Abstract
Standard treatments for advanced high‐grade serous ovarian carcinomas (HGSOCs) show significant side‐effects and provide only short‐term survival benefits due to disease recurrence. Thus, identification of novel prognostic and predictive biomarkers is urgently needed. We have used 42 paraffin‐embedded HGSOCs, to evaluate the utility of DNA copy number alterations, as potential predictors of clinical outcome. Copy number‐based unsupervised clustering stratified HGSOCs into two clusters of different immunohistopathological features and survival outcome (HR = 0.15, 95%CI = 0.03–0.81; Padj = 0.03). We found that loss at 6q24.2–26 was significantly associated with the cluster of longer survival independently from other confounding factors (HR = 0.06, 95%CI = 0.01–0.43, Padj = 0.005). The prognostic value of this deletion was validated in two independent series, one consisting of 36 HGSOCs analyzed by fluorescent in situ hybridization (P = 0.04) and another comprised of 411 HGSOCs from the Cancer Genome Atlas study (TCGA) (HR = 0.67, 95%CI = 0.48–0.93, Padj = 0.019). In addition, we confirmed the association of low expression of the genes from the region with longer survival in 799 HGSOCs (HR = 0.74, 95%CI = 0.61–0.90, log‐rank P = 0.002) and 675 high‐FIGO stage HGSOCs (HR = 0.76, 95%CI = 0.61–0.96, log‐rank P = 0.02) available from the online tool KM‐plotter. Finally, by integrating copy number, RNAseq and survival data of 296 HGSOCs from TCGA we propose a few candidate genes that can potentially explain the association. Altogether our findings indicate that the 6q24.2–26 deletion is an independent marker of favorable outcome in HGSOCs with potential clinical value as it can be analyzed by FISH on tumor sections and guide the selection of patients towards more conservative therapeutic strategies in order to reduce side‐effects and improve quality of life.
Keywords: HGSOC, CGH, Survival, 6q24.2–26 deletion, Prognosis
Highlights
DNA copy number‐based clustering stratifies HGSOCs into groups of different outcome.
Deletion at 6q24.2‐26 predicts longer survival of HGSOC patients.
Prognostic value of the 6q24.2–26 deletion is independent of known factors.
Validation in independent series confirmed prognostic utility of the 6q24.2–26 loss.
A combination of lost genes within 6q24.2–26 region may explain the association.
1. Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy and fifth leading cause of cancer‐related death in Western countries (Ferlay et al., 2008). High‐grade serous ovarian carcinomas (HGSOCs), the most common and aggressive ovarian cancer subtype, account for the majority (60–80%) of EOC deaths (Cannistra, 2004). HGSOCs show a very poor prognosis due to late stage at presentation and the development of chemoresistance (Seidman et al., 2004). In spite of high rates (∼80%) of initial response to platinum‐based treatment, the majority of patients relapse (Piccart et al., 2001). Although, over recent decades treatment has advanced significantly thanks to improved surgical techniques and chemotherapy regimens, the 5‐year survival rate has remained relatively unchanged (between 35 and 40%) (Berns and Bowtell, 2012; Siegel et al., 2013). It is therefore essential to improve our understanding of the molecular events underlying the pathogenesis of HGSOCs in order to develop better prognostic and predictive markers. Given the fact that the presence of widespread DNA copy number changes is a hallmark of HGSOCs (Bowtell, 2010; TCGA, 2011), such alterations may serve as relevant markers for predicting prognosis, progression and drug sensitivity.
Comparative genomic hybridization (CGH) has been the most widely used method for the global assessment of DNA copy number alterations (CNAs). To date, there have been several studies utilizing either conventional metaphase chromosome‐based CGH (Bruchim et al., 2009; Ramus et al., 2003), or array‐based high‐resolution techniques to identify the landscape of copy number events in ovarian cancer (Leunen et al., 2009; TCGA, 2011). Among the most frequently reported gained regions are 1q, 3q, 8q and 20q, while common lost regions include 4q, 5q, 6q, 8p, 17p, 18q and 22q (Bruchim et al., 2009; Gorringe and Campbell, 2009; Ramus et al., 2003). Especially interesting are losses and rearrangements at the long arm of chromosome 6, which have been recurrently described not only in ovarian cancer (Foulkes et al., 1993; Orphanos et al., 1995; Saito et al., 1992), but also in other types of carcinomas and in non‐epithelial tumors including melanoma and hematological and central nervous system malignancies (Burkhardt et al., 2006; Guo et al., 2011; Li et al., 2013; Nelson et al., 2008; Theile et al., 1996; Vajdic et al., 2003).
In particular, loss at 6q24–27 has been extensively studied for its potential role in tumor suppression (Hayashi et al., 2012; Sun et al., 2003) and some candidate genes have been proposed such as PLAGL1 (Abdollahi et al., 2003), GRM1, SOD2 (Shridhar et al., 1999), SASH1 (Zeller et al., 2003) or Parkin (Denison et al., 2003). However, few studies so far have aimed to define specific DNA copy number markers that may have clinical relevance in predicting outcome in ovarian cancer (Baumbusch et al., 2013; Bruchim et al., 2009; Engler et al., 2012; Wang et al., 2012; Yamamoto et al., 2009). Most studies that have focused on the assessment of specific alterations have been limited by the absence of independent copy number replication datasets (Bruchim et al., 2009; Yamamoto et al., 2009). Other studies including independent validation series have been mainly focused on describing general features (ie. genomic instability or LOH profiles) which are more difficult to implement in the clinic than distinct individual changes (Baumbusch et al., 2013; Wang et al., 2012).
In our study, we used a series of familial and sporadic HGSOCs, whose DNA copy number profiles had been previously characterized (Kamieniak et al., 2013). We performed a DNA copy number‐based unsupervised clustering that revealed two main groups of tumors of distinct immunohistopathological features and clinical outcome. We further identified a single region of deletion at 6q24.2–26 that differentiated both clusters and that was subsequently found to be associated with longer survival. We used different independent datasets to validate the prognostic value of this deletion at the DNA copy number (N = 447) and at the expression level (N = 799) and finally propose some candidate genes that may potentially explain the association.
2. Materials and methods
2.1. Patients and tumors
A series of 42 formalin‐fixed, paraffin‐embedded (FFPE) HGSOCs previously characterized using high‐resolution array CGH (4 × 180 K, Agilent Technologies, Palo Alto, CA) assuring >80% tumoral cellularity (Kamieniak et al., 2013) was included in the present study. Since the previous study (Kamieniak et al., 2013) aimed to characterize copy number changes in hereditary ovarian tumors and to describe similarities and differences with sporadic tumors the series comprised 30 familial and 12 sporadic HGSOCs. Inclusion of hereditary and sporadic cases in the current study allowed us to evaluate associations between the BRCA status and potential DNA copy number alterations predicting clinical outcome. Familial tumors were obtained from index‐cases from high‐risk breast and ovarian cancer families and corresponded to 13 BRCA1 mutation carriers (BRCA1 tumors), 5 BRCA2 mutation carriers (BRCA2 tumors) and 12 patients without mutations in neither of those genes (BRCAX tumors). Families were ascertained at Spanish hospitals and at the Spanish National Cancer Research Center (CNIO) and fulfilled one of the following criteria in order to be selected for the present study: (a) at least two cases of ovarian cancer in the same family line; (b) at least one case of ovarian cancer and at least one case of breast cancer in the same family line; (c) at least one woman with both breast and ovarian cancer; (d) at least one woman with bilateral ovarian cancer. The index case of each family was screened for mutations in the BRCA1 and BRCA2 genes by a combination of denaturing high performance liquid chromatography (DHPLC) and sequencing. Details about the screening of germline mutations in the BRCA1 and BRCA2 genes can be found elsewhere (Kamieniak et al., 2013). The 12 sporadic tumors (with no reported first or second degree relative with breast or ovarian cancer) were obtained from one hospital (H. Virgen del Rocio, Seville). Patients whose tumors were included in the discovery series (N = 42) and in the validation series characterized using tissue microarrays (N = 36 HGSOCs) underwent surgical intervention (following diagnosis) in the years 1990–2008 and 1991‐2010, respectively. Surgical resections were classified as optimal (less than or equal 1 cm diameter of residual tumor) or suboptimal (greater than 1 cm diameter of residual tumor). Patients were treated according to a standardized protocol with a combination of taxane and platinum agents. The length of overall survival (OS) was defined from the date of primary surgery to the date of patient death, while progression‐free survival (PFS) was calculated from the date of primary surgery to the date of disease progression, defined as an increase in CA125, or radiological or surgical evidence of relapse. For both analyses, time was censored at the date of last follow‐up.
Additional information about the clinicohistopathological features of the series is shown in Supplementary Table 1. The study was approved by the research ethics committees from each of the participating centers and all patients gave informed consent.
Tumors were blindly reviewed by two pathologists (I.M‐R. and J.P.) and classified histopathologically by evaluation of immunohistochemical markers such as Wilms Tumor protein (WT1), tumor protein p53 (TP53), estrogen receptor (ESR), progesterone receptor (PGR) and cyclin‐dependent kinase inhibitor 2A (p16) (CDKN2A) (Kalloger et al., 2011; Kobel et al., 2009) to assure proper identifications of serous histotype. Grading of the serous tumors was assessed according to two‐tier M.D. Anderson Cancer Center (MDACC) system (Malpica et al., 2004).
2.2. Independent validation series
Two independent series were used to validate associations found between DNA copy number changes and patients' outcome. First series consisted of 36 HGSOCs among which 25 were sporadic and 11 were familial cases. Almost 70% of the series was represented by high‐FIGO stage tumors (III and IV). The second validation series was composed of 411 HGSOCs from The Cancer Genome Atlas (TCGA) study with publically available DNA copy number and clinical data (TCGA, 2011). It included mainly sporadic (91%), high‐grade (87%), and high (III and IV) FIGO stage (95%) serous adenocarcinomas.
In addition, 799 HGSOCs, among which 675 were high‐FIGO stage tumors, from KM‐plotter (Gyorffy et al., 2012) were used for validation at the gene expression level.
2.3. Immunohistochemistry
Staining pattern of 33 proteins was assessed by two pathologists (I.M‐R. and J.P.) on 4 TMAs containing 170 EOCs. Evaluated proteins were involved in the following cellular processes: hormone signaling (ER, PR and AR), proliferation (topoisomerase IIα and Ki‐67), cell cycle (cyclin D1, cyclin E, p21, p27, p16, p53 and Rb), apoptosis (BCL‐XL, Bcl‐2 and survivin), cell adhesion (E‐cadherin, β‐catenin), tumor progression (KLK7, KLK6, EMA, MMP7 and PIK3CA), angiogenesis (VEGF), signaling (HER2, C‐KIT and EGFR) or DNA repair (ERCC1, XPG, XPF, RAD50, RAD51 and CHEK2). The antibodies, dilutions, suppliers, visualization systems, immunostainers and scoring used are shown in Supplementary Table 2. Between 100 and 150 cells per core were scored to determine the percentage of positive nuclei, cytoplasm, or membranes, depending on the marker. Nuclear staining was evaluated for estrogen receptor (ER), progesterone receptor (PR), androgen receptor (AR), p53, Ki‐67, cyclins D1 and E, p27, p21, Rb, topoisomerase IIα, survivin, RAD50, RAD51, XPF, XPG, CHEK2, ERCC1, EGFR, metalloproteinase 7 (MMP7), kallikrein 7 (KLK7), E‐cadherin, β‐catenin and PIK3CA. Cytoplasmic staining was assessed for p16, BCL‐XL, Bcl‐2, survivin, kallikrein 6 (KLK6) and KLK7, EMA, VEGF, C‐Kit, MMP7, EGFR, e‐cadherin, β‐catenin, RAD51 and PIK3CA. Membrane staining was evaluated for HER2, EGFR, e‐cadherin and β‐catenin. The thresholds to determine over‐expression of each marker were established based on literature (Supplementary Table 2) as described elsewhere (Bali et al., 2004; Brun et al., 2008; Honrado et al., 2005; Ni et al., 2004; Rosen et al., 2006; Schindlbeck et al., 2007; Schmandt et al., 2003; Tangjitgamol et al., 2009; Xia et al., 2009). The percentage of stained nuclei, independent of the intensity, was scored for ER, PR, AR, Ki‐67, p53, cyclinD1, cyclinE, p27, p21, Rb, RAD51, XPF, XPG, CHEK2, ERCC1, EGFR, MMP7, KLK7. For EGFR, E‐cadherin and β‐catenin expression, the percentage of cells with membrane staining and staining intensity was determined.
2.4. Definition of regions differentiating DNA copy number‐based clusters
Called CGH data for 42 HGSOCs, preprocessed as described previously (Kamieniak et al., 2013) were subjected to unsupervised hierarchical clustering using total linkage and overall similarity algorithms implemented in WECCA (Weighted Clustering of Called aCGH Data) R package (Van Wieringen et al., 2008). The cut‐off of two clusters was chosen by plotting a ratio of the within‐cluster similarity and the between‐cluster similarity (a statistics that evaluates the effect of a new cluster split). Two was shown to be the optimal cut‐off and optimal number of clusters.
The DNA copy number alterations that best distinguished the generated clusters were defined in Nexus Copy Number v5.1 (BioDiscovery, Inc; El Segundo, CA) using “comparisons” option, minimal frequency difference between both clusters of 35% and Fisher Exact Test with significant p‐value lower than 0.05. Benjamini–Hochberg was used for multiple testing correction and FDR <0.05 was considered significant. The boundaries of the defined regions were based on the similar frequency of the alteration in a given cluster.
2.5. Association of defined regions with survival
Defined copy number alterations were evaluated for their association with overall survival (OS) using univariate log‐rank. Multivariate analysis, including all the significant variables (P‐value <0.05) in the univariate analysis, was performed with the Cox regression model. In addition, the association of the regions with OS was further proved on adjustment for other potentially confounding factor, even if not statistically significant in our series (such as residual tumor, age of diagnosis, BRCA mutation status) producing consistent results.
2.6. Validation by fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization (FISH) was performed on TMA sections according to Vysis's protocol (Vysis, Downers Grove, IL, USA). The test probe mapping to the 6q25.1 (157,099,063–157,530,401) region of deletion was composed of three BACs (RP11‐608N7, RP11‐68I24 and RP11‐100N9) and was labeled by nick translation with dUTP‐SpectrumOrange (Abbott Molecular, IL, USA). The reference probe targeting 6p21 (43,490,072–43,543,812) consisted of two BACs (RP11‐410B13 and RP11‐107P14) and was labeled with dUTP‐SpectrumGreen (Abbott Molecular, IL, USA). BACs were obtained from the BACPAC Resource Center (BPRC) at the Children's Hospital Oakland Research Institute (Oakland, CA). Briefly, probes were blocked with Cot‐1 Human DNA (Roche Diagnostics GmbH, Mannheim, Germany) to suppress repetitive sequences. Probe specificity was confirmed on normal peripheral blood metaphase cells. FISH analysis was performed by two investigators (M.G. and M.K.) who had no prior knowledge of the genetic, clinical, or immunohistochemical features of the tumors. On average 5 (3–7) high‐power fields with well defined‐nuclei were analyzed per each sample (always in duplicates). Deletion was considered as positive, when at least 100 cells/per tissue core showed one red signal less than green in the same nuclei. The deletion status in these tumors was then analyzed for association with patient's outcome.
2.7. Validation at the global gene expression level
The association of the 6q24.2–q26 deletion with survival was validated at the gene expression level using KM‐plotter (Gyorffy et al., 2012) with the JetSet probset. The KM‐plotter is an online tool that allows the assessment of the prognostic value of the expression levels of microarray‐quantified genes in ovarian cancer patients. The current database is set up using gene expression data and survival information of 1436 ovarian cancer patients downloaded from Gene Expression Omnibus and The Cancer Genome Atlas (10 different datasets). The association with overall survival was assessed in 799 HGSOCs (grade 3) and in 675 high‐FIGO HGSOCs (FIGO III and IV, grade 3). The mean expression of all the genes from the 6q24.2–q26 region with available data in the KM‐plotter (53 out of 85 with validated RefSeq, as for May 2014) was used and the data was dichotomized at the automatically selected, best fitted cut‐off into higher and lower expressing groups. Kaplan–Meier and log‐rank tests were used to characterize the distribution and estimate the outcomes. In addition, Cox proportional hazard model was used to estimate the hazard ratios and 95% Confidence Intervals.
2.8. Definition of candidate genes at 6q24–q26
In order to propose individual candidate genes that might explain the observed association with patient survival, we first identified those genes in the 6q24–26 region whose loss had an impact on expression. To this end we used 232 tumors from TCGA ovarian study for whose copy number and expression data were available and assessed a total of 81 genes (out of 85 in the region with validated RefSeq) localized at 6q24.2–q26 for whom RNAseq data (RPKM) was available. The Wilcoxon rank test was used to compare the expression values of each gene between tumors with normal copy number status and tumors with genomic loss at each locus. Significant genes were then evaluated for potential association between their expression levels and patient survival.
Before running survival analysis, RPKM values were normalized, by subtracting the mean of all samples and dividing by the standard deviation, in order to allow a direct comparison of hazard ratios between genes. Next, the normalized gene expression values for each of 296 HGSOCs from TCGA study (for whom clinical and RPKM data were available) were included as an explanatory variable in Cox regression models, together with cofactors significantly associated with survival in the series (FIGO stage, age of diagnosis and BRCA mutation status).
2.9. Statistical analyses
Statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL). Comparison of continuous variables was assessed using a two‐tailed Student's t‐test for variables with approximate normal distribution (as determined by Kolmogorov–Smirnov test), or by Mann–Whitney test otherwise. For categorical data (FIGO stage, BRCA1/2 mutation status) Fisher's Exact Test was used. All tests were two‐sided and P‐value <0.05 was considered statistically significant. Estimation of survival time distribution was performed using Kaplan–Meier method and statistical differences between survival curves were assessed with log‐rank test if the proportional hazard assumption was true or Gehan–Breslow–Wilcoxon otherwise. Univariate Cox regression was used to estimate Hazard ratios (HR) and 95% confidence intervals (95% CI). To adjust for confounding factors, for each tumor series we created a multivariate Cox proportional hazards regression model including all possibly confounding variables: FIGO stage, residual tumor, age of diagnosis, familial status and BRCA mutation status. In the final model, with tested variable, all covariables with P‐values <0.05 were included. The prognostic value of 6q24–26 deletion was tested (in discovery and validation sets) by comparing patients positive for the deletion (having at least 90% of the region lost) versus all the others. All statistical tests were two‐sided and nominal P‐values less than 0.05 were considered statistically significant.
3. Results
3.1. Immunohistochemical characterization of DNA copy number‐based tumor clusters
To determine whether DNA copy number changes may define novel ovarian tumor subtypes with distinct biology we performed DNA copy number‐based unsupervised hierarchical clustering of 42 HGSOCs (Figure 1). This analysis rendered two main clusters, A and B. Cluster B, with greater amount of genome lost (as defined by a number and length of lost regions), exhibited some enrichment in tumors from BRCA1/2 mutation carriers, but did not differ significantly from cluster A with regard to BRCA mutation status or FIGO stage. As for immunohistochemical markers, cluster B showed a significantly higher immunostaining of survivin and marginally higher expression of progesterone receptor, whereas greater expression of CCNE1 was significantly associated with cluster A (Table 1).
Figure 1.
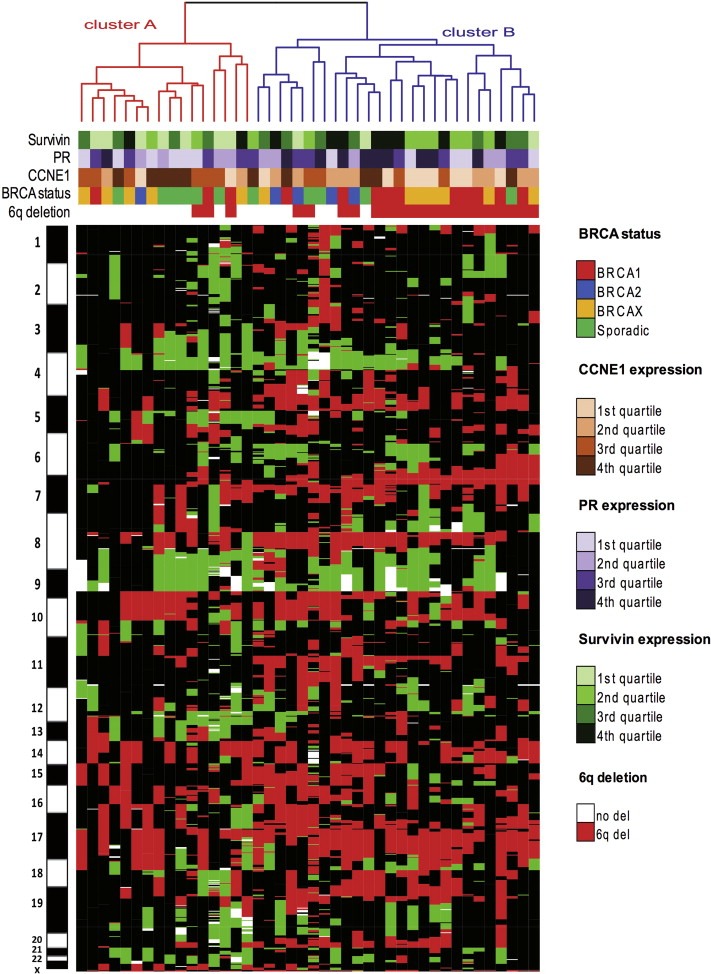
DNA copy number‐based unsupervised hierarchical clustering of 42 high‐grade serous ovarian carcinomas (HGSOCs). Each column represents a tumor sample and each row corresponds to DNA copy number changes mapped according to chromosomal location. Colors correspond to different copy number categories: red, loss; green, gain; white, amplification; black, lack of copy number changes. Dendrogram highlights the division of the samples into two main clusters. Hereditary or sporadic condition of tumors, immunohistopathological features and the 6q deletion status are represented by color labels shown below the dendrogram.
Table 1.
Correlation of the clusters defined by unsupervised hierarchical clustering of 42 HGSOC with immunohistopathological and genomic instability features.
| HGSOC | Cluster A | Cluster B | P‐value | ||
|---|---|---|---|---|---|
| n = 16 | n = 26 | ||||
| BRCA status , n (%) | |||||
| Familial | 9 | (56) | 21 | (81) | |
| BRCA1 | 3 | 10 | |||
| BRCA2 | 1 | 4 | 0.25 | ||
| BRCAX | 5 | 7 | |||
| Sporadic | 7 | (44) | 5 | (19) | |
| FIGO stage, n (%) | |||||
| Low (I,II) | 1 | (6.25) | 4 | (15.4) | |
| High (III,IV) | 10 | (62.5) | 16 | (61.5) | 0.62 |
| NA | 5 | (31.25) | 6 | (23.1) | |
| Genomic instability | |||||
| Median number per tumor (95% CI) | |||||
| Amplifications | 4 | (0.9–15) | 3 | (2–3.9) | 0.10 |
| Gains | 20 | (13–38) | 18 | (14.7–26) | 0.95 |
| Losses | 20 | (16–30) | 32 | (29.5–38.8) | 0.004 |
| Homozygous deletions | 2 | (0.4–6.6) | 4 | (3.3–8.9) | 0.02 |
| Median length per tumor b (95% CI) | |||||
| Gains | 393.6 | (268–545) | 269.3 | (225.6.7–354.7) | 0.12 |
| Losses | 641.5 | (445–692) | 727.1 | (658.4–841) | 0.019 a |
| Immunohistochemical markers | |||||
| Median expression c (95% CI) | |||||
| Nuclear Survivin | 14 | (8‐22) | 22 | (17.3‐29) | 0.029 |
| PR | 3 | (1‐20.5) | 21 | (17‐43) | 0.05 |
| CCNE1 | 74 | (63‐79) | 59 | (47‐64) | 0.009 a |
For categorical variables (BRCA status, FIGO stage) Fisher Exact Test was used; All the variables expressed by continuous values were compared using atwo‐tailed Student's t‐test (variables with approximate normal distribution), or by Mann–Whitney test (otherwise); P‐values <0.05 were considered statistically significant and shown in bold and italics. bExpressed in Megabases. cThe percentage of stained nuclei, independent of the intensity.
3.2. DNA copy number‐based tumor clusters and survival
We also aimed to determine whether groups of ovarian tumors defined according to their copy number features differed in terms of patients' prognosis. Association of the DNA copy number‐defined clusters with OS revealed a significant association of cluster B with longer survival (P = 0.003, Figure 2A). This association remained significant (HR = 0.15, 95%CI = 0.03‐0.81; P adj = 0.03) (Table 2) on adjustment for the covariate significantly associated with survival in this series ‐FIGO stage (Supplementary Table 3)‐ and on adjustment for other potentially confounding factors (residual tumor and age of diagnosis). To minimize the effect of non‐ovarian cancer‐related deaths we confirmed the association after censoring the data five years after diagnosis (Table 2).
Figure 2.
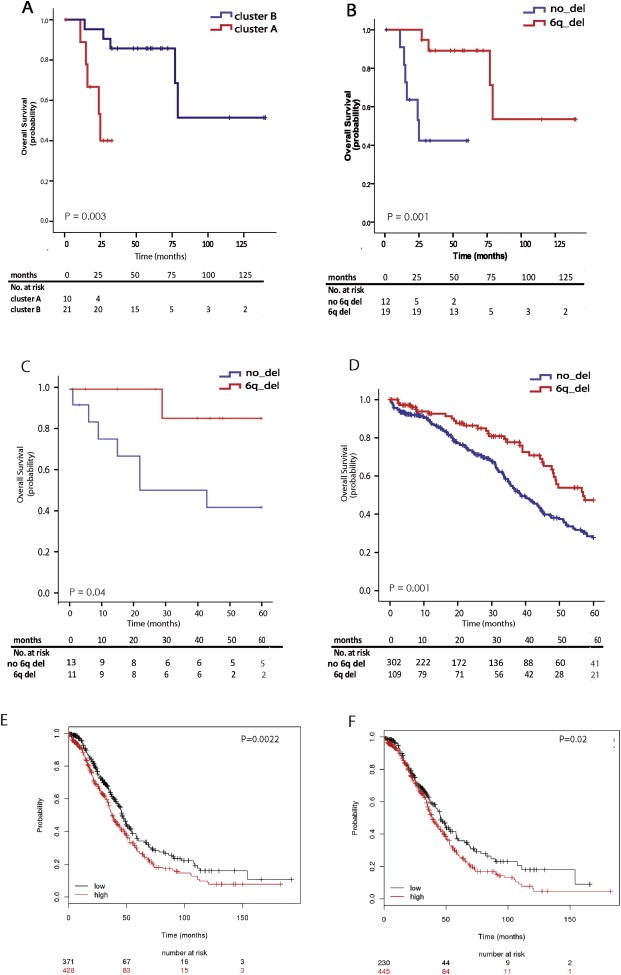
Kaplan–Meier survival curves. Overall Survival of HGSOCs from the discovery series according to (A) clusters defined by DNA copy number‐based unsupervised analysis (log‐rank P = 0.003) and (B) the presence of the 6q24.2–26 deletion (log‐rank P = 0.001). Validation of the association observed between the presence of the 6q24.2–26 deletion and improved 5‐year survival in two independent series from (C) sporadic HGSOCs where the deletion was evaluated by Fluorescence in situ Hybridization (log‐rank P = 0.04) and (D) HGSOC patients from the TCGA study (log‐rank test P = 0.001). Lower expression of the genes within 6q24.2–26 region predicts longer Overall Survival in (E) 799 HGSOCs (log‐rank P = 0.002) and in (F) 675 High‐FIGO HGSOC carcinomas (log‐rank P = 0.02), as shown using an online tool KM‐plotter from publicly‐available microarray data (Gyorffy et al., 2012).
Table 2.
Multivariate Cox regression model of prognostic factors for overall and 5‐yrs survival.
| HGSOCs | Comparison | P‐value | HR | HR (95% CI) | P adj | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Overall Survival | ||||||
| cluster B vs A | 0.003 | 0.15 | 0.03 | 0.81 | 0.03 | |
| FIGO stage | 8.32 | 1.55 | 44.61 | 0.01 | ||
| 6q24.2–6q26 | 0.001 | 0.06 | 0.01 | 0.43 | 0.005 | |
| FIGO stage | 10.80 | 1.67 | 69.70 | 0.01 | ||
| 5‐yrs survival | ||||||
| cluster B vs A | 0.004 | 0.15 | 0.03 | 0.81 | 0.03 | |
| FIGO stage | 8.29 | 1.54 | 44.75 | 0.01 | ||
| 6q24.2–6q26 | 0.002 | 0.06 | 0.01 | 0.43 | 0.005 | |
| FIGO stage | 10.79 | 1.67 | 69.81 | 0.01 | ||
P‐values in the univariate analysis calculated with log‐rank test; Padj as calculated in multivariate analysis with Cox regression model with Overall Survival (OS) and 5‐year survival as endpoints. FIGO stage (high, ≥III; low, I&II), HR, Hazard Ratio; CI, Confidence Interval, significant P‐values (<0.05) for the tested variable are highlighted in bold and italics.
Since ovarian cancer patients with germline BRCA1/2 mutations have been shown to present better survival (Alsop et al., 2012; Bolton et al., 2012) and cluster B exhibited a non‐significant enrichment in tumors from mutation carriers (Table 1), we also adjusted for this factor (in addition to FIGO stage). The association remained statistically significant (HR = 0.15, 95%CI=0.027–0.82, P adj = 0.028) indicating that BRCA1/2 mutation status would not explain the better survival of patients from the higher genomic instability cluster. In addition, we verified that the association was also significant on further adjustment for familial status (HR = 0.14, 95%CI = 0.03–0.72; P adj = 0.02). The reason for this adjustment is that some of the BRCAX familial cases (negative for mutations in the BRCA1/2 genes) might still harbor germline mutations in other BRCA‐Fanconi Anemia pathway genes, thus mimicking the phenotypes of BRCA1/2 germline carriers. Moreover the association was proved significant on further adjustment for potential confounders, that showed different distribution between the clusters and have been previously associated with clinical outcome in HGSOCs, such as progesterone receptor (HR = 0.13, 95%CI=0. 021–0.79, P adj = 0.027) and CCNE1 expression (HR = 0.045, 95%CI=0. 003–0.68, P adj = 0.025).
We next sought to define specific copy number changes that might explain the observed association. To this end we defined 32 the regions that differentiated the clusters (at least 35% frequency difference, P‐value <0.05). Out of these regions, six passed the multiple testing correction (FDR <0.05) and were subsequently tested for their association with survival (Table 3). Only a deletion at 6q24.2–6q26 (145,593,087–162,867,181), more frequent in cluster B, was found to be significantly associated with longer survival (P = 0.001) (Table 2 and Figure 2B). This association remained significant after adjustment for FIGO stage (HR = 0.06, 95%CI=0.01–0.43, P adj = 0.005) (Table 2) and on adjustment for other potentially confounding factors (age of diagnosis, residual tumor, BRCA1/2 mutation status and PR expression) and also at 5‐year survival endpoint (Table 2).
Table 3.
List of regions differentiating the clusters defined in HGSOCs.
| Cytoband | Start [bp] | End [bp] | Size [bp] | Nr of genes | Frequency in cluster A [%] | Frequency in cluster B [%] | Difference | Cancer gene census | P | HR | 95% CI | P adj | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6q24.2 – q26 | 145,593,087 | 162,867,181 | 17,274,094 | 169 | 14.29 | 75.55 | 61.26 | 0.001 | 0.06 | 0.01 | 0.43 | 0.005 | |
| 11p15.5–15.3 | 0 | 12,467,187 | 12,467,187 | 393 | 9.56 | 67.65 | 58.09 | HRAS CARS NUP98 LMO1 | 0.07 | 0.30 | 0.06 | 1.41 | 0.13 |
| 16q21 | 62,541,693 | 64,974,691 | 2,432,998 | 4 | 25.00 | 76.92 | 51.92 | CDH11 | 0.71 | 0.62 | 0.15 | 2.60 | 0.51 |
| 19q13.31 – q13.32 | 48,908,403 | 51,085,851 | 2,177,448 | 73 | 1.56 | 51.92 | 50.36 | BCL3 CBLC ERCC2 | 0.17 | 0.31 | 0.04 | 2.34 | 0.26 |
| 22q12.3 – q13.1 | 35,051,869 | 37,251,402 | 2,199,533 | 110 | 25.00 | 78.85 | 53.85 | MYH9 | 0.56 | 0.25 | 0.04 | 1.56 | 0.14 |
| 22q13.2 – q13.31 | 39,940,439 | 46,673,931 | 6,733,492 | 105 | 26.25 | 80.77 | 54.52 | 0.72 | 0.53 | 0.08 | 3.30 | 0.49 | |
Regions differentiating clusters (Clusters A and B) in the high‐grade serous ovarian carcinomas (HGSOCs) defined to be at least 35% more frequent in one of the clusters and significant after correction for multiple testing (FDR <0.05); all the regions were tested for the association with overall survival (OS), highlighted regions were significantly associated with better prognosis (P < 0.05), P‐values‐calculated in the univariate analysis with the log‐rank test; Padj as calculated in multivariate Cox regression model in which significant covariates were included; HR, Hazard Ratio; CI, Confidence Intervals.
3.3. Validation of the prognostic value of 6q24.2–q26 in an independent ovarian cancer series by FISH analysis
In order to validate our findings we performed FISH analysis using a 6q25.1 probe (Figure 3) in an independent series of 36 HGSOCs. Twenty‐eight tumors were successfully hybridized. The deletion was detected in 54% of successfully hybridized cases and was associated with significantly better overall survival (P = 0.04) and 5yrs survival (P = 0.03). Since none of the potential confounding factors were significant in this series, the multivariate cox was not carried out, however in order to minimize the influence of the BRCA mutation and familial status the 5yrs prognostic value was confirmed in sporadic cases alone (HR = 0.15, 95%CI = 0.19–1.2, P = 0.04) (Figure 2C).
Figure 3.
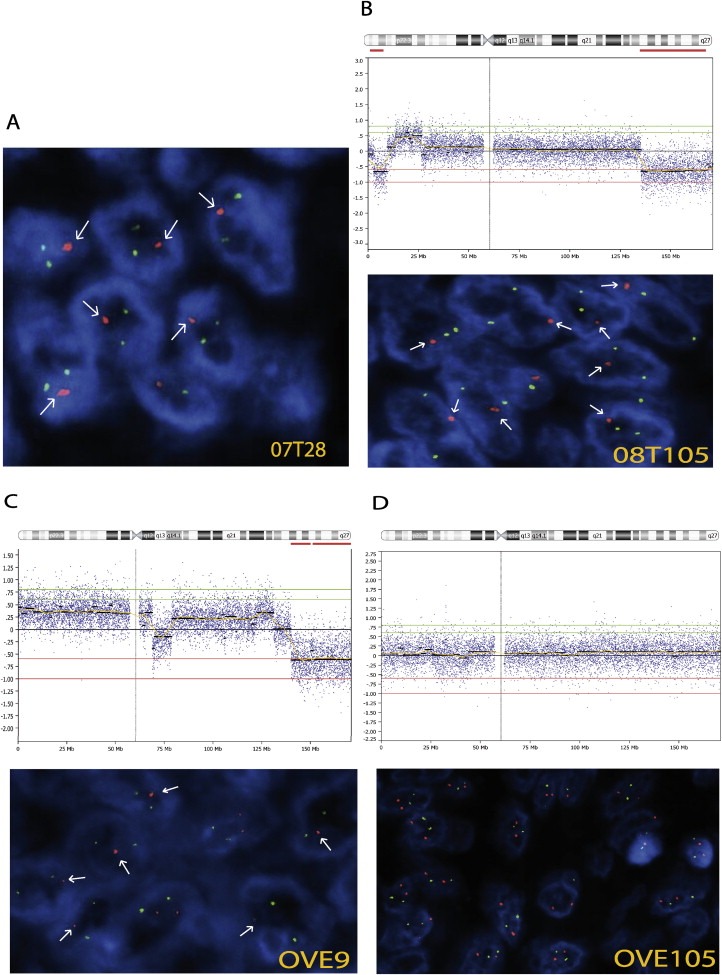
Fluorescence in situ Hybridization (FISH) on tumor sections. Evaluation of the 6q24.2–26 deletion by Fluorescence in situ Hybridization (FISH) on paraffin‐embedded tissue sections. Test (red) and reference (green) probes mapped to 6q25.1 and 6p21, respectively. Presence of one single red signal is indicated with an arrow. (A) Tumor from validation series showing deletion at 6q25.1. (B, C, D) Chromosome 6 array‐CGH profiles (top panels) of tumors from the discovery series and corresponding FISH analysis (lower panels) confirming the presence of the deletion (B, C) and normal DNA copy number at this locus (D). Magnification: 100×.
3.4. Validation of the prognostic value of the 6q24–26 deletion in the TCGA ovarian cancer series
We aimed to further validate the prognostic value of the 6q24.2–q26 deletion in a larger homogeneous group of 411 HGSOCs from TCGA ovarian cancer study (TCGA, 2011). Normalized log2 ratios from 1 M Agilent Sure Print Human Microarray platform were downloaded from TCGA website (https://tcga‐data.nci.nih.gov/tcga/dataAccessMatrix.htm) and subjected to the segmentation and calling algorithms using CGHcall R package and following the pipeline applied for our discovery series (Kamieniak et al., 2013). The previously defined region of loss at 6q24.2–q26 (as described in Section 2.4) was also called as “deleted” in more than 35% of TCGA series. Tumors were considered deletion‐positive if at least 90% of the defined region (6q24.2–q26, 145,593,087–162,867,181) was lost (regardless the exact location of the lost segment within the region boundaries). The deletion was associated with overall (P = 0.002) and 5yrs survival advantage (P = 0.001) (Figure 2D) in the univariate analysis. The association remained significant after adjustment for significant confounders such as FIGO stage, BRCA1/2 mutation status and age at diagnosis for OS (HR = 0.67, 95%CI = 0.48–0.93, P adj = 0.019) and also for 5‐years time point (HR = 0.61, 95%CI = 0.41–0.89, P adj = 0.010). Further adjustment for debulking status produced consistent results.
3.5. Expression of genes at 6q24.2–q26 and survival
Assuming that copy number status has an impact on mRNA level, we evaluated the prognostic value of the deletion at the gene expression level using KM‐plotter (Gyorffy et al., 2012), which integrates gene expression and clinical data from 10 different data sets for 1436 EOCs patients. We found that low mean expression of all the genes within the 6q24.2–26 region with available data in the KM‐plotter tool (53 out of 85 RefSeq genes) was associated with longer OS in 799 HGSOCs (HR = 0.74, 95%CI = 0.61–0.90, log‐rank P = 0.002) (Figure 2E) and on limiting the analysis to 675 high‐FIGO stage HGSOCs (HR = 0.76, 95%CI = 0.61–0.96, log‐rank P = 0.02) (Figure 2F). Similar results were obtained following censoring of survival data at 5‐years endpoint. In addition, to account for the effect of confounding factors, the association was confirmed separately in stratified groups according to debulking status (optimally and suboptimally debulked tumors).
In order to propose individual genes that might explain the observed association, we selected only those whose copy number status had an impact on the expression level. To address this, we used TCGA study, as it allowed us to combine copy number and expression data for the majority of the genes from the region (81 out of 85 RefSeq genes), which was not possible with KM‐plotter. By using 232 HGSOCs with accessible copy number and RNAseq data from TCGA study we found that 76% of the examined genes in the region (62 out of 81) were significantly down‐regulated when lost (FDR <0.05) (Supplementary Figure 1). Of these, multivariate Cox regression analysis identified four genes whose downregulation was significantly associated (P adj < 0.05) with better survival independently of known prognostic factors (FIGO stage, age at diagnosis, and BRCA1/2 mutation status) and nine additional genes that showed borderline associations (P adj < 0.15) (Table 4).
Table 4.
List of the genes from the region whose expression was associated with overall survival.
| Gene symbol | Gene name | HR | HR (95% CI) | Padj | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| SLC22A2 | solute carrier family 22, member 2 | 1.34 | 1.17 | 1.54 | <0.0001 |
| ARID1B | AT rich interactive doma in 1B (SWI1‐like) | 1.18 | 1.02 | 1.36 | 0.029 |
| SLC22A3 | solute carrier family 22, member 3 | 1.17 | 1.01 | 1.36 | 0.040 |
| SAMD5 | sterile alpha motif domain containing 5 | 1.17 | 1.01 | 1.37 | 0.043 |
| GRM1 | glutamate receptor, metabotropic 1 | 1.16 | 0.99 | 1.37 | 0.07 |
| TAB2 | TGF‐beta activated kinase 1/MAP3K7 binding protein 2 | 1.13 | 0.98 | 1.30 | 0.08 |
| PPIL4 | peptidylprolyl isomerase (cyclophilin)‐like 4 | 1.15 | 0.98 | 1.35 | 0.08 |
| TIAM2 | T‐cell lymphoma invasion and metastasis 2 | 1.13 | 0.98 | 1.32 | 0.09 |
| AKAP12 | A kinase (PRKA) anchor protein 12 | 1.13 | 0.98 | 1.31 | 0.09 |
| GTF2H5 | general transcription factor IIH, polypeptide 5 | 1.15 | 0.98 | 1.36 | 0.10 |
| ULBP1 | UL16 binding protein 1 | 1.12 | 0.98 | 1.29 | 0.10 |
| SASH1 | SAM and SH3 domain containing 1 | 1.11 | 0.98 | 1.26 | 0.10 |
| SHPRH | SNF2 histone linker PHD RING helicase | 1.13 | 0.08 | 1.48 | 0.14 |
P adj ‐P‐values calculated with Cox proportional hazard model for each gene individually adjusting for cofactors (FIGO stage, age of diagnosis and BRCA1/2 mutation status), significant P‐values (<0.05) are highlighted in italics and significant genes in bold; HR, Hazard Ratio, CI, Confidence Interval.
4. Discussion
HGSOCs remain a major cause of gynecological‐related deaths and the identification of novel molecular markers that may explain the different clinical behavior of HGSOC patients is of critical importance. Novel prognostic and predictive markers may not only assist in treatment decision‐making, but may eventually lead to the development of more effective therapies. DNA copy number alterations have been shown to be a hallmark of HGSOCs (TCGA, 2011) and might be potentially used as molecular predictors of patient outcomes. In this study, we have investigated whether stratification of HGSOCs on the basis of DNA copy number may delineate novel categories of tumors with different underlying biology, as defined by a distinct immunostaining pattern and more importantly, by a different clinical outcome.
Consistent with previous results from our group (Kamieniak et al., 2013) the present study indicates that DNA copy number‐based clustering would not stratify HGSOCs according to their sporadic or familial condition. However, higher extent of genome loss seems to define a cluster of HGSOCs enriched in BRCA1/2 mutation carriers (and possibly BRCA‐like sporadic tumors) and characterized by the features associated with those BRCA1/2 tumors, such as: greater expression of progesterone receptor (Munoz‐Repeto et al., 2013), higher proliferation rate as defined here by survivin expression (Fields et al., 2004) and mutual exclusivity with CCNE1 amplification (TCGA, 2011) as reflected here by immunohistochemical staining.
Besides differences in the immunohistopathological features, the DNA copy number‐defined tumor groups differed also regarding their survival outcome. Importantly, this difference was statistically significant after accounting for all specific clinicopatholigcal features, that have a potential or already well defined effect on survival of ovarian cancer patients. Thus, the association of one of the clusters with better survival was maintained significant after adjustment for: BRCA1/2 mutation status, that is an established marker of improved overall survival and better response to platinum‐based therapy (Alsop et al., 2012; Bolton et al., 2012); expression of progesterone receptor that has been associated with longer survival in HGSOCs (Sieh et al., 2013) and also after adjustment for CCNE1 expression, although its association with worse clinical outcome is not certain, once its inverse correlation with BRCA1/2 mutation status is considered (TCGA, 2011).
Examination of distinct features characterizing the cluster of better survival led us to define a specific copy number loss at 6q24.2–26 that was the alteration the most significantly associated with this cluster and the only one associated with better survival. This result was replicated in two independent series and the prognostic value of this loss was confirmed after adjustment, for all the other variables, among them for BRCA1/2 mutation status. Therefore, the association between the 6q24.2–26 loss and outcome appears to be driven by mechanisms other than those proposed to mediate the survival advantage of BRCA1/2 mutation carriers (Alsop et al., 2012; Bolton et al., 2012).
The long arm of chromosome 6 is frequently altered in many human malignances and loss of 6q24–26 has been studied as potential location for genes with a tumor suppressor role (Hayashi et al., 2012; Sun et al., 2003). However, only a very few reports have associated this loss with clinical outcome, reaching contradictory results depending on the cancer type. In most of them the deletion was associated with poor prognosis and tumor recurrence (Cui et al., 2011; Fischer et al., 2004; Letessier et al., 2007; Schwaenen et al., 2009), however there were also a few studies that indicated the favorable outcome (Dalsass et al., 2013; Monoranu et al., 2008; Pfister et al., 2009). Such inconsistent findings may be related to small study size, heterogeneity of the tumors and most importantly lack of independent validation series (Bruchim et al., 2009; Yamamoto et al., 2009). In the TCGA study the only 6q loss defined among the 50 focal losses was 6q27, but not 6q24.2–26. This might be due to different methodology and composition of the tumor series, but more importantly, 6q24.2–26 in our series was not defined based on recurrence in HGSOCs, but from the comparison of genomic alterations between tumor clusters, that were found to be associated with survival. Of interest, in a recent high‐throughput data‐based study, a regional loss at 6q15–q27 was defined among 8 copy number losses that significantly affected candidate pathways associated with chemo‐response in epithelial ovarian cancer (Gonzalez Bosquet et al., 2014).
We showed that 6q24.2–26 loss has an impact on gene expression, as evidenced by downregulation of 76% genes from the lost region and significant association of low expression of the genes from the region with longer survival. We also pointed several candidate genes that might explain the survival association.
One of the candidate genes is the DNA‐binding subunit of SWI/SNF chromatin remodeling complex ARID1B. The presence of this gene in the complex is mutually exclusive with ARID1A, which has been found to be mutated in clear cell and endometrioid ovarian carcinomas (Wiegand et al., 2010). Among other processes, the SWI/SNF complexes are involved in double‐strand break repair (Ogiwara et al., 2011; Park et al., 2009) and downregulation of some of their key components has been shown to modulate cisplatin cytotoxicity (Kothandapani et al., 2012). Therefore, it might be plausible that compromised expression of ARID1B due to the 6q24.2–26 deletion could enhance cancer cells sensitization to cytotoxic drugs and contribute to explain the association found in the present study. Similar effect might be attributed to reduced expression of other DNA‐repair gene found in the region, such as the General Transcription factor IIH Polypeptide 5 (GTF2H5), which has an essential role in nucleotide excision repair (Theil et al., 2013). In addition, loss of the SNF2 histone linker PHD RING helicase (SHPRH) that plays a crucial role in an error‐free repair of stalled replication forks, caused by DNA damage, may also sensitize cells to genotoxic stress (Motegi et al., 2008).
An individual loss of the above mentioned, or of other genes in the deleted region, might indeed have an impact on patient survival; however, it seems likely that it is not just a single gene that needs to be deleted to influence tumor progression, but rather a combination of them. Moreover we should not overlook the role of non‐coding DNA fragments such as regulatory elements, miRNAs or other functionally important sequences that might be completely lost within the region or whose function might be disrupted at its boundaries. Also, the possibility that the deletion might cause dysregulation of flanking genes cannot be excluded.
5. Conclusions
In this study, we have demonstrated in different tumor series, at DNA copy number and at the gene expression level, that the 6q24–26 deletion is an independent marker of favorable outcome in HGSOCs. These findings have potentially relevant clinical value, as this marker could help to guide the selection of patients, whose favorable prognosis would support the use of new treatment regimens focused on improving tolerability without jeopardizing efficacy. Importantly, DNA‐based markers that can be analyzed by FISH, as we have demonstrated for this deletion, are particularly suitable for routine clinical applications due to their robustness and suitability for use with paraffin sections. In addition, the deletion, together with other emerging prognostic and predictive biomarkers in ovarian cancer, could be used to better balance patients between treatment arms in clinical trials. Future research should be dedicated to the prospective validation of this marker and further characterization of tumors that carry the deletion, as well as of defining the role of the genes in the region. This may not only offer insights into tumor biology, but may eventually lead to the development of effective targeted therapies.
Conflict of interests
The authors declare that they have no conflict of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary Figure 1 Association between copy number and gene expression level. Association between copy number status of 81 genes from the 6q24.2–26 region and their expression level (based on RPKM values) from TCGA studies. Fold change (log‐ratio) in gene expression between cases with loss vs. normal copy number status at this locus on the y‐axis; genomic location on the 6q chromosome on the x‐axis; significant changes (FDR < 0.05) are marked in red.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
This study was financially supported by the Fondo de Investigación Sanitaria (FIS), Instituto de Salud Carlos III (grants PS09/01094 and PI12/01319). MJG is recipient of a research contract from the Instituto de Salud Carlos III of the Ministerio Español de Sanidad y Consumo (Miguel Servet tipo II Program, CPII 13/00047). MMK and KI have financial support from the Fundación La Caixa. DR and KI are supported by grant BIO2012‐40205. LP‐A, JCC and JP are recipients of financial support from Red Temática de Investigación Cooperativa en Cáncer (RTICC) (grants RD12/0036/0028, RD12/0036/0037 and RD12/0036/0064).
We want to thank to María del Carmen González‐Neira and Victoria Fernández (from the CNIO Human Cancer Genetics group) and to the CNIO Tumor Bank Network, in particular to María Jesús Artiga, for exceptional assistance in the retrieval of clinical data. We also thank the CNIO Histology and Immunohistochemistry Core Unit, especially Lydia Sánchez for excellent support. We are very grateful to Miguel Vázquez from the CNIO Structural Computational Biology Group for helpful discussion and assistance with data management. We acknowledge the contribution of pathologist Beatriz López Martínez Bernal from Instituto de Investigación Sanitaria Gregorio Marañón who provided some of the tissue samples used in this study. Special thanks to the ASACO association of ovarian cancer patients for their support and enthusiastic collaboration. Finally, we express our gratitude to Professor Ahmed Ahmed for critically reviewing the manuscript and for his valuable comments.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.09.010.
Kamieniak Marta M., Rico Daniel, Milne Roger L., Muñoz-Repeto Ivan, Ibáñez Kristina, Grillo Miguel A., Domingo Samuel, Borrego Salud, Cazorla Alicia, García-Bueno José M., Hernando Susana, García-Donas Jesús, Hernández-Agudo Elena, y Cajal Teresa Ramón, Robles-Díaz Luis, Márquez-Rodas Ivan, Cusidó Maite, Sáez Raquel, Lacambra-Calvet Carmen, Osorio Ana, Urioste Miguel, Cigudosa Juan C., Paz-Ares Luis, Palacios José, Benítez Javier, García María J., (2015), Deletion at 6q24.2–26 predicts longer survival of high‐grade serous epithelial ovarian cancer patients, Molecular Oncology, 9, doi: 10.1016/j.molonc.2014.09.010.
References
- Abdollahi, A. , Pisarcik, D. , Roberts, D. , Weinstein, J. , Cairns, P. , Hamilton, T.C. , 2003. LOT1 (PLAGL1/ZAC1), the candidate tumor suppressor gene at chromosome 6q24-25, is epigenetically regulated in cancer. J. Biol. Chem. 278, 6041–6049. [DOI] [PubMed] [Google Scholar]
- Alsop, K. , Fereday, S. , Meldrum, C. , Defazio, A. , Emmanuel, C. , George, J. , Dobrovic, A. , Birrer, M.J. , Webb, P.M. , Stewart, C. , Friedlander, M. , Fox, S. , Bowtell, D. , Mitchell, G. , 2012. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian Cancer: a report from the Australian ovarian Cancer study group. J. Clin. Oncol. 30, 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali, A. , O'Brien, P.M. , Edwards, L.S. , Sutherland, R.L. , Hacker, N.F. , Henshall, S.M. , 2004. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin. Cancer Res. 10, 5168–5177. [DOI] [PubMed] [Google Scholar]
- Baumbusch, L.O. , Helland, A. , Wang, Y. , Liestol, K. , Schaner, M.E. , Holm, R. , Etemadmoghadam, D. , Alsop, K. , Brown, P. , Mitchell, G. , Fereday, S. , DeFazio, A. , Bowtell, D.D. , Kristensen, G.B. , Lingjaerde, O.C. , Borresen-Dale, A.L. , 2013. High levels of genomic aberrations in serous ovarian cancers are associated with better survival. PLoS One. 8, e54356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns, E.M. , Bowtell, D.D. , 2012. The changing view of high-grade serous ovarian cancer. Cancer Res. 72, 2701–2704. [DOI] [PubMed] [Google Scholar]
- Bolton, K.L. , Chenevix-Trench, G. , Goh, C. , Sadetzki, S. , Ramus, S.J. , Karlan, B.Y. , Lambrechts, D. , Despierre, E. , Barrowdale, D. , McGuffog, L. , Healey, S. , Easton, D.F. , Sinilnikova, O. , Benitez, J. , Garcia, M.J. , Neuhausen, S. , Gail, M.H. , Hartge, P. , Peock, S. , Frost, D. , Evans, D.G. , Eeles, R. , Godwin, A.K. , Daly, M.B. , Kwong, A. , Ma, E.S. , Lazaro, C. , Blanco, I. , Montagna, M. , D'Andrea, E. , Nicoletto, M.O. , Johnatty, S.E. , Kjaer, S.K. , Jensen, A. , Hogdall, E. , Goode, E.L. , Fridley, B.L. , Loud, J.T. , Greene, M.H. , Mai, P.L. , Chetrit, A. , Lubin, F. , Hirsh-Yechezkel, G. , Glendon, G. , Andrulis, I.L. , Toland, A.E. , Senter, L. , Gore, M.E. , Gourley, C. , Michie, C.O. , Song, H. , Tyrer, J. , Whittemore, A.S. , McGuire, V. , Sieh, W. , Kristoffersson, U. , Olsson, H. , Borg, A. , Levine, D.A. , Steele, L. , Beattie, M.S. , Chan, S. , Nussbaum, R.L. , Moysich, K.B. , Gross, J. , Cass, I. , Walsh, C. , Li, A.J. , Leuchter, R. , Gordon, O. , Garcia-Closas, M. , Gayther, S.A. , Chanock, S.J. , Antoniou, A.C. , Pharoah, P.D. , 2012. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 307, 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell, D.D. , 2010. The genesis and evolution of high-grade serous ovarian cancer. Nat. Rev. Cancer. 10, 803–808. [DOI] [PubMed] [Google Scholar]
- Bruchim, I. , Israeli, O. , Mahmud, S.M. , Aviram-Goldring, A. , Rienstein, S. , Friedman, E. , Ben-Baruch, G. , Gotlieb, W.H. , 2009. Genetic alterations detected by comparative genomic hybridization and recurrence rate in epithelial ovarian carcinoma. Cancer Genet. Cytogenet. 190, 66–70. [DOI] [PubMed] [Google Scholar]
- Brun, J.L. , Cortez, A. , Commo, F. , Uzan, S. , Rouzier, R. , Darai, E. , 2008. Serous and mucinous ovarian tumors express different profiles of MMP-2, -7, -9, MT1-MMP, and TIMP-1 and -2. Int. J. Oncol. 33, 1239–1246. [PubMed] [Google Scholar]
- Burkhardt, B. , Bruch, J. , Zimmermann, M. , Strauch, K. , Parwaresch, R. , Ludwig, W.D. , Harder, L. , Schlegelberger, B. , Mueller, F. , Harbott, J. , Reiter, A. , 2006. Loss of heterozygosity on chromosome 6q14-q24 is associated with poor outcome in children and adolescents with T-cell lymphoblastic lymphoma. Leukemia. 20, 1422–1429. [DOI] [PubMed] [Google Scholar]
- Cannistra, S.A. , 2004. Cancer of the ovary. N. Engl. J. Med. 351, 2519–2529. [DOI] [PubMed] [Google Scholar]
- Cui, R. , Okada, Y. , Jang, S.G. , Ku, J.L. , Park, J.G. , Kamatani, Y. , Hosono, N. , Tsunoda, T. , Kumar, V. , Tanikawa, C. , Kamatani, N. , Yamada, R. , Kubo, M. , Nakamura, Y. , Matsuda, K. , 2011. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 60, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsass, A. , Mestichelli, F. , Ruggeri, M. , Gaspari, P. , Pezzoni, V. , Vagnoni, D. , Angelini, M. , Angelini, S. , Bigazzi, C. , Falcioni, S. , Troiani, E. , Alesiani, F. , Catarini, M. , Attolico, I. , Scortechini, I. , Discepoli, G. , Galieni, P. , 2013. 6q deletion detected by fluorescence in situ hybridization using bacterial artificial chromosome (BAC) in chronic lymphocytic leukemia. Eur. J. Haematol. 91, 10–19. [DOI] [PubMed] [Google Scholar]
- Denison, S.R. , Wang, F. , Becker, N.A. , Schule, B. , Kock, N. , Phillips, L.A. , Klein, C. , Smith, D.I. , 2003. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 22, 8370–8378. [DOI] [PubMed] [Google Scholar]
- Engler, D.A. , Gupta, S. , Growdon, W.B. , Drapkin, R.I. , Nitta, M. , Sergent, P.A. , Allred, S.F. , Gross, J. , Deavers, M.T. , Kuo, W.L. , Karlan, B.Y. , Rueda, B.R. , Orsulic, S. , Gershenson, D.M. , Birrer, M.J. , Gray, J.W. , Mohapatra, G. , 2012. Genome wide DNA copy number analysis of serous type ovarian carcinomas identifies genetic markers predictive of clinical outcome. PLoS One. 7, e30996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay, J. , Shin, H.R. , Bray, F. , Forman, D. , Mathers, C. , Parkin, D.M. , 2008. Estimates of worldwide burden of cancer in 2008: GLOBOCAN. Int. J. Cancer. 127, 2893–2917. [DOI] [PubMed] [Google Scholar]
- Fields, A.C. , Cotsonis, G. , Sexton, D. , Santoianni, R. , Cohen, C. , 2004. Survivin expression in hepatocellular carcinoma: correlation with proliferation, prognostic parameters, and outcome. Mod. Pathol.17, 1378–1385. [DOI] [PubMed] [Google Scholar]
- Fischer, T.C. , Gellrich, S. , Muche, J.M. , Sherev, T. , Audring, H. , Neitzel, H. , Walden, P. , Sterry, W. , Tonnies, H. , 2004. Genomic aberrations and survival in cutaneous T cell lymphomas. J. Invest. Dermatol.122, 579–586. [DOI] [PubMed] [Google Scholar]
- Foulkes, W.D. , Campbell, I.G. , Stamp, G.W. , Trowsdale, J. , 1993. Loss of heterozygosity and amplification on chromosome 11q in human ovarian cancer. Br. J. Cancer. 67, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Bosquet, J. , Marchion, D.C. , Chon, H. , Lancaster, J.M. , Chanock, S. , 2014. Analysis of chemotherapeutic response in ovarian cancers using publically available high-throughput data. Cancer Res.74, 3902–3912. [DOI] [PubMed] [Google Scholar]
- Gorringe, K.L. , Campbell, I.G. , 2009. Large-scale genomic analysis of ovarian carcinomas. Mol. Oncol.3, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Yanna, Ma, X. , An, J. , Shang, Y. , Huang, Q. , Yang, H. , Chen, Z. , Xing, J. , 2011. A meta-analysis of array-CGH studies implicates antiviral immunity pathways in the development of hepatocellular carcinoma. PLoS One. 6, e28404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy, B. , Lanczky, A. , Szallasi, Z. , 2012. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 19, 197–208. [DOI] [PubMed] [Google Scholar]
- Hayashi, M. , Nomoto, S. , Kanda, M. , Okamura, Y. , Nishikawa, Y. , Yamada, S. , Fujii, T. , Sugimoto, H. , Takeda, S. , Kodera, Y. , 2012. Identification of the A kinase anchor protein 12 (AKAP12) gene as a candidate tumor suppressor of hepatocellular carcinoma. J. Surg. Oncol.105, 381–386. [DOI] [PubMed] [Google Scholar]
- Honrado, E. , Osorio, A. , Palacios, J. , Milne, R.L. , Sanchez, L. , Diez, O. , Cazorla, A. , Syrjakoski, K. , Huntsman, D. , Heikkila, P. , Lerma, E. , Kallioniemi, A. , Rivas, C. , Foulkes, W.D. , Nevanlinna, H. , Benitez, J. , 2005. Immunohistochemical expression of DNA repair proteins in familial breast cancer differentiate BRCA2-associated tumors. J. Clin. Oncol.23, 7503–7511. [DOI] [PubMed] [Google Scholar]
- Kalloger, S.E. , Kobel, M. , Leung, S. , Mehl, E. , Gao, D. , Marcon, K.M. , Chow, C. , Clarke, B.A. , Huntsman, D.G. , Gilks, C.B. , 2011. Calculator for ovarian carcinoma subtype prediction. Mod. Pathol.24, 512–521. [DOI] [PubMed] [Google Scholar]
- Kamieniak, M.M. , Munoz-Repeto, I. , Rico, D. , Osorio, A. , Urioste, M. , Garcia-Donas, J. , Hernando, S. , Robles-Diaz, L. , Ramon, Y.C.T. , Cazorla, A. , Saez, R. , Garcia-Bueno, J.M. , Domingo, S. , Borrego, S. , Palacios, J. , van de Wiel, M.A. , Ylstra, B. , Benitez, J. , Garcia, M.J. , 2013. DNA copy number profiling reveals extensive genomic loss in hereditary BRCA1 and BRCA2 ovarian carcinomas. Br. J. Cancer. 108, 1732–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel, M. , Kalloger, S.E. , Carrick, J. , Huntsman, D. , Asad, H. , Oliva, E. , Ewanowich, C.A. , Soslow, R.A. , Gilks, C.B. , 2009. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am. J. Surg. Pathol.33, 14–21. [DOI] [PubMed] [Google Scholar]
- Kothandapani, A. , Gopalakrishnan, K. , Kahali, B. , Reisman, D. , Patrick, S.M. , 2012. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp. Cell Res.318, 1973–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier, A. , Garrido-Urbani, S. , Ginestier, C. , Fournier, G. , Esterni, B. , Monville, F. , Adelaide, J. , Geneix, J. , Xerri, L. , Dubreuil, P. , Viens, P. , Charafe-Jauffret, E. , Jacquemier, J. , Birnbaum, D. , Lopez, M. , Chaffanet, M. , 2007. Correlated break at PARK2/FRA6E and loss of AF-6/Afadin protein expression are associated with poor outcome in breast cancer. Oncogene. 26, 298–307. [DOI] [PubMed] [Google Scholar]
- Leunen, K. , Gevaert, O. , Daemen, A. , Vanspauwen, V. , Michils, G. , De Moor, B. , Moerman, P. , Vergote, I. , Legius, E. , 2009. Recurrent copy number alterations in BRCA1-mutated ovarian tumors alter biological pathways. Hum. Mutat.30, 1693–1702. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, D. , Wang, L. , Yu, J. , Du, D. , Chen, Y. , Gao, P. , Wang, D.M. , Zhang, F. , Fu, S. , 2013. Distinct genomic aberrations between low-grade and high-grade gliomas of Chinese patients. PLoS One. 8, e57168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica, A. , Deavers, M.T. , Lu, K. , Bodurka, D.C. , Atkinson, E.N. , Gershenson, D.M. , Silva, E.G. , 2004. Grading ovarian serous carcinoma using a two-tier system. Am. J. Surg. Pathol.28, 496–504. [DOI] [PubMed] [Google Scholar]
- Monoranu, C.M. , Huang, B. , Zangen, I.L. , Rutkowski, S. , Vince, G.H. , Gerber, N.U. , Puppe, B. , Roggendorf, W. , 2008. Correlation between 6q25.3 deletion status and survival in pediatric intracranial ependymomas. Cancer Genet. Cytogenet.182, 18–26. [DOI] [PubMed] [Google Scholar]
- Motegi, A. , Liaw, H.J. , Lee, K.Y. , Roest, H.P. , Maas, A. , Wu, X. , Moinova, H. , Markowitz, S.D. , Ding, H. , Hoeijmakers, J.H. , Myung, K. , 2008. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U S A. 105, 12411–12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Repeto, I. , Garcia, M.J. , Kamieniak, M. , Ramon, Y.C.T. , Domingo, S. , Cazorla, A. , Garcia Donas, J. , Hernando Polo, S. , Garcia Sagredo, J.M. , Hernandez, E. , Lacambra, C. , Saez, R. , Robles, L. , Borrego, S. , Prat, J. , Palacios, J. , Benitez, J. , 2013. Phenotypic characterization of hereditary epithelial ovarian cancer based on a tissue microarray study. Histol. Histopathol.28, 133–144. [DOI] [PubMed] [Google Scholar]
- Nelson, M. , Horsman, D.E. , Weisenburger, D.D. , Gascoyne, R.D. , Dave, B.J. , Loberiza, F.R. , Ludkovski, O. , Savage, K.J. , Armitage, J.O. , Sanger, W.G. , 2008. Cytogenetic abnormalities and clinical correlations in peripheral T-cell lymphoma. Br. J. Haematol.141, 461–469. [DOI] [PubMed] [Google Scholar]
- Ni, X. , Zhang, W. , Huang, K.C. , Wang, Y. , Ng, S.K. , Mok, S.C. , Berkowitz, R.S. , Ng, S.W. , 2004. Characterisation of human kallikrein 6/protease M expression in ovarian cancer. Br. J. Cancer. 91, 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara, H. , Ui, A. , Otsuka, A. , Satoh, H. , Yokomi, I. , Nakajima, S. , Yasui, A. , Yokota, J. , Kohno, T. , 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 30, 2135–2146. [DOI] [PubMed] [Google Scholar]
- Orphanos, V. , McGown, G. , Hey, Y. , Thorncroft, M. , Santibanez-Koref, M. , Russell, S.E. , Hickey, I. , Atkinson, R.J. , Boyle, J.M. , 1995. Allelic imbalance of chromosome 6q in ovarian tumours. Br. J. Cancer. 71, 666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.H. , Park, E.J. , Hur, S.K. , Kim, S. , Kwon, J. , 2009. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst). 8, 29–39. [DOI] [PubMed] [Google Scholar]
- Pfister, S. , Remke, M. , Benner, A. , Mendrzyk, F. , Toedt, G. , Felsberg, J. , Wittmann, A. , Devens, F. , Gerber, N.U. , Joos, S. , Kulozik, A. , Reifenberger, G. , Rutkowski, S. , Wiestler, O.D. , Radlwimmer, B. , Scheurlen, W. , Lichter, P. , Korshunov, A. , 2009. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J. Clin. Oncol.27, 1627–1636. [DOI] [PubMed] [Google Scholar]
- Piccart, M.J. , Lamb, H. , Vermorken, J.B. , 2001. Current and future potential roles of the platinum drugs in the treatment of ovarian cancer. Ann. Oncol.12, 1195–1203. [DOI] [PubMed] [Google Scholar]
- Ramus, S.J. , Pharoah, P.D. , Harrington, P. , Pye, C. , Werness, B. , Bobrow, L. , Ayhan, A. , Wells, D. , Fishman, A. , Gore, M. , DiCioccio, R.A. , Piver, M.S. , Whittemore, A.S. , Ponder, B.A. , Gayther, S.A. , 2003. BRCA1/2 mutation status influences somatic genetic progression in inherited and sporadic epithelial ovarian cancer cases. Cancer Res.63, 417–423. [PubMed] [Google Scholar]
- Rosen, E.M. , Fan, S. , Ma, Y. , 2006. BRCA1 regulation of transcription. Cancer Lett.236, 175–185. [DOI] [PubMed] [Google Scholar]
- Saito, S. , Saito, H. , Koi, S. , Sagae, S. , Kudo, R. , Saito, J. , Noda, K. , Nakamura, Y. , 1992. Fine-scale deletion mapping of the distal long arm of chromosome 6 in 70 human ovarian cancers. Cancer Res.52, 5815–5817. [PubMed] [Google Scholar]
- Schindlbeck, C. , Hantschmann, P. , Zerzer, M. , Jahns, B. , Rjosk, D. , Janni, W. , Rack, B. , Sommer, H. , Friese, K. , 2007. Prognostic impact of KI67, p53, human epithelial growth factor receptor 2, topoisomerase IIalpha, epidermal growth factor receptor, and nm23 expression of ovarian carcinomas and disseminated tumor cells in the bone marrow. Int. J. Gynecol. Cancer. 17, 1047–1055. [DOI] [PubMed] [Google Scholar]
- Schmandt, R.E. , Broaddus, R. , Lu, K.H. , Shvartsman, H. , Thornton, A. , Malpica, A. , Sun, C. , Bodurka, D.C. , Gershenson, D.M. , 2003. Expression of c-ABL, c-KIT, and platelet-derived growth factor receptor-beta in ovarian serous carcinoma and normal ovarian surface epithelium. Cancer. 98, 758–764. [DOI] [PubMed] [Google Scholar]
- Schwaenen, C. , Viardot, A. , Berger, H. , Barth, T.F. , Bentink, S. , Dohner, H. , Enz, M. , Feller, A.C. , Hansmann, M.L. , Hummel, M. , Kestler, H.A. , Klapper, W. , Kreuz, M. , Lenze, D. , Loeffler, M. , Moller, P. , Muller-Hermelink, H.K. , Ott, G. , Rosolowski, M. , Rosenwald, A. , Ruf, S. , Siebert, R. , Spang, R. , Stein, H. , Truemper, L. , Lichter, P. , Bentz, M. , Wessendorf, S. , 2009. Microarray-based genomic profiling reveals novel genomic aberrations in follicular lymphoma which associate with patient survival and gene expression status. Genes Chromosomes Cancer. 48, 39–54. [DOI] [PubMed] [Google Scholar]
- Seidman, J.D. , Horkayne-Szakaly, I. , Haiba, M. , Boice, C.R. , Kurman, R.J. , Ronnett, B.M. , 2004. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int. J. Gynecol. Pathol.23, 41–44. [DOI] [PubMed] [Google Scholar]
- Shridhar, V. , Staub, J. , Huntley, B. , Cliby, W. , Jenkins, R. , Pass, H.I. , Hartmann, L. , Smith, D.I. , 1999. A novel region of deletion on chromosome 6q23.3 spanning less than 500 Kb in high grade invasive epithelial ovarian cancer. Oncogene. 18, 3913–3918. [DOI] [PubMed] [Google Scholar]
- Siegel, R. , Naishadham, D. , Jemal, A. , 2013. Cancer statistics, 2013. CA Cancer J. Clin.63, 11–30. [DOI] [PubMed] [Google Scholar]
- Sieh, W. , Kobel, M. , Longacre, T.A. , Bowtell, D.D. , Defazio, A. , Goodman, M.T. , Hogdall, E. , Deen, S. , Wentzensen, N. , Moysich, K.B. , Brenton, J.D. , Clarke, B.A. , Menon, U. , Gilks, C.B. , Kim, A. , Madore, J. , Fereday, S. , George, J. , Galletta, L. , Lurie, G. , Wilkens, L.R. , Carney, M.E. , Thompson, P.J. , Matsuno, R.K. , Kjaer, S.K. , Jensen, A. , Hogdall, C. , Kalli, K.R. , Fridley, B.L. , Keeney, G.L. , Vierkant, R.A. , Cunningham, J.M. , Brinton, L.A. , Yang, H.P. , Sherman, M.E. , Garcia-Closas, M. , Lissowska, J. , Odunsi, K. , Morrison, C. , Lele, S. , Bshara, W. , Sucheston, L. , Jimenez-Linan, M. , Driver, K. , Alsop, J. , Mack, M. , McGuire, V. , Rothstein, J.H. , Rosen, B.P. , Bernardini, M.Q. , Mackay, H. , Oza, A. , Wozniak, E.L. , Benjamin, E. , Gentry-Maharaj, A. , Gayther, S.A. , Tinker, A.V. , Prentice, L.M. , Chow, C. , Anglesio, M.S. , Johnatty, S.E. , Chenevix-Trench, G. , Whittemore, A.S. , Pharoah, P.D. , Goode, E.L. , Huntsman, D.G. , Ramus, S.J. , 2013. Hormone-receptor expression and ovarian cancer survival: an ovarian tumor tissue analysis consortium study. Lancet Oncol.14, 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H.S. , Su, I.J. , Lin, Y.C. , Chen, J.S. , Fang, S.Y. , 2003. A 2.6 Mb interval on chromosome 6q25.2-q25.3 is commonly deleted in human nasal natural killer/T-cell lymphoma. Br. J. Haematol.122, 590–599. [DOI] [PubMed] [Google Scholar]
- Tangjitgamol, S. , Manusirivithaya, S. , Khunnarong, J. , Jesadapatarakul, S. , Tanwanich, S. , 2009. Expressions of estrogen and progesterone receptors in epithelial ovarian cancer: a clinicopathologic study. Int. J. Gynecol. Cancer. 19, 620–627. [DOI] [PubMed] [Google Scholar]
- TCGA, 2011. Integrated genomic analyses of ovarian carcinoma. Nature. 474, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil, A.F. , Nonnekens, J. , Steurer, B. , Mari, P.O. , de Wit, J. , Lemaitre, C. , Marteijn, J.A. , Raams, A. , Maas, A. , Vermeij, M. , Essers, J. , Hoeijmakers, J.H. , Giglia-Mari, G. , Vermeulen, W. , 2013. Disruption of TTDA results in complete nucleotide excision repair deficiency and embryonic lethality. PLos Genet.9, e1003431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theile, M. , Seitz, S. , Arnold, W. , Jandrig, B. , Frege, R. , Schlag, P.M. , Haensch, W. , Guski, H. , Winzer, K.J. , Barrett, J.C. , Scherneck, S. , 1996. A defined chromosome 6q fragment (at D6S310) harbors a putative tumor suppressor gene for breast cancer. Oncogene. 13, 677–685. [PubMed] [Google Scholar]
- Vajdic, C.M. , Hutchins, A.M. , Kricker, A. , Aitken, J.F. , Armstrong, B.K. , Hayward, N.K. , Armes, J.E. , 2003. Chromosomal gains and losses in ocular melanoma detected by comparative genomic hybridization in an Australian population-based study. Cancer Genet. Cytogenet.144, 12–17. [DOI] [PubMed] [Google Scholar]
- Van Wieringen, W.N. , Van De Wiel, M.A. , Ylstra, B. , 2008. Weighted clustering of called array CGH data. Biostatistics. 9, 484–500. [DOI] [PubMed] [Google Scholar]
- Wang, Z.C. , Birkbak, N.J. , Culhane, A. , Drapkin, R.I. , Fatima, A. , Tian, R. , Schwede, M. , Alsop, K. , Daniels, K.E. , Piao, H. , Liu, J. , Etemadmoghadam, D. , Miron, A. , Salvesen, H.B. , Mitchell, G. , Defazio, A. , Quackenbush, J. , Berkowitz, R.S. , Iglehart, J.D. , Bowtell, D.D. , Matulonis, U.A. , 2012. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin. Cancer Res.18, 5806–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand, K.C. , Shah, S.P. , Al-Agha, O.M. , Zhao, Y. , Tse, K. , Zeng, T. , Senz, J. , McConechy, M.K. , Anglesio, M.S. , Kalloger, S.E. , Yang, W. , Heravi-Moussavi, A. , Giuliany, R. , Chow, C. , Fee, J. , Zayed, A. , Prentice, L. , Melnyk, N. , Turashvili, G. , Delaney, A.D. , Madore, J. , Yip, S. , McPherson, A.W. , Ha, G. , Bell, L. , Fereday, S. , Tam, A. , Galletta, L. , Tonin, P.N. , Provencher, D. , Miller, D. , Jones, S.J. , Moore, R.A. , Morin, G.B. , Oloumi, A. , Boyd, N. , Aparicio, S.A. , Shih, Ie.M. , Mes-Masson, A.M. , Bowtell, D.D. , Hirst, M. , Gilks, B. , Marra, M.A. , Huntsman, D.G. , 2010. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med.363, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, W. , Wei, Y. , Du, Y. , Liu, J. , Chang, B. , Yu, Y.L. , Huo, L.F. , Miller, S. , Hung, M.C. , 2009. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol. Carcinog.48, 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S. , Tsuda, H. , Honda, K. , Onozato, K. , Takano, M. , Tamai, S. , Imoto, I. , Inazawa, J. , Yamada, T. , Matsubara, O. , 2009. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod. Pathol.22, 499–507. [DOI] [PubMed] [Google Scholar]
- Zeller, C. , Hinzmann, B. , Seitz, S. , Prokoph, H. , Burkhard-Goettges, E. , Fischer, J. , Jandrig, B. , Schwarz, L.E. , Rosenthal, A. , Scherneck, S. , 2003. SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene. 22, 2972–2983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary Figure 1 Association between copy number and gene expression level. Association between copy number status of 81 genes from the 6q24.2–26 region and their expression level (based on RPKM values) from TCGA studies. Fold change (log‐ratio) in gene expression between cases with loss vs. normal copy number status at this locus on the y‐axis; genomic location on the 6q chromosome on the x‐axis; significant changes (FDR < 0.05) are marked in red.
Supplementary data
Supplementary data
Supplementary data
Supplementary data


