how the mutations of the missense‐nonDBM class modulate cell responses to cisplatin‐based cytotoxic treatments.
It should be noted that some of the mutations classified as missense‐DBM may exert a dual effect by abrogating critical contacts between p53 protein and target DNA and by disrupting the overall structure of the protein. Thus, this mutation class may actually contain mutations that are functionally equivalent to missense‐nonDBM, that is, turning the protein into a mutant with altered protein architecture. However, in the predictive analysis, missense‐DBM mutant were not distinguishable from non‐missense mutants. Therefore, our analysis does not support the hypothesis that further sub‐classification of missense‐DBM mutants in sub‐groups according to suspected structure/function effect may improve the predictive value of TP53 mutations in IALT‐Bio.
Many previous studies have shown a strong but incomplete correlation between TP53 mutation and p53 protein accumulation detected by immuno‐histochemistry (Scoccianti et al., 2012; Tsao et al., 2007). Such a strong but incomplete correlation was also observed in the IALT‐Bio series (p < 0.001). Nevertheless, a substantial number of immuno‐positive cases did not show detectable mutation (45%) and 40% of the cases with mutations did not show high p53 protein accumulation. Immuno‐positive cases without mutant TP53 may result from multiple mechanisms, including accumulation of wild‐type p53 in an active or inactive form in tumors that may carry mutations in factors that regulate p53 function. In the IALT‐Bio series, it is also possible that a small proportion of these immune‐positive cases may represent tumors with mutant TP53 occurring in regions of the gene that have not been analyzed. In contrast, immune‐negative tumors with mutant TP53 may be caused by nonsense or frameshift mutations (predicted to result in a null‐p53 phenotype). Thus, p53 immuno‐reactivity cannot be regarded as a surrogate for TP53 gene mutation and should be considered as a biomarker with distinct biological and clinical significance as compared to TP53 mutations. In the IALT‐Bio series, positive p53 immuno‐histochemistry was neither prognostic nor predictive. However, taken together, patients with tumors containing TP53 mutation and/or positive p53 immunohistochemistry had a tendency for borderline significant worse OS than patients with wild‐type TP53 and no detectable p53 protein.
Another trial of cisplatin‐based therapy against placebo, the JBR.10 trial, has reported that TP53 mutations were not predictive of treatment outcomes with an interaction test p = 0.65) (HR = 0.78; 95% CI [0.46–1.32] for 124 patients with mutations and HR = 0.67; 95% CI, [0.46 to 0.98]; p = 0.04 for the 273 patients with wild‐type TP53) The test for interaction between TP53 status and treatment was non‐significant. Furthermore, at a significance threshold of p ≤ 0.05, p53 protein expression emerged as prognostic marker of shortened survival (p = 0.02) and as predictor for a differentially greater benefit of chemotherapy (p = 0.03). The main differences between the set‐up of JBR.10 and of IALT‐Bio are that the latter trial included a larger proportion of advanced stage patients and of adenocarcinoma than JBR.10. Although these co‐variables have been taken into account in the analysis of IALT‐Bio, it is possible that the differences observed in the significance of p53 accumulation may be caused by differences in patient's recruitment. Alternatively, this discrepancy may be due to inter‐laboratory variations in assessing p53 immuno‐positive status.
In conclusion, the main strength of the present study is that it is the largest to date, developed within a strictly controlled randomized trial and using stringent laboratory controls as well as statistical methods. Its weaknesses are the use of archival paraffin specimens that precluded the exhaustive analysis of TP53 mutation status, as well as the fact that other genetic variations at the TP53 locus have not been analysed (including in particular loss of the wild‐type allele in tumors that contain a TP53 mutation). Furthermore, the present study is limited to 524 samples, taking into account the fact that 259 samples had to be excludes from the present study because of incomplete or unconfirmed analysis of exons 4 to 8. The original design of IALT Bio was based on the analysis of a larger set of samples in order to ensure reasonable power, especially for detecting interactions between treatment effect and marker status. Therefore, our lack of significant results for the predictive study may be due to lack of power due to the loss for analysis of a total of 259 samples.
Overall, the results show that TP53 mutations have no significant prognostic or predictive value for response to cisplatin‐based chemotherapy, but underline a tendency for patients with mutations to show more adverse events in the treatment group as compared to the observation group. These findings should prompt further investigation on the identification of a subset of mutations that may exert gain‐of‐function effects associated with detrimental responses to chemotherapy.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgments
The assistance of M Olivier in classifying mutations is acknowledged, as well as the technical support of Mrs G Martel Planche and S Villar for mutation analysis.
Grant support: The study was supported by Grants No. 2015 from Association pour la Recherche sur le Cancer and No. 95009 from Programme Hospitalier de Recherche Clinique. Also supported by Association pour la Recherche sur le Cancer Pulmonaire, Fédération Nationale des Groupements des Entreprises Françaises dans la Lutte contre le Cancer, Ligue Nationale Contre le Cancer, Eli Lilly France, Swiss Group for Clinical Cancer Research, University Paris‐Sud, EU FP7 program 2007–2013 CURELUNG HEALTH F2‐2010‐258677, and Programme National d'Expertise Spécialisée sur le Cancer du Poumon (PNES‐LUNG), Institut National du Cancer, France. XM was recipient of a fellowship of the International Agency for Research on Cancer.
Database linking: IARC TP53 database: http://p53.iarc.fr/
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.12.015.
Ma Xiaoli, Rousseau Vanessa, Sun Haiji, Lantuejoul Sylvie, Filipits Martin, Pirker Robert, Popper Helmut, Mendiboure Jean, Vataire Anne-Lise, Le Chevalier Thierry, Soria Jean Charles, Brambilla Elisabeth, Dunant Ariane and Hainaut Pierre, (2014), Significance of TP53 mutations as predictive markers of adjuvant cisplatin‐based chemotherapy in completely resected non‐small‐cell lung cancer, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.12.015.
References
- Arriagada, R. , Auperin, A. , Burdett, S. , Higgins, J.P. , Johnson, D.H. , Le Chevalier, T. , Le Pechoux, C. , Parmar, M.K. , Pignon, J.P. , Souhami, R.L. , Stephens, R.J. , Stewart, L.A. , Tierney, J.F. , Tribodet, H. , van Meerbeeck, J. , 2010. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet. 375, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada, R. , Bergman, B. , Dunant, A. , Le Chevalier, T. , Pignon, J.P. , Vansteenkiste, J. , 2004. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N. Engl. J. Med.. 350, 351–360. [DOI] [PubMed] [Google Scholar]
- Arriagada, R. , Dunant, A. , Pignon, J.P. , Bergman, B. , Chabowski, M. , Grunenwald, D. , Kozlowski, M. , Le Pechoux, C. , Pirker, R. , Pinel, M.I. , Tarayre, M. , Le Chevalier, T. , 2010. Long-term results of the International adjuvant lung cancer trial evaluating adjuvant cisplatin-based chemotherapy in resected lung cancer. J. Clin. Oncol.. 28, 35–42. [DOI] [PubMed] [Google Scholar]
- Bepler, G. , Olaussen, K.A. , Vataire, A.L. , Soria, J.C. , Zheng, Z. , Dunant, A. , Pignon, J.P. , Schell, M.J. , Fouret, P. , Pirker, R. , Filipits, M. , Brambilla, E. , 2011. Ercc1 and Rrm1 in the International adjuvant lung trial by automated quantitative in situ analysis. Am. J. Pathol.. 178, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla, E. , Lantuejoul, S. , Dunant, A. , Popper, H. , Ribes, P. , Andre, F. , Filipits, M. , Pirker, R. , Soria, J.C. , Le Chevalier, T. , 2005. Ialt (International adjuvant lung cancer trial): quality assessment and histopathological review according to the who 2004 classification and Assessment of prognostic and predictive role of pathological criteria. Lung Cancer. 49, S2–S44. [Google Scholar]
- Bray, F. , Ren, J.S. , Masuyer, E. , Ferlay, J. , 2013. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer. 132, 1133–1145. [DOI] [PubMed] [Google Scholar]
- Douillard, J.Y. , Rosell, R. , De Lena, M. , Carpagnano, F. , Ramlau, R. , Gonzales-Larriba, J.L. , Grodzki, T. , Pereira, J.R. , Le Groumellec, A. , Lorusso, V. , Clary, C. , Torres, A.J. , Dahabreh, J. , Souquet, P.J. , Astudillo, J. , Fournel, P. , Artal-Cortes, A. , Jassem, J. , Koubkova, L. , His, P. , Riggi, M. , Hurteloup, P. , 2006. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage Ib-Iiia non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [Anita]): a randomised controlled trial. Lancet Oncol.. 7, 719–727. [DOI] [PubMed] [Google Scholar]
- Dunant, A. , Pignon, J.P. , Le Chevalier, T. , 2005. Adjuvant chemotherapy for non-small cell lung cancer: contribution of the International adjuvant lung trial. Clin. Cancer Res.. 11, 5017s–5021s. [DOI] [PubMed] [Google Scholar]
- Farjah, F. , Flum, D.R. , Ramsey, S.D. , Heagerty, P.J. , Symons, R.G. , Wood, D.E. , 2009. Multi-modality mediastinal staging for lung cancer among medicare beneficiaries. J. Thorac. Oncol.. 4, 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipits, M. , Pirker, R. , Dunant, A. , Lantuejoul, S. , Schmid, K. , Huynh, A. , Haddad, V. , Andre, F. , Stahel, R. , Pignon, J.P. , Soria, J.C. , Popper, H.H. , Le Chevalier, T. , Brambilla, E. , 2007. Cell Cycle regulators and outcome of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer: the International adjuvant lung cancer trial biologic program. J. Clin. Oncol.. 25, 2735–2740. [DOI] [PubMed] [Google Scholar]
- Goldstein, I. , Marcel, V. , Olivier, M. , Oren, M. , Rotter, V. , Hainaut, P. , 2011. Understanding wild-type and mutant P53 activities in human cancer: new landmarks on the way to targeted therapies. Cancer Gene Ther.. 18, 2–11. [DOI] [PubMed] [Google Scholar]
- Goldstraw, P. , Ball, D. , Jett, J.R. , Le Chevalier, T. , Lim, E. , Nicholson, A.G. , Shepherd, F.A. , 2011. Non-small-cell lung cancer. Lancet. 378, 1727–1740. [DOI] [PubMed] [Google Scholar]
- Hainaut, P. , Olivier, M. , Pfeifer, G.P. , 2001. TP53 mutation spectrum in lung cancers and mutagenic signature of components of tobacco smoke: lessons from the iarc TP53 mutation database. Mutagenesis. 16, 551–553. [DOI] [PubMed] [Google Scholar]
- Hollstein, M. , Hainaut, P. , 2010. Massively regulated genes: the example of TP53. J. Pathol.. 220, 164–173. [DOI] [PubMed] [Google Scholar]
- Mathe, E. , Olivier, M. , Kato, S. , Ishioka, C. , Hainaut, P. , Tavtigian, S.V. , 2006. Computational approaches for predicting the biological effect of P53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res.. 34, 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe, E. , Olivier, M. , Kato, S. , Ishioka, C. , Vaisman, I. , Hainaut, P. , 2006. Predicting the transactivation activity of P53 missense mutants using a four-body potential score derived from delaunay tessellations. Hum. Mutat.. 27, 163–172. [DOI] [PubMed] [Google Scholar]
- Olaussen, K.A. , Dunant, A. , Fouret, P. , Brambilla, E. , Andre, F. , Haddad, V. , Taranchon, E. , Filipits, M. , Pirker, R. , Popper, H.H. , Stahel, R. , Sabatier, L. , Pignon, J.P. , Tursz, T. , Le Chevalier, T. , Soria, J.C. , 2006. DNA repair by Ercc1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med.. 355, 983–991. [DOI] [PubMed] [Google Scholar]
- Olivier, M. , Langerod, A. , Carrieri, P. , Bergh, J. , Klaar, S. , Eyfjord, J. , Theillet, C. , Rodriguez, C. , Lidereau, R. , Bieche, I. , Varley, J. , Bignon, Y. , Uhrhammer, N. , Winqvist, R. , Jukkola-Vuorinen, A. , Niederacher, D. , Kato, S. , Ishioka, C. , Hainaut, P. , Borresen-Dale, A.L. , 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res.. 12, 1157–1167. [DOI] [PubMed] [Google Scholar]
- Oren, M. , Rotter, V. , 2010. Mutant P53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol.. 2, a001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean, A. , Achatz, M.I. , Borresen-Dale, A.L. , Hainaut, P. , Olivier, M. , 2007. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 26, 2157–2165. [DOI] [PubMed] [Google Scholar]
- Pfeifer, G.P. , Denissenko, M.F. , Olivier, M. , Tretyakova, N. , Hecht, S.S. , Hainaut, P. , 2002. Tobacco smoke carcinogens, DNA damage and P53 mutations in smoking-associated cancers. Oncogene. 21, 7435–7451. [DOI] [PubMed] [Google Scholar]
- Pierceall, W.E. , Olaussen, K.A. , Rousseau, V. , Brambilla, E. , Sprott, K.M. , Andre, F. , Pignon, J.P. , Le Chevalier, T. , Pirker, R. , Jiang, C. , Filipits, M. , Chen, Y. , Kutok, J.L. , Weaver, D.T. , Ward, B.E. , Soria, J.C. , 2012. Cisplatin benefit is predicted by immunohistochemical analysis of DNA repair proteins in squamous cell carcinoma but not adenocarcinoma: theranostic modeling by Nsclc constituent histological subclasses. Ann. Oncol.. 23, 2245–2252. [DOI] [PubMed] [Google Scholar]
- Pignon, J.P. , Tribodet, H. , Scagliotti, G.V. , Douillard, J.Y. , Shepherd, F.A. , Stephens, R.J. , Dunant, A. , Torri, V. , Rosell, R. , Seymour, L. , Spiro, S.G. , Rolland, E. , Fossati, R. , Aubert, D. , Ding, K. , Waller, D. , Le Chevalier, T. , 2008. Lung adjuvant cisplatin evaluation: a pooled analysis by the Lace Collaborative group. J. Clin. Oncol.. 26, 3552–3559. [DOI] [PubMed] [Google Scholar]
- Sano, D. , Xie, T.X. , Ow, T.J. , Zhao, M. , Pickering, C.R. , Zhou, G. , Sandulache, V.C. , Wheeler, D.A. , Gibbs, R.A. , Caulin, C. , Myers, J.N. , 2011. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin. Cancer Res.. 17, 6658–6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoccianti, C. , Vesin, A. , Martel, G. , Olivier, M. , Brambilla, E. , Timsit, J.F. , Tavecchio, L. , Brambilla, C. , Field, J.K. , Hainaut, P. , 2012. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: the EUELC cohort. Eur. Respir. J.. 40, 177–184. [DOI] [PubMed] [Google Scholar]
- Skinner, H.D. , Sandulache, V.C. , Ow, T.J. , Meyn, R.E. , Yordy, J.S. , Beadle, B.M. , Fitzgerald, A.L. , Giri, U. , Ang, K.K. , Myers, J.N. , 2012. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin. Cancer Res.. 18, 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, H. , Madar, S. , Rotter, V. , 2011. Mutant P53 gain of function is Interwoven into the hallmarks of cancer. J. Pathol.. 225, 475–478. [DOI] [PubMed] [Google Scholar]
- Steels, E. , Paesmans, M. , Berghmans, T. , Branle, F. , Lemaitre, F. , Mascaux, C. , Meert, A.P. , Vallot, F. , Lafitte, J.J. , Sculier, J.P. , 2001. Role of P53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur. Respir. J.. 18, 705–719. [DOI] [PubMed] [Google Scholar]
- Strauss, G.M. , Herndon, J.E.n , Maddaus, M.A. , Johnstone, D.W. , Johnson, E.A. , Harpole, D.H. , Gillenwater, H.H. , Watson, D.M. , Sugarbaker, D.J. , Schilsky, R.L. , Vokes, E.E. , Green, M.R. , 2008. Adjuvant paclitaxel plus carboplatin compared with observation in stage Ib non-small-cell lung cancer: Calgb 9633 with the cancer and leukemia group B, radiation therapy oncology group, and North Central cancer treatment group study groups. J. Clin. Oncol.. 26, 5043–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao, M.S. , Aviel-Ronen, S. , Ding, K. , Lau, D. , Liu, N. , Sakurada, A. , Whitehead, M. , Zhu, C.Q. , Livingston, R. , Johnson, D.H. , Rigas, J. , Seymour, L. , Winton, T. , Shepherd, F.A. , 2007. Prognostic and predictive importance of P53 and Ras for adjuvant chemotherapy in non small-cell lung cancer. J. Clin. Oncol.. 25, 5240–5247. [DOI] [PubMed] [Google Scholar]
- Winton, T. , Livingston, R. , Johnson, D. , Rigas, J. , Johnston, M. , Butts, C. , Cormier, Y. , Goss, G. , Inculet, R. , Vallieres, E. , Fry, W. , Bethune, D. , Ayoub, J. , Ding, K. , Seymour, L. , Graham, B. , Tsao, M.S. , Gandara, D. , Kesler, K. , Demmy, T. , Shepherd, F. , 2005. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N. Engl. J. Med.. 352, 2589–2597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


