Abstract
Glioblastoma (GBM) is one of the most common and aggressive primary brain tumors in adults. Deregulated expression of microRNAs (miRNAs) has been associated with GBM progression through alterations in either oncogenic or tumor suppressor targets. Here, we elucidated the function and the possible molecular mechanisms of miR‐449a in human GBM cell lines and tumor specimens‐derived glioblastoma stem cells (GSCs). Quantitative real‐time PCR demonstrated that miR‐449a was down‐regulated in human GBM cell lines and GSCs. Functionally, miR‐449a acted as a tumor suppressor by reducing cell proliferation, migration and invasion as well as inducing apoptosis in human GBM cell lines and GSCs. Myc‐associated zinc‐finger protein (MAZ) was identified as a direct target of miR‐449a, mediating these tumor‐suppressive effects, demonstrated by Western blot assay and luciferase assays. Moreover, over‐expression of miR‐449a inhibited the expression of Podoplanin (PDPN) by down‐regulating MAZ which could positively control the promoter activities via binding to the promoter of PDPN, demonstrated by luciferase assays and chromatin immunoprecipitation assays. Further, the PI3K/AKT pathway was blocked when MAZ was down‐regulated by miR‐449a. This process was coincided with the up‐regulation of apoptotic proteins and the down‐regulation of anti‐apoptotic proteins, MMP2 and MMP9. Furthermore, nude mice carrying over‐expressed miR‐449a combined with knockdown MAZ tumors produced the smallest tumors and the highest survival. These results elucidated a novel molecular mechanism of GBM progression, and may thus suggest a promising application for GBM treatment.
Keywords: MicroRNAs, MiR‐449a, Glioblastoma, Stem cells, MAZ, PDPN
Highlights
MiR‐449a expression was down‐regulated in human GBM cell lines and GSCs.
MiR‐449a reduced proliferation, migration and invasion, and induced apoptosis.
MAZ was identified as a direct target of miR‐449a.
MAZ up‐regulated the promoter activities and bound to the PDPN promoters.
The mechanisms were the down‐regulation of PI3K/AKT pathway and PDPN expression.
1. Introduction
Glioblastoma (GBM) is one of the most common and aggressive primary brain tumors in adults (Ostrom et al., 2013). Despite advances in clinical therapies and technologies, the outcome of patients diagnosed with GBM has shown only marginal improvement over the past several decades with a median survival of only 15 months (Ohgaki, 2009; Stupp et al., 2007). The deadly nature of GBM originated from its rapid diffusely infiltrative growth and the inherent complexity of the tumor (Furnari et al., 2007). Glioblastoma stem cells (GSCs) are a neoplastic subpopulation within GBMs that is highly tumorigenic and responsible for therapy resistance and poor survival rate (Bao et al., 2006; Chen et al., 2012b; Hadjipanayis and Van Meir, 2009; Liu et al., 2014a; Zhou et al., 2009). GSCs have capabilities of neurosphere formation, self‐renewal, multipotency of differentiation and initiation of tumors (Alcantara Llaguno et al., 2009, 2006, 2003, 2004). Therefore, researches on the molecular mechanisms relating the oncogenic effects of GSCs become urgent missions, which may improve the GBM treatment.
MicroRNAs (miRNAs) are small non‐coding RNAs that regulate gene expression by targeting mRNA for deregulation or translational repression (Bartel, 2004). MiRNAs are involved in both physiological and pathological events including proliferation, migration, invasion and apoptosis (Calin and Croce, 2006; Croce, 2011; Pasquinelli et al., 2005). There was ample evidence that miRNAs have regulatory functions in GBM progression (Kim et al., 2011). MiR‐663 was found to inhibit the proliferation and invasion of glioblastoma cells in vitro and in vivo by directly targeting PIK3CD, and predicted better prognosis in human GBM (Shi et al., 2014). However, miR‐148a exerted oncogenic effects by regulating BIM and MIG6, and inversely correlates with patient survival (Kim et al., 2014). In addition, a number of studies have highlighted the important roles of miRNAs in GSCs (Brower et al., 2014). MiR‐34a have been shown to be down‐regulated in GSCs, and suppressed cell proliferation and tumor growth by targeting Akt and Wnt signaling pathways (Guessous et al., 2010; Rathod et al., 2014). However, miR‐330 enhanced the malignant behaviors of GSCs by targeting SH3GL2 via the activation of ERK and PI3K/AKT signaling pathways (Yao et al., 2014). Taken together, aberrant expressions of miRNAs and its specific target gene in GBM cells and GSCs were associated with the initiation and development of GBM. However, the miRNAs and their mechanisms in the regulation of GBM and GSCs still need to be well documented.
MiR‐449a has shown tumor suppressive effects in several types of cancer, including prostatic carcinoma (Noonan et al., 2009), lung cancer (Ren et al., 2014) and endometrial cancer (Ye et al., 2014). MiR‐449a also acted as a tumor suppressor in hepatocellular carcinoma cells by targeting c‐MET, promoting apoptosis and reducing proliferation (Buurman et al., 2012). In addition, miR‐449a directly targeted lymphoid enhancer‐binding factor‐1 which in turn reduces Sox 9 expression leading to the proper regulation of the differentiation and chondrogenesis of human bone marrow‐derived mesenchymal stem cells (Paik et al., 2012). These evidences showed that miR‐449a played a tumor‐suppressive role in most cancers even in the stem cells, but whether miR‐449a exerts tumor‐suppressive functions in human GBM and GSCs needs to be explored urgently.
In this study, we provided functional evidence that miR‐449a exerted tumor‐suppressive functions in human GBM cell lines and tumor specimens‐derived GSCs. Mechanistically, over‐expression of miR‐449a impaired the oncogenic abilities by targeting Myc‐associated zinc‐finger protein (MAZ), and additionally lead to the down‐regulation of Podoplanin (PDPN) and the inactivation of PI3K/AKT pathway. These results elucidated a novel molecular mechanism of GBM progression, and may thus suggest a promising application for GBM treatment.
2. Materials and methods
2.1. Cell culture and human tissue specimens
Human GBM cell lines (U87 and U251) and human embryonic kidney (HEK) 293T cells were obtained from Shanghai Institutes for Biological Sciences Cell Resource Center, grown in Dulbecco's Modified Eagle Medium (DMEM) of high glucose with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA). Human normal astrocytes were purchased from the ScienCell Research Laboratories (Carlsbad, CA, USA), cultured according to the instruction of the manufacturer. All cells were incubated in a humidified air at 37 °C with 5% CO2. Glioma tissues and normal brain tissues (NBTs) were obtained from patients at the Shengjing Hospital of China Medical University. After surgical resection, part of the fresh glioma tissues were sent for routine neuropathological evaluation, and the other parts were frozen in liquid nitrogen or used for subsequent experiments. Informed consent was obtained from all patients and the study was approved by the Ethics Committee of Shengjing Hospital of China Medical University. Grading of tumors was identified according to WHO classification by neuropathologists.
2.2. Isolation and identification of GSCs
GSCs were isolated from GBM tissues (GBMs) as described previously (Bao et al., 2006; Singh et al., 2003), and were resuspended in DMEM/F‐12 medium (Life Technologies Corporation, Grand Island, NY, USA) supplemented with basic fibroblast growth factor (bFGF, 20 ng/ml, Life Technologies Corporation, Carlsbad, CA, USA), epidermal growth factor (EGF, 20 ng/ml, Life Technologies Corporation, Gaithersburg, MD, USA) and 2% B27 (50×, Life Technologies Corporation, Grand Island, NY, USA). Sphere cells were dissociated and planted in 96‐well plates for the limiting dilution assay and primary sphere formation assay, as described by Singh et al. (Singh et al., 2003). For differentiation assay, sphere cells were plated onto glass coverslips coated with poly‐L‐ornithine (BD Biosciences, Franklin Lakes, NJ, USA) in medium with 10% FBS. For immunostaining of undifferentiated spheres, cells were stained with antibodies against Nestin and CD133 (also known as prominin‐1) (1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA). For immunostaining of differentiated spheres, cells were stained with antibodies against GFAP (1:100, Abcam, Cambridge, MA, USA) and beta‐tubulin III (1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA). The primary antibodies complexes were visualized using anti‐rabbit Alexa Fluor 488 and anti‐mouse Alexa Fluor 555 secondary antibodies (Beyotime Institute of Biotechnology, Jiangsu, China). Nuclei were counterstained using 4′, 6‐diamidino‐2‐phenylindole (DAPI).
2.3. Reverse transcription and quantitative real‐time PCR (qRT‐PCR)
Total RNA was isolated from cells using Trizol reagent (Life Technologies Corporation, Carlsbad, CA, USA). We used TaqMan MicroRNA Reverse Transcription kit and High Capacity cDNA Reverse Transcription Kit for miRNA and mRNA reverse transcription, respectively (Applied Biosystems, Foster City, CA, USA). Quantitative real‐time PCR (qRT‐PCR) was conducted using TaqMan Universal Master Mix II with TaqMan microRNA assays of miR‐449a and U6 or TaqMan gene expression assays of MAZ and GAPDH (Applied Biosystems, Foster City, CA, USA). U6 and GAPDH were used as endogenous controls for miRNA and gene expressions, respectively. Expression were normalized to endogenous controls and fold changes were calculated using relative quantification (2−ΔΔCt) method.
2.4. Cell transfections
MiR‐449a agomir, miR‐449a antagomir and their respective non‐targeting sequence (negative control, NC) were synthesized (GenePharma, Shanghai, China). U87, U251 and GSCs were transfected with miR‐449a agomir (pre‐miR‐449a), miR‐449a antagomir (anti‐miR‐449a) or their respective NC using Lipofectamine 2000 reagent (Life Technologies Corporation, Carlsbad, CA, USA). The transfected efficacy was evaluated by qRT‐PCR, and the high transfection efficacy of these could sustain 7 days from 48 h post‐transfection, thus 72 h post‐transfection was considered as the harvested time in the subsequent experiments.
2.4.1. Lentiviral vector construction and infection
Human MAZ coding sequence (CDS) was ligated into the LV5‐CMV‐GFP‐EF1a‐Puro lentiviral vector (GenePharma, Shanghai, China) to generate LV5‐CMV‐GFP‐EF1a‐Puro‐MAZ lentiviral vector. Lentivirus was harvested 48 h after the LV5‐CMV‐GFP‐EF1a‐Puro‐MAZ lentiviral vectors or the empty lentiviral vectors (negative control, NC) co‐transfection with the packaging vectors into HEK 293T cells using Lipofectamine 2000. Cells were then infected with the lentivirus. GFP‐positive cells were pooled as MAZ and MAZ‐NC, and then used for subsequent assays. These stable expressing cells co‐transfected with pre‐miR‐449a (or pre‐NC) were divided into 5 groups: control group, pre‐NC + MAZ‐NC group (MAZ‐NC stable expressing cells co‐transfected with pre‐NC), pre‐miR‐449a + MAZ‐NC group (MAZ‐NC stable expressing cells co‐transfected with pre‐miR‐449a), pre‐NC + MAZ group (MAZ stable expressing cells co‐transfected with pre‐NC) and pre‐miR‐449a + MAZ group (MAZ stable expressing cells co‐transfected with pre‐miR‐449a).
2.5. Cell proliferation assay
Cells were seeded in 96‐well plates. After cells were transfected 72 h, 20 μl of Cell Counting Kit‐8 (Beyotime Institute of Biotechnology, Jiangsu, China) was added into each well. The plate was allowed to stand for 2 h at 37 °C and the absorbance at 450 nm was recorded.
2.6. Quantization of apoptosis by flow cytometry
Apoptosis was assessed using ApoScreen Annexin V Apoptosis detection kit (SouthernBiotech, Birmingham, AL, USA). Cells were harvested and stained with Annexin V‐PE and 7‐AAD according to the manufacturer's instructions. Cell samples were analyzed on flow cytometry (FACScan, BD Biosciences) and apoptotic fractions were determined.
2.7. Cell migration and invasion assay
Cells were resuspended in 100 μl serum‐free medium and placed in the upper chamber (or pre‐coated with 500 ng/μl Matrigel solution (BD, Franklin Lakes, NJ, USA) for cell invasion assay) of the 24‐well insert with 8 μm pore size. 600 μl of 10% FBS medium was placed in the lower chamber. After incubation for 48 h, the cells on the upper membrane surface were mechanically removed. Cells that had migrated or invaded to the lower side of membrane were fixed and stained with 20% Giemsa. Cells were counted under a microscope in five randomly chosen fields and photographs were taken.
2.8. Western blot analysis
Cells were lysed in RIPA buffer on ice and total proteins were further analyzed by SDS–PAGE and electrophoretically transferred to polyvinylidene difluoride membranes. Non‐specific bindings were blocked by incubating membranes in Tris‐buffered saline‐Tween (TBST) containing 5% nonfat milk for 2 h at room temperature. The membranes were then incubated with primary antibodies against MAZ, PDPN (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA), PI3K, p‐PI3K, AKT, p‐AKT (1:1000, Cell Signaling Technology, Beverly, MA, USA), XIAP, Bcl‐2, Caspase‐3, TRAIL (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and GAPDH (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA, USA). After this, the membranes were incubated with horseradish peroxidase conjugated secondary antibody. Immunoblots were visualized by ECL chemiluminescent detection system. The blots were scanned and the integrated density value (IDV) was measured on FluorChem 2.0 software.
2.9. Reporter vectors constructs and luciferase assays
The 3′‐UTR fragments of MAZ gene and its mutant of the predicted miR‐449a binding sites were subcloned into a pmirGLO Dual‐luciferase miRNA Target Expression Vector to form MAZ‐3′UTR‐Wt and MAZ‐3′UTR‐Mut (GenePharma, Shanghai, China), respectively. Cells were co‐transfected with MAZ‐3′UTR‐Wt (or MAZ‐3′UTR‐Mut) and pre‐NC (or pre‐miR‐449a). After transfection, cells were harvested and lysed for luciferase assays using the Dual‐Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Promoter activities were measured to determine the responsive MAZ‐binding sites in the human PDPN promoter using Dual‐Luciferase Reporter Assay System, as previously described (Ma et al., 2013). Different promoter fragments of PDPN and human full‐length MAZ were subcloned into pGL3‐Basic vector (Promega, Madison, WI, USA) and pEX3 vector (GenePharma, Shanghai, China), respectively. The firefly luciferase activity was normalized to renilla luciferase activity for each individual analysis.
2.10. Chromatin immunoprecipitation (ChIP) assay
Simple ChIP Enzymatic Chromatin IP Kit (Cell signaling Technology, Danvers, Massachusetts, USA) was used for ChIP assays according to the manufacturer's protocol, as described previously (Ma et al., 2013). Cells were cross‐linked with 1% formaldehyde and collected in lysis buffer. 2% lysates were used as an input control and the remaining lysates were immunoprecipitated with normal rabbit IgG and MAZ antibody. DNA was extracted for PCR amplification of the following DNA fragments: putative binding site 1 using the primers 5′‐TGCGGCCCTGTAACTTTAA‐3′ and 5′‐CGAGCGGAGTCAGCATTTATC‐3′, yielding a 155 bp product; putative binding site 2 using the primers 5′‐CTCCAGCGACACTGGTTTA‐3′ and 5′‐GCTCCACATTGCGTGAA‐3′, yielding a 181 bp product; control using the primers 5′‐CCCTTACCTATTGAATCAGCAC‐3′ and 5′‐GAACACGCACTATTTCCTTT‐3′, yielding a 171 bp product.
2.11. Tumor xenografts in nude mice
For the in vivo study, the stable expressing cells were used. Lentivirus encoding pre‐miR‐449a was generated using pLenti6.3/V5–DEST Gateway Vector Kit (Life Technologies Corporation, Carlsbad, CA, USA). The pre‐miR‐449a and short‐hairpin RNA targeting human MAZ were ligated into the pLenti6.3/V5–DEST vector and LV3‐CMV‐GFP‐Puro vector (GenePharma, Shanghai, China), respectively. And then those generated pLenti6.3/V5–DEST‐miR‐449a and LV3‐CMV‐GFP‐Puro‐shMAZ vectors. Lentivirus was generated in 293FT cells using the ViraPower Packaging Mix. After infection, the stable expressing cells of miR‐449a and shMAZ were picked. The lentiviruses of pre‐miR‐449a were transduced in shMAZ stable expressing cells to generate miR‐449a + shMAZ stable expressing cells.
Four‐week old BALB/c athymic nude mice were purchased from Cancer Institute of the Chinese Academy of Medical Science. Animal handling and experimental procedures were in accordance with the Guide for the Care and Use of Laboratory Animals, and approved by the Animal Care Committee of the Shengjing Hospital. For subcutaneous implantation, 3 × 105 cells were subcutaneously inoculated in the right flanks of the mice. Tumor volume was measured every five days and calculated by the formula: volume (mm3) = length × width2/2. For survival analysis in orthotopic inoculations, 3 × 105 cells were stereotactically implanted into the right striatum of the mice. The number of survived nude mice was recorded and survival analysis was performed using Kaplan–Meier survival curve.
2.12. Statistical analysis
Data are presented as mean ± standard deviation (SD). Differences were evaluated by SPSS 18.0 statistical software with the Student's t‐test or one‐way ANOVA. Differences were considered to be significant when P < 0.05.
3. Results
3.1. Isolation and identification of GSCs
As shown in Figure 1A‐a, cells isolated from GBMs were cultured in the serum‐free medium and formed cell spheres. In order to test the self‐renewing ability, the spheres were harvested and a second round sphere‐forming assay was performed. Sphere generated again from a single cell (Figure 1A‐b). The stain of Nestin and CD133 proved that most cells within spheres were positive to neural stem cell lineage markers on membranes (Figure 1B). Cell spheres were differentiated and stained for GFAP and beta‐tubulin‐III lineage markers suggesting that there were typical morphological differentiation towards astrocytic and neuronal lineages (Figure 1C).
Figure 1.
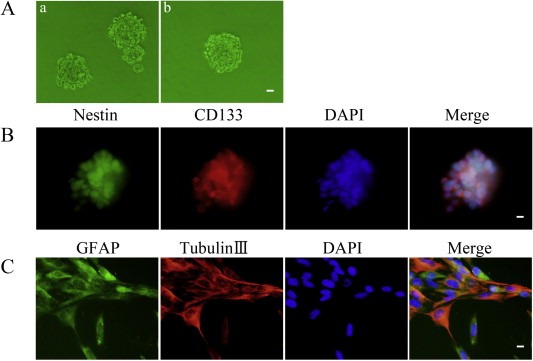
Isolation and identification of GSCs. (A) a: Cells formed spheres in serum‐free medium. b: a single cell formed spheres again by a second round sphere‐forming assay. (B) Cell spheres stained for Nestin (green) and CD133 (red) by immunofluorescence analysis. (C) Cell spheres were differentiated and then stained for GFAP (green) and beta‐tubulin III (red) by immunofluorescence analysis. Nuclei (blue) were labeled with DAPI. Images are representative of independent experiments (n = 5). Scale bars represent 20 μm.
3.2. MiR‐449a expression was down‐regulated in human GBM cell lines and GSCs
The expression levels of miR‐449a were evaluated by qRT‐PCR analysis. We first evaluated miR‐449a expression in NBTs and human glioma tissues of different grades (8 tissues in each group). The results showed that miR‐449a was negatively correlated with histopathological grade in human glioma tissues. Compared with NBTs, miR‐449a expression was reduced to 0.79‐, 0.64‐, 0.39‐ and 0.15‐fold in Grade I, Grade II, Grade III and Grade IV, respectively. MiR‐449a expression was significantly reduced in Grade IV glioma tissues as compared to NBTs and glioma tissues of other grades (Figure 2A). Moreover, we also evaluated miR‐449a expression in NBTs, GBMs and GSCs (5 tissues in each group), and found that miR‐449a expression was significantly lower in GBMs and GSCs than in NBTs (P < 0.01), and was significantly lower in GSCs than in GBMs (P < 0.05) (Figure 2B). This was similar to the suppressed expression level of miR‐449a in human GBM cell lines U87 and U251 (P < 0.01) (Figure 2C). These results suggested that the reduced miR‐449a expression may be critically involved in GBM development.
Figure 2.
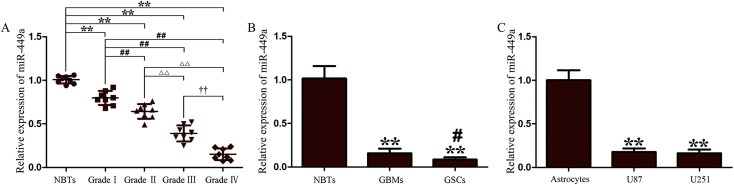
MiR‐449a expression in human glioma tissues, GSCs and GBM cell lines. (A) MiR‐449a expression in normal brain tissues (NBTs) and human glioma tissues of different grades. Data are presented as the mean ± SD (n = 8, each group). ∗∗P < 0.01 vs. NBTs group, ##P < 0.01 vs. Grade I group, △△P < 0.01 vs. Grade II group and ††P < 0.01 vs. Grade III group. (B) MiR‐449a expression in NBTs, GBM tissues (GBMs) and glioblastoma stem cells (GSCs). Data are presented as the mean ± SD (n = 5, each group). **P < 0.01 vs. NBTs group, #P < 0.05 vs. GBMs group. (C) MiR‐449a expression in human normal astrocytes and GBM cell lines (U87 and U251). Data are presented as the mean ± SD (n = 5, each group). **P < 0.01 vs. Astrocytes group.
3.3. MiR‐449a was a tumor suppressor in human GBM cell lines and GSCs
To better understand the functional consequences of reduced miR‐449a in GBM progression, we next assessed the effects of miR‐449a over‐expression and inhibition on cell proliferation, apoptosis, migration and invasion in GBM cell lines and GSCs. The high transfection efficacy of pre‐miR‐449a and anti‐miR‐449a could sustain 7 days from 48 h post‐transfection. The transfected efficacy at 72 h post‐transfection was shown in Figure 3A. As shown in Figure 3B, miR‐449a over‐expression reduced the proliferation of U87, U251 and GSCs, whereas miR‐449a inhibition promoted the proliferation of these cells. Similarly, over‐expression of miR‐449a led to a significant induction of apoptosis, whereas inhibition of miR‐449a led to a significant suppression of apoptosis in U87, U251 and GSCs (Figure 3C). As shown in Figure 3D and E, over‐expression of miR‐449a significantly decreased the migration and invasion of U87, U251 and GSCs, whereas inhibition of miR‐449a significantly increased the migration and invasion of these cells. Taken together, these results demonstrated the tumor‐suppressive functions of miR‐449a in human GBM cell lines and GSCs.
Figure 3.
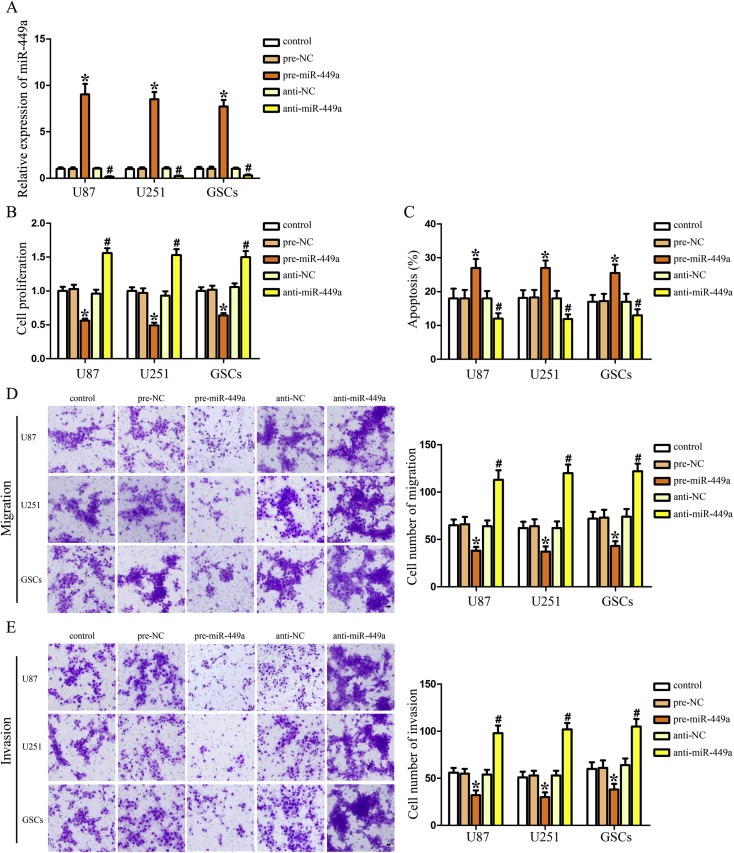
MiR‐449a was a tumor suppressor in human GBM cell lines and GSCs. (A) Relative expression of miR‐449a after cells transfected with pre‐miR‐449a and anti‐miR‐449a. (B) Effect of miR‐449a on the proliferation of U87, U251 and GSCs. (C) Effect of miR‐449a on the apoptosis of U87, U251 and GSCs. Effect of miR‐449a on cell migration (D) and invasion (E) of U87, U251 and GSCs. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. pre‐NC group, #P < 0.05 vs. anti‐NC group. Scale bars represent 20 μm.
3.4. MiR‐449a inhibited the expression of MAZ by targeting its 3′‐UTR
The bioinformatics databases (TargetScan and RNAhybrid) were used to identify potential targets of miR‐449a. To experimentally verify these potential targets, cells transfected with pre‐miR‐449a or anti‐miR‐449a were assessed the mRNA and protein expression levels of targets by qRT‐PCR and Western blot, respectively. MAZ was identified as the target of miR‐449a. We found that over‐expression of miR‐449a decreased the mRNA and protein expression of MAZ, whereas inhibition of miR‐449a increased the mRNA and protein expression of MAZ in U87, U251 and GSCs (Figure 4A and B). These results indicated that miR‐449a could inhibit MAZ expression in human GBM cell lines and GSCs. As shown in Figure 4C, MAZ expression was higher in GBMs and GSCs than in NBTs, and was higher in GSCs than in GBMs. This was similar to the increased MAZ expression in human GBM cell lines U87 and U251 (Figure 4D). These results revealed that MAZ might play an oncogenic role in GBM development.
Figure 4.
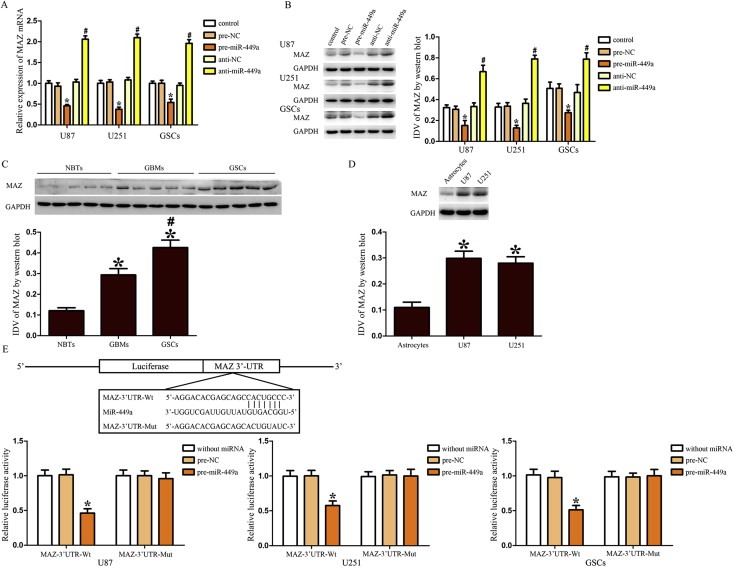
Over‐expressed miR‐449a inhibited the expression of MAZ by targeting its 3′‐UTR. (A) Effect of miR‐449a on the mRNA expression of MAZ in U87, U251 and GSCs. (B) Effect of miR‐449a on the protein expression of MAZ in U87, U251 and GSCs. *P < 0.05 vs. pre‐NC group, #P < 0.05 vs. anti‐NC group. (C) MAZ expression in NBTs, GBMs and GSCs. *P < 0.05 vs. NBTs group, #P < 0.05 vs. GBMs group. (D) MAZ expression in human normal astrocytes and GBM cell lines (U87 and U251). *P < 0.05 vs. Astrocytes group. (E) The predicted miR‐449a binding sites in the 3′‐UTR region of MAZ (MAZ‐3′UTR‐Wt) and the designed mutant sequence (MAZ‐3′UTR‐Mut) were indicated. Cells were co‐transfected with MAZ‐3′UTR‐Wt (or MAZ‐3′UTR‐Mut) and the indicated miRNAs, and then the luciferase assay was conducted. Data are presented as the mean ± SD (n = 5, each group). ∗P < 0.05 vs. MAZ‐3′UTR‐Wt + pre‐NC group.
Using TargetScan 6.2, MAZ was predicted to harbor one putative miR‐449a binding site in the 3′‐UTR. To elucidate whether MAZ was a functional target of miR‐449a, we cloned a reporter plasmid containing the wide‐type 3′‐UTR of MAZ (MAZ‐3′UTR‐Wt). The seeds for miR‐449a to MAZ 3′‐UTR were indicated. In the U87 cell line, co‐transfection of pre‐miR‐449a and MAZ‐3′UTR‐Wt decreased the luciferase activity, whereas co‐transfection of pre‐NC and MAZ‐3′UTR‐Wt did not change the luciferase activity. These results suggested that miR‐449a targeted the 3′‐UTR of MAZ. In parallel, the reporter plasmid mutated the miR‐449a binding sites were constructed (MAZ‐3′UTR‐Mut). Co‐transfection of pre‐miR‐449a and MAZ‐3′UTR‐Mut did not change the luciferase activity in U87 cells. In addition, the same changes were also observed in U251 and GSCs. These results suggested that MAZ was a direct target of miR‐449a with the specific binding site in human GBM cell lines and GSCs (Figure 4E).
3.5. MAZ mediated the tumor‐suppressive effects of miR‐449a on human GBM cell lines and GSCs
To determine whether the tumor‐suppressive effects of miR‐449a were mediated by MAZ, MAZ down‐regulation by pre‐miR‐449a was rescued using MAZ prior to the assessment of the cell proliferation, apoptosis, migration and invasion. As shown in Figure 5A, over‐expression of miR‐449a reduced the proliferation of U87, U251 and GSCs, whereas over‐expression of MAZ promoted the proliferation of these cells. MAZ over‐expression rescued the inhibitory effect of miR‐449a over‐expression on the proliferation of U87, U251 and GSCs. Similar to earlier results, over‐expression of miR‐449a led to a significant induction of apoptosis, whereas over‐expression of MAZ led to a significant suppression of apoptosis in U87, U251 and GSCs. MAZ over‐expression rescued the increased apoptosis induced by miR‐449a over‐expression in U87, U251 and GSCs (Figure 5B). In addition, over‐expression of miR‐449a inhibited the migration and invasion of U87, U251 and GSCs, whereas over‐expression of MAZ increased the migration and invasion of these cells. MAZ over‐expression rescued the inhibitory effect of miR‐449a over‐expression on cell migration and invasion of U87, U251 and GSCs (Figure 5C and D). The above data have shown that the tumor‐suppressive effects of miR‐449a were mediated by MAZ in human GBM cell lines and GSCs.
Figure 5.
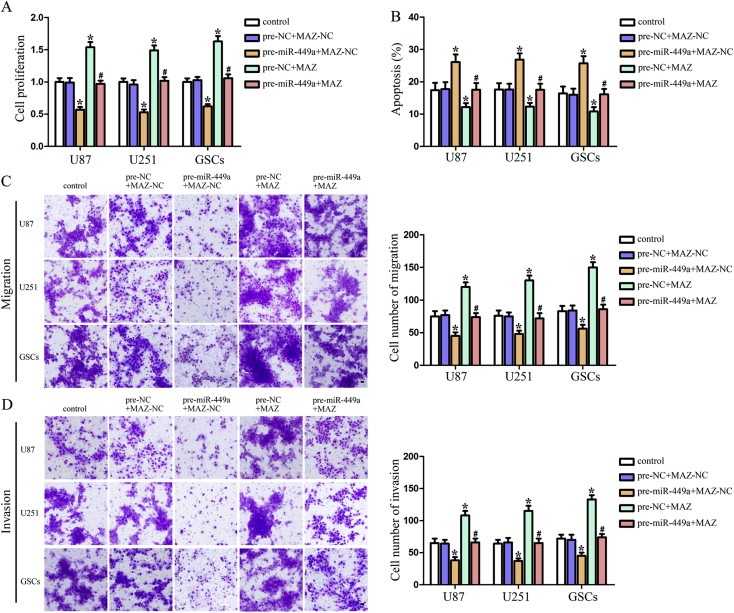
MAZ mediated the tumor‐suppressive effects of miR‐449a on human GBM cells and GSCs. (A) CCK8 assay to evaluate the effect of miR‐449a and MAZ on the proliferation in U87, U251 and GSCs. (B) Flow cytometry analysis to evaluate the effect of miR‐449a and MAZ on the cell apoptosis in U87, U251 and GSCs. Quantification of cell migration (C) and invasion (D) to evaluate the effect of miR‐449a and MAZ on the cell migration and invasion in U87, U251 and GSCs. Representative images and accompanying statistical plots were presented. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. pre‐NC + MAZ‐NC group, #P < 0.05 vs. pre‐miR‐449a + MAZ‐NC group. Scale bars represent 20 μm.
3.6. Over‐expression of miR‐449a inhibited the expression of PDPN by down‐regulating MAZ
Silencing of Podoplanin (PDPN) expression lead to the reduced proliferation and migration of glioma cells, suggesting a functional role of PDPN in glioma progression and malignancy (Peterziel et al., 2012). To further determine the underlying mechanisms of the tumor‐suppressive effects of miR‐449a, we focused on PDPN. To clarify whether PDPN was involved in the tumor‐suppressive effects of miR‐449a, the expression levels of PDPN were assessed after cells transfected with pre‐miR‐449a or anti‐miR‐449a. In U87, U251 and GSCs, miR‐449a over‐expression reduced PDPN expression, whereas miR‐449a inhibition increased PDPN expression (Figure 6A). However, the 3′‐UTR of PDPN was predicted to harbor none putative miR‐449a binding site, suggesting that it was not directly regulated by miR‐449a. Additionally, through analyzing the DNA sequence of the PDPN promoter, two putative MAZ binding sites were found, which suggested that PDPN might be a target gene of MAZ. To clarify whether miR‐449a inhibiting the expression of PDPN was mediated by MAZ, MAZ down‐regulation by pre‐miR‐449a was rescued using MAZ prior to the assessment of the expression of MAZ and PDPN. As shown in Figure 6B, over‐expression of miR‐449a inhibited the MAZ expression and meanwhile reduced the expression of PDPN, whereas over‐expression of MAZ increased PDPN expression. MAZ over‐expression rescued the inhibitory effect of miR‐449a over‐expression on the expression of PDPN in U87, U251 and GSCs. These results indicated that miR‐449a inhibited the expression of PDPN by down‐regulating MAZ.
Figure 6.
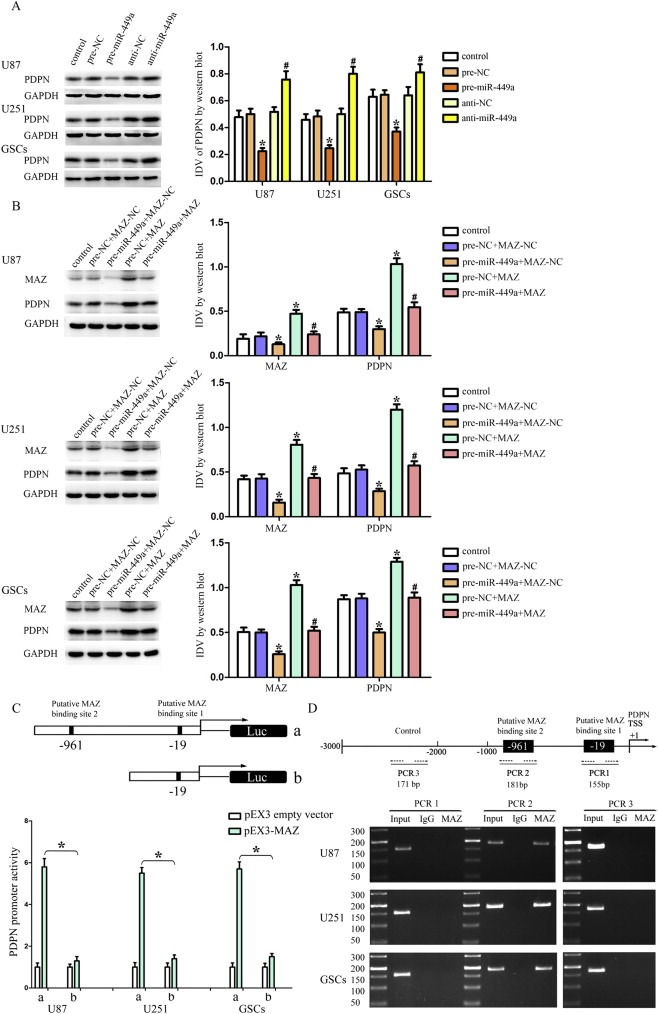
Over‐expression of miR‐449a inhibited PDPN expression by down‐regulating MAZ. (A) Western blot analysis of the PDPN expression regulated by miR‐449a in U87, U251 and GSCs. *P < 0.05 vs. pre‐NC group, #P < 0.05 vs. anti‐NC group. (B) Western blot analysis of the PDPN expression regulated by miR‐449a and MAZ in U87, U251 and GSCs. *P < 0.05 vs. pre‐NC + MAZ‐NC group, #P < 0.05 vs. pre‐miR‐449a + MAZ‐NC group. (C) MAZ on promoter activities of PDPN in U87, U251 and GSCs. The Y‐bar shows the constructed plasmid activity after normalization with the co‐transfected reference vector (pRL‐TK), and relative to the activity of pEX3 empty vector, which the activity was set to 1. X‐bar shows the position of the deletions on the promoter of PDPN. (D) MAZ bound to the promoters of PDPN in U87, U251 and GSCs. Transcription start site (TSS) was designated as +1. Putative MAZ binding sites are indicated. Immunoprecipitated DNA was amplified by PCR. Normal rabbit IgG was used as a negative control.
To test whether MAZ is required for the promoter activity of PDPN, a series of constructs were generated and luciferase assays were performed. MAZ recognized the GA box motif and GC‐rich sequences which is element of promoters of target genes (Cogoi et al., 2010; Himeda et al., 2008; Parks and Shenk, 1996). The sequences of PDPN promoter and the position of transcription start site (TSS) were set according to DBTSS HOME. By analyzing these DNA sequence in the 1000 bp region upstream and its 100 bp downstream sequence of the TSS, two putative binding sites of MAZ were confirmed. Wild‐type, deletion construct and putative MAZ binding sites were indicated. Deletion of the putative MAZ binding site 2 (‐961 site region) significantly decreased the promoter activities of PDPN in U87, U251 and GSCs, suggesting that important functional elements which were necessary for high PDPN promoter activity were resided in the ‐961 site region (Figure 6C). To further determine whether MAZ directly associated with the PDPN promoters in U87, U251 and GSCs, ChIP assays were performed. As a negative control, PCR was conducted to amplify the 1000 bp upstream region of the putative MAZ binding site that was not expected to associate with MAZ. Our results showed that there was an interaction of MAZ with putative binding site 2 of PDPN. There was no interaction of MAZ with the control region (Figure 6D). These results demonstrated that MAZ could up‐regulate the promoter activities and bind to the PDPN promoters in human GBM cell lines and GSCs.
3.7. Over‐expression of miR‐449a blocked the PI3K/AKT pathway by down‐regulating MAZ
The activity of PI3K/AKT pathway was detected by Western blot assays. As shown in Figure 7A, over‐expression of miR‐449a inhibited the p‐PI3K and p‐AKT expression, whereas over‐expression of MAZ increased the p‐PI3K and p‐AKT expression in U87 cells. MAZ over‐expression rescued the inhibitory effect of miR‐449a over‐expression on this pathways. The same changes were also observed in U251 and GSCs (Figure 7B and C). These results demonstrated that over‐expressed miR‐449a could block the PI3K/AKT pathway by down‐regulating MAZ.
Figure 7.
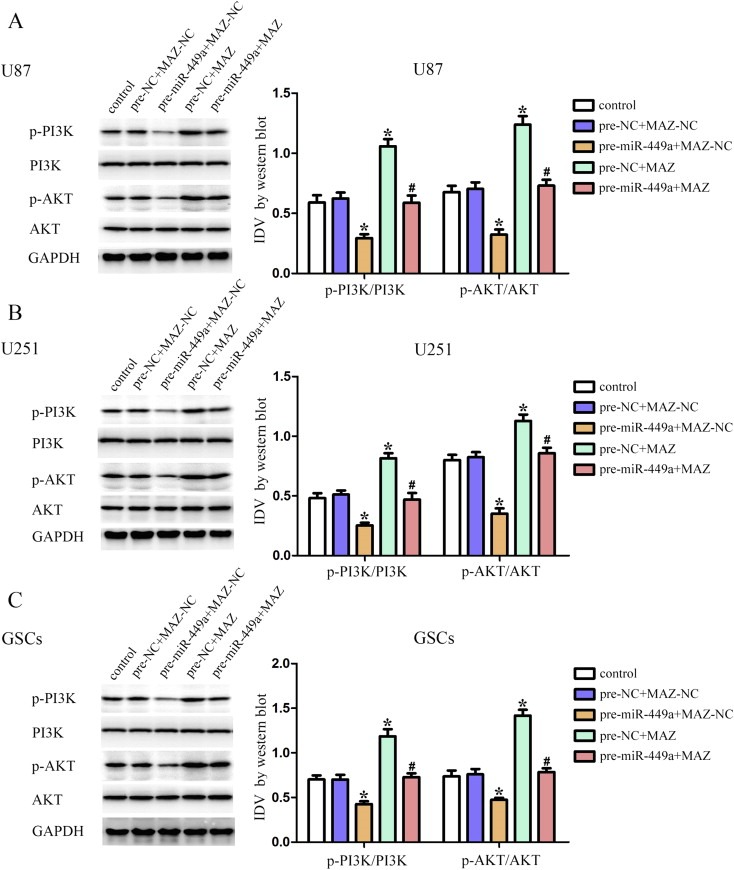
Over‐expression of miR‐449a blocked the PI3K/AKT pathway by down‐regulating MAZ. Western blot analysis of the PI3K/AKT pathway regulated by miR‐449a and MAZ in U87 (A), U251 (B) and GSCs (C). Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. pre‐NC + MAZ‐NC group, #P < 0.05 vs. pre‐miR‐449a + MAZ‐NC group.
3.8. Over‐expression of miR‐449a up‐regulated the apoptotic protein expression, and down‐regulated the expression of anti‐apoptotic proteins, MMP2 and MMP9 by down‐regulating MAZ
To determine whether anti‐apoptotic proteins, apoptotic proteins, MMP2 and MMP9 were involved in the tumor‐suppressive effects induced by the over‐expression of miR‐449a via down‐regulating of MAZ, the expression of anti‐apoptotic proteins, apoptotic proteins, MMP2 and MMP9 in U87, U251 and GSCs were measured by Western blot. As shown in Figure 8A, the analysis of anti‐apoptotic protein showed that over‐expression of miR‐449a inhibited the expression of XIAP and Bcl‐2, whereas over‐expression of MAZ increased the expression of XIAP and Bcl‐2. MAZ over‐expression rescued the inhibitory effect of miR‐449a over‐expression on the expression of anti‐apoptotic proteins in U87, U251 and GSCs. The analysis of apoptotic protein Capase‐3 and TRAIL showed a contrary result. These results suggested that over‐expression of miR‐449a could induce apoptosis by regulating the expression of anti‐apoptotic proteins and apoptotic proteins via down‐regulating MAZ. As shown in Figure 8B, over‐expression of miR‐449a inhibited the expression of MMP‐2 and MMP‐9, whereas over‐expression of MAZ increased the expression of MMP‐2 and MMP‐9. MAZ over‐expression rescued the inhibitory effect of miR‐449a over‐expression on the expression of MMP‐2 and MMP‐9 in U87, U251 and GSCs. The above results were summarized in Table 1. These results indicated that over‐expression of miR‐449a could inhibit the expression of MMP2 and MMP9 by down‐regulating MAZ.
Figure 8.
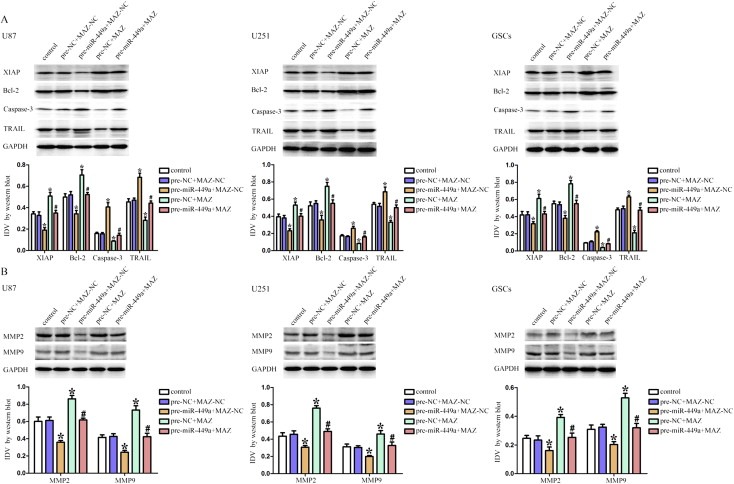
Over‐expression of miR‐449a up‐regulated the apoptotic protein expression, and down‐regulated the expression of anti‐apoptotic proteins, MMP2 and MMP9 by down‐regulating MAZ. (A) Western blot analysis of anti‐apoptotic proteins (XIAP and Bcl‐2) and apoptotic proteins (Capase‐3 and TRAIL) regulated by miR‐449a and MAZ in U87, U251 and GSCs. (B) Western blot analysis of MMP‐2 and MMP‐9 regulated by miR‐449a and MAZ in U87, U251 and GSCs. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05 vs. pre‐NC + MAZ‐NC group, #P < 0.05 vs. pre‐miR‐449a + MAZ‐NC group.
Table 1.
Over‐expression of miR‐449a up‐regulated the apoptotic protein expression, and down‐regulated the expression of anti‐apoptotic proteins, MMP2 and MMP9 by down‐regulating MAZ.
| Category | Protein name | Control groups | Pre‐NC + MAZ‐NC groups | Pre‐miR‐449a + MAZ‐NC groups | Pre‐NC + MAZ groups | Pre‐miR‐449a + MAZ groups |
|---|---|---|---|---|---|---|
| Anti‐apoptotic proteins | XIAP | – | – | Decreased | Increased | Rescued |
| Bcl‐2 | – | – | Decreased | Increased | Rescued | |
| Apoptotic proteins | Capase‐3 | – | – | Increased | Decreased | Rescued |
| TRAIL | – | – | Increased | Decreased | Rescued | |
| Cell migration and invasion related proteins | MMP2 | – | – | Decreased | Increased | Rescued |
| MMP9 | – | – | Decreased | Increased | Rescued |
3.9. Over‐expressed miR‐449a combined with knockdown MAZ suppressed tumor growth and had high survival in nude mice
As shown in Figure 9A, B and C, the in vivo studies showed that miR‐449a over‐expression, MAZ inhibition or miR‐449a over‐expression combined with MAZ inhibition produced smaller tumors than control. MiR‐449a over‐expression combined with MAZ inhibition resulted in the smallest tumor volume. As shown in Figure 9D, the survival analysis showed that miR‐449a over‐expression, MAZ inhibition or miR‐449a over‐expression combined with MAZ inhibition produced longer survival than control. MiR‐449a over‐expression combined with MAZ inhibition produced the longest survival. These data showed that nude mice carrying GBM cell lines and GSCs which were over‐expressed miR‐449a combined with knockdown of MAZ produced the smallest tumors and had the highest survival.
Figure 9.
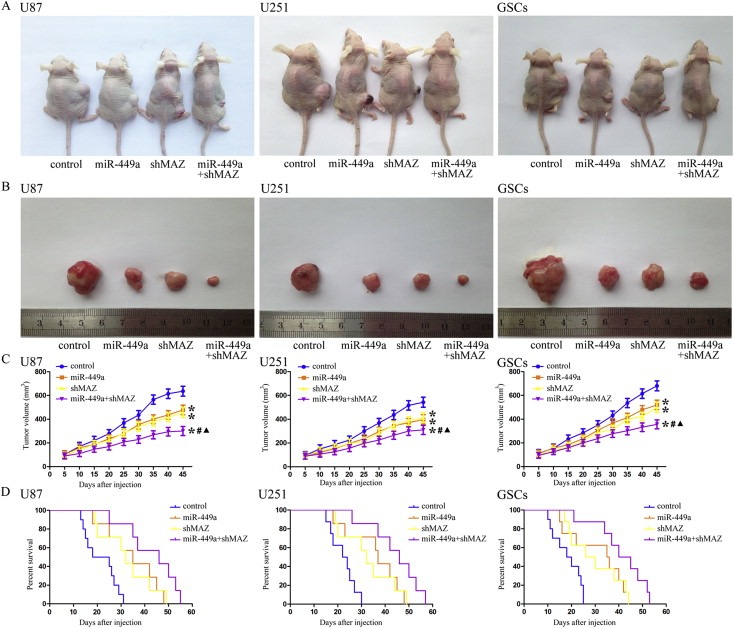
In vivo tumor xenografts study. (A) The stable expressing cells were used for the in vivo study. The nude mice carrying tumors from respective groups were shown. (B) The sample tumor from respective group was shown. (C) Tumor growth curves in nude mice. Tumor volume was calculated every five days after injection. *P < 0.05 vs. control group, #P < 0.05 vs. miR‐449a group, ▲P < 0.05 vs. shMAZ group. (D) The survival curves of nude mice injected into the right striatum (n = 15). P < 0.05 (miR‐449a or shMAZ group vs. control group), P < 0.01 (miR‐449a + shMAZ group vs. control group).
4. Discussion
In this study, we demonstrated that miR‐449a had a low expression level in human GBM cell lines and GSCs. Over‐expression of miR‐449a inhibited cell proliferation, migration and invasion as well as inducing apoptosis in these cells. In addition, MAZ was identified as a direct target of miR‐449a and mediated these tumor‐suppressive effects by miR‐449a. Furthermore, over‐expression of miR‐449a inhibited the expression of PDPN by down‐regulating MAZ which could positively control the promoter activities via binding to the promoter of PDPN. Further, PI3K/AKT pathway was blocked when MAZ was down‐regulated by miR‐449a. This process was coincided with the up‐regulation of apoptotic proteins and the down‐regulation of anti‐apoptotic proteins, MMP2 and MMP9. The in vivo studies showed that nude mice carrying over‐expressed miR‐449a combined with knockdown MAZ cells produced the smallest tumors and had the highest survival.
Our present data indicated that miR‐449a was negatively correlated with histopathological grade in human glioma tissues and had a low expression level in GBM cell lines (U87 and U251) and GSCs, suggesting that the reduced miR‐449a expression may be critically involved in GBM development. To better understand the functional consequences of reduced miR‐449a in GBM progression, we next assessed the effects of miR‐449a over‐expression and inhibition on cell proliferation, apoptosis, migration and invasion in human GBM cell lines and GSCs. Our results have shown that over‐expression of miR‐449a inhibited the cell proliferation, migration and invasion as well as inducing apoptosis in U87, U251 and GSCs. Taken together, miR‐449a acted as a tumor suppressor to inhibit the growth and infiltration of U87, U251 and GSCs, which may have therapeutic potential in GBM treatment. Previous studies also showed the same tumor‐suppressive function of miR‐449a in bladder cancer, gastric adenocarcinoma, retinoblastoma and non‐small cell lung cancer (Chen et al., 2012a; Luo et al., 2013; Martin et al., 2013; Wei et al., 2013). Our results for the first time showed that miR‐449a acted as a tumor suppressor in human GBM cell lines and GSCs, but the underlying mechanisms still need to be investigated.
Myc‐associated zinc‐finger protein (MAZ) was confirmed as the target of miR‐449a in our study. MAZ is a zinc‐finger protein expressed ubiquitously in different human tissues (Song et al., 1998), and has been discovered to regulate a variety of genes including c‐myc, insulin and serotonin receptors (Bossone et al., 1992; Kennedy and Rutter, 1992; Parks and Shenk, 1996), which could regulate many pathways involved in cell proliferation, apoptosis, differentiation and development (Cogoi et al., 2013; Jiao et al., 2013; Smits et al., 2012). Our results showed that MAZ expression was higher in human GBM cell lines than in astrocytes, and was higher in GSCs than in NBTs and GBMs, respectively. These results suggested that MAZ played an oncogenic role in GBM. This was consistent with the previous studies showing that MAZ was over‐expressed in breast cancer and hepatocellular carcinoma (Dudas et al., 2008; Wang et al., 2008), and down‐regulation of MAZ suppressed cell growth and induced apoptosis by inhibition of PPARgamma1 in breast cancer cells (Zaytseva et al., 2008). Mechanistically, MAZ was identified as a direct target of miR‐449a by luciferase assay, suggesting that miR‐449a inhibited the expression of MAZ by targeting its 3′‐UTRs. Furthermore, our results showed that over‐expression of miR‐449a inhibited cell proliferation, migration and invasion as well as inducing apoptosis in U87, U251 and GSCs by down‐regulating MAZ, suggesting that MAZ mediated the tumor‐suppressive effects of miR‐449a. What's more, the in vivo studies showed that nude mice carrying over‐expressed miR‐449a combined with knockdown MAZ cells produced the smallest tumors and highest survival, suggesting that these were likely to achieve synergistic anti‐tumor effects. Collectively, miR‐449a acted as a tumor suppressor by down‐regulating MAZ in human GBM cell lines and GSCs, but the further mechanisms under the MAZ were largely unknown.
Podoplanin (PDPN) is an identified platelet aggregation‐inducing factor of cancer cells (Kaneko et al., 2006). Ectopic expression of PDPN was observed in various cancer cells such as colorectal tumors (Kato et al., 2003), testicular germ cell tumors (Kato et al., 2004), lung squamous cell carcinoma (Kato et al., 2005), malignant astrocytic tumors (Mishima et al., 2006) and glioblastoma (Cortez et al., 2010). PDPN was also known as a specific marker of lymphatic endothelium (Breiteneder‐Geleff et al., 1999), and it has been reported that expression of PDPN by stromal cancer‐associated fibroblasts was a prognostic indicator in various types of cancer (Shindo et al., 2013). Silencing of PDPN expression leads to the reduced proliferation and migration of glioma cells, suggesting a functional role of PDPN in glioma progression and malignancy (Peterziel et al., 2012). In addition, PDPN as a molecular marker was associated with the clinical outcome of glioma patients (Ernst et al., 2009). Anti‐PDPN immunotoxin had highly cytotoxic to glioblastoma and medulloblastoma and increased the survival days when the intracranial tumor model treated with it (Chandramohan et al., 2013). In our study, miR‐449a over‐expression inhibited PDPN expression, whereas miR‐449a inhibition increased PDPN expression in U87, U251 and GSCs, suggesting that it was involved in the tumor‐suppressive effects of miR‐449a. Furthermore, as there are two putative MAZ binding sites in the PDPN promoter, we determined whether miR‐449a could inhibit the PDPN expression by down‐regulating MAZ. Our results showed that over‐expression of miR‐449a inhibited the expression of PDPN by down‐regulating MAZ. Further, the luciferase assay and ChIP assay were conducted to clarify the potential mechanisms of the regulation of PDPN by MAZ. The results indicated that MAZ could up‐regulate the promoter activities and bind to the PDPN promoters in human GBM cell lines and GSCs.
The activation of PI3K/AKT pathway participated in the glioma progression and various biological effects including proliferation, migration, invasion and apoptosis (Liu et al., 2014b; Wang et al., 2014). In addition, previous study found that the increased PDPN expression in human GBM was caused by the activation of the PI3K‐AKT‐AP‐1 signaling pathway. Thus the detection of PI3K/AKT signal pathway was carried out to illustrate the mechanisms of the MAZ‐dependent tumor suppressive effects of miR‐449a in U87, U251 and GSCs. Our results showed that over‐expression of miR‐449a blocked the PI3K/AKT pathway by down‐regulating MAZ. In addition, our results also showed that over‐expression of miR‐449a inhibited the expression of PDPN by down‐regulating MAZ. The functional study of PDPN showed that the increased PDPN expression in human GBM was caused by the activation of the PI3K‐AKT‐AP‐1 signaling pathway (Peterziel et al., 2012). Combined with this previous study, our present study indicated that the inactivation of PI3K/AKT pathway might contribute to the reduced expression of PDPN, which was also found to be directly regulated by MAZ. Thus, the reduced expression of MAZ might attenuate the oncogenic function of human GBM cell lines and GSCs through directly down‐regulating PDPN or the inactivating PI3K/AKT pathway. However, the regulation between MAZ and PI3K/AKT pathway need to be further investigated. Previous studies showed that the activation of PI3K/AKT signaling pathway could stimulate the XIAP and Bcl‐2 expression, whereas inhibited this pathway lead to TRAIL‐induced apoptosis which resulted in the activation of caspase‐8 and ‐10, inducing apoptosis by subsequent activation of the executioner caspase‐3 (Gogineni et al., 2012; Johnstone et al., 2008; Zhang et al., 2014). To determine whether anti‐apoptotic proteins and apoptotic proteins were involved in the increased apoptosis induced by the over‐expression of miR‐449a via down‐regulating MAZ, the expression of anti‐apoptotic proteins and apoptotic proteins in U87, U251 and GSCs were measured. Our results showed that over‐expression of miR‐449a inhibited the expression of anti‐apoptotic proteins (XIAP and Bcl‐2) and increased the expression of apoptotic proteins (Capase‐3 and TRAIL) by down‐regulating MAZ. Previous studies found that the expression of MMP2 and MMP9 were highly increased upon GBM progression, and MMP2 and MMP9 played critical roles in GBM cell migration and invasion (Roy et al., 2009; Song et al., 2009). Moreover, PDPN was found to have a marked influence on cancer cell migration and invasion in association with the expression of MMP2 and MMP‐9 (Inoue et al., 2012; Shindo et al., 2013). PI3K/AKT pathway could also influence on cell migration and invasion by regulating the expression of MMP2 and MMP‐9 (Birner et al., 2014; Mao et al., 2013). In order to determine whether MMP2 and MMP9 were involved in the reduced cell migration and invasion induced by the over‐expression of miR‐449a via down‐regulating MAZ, the detection of MMP2 and MMP‐9 was carried out. Our results indicated that over‐expression of miR‐449a inhibited the expression of MMP2 and MMP9 by down‐regulating MAZ. Collectively, over‐expression of miR‐449a inhibited MAZ expression, and further directly reduced the PDPN expression leading to the decreased expression of MMP‐2 and MMP‐9, or further indirectly reduced PDPN expression which was caused by the inactivation of PI3K/AKT pathway, resulting in the decreased expression of anti‐apoptotic proteins, the increased expression of apoptotic proteins as well as the decreased expression of MMP‐2 and MMP‐9 in human GBM cell lines and GSCs. However, recent studies showed that miR‐449a could inhibit the Bcl‐2 expression by targeting its 3′‐UTR in human gastric adenocarcinoma (Wei et al., 2013) and MAZ could induce MMP‐9 expression in chondrocyte and synoviocyte cells by stimulating MMP‐9 promoter (Ray et al., 2005). Our study could not rule out the possibility that miR‐449a and MAZ can directly regulate the anti‐apoptotic proteins and apoptotic proteins as well as MMP‐2 and MMP‐9 which needs to be further investigated. The mechanism underlying tumor suppression of human GBM cell lines and GSCs by miR‐449a is schematically presented in Figure 10.
Figure 10.
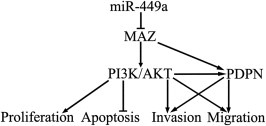
The schematic cartoon of the mechanism underlying tumor suppression of human GBM cell lines and GSCs by miR‐449a.
In conclusion, our study revealed that miR‐449a inhibited the cell proliferation, migration and invasion as well as inducing apoptosis by directly targeting MAZ in human GBM cell lines and GSCs. Mechanistically, the down‐regulating MAZ inhibited PDPN expression and dampened the activation of the PI3K/AKT signaling pathway. More importantly, miR‐449a/MAZ may represent a promising therapeutic targets for the treatment of human glioblastoma.
Acknowledgments
This work is supported by grants from the Natural Science Foundation of China (81172197, 81171131, 81272564, 81272795, 81372484 and 81372682), Shenyang Science and Technology Plan Projects (nos. F11‐264‐1‐15, F12‐277‐1‐05, F13‐318‐1‐16, F13‐318‐1‐19 and F13‐220‐9‐15), and Outstanding Scientific Fund of Shengjing Hospital (No. 201304).
Yao Yilong, Ma Jun, Xue Yixue, Wang Ping, Li Zhen, Li Zhiqing, Hu Yi, Shang Xiuli, Liu Yunhui, (2015), MiR‐449a exerts tumor‐suppressive functions in human glioblastoma by targeting Myc‐associated zinc‐finger protein, Molecular Oncology, 9, doi: 10.1016/j.molonc.2014.11.003.
References
- Alcantara Llaguno, S.R. , Chen, J. , Parada, L.F. , 2009. Signaling in malignant astrocytomas: role of neural stem cells and its therapeutic implications. Clin. Cancer Res. 15, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, S. , Wu, Q. , McLendon, R.E. , Hao, Y. , Shi, Q. , Hjelmeland, A.B. , Dewhirst, M.W. , Bigner, D.D. , Rich, J.N. , 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444, 756–760. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. , 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Birner, P. , Pusch, S. , Christov, C. , Mihaylova, S. , Toumangelova-Uzeir, K. , Natchev, S. , Schoppmann, S.F. , Tchorbanov, A. , Streubel, B. , Tuettenberg, J. , Guentchev, M. , 2014. Mutant IDH1 inhibits PI3K/Akt signaling in human glioma. Cancer. 120, 2240–2447. [DOI] [PubMed] [Google Scholar]
- Bossone, S.A. , Asselin, C. , Patel, A.J. , Marcu, K.B. , 1992. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. U. S. A. 89, 7452–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiteneder-Geleff, S. , Soleiman, A. , Kowalski, H. , Horvat, R. , Amann, G. , Kriehuber, E. , Diem, K. , Weninger, W. , Tschachler, E. , Alitalo, K. , Kerjaschki, D. , 1999. Angiosarcomas express mixed endothelial phenotypes of blood and lymphatic capillaries: podoplanin as a specific marker for lymphatic endothelium. Am. J. Pathol. 154, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower, J.V. , Clark, P.A. , Lyon, W. , Kuo, J.S. , 2014. MicroRNAs in cancer: glioblastoma and glioblastoma cancer stem cells. Neurochem. Int. 77, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buurman, R. , Gurlevik, E. , Schaffer, V. , Eilers, M. , Sandbothe, M. , Kreipe, H. , Wilkens, L. , Schlegelberger, B. , Kuhnel, F. , Skawran, B. , 2012. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 143, 811–820. e811–815 [DOI] [PubMed] [Google Scholar]
- Calin, G.A. , Croce, C.M. , 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6, 857–866. [DOI] [PubMed] [Google Scholar]
- Chandramohan, V. , Bao, X. , Kato Kaneko, M. , Kato, Y. , Keir, S.T. , Szafranski, S.E. , Kuan, C.T. , Pastan, I.H. , Bigner, D.D. , 2013. Recombinant anti-podoplanin (NZ-1) immunotoxin for the treatment of malignant brain tumors. Int. J. Cancer. 132, 2339–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Lin, Y.W. , Mao, Y.Q. , Wu, J. , Liu, Y.F. , Zheng, X.Y. , Xie, L.P. , 2012. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett. 320, 40–47. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Li, Y. , Yu, T.S. , McKay, R.M. , Burns, D.K. , Kernie, S.G. , Parada, L.F. , 2012. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 488, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi, S. , Paramasivam, M. , Membrino, A. , Yokoyama, K.K. , Xodo, L.E. , 2010. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 285, 22003–22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi, S. , Zorzet, S. , Rapozzi, V. , Geci, I. , Pedersen, E.B. , Xodo, L.E. , 2013. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 41, 4049–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, M.A. , Nicoloso, M.S. , Shimizu, M. , Rossi, S. , Gopisetty, G. , Molina, J.R. , Carlotti, C. , Tirapelli, D. , Neder, L. , Brassesco, M.S. , Scrideli, C.A. , Tone, L.G. , Georgescu, M.M. , Zhang, W. , Puduvalli, V. , Calin, G.A. , 2010. miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer. 49, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce, C.M. , 2011. miRNAs in the spotlight: understanding cancer gene dependency. Nat. Med. 17, 935–936. [DOI] [PubMed] [Google Scholar]
- Dudas, J. , Mansuroglu, T. , Moriconi, F. , Haller, F. , Wilting, J. , Lorf, T. , Fuzesi, L. , Ramadori, G. , 2008. Altered regulation of Prox1-gene-expression in liver tumors. BMC Cancer. 8, 92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, A. , Hofmann, S. , Ahmadi, R. , Becker, N. , Korshunov, A. , Engel, F. , Hartmann, C. , Felsberg, J. , Sabel, M. , Peterziel, H. , Durchdewald, M. , Hess, J. , Barbus, S. , Campos, B. , Starzinski-Powitz, A. , Unterberg, A. , Reifenberger, G. , Lichter, P. , Herold-Mende, C. , Radlwimmer, B. , 2009. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin. Cancer Res. 15, 6541–6550. [DOI] [PubMed] [Google Scholar]
- Furnari, F.B. , Fenton, T. , Bachoo, R.M. , Mukasa, A. , Stommel, J.M. , Stegh, A. , Hahn, W.C. , Ligon, K.L. , Louis, D.N. , Brennan, C. , Chin, L. , DePinho, R.A. , Cavenee, W.K. , 2007. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 21, 2683–2710. [DOI] [PubMed] [Google Scholar]
- Gogineni, V.R. , Gupta, R. , Nalla, A.K. , Velpula, K.K. , Rao, J.S. , 2012. uPAR and cathepsin B shRNA impedes TGF-beta1-driven proliferation and invasion of meningioma cells in a XIAP-dependent pathway. Cell Death Dis. 3, e439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous, F. , Zhang, Y. , Kofman, A. , Catania, A. , Li, Y. , Schiff, D. , Purow, B. , Abounader, R. , 2010. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 9, 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjipanayis, C.G. , Van Meir, E.G. , 2009. Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol. Med. 15, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeda, C.L. , Ranish, J.A. , Hauschka, S.D. , 2008. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol. Cell. Biol. 28, 6521–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H. , Miyazaki, Y. , Kikuchi, K. , Yoshida, N. , Ide, F. , Ohmori, Y. , Tomomura, A. , Sakashita, H. , Kusama, K. , 2012. Podoplanin promotes cell migration via the EGF-Src-Cas pathway in oral squamous cell carcinoma cell lines. J. Oral Sci. 54, 241–250. [DOI] [PubMed] [Google Scholar]
- Jiao, L. , Li, Y. , Shen, D. , Xu, C. , Wang, L. , Huang, G. , Chen, L. , Yang, Y. , Yang, C. , Yu, Y. , Sun, Y. , 2013. The prostate cancer-up-regulated Myc-associated zinc-finger protein (MAZ) modulates proliferation and metastasis through reciprocal regulation of androgen receptor. Med. Oncol. 30, 570 [DOI] [PubMed] [Google Scholar]
- Johnstone, R.W. , Frew, A.J. , Smyth, M.J. , 2008. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer. 8, 782–798. [DOI] [PubMed] [Google Scholar]
- Kaneko, M.K. , Kato, Y. , Kitano, T. , Osawa, M. , 2006. Conservation of a platelet activating domain of Aggrus/podoplanin as a platelet aggregation-inducing factor. Gene. 378, 52–57. [DOI] [PubMed] [Google Scholar]
- Kato, Y. , Fujita, N. , Kunita, A. , Sato, S. , Kaneko, M. , Osawa, M. , Tsuruo, T. , 2003. Molecular identification of Aggrus/T1alpha as a platelet aggregation-inducing factor expressed in colorectal tumors. J. Biol. Chem. 278, 51599–51605. [DOI] [PubMed] [Google Scholar]
- Kato, Y. , Kaneko, M. , Sata, M. , Fujita, N. , Tsuruo, T. , Osawa, M. , 2005. Enhanced expression of Aggrus (T1alpha/podoplanin), a platelet-aggregation-inducing factor in lung squamous cell carcinoma. Tumour Biol. 26, 195–200. [DOI] [PubMed] [Google Scholar]
- Kato, Y. , Sasagawa, I. , Kaneko, M. , Osawa, M. , Fujita, N. , Tsuruo, T. , 2004. Aggrus: a diagnostic marker that distinguishes seminoma from embryonal carcinoma in testicular germ cell tumors. Oncogene. 23, 8552–8556. [DOI] [PubMed] [Google Scholar]
- Kennedy, G.C. , Rutter, W.J. , 1992. Pur-1, a zinc-finger protein that binds to purine-rich sequences, transactivates an insulin promoter in heterologous cells. Proc. Natl. Acad. Sci. U. S. A. 89, 11498–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Zhang, Y. , Skalski, M. , Hayes, J. , Kefas, B. , Schiff, D. , Purow, B. , Parsons, S. , Lawler, S. , Abounader, R. , 2014. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 74, 1541–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.M. , Huang, W. , Park, R. , Park, P.J. , Johnson, M.D. , 2011. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 71, 3387–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Kotliarova, S. , Kotliarov, Y. , Li, A. , Su, Q. , Donin, N.M. , Pastorino, S. , Purow, B.W. , Christopher, N. , Zhang, W. , Park, J.K. , Fine, H.A. , 2006. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 9, 391–403. [DOI] [PubMed] [Google Scholar]
- Liu, H.L. , Fan, C.H. , Ting, C.Y. , Yeh, C.K. , 2014. Combining microbubbles and ultrasound for drug delivery to brain tumors: current progress and overview. Theranostics. 4, 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Jiang, Z. , Huang, J. , Huang, S. , Li, Y. , Yu, S. , Yu, S. , Liu, X. , 2014. miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int. J. Oncol. 44, 1571–1580. [DOI] [PubMed] [Google Scholar]
- Luo, W. , Huang, B. , Li, Z. , Li, H. , Sun, L. , Zhang, Q. , Qiu, X. , Wang, E. , 2013. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS One. 8, e64759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , Wang, P. , Liu, Y. , Zhao, L. , Li, Z. , Xue, Y. , 2014. Kruppel-like factor 4 regulates blood-tumor barrier permeability via ZO-1, occludin and claudin-5. J. Cell. Physiol. 229, 916–926. [DOI] [PubMed] [Google Scholar]
- Mao, F. , Wang, B. , Xiao, Q. , Xi, G. , Sun, W. , Zhang, H. , Ye, F. , Wan, F. , Guo, D. , Lei, T. , Chen, X. , 2013. A role for LRIG1 in the regulation of malignant glioma aggressiveness. Int. J. Oncol. 42, 1081–1087. [DOI] [PubMed] [Google Scholar]
- Martin, A. , Jones, A. , Bryar, P.J. , Mets, M. , Weinstein, J. , Zhang, G. , Laurie, N.A. , 2013. MicroRNAs-449a and -449b exhibit tumor suppressive effects in retinoblastoma. Biochem. Biophys. Res. Commun. 440, 599–603. [DOI] [PubMed] [Google Scholar]
- Mishima, K. , Kato, Y. , Kaneko, M.K. , Nishikawa, R. , Hirose, T. , Matsutani, M. , 2006. Increased expression of podoplanin in malignant astrocytic tumors as a novel molecular marker of malignant progression. Acta Neuropathol. 111, 483–488. [DOI] [PubMed] [Google Scholar]
- Noonan, E.J. , Place, R.F. , Pookot, D. , Basak, S. , Whitson, J.M. , Hirata, H. , Giardina, C. , Dahiya, R. , 2009. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 28, 1714–1724. [DOI] [PubMed] [Google Scholar]
- Ohgaki, H. , 2009. Epidemiology of brain tumors. Methods Mol. Biol. 472, 323–342. [DOI] [PubMed] [Google Scholar]
- Ostrom, Q.T. , Gittleman, H. , Farah, P. , Ondracek, A. , Chen, Y. , Wolinsky, Y. , Stroup, N.E. , Kruchko, C. , Barnholtz-Sloan, J.S. , 2013. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol. 15, (Suppl 2) ii1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, S. , Jung, H.S. , Lee, S. , Yoon, D.S. , Park, M.S. , Lee, J.W. , 2012. miR-449a regulates the chondrogenesis of human mesenchymal stem cells through direct targeting of lymphoid enhancer-binding factor-1. Stem Cells Dev. 21, 3298–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, C.L. , Shenk, T. , 1996. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. J. Biol. Chem. 271, 4417–4430. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E. , Hunter, S. , Bracht, J. , 2005. MicroRNAs: a developing story. Curr. Opin. Genet. Dev. 15, 200–205. [DOI] [PubMed] [Google Scholar]
- Peterziel, H. , Muller, J. , Danner, A. , Barbus, S. , Liu, H.K. , Radlwimmer, B. , Pietsch, T. , Lichter, P. , Schutz, G. , Hess, J. , Angel, P. , 2012. Expression of podoplanin in human astrocytic brain tumors is controlled by the PI3K-AKT-AP-1 signaling pathway and promoter methylation. Neuro Oncol. 14, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod, S.S. , Rani, S.B. , Khan, M. , Muzumdar, D. , Shiras, A. , 2014. Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio. 4, 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, A. , Bal, B.S. , Ray, B.K. , 2005. Transcriptional induction of matrix metalloproteinase-9 in the chondrocyte and synoviocyte cells is regulated via a novel mechanism: evidence for functional cooperation between serum amyloid A-activating factor-1 and AP-1. J. Immunol. 175, 4039–4048. [DOI] [PubMed] [Google Scholar]
- Ren, X.S. , Yin, M.H. , Zhang, X. , Wang, Z. , Feng, S.P. , Wang, G.X. , Luo, Y.J. , Liang, P.Z. , Yang, X.Q. , He, J.X. , Zhang, B.L. , 2014. Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett. 344, 195–203. [DOI] [PubMed] [Google Scholar]
- Roy, R. , Yang, J. , Moses, M.A. , 2009. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 27, 5287–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Chen, C. , Zhang, X. , Liu, Q. , Xu, J.L. , Zhang, H.R. , Yao, X.H. , Jiang, T. , He, Z.C. , Ren, Y. , Cui, W. , Xu, C. , Liu, L. , Cui, Y.H. , Yu, S.Z. , Ping, Y.F. , Bian, X.W. , 2014. Primate-specific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clin. Cancer Res. 20, 1803–1813. [DOI] [PubMed] [Google Scholar]
- Shindo, K. , Aishima, S. , Ohuchida, K. , Fujiwara, K. , Fujino, M. , Mizuuchi, Y. , Hattori, M. , Mizumoto, K. , Tanaka, M. , Oda, Y. , 2013. Podoplanin expression in cancer-associated fibroblasts enhances tumor progression of invasive ductal carcinoma of the pancreas. Mol. Cancer. 12, 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K. , Clarke, I.D. , Terasaki, M. , Bonn, V.E. , Hawkins, C. , Squire, J. , Dirks, P.B. , 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63, 5821–5828. [PubMed] [Google Scholar]
- Singh, S.K. , Hawkins, C. , Clarke, I.D. , Squire, J.A. , Bayani, J. , Hide, T. , Henkelman, R.M. , Cusimano, M.D. , Dirks, P.B. , 2004. Identification of human brain tumour initiating cells. Nature. 432, 396–401. [DOI] [PubMed] [Google Scholar]
- Smits, M. , Wurdinger, T. , van het Hof, B. , Drexhage, J.A. , Geerts, D. , Wesseling, P. , Noske, D.P. , Vandertop, W.P. , de Vries, H.E. , Reijerkerk, A. , 2012. Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB J. 26, 2639–2647. [DOI] [PubMed] [Google Scholar]
- Song, H. , Li, Y. , Lee, J. , Schwartz, A.L. , Bu, G. , 2009. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res. 69, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Murakami, H. , Tsutsui, H. , Tang, X. , Matsumura, M. , Itakura, K. , Kanazawa, I. , Sun, K. , Yokoyama, K.K. , 1998. Genomic organization and expression of a human gene for Myc-associated zinc finger protein (MAZ). J. Biol. Chem. 273, 20603–20614. [DOI] [PubMed] [Google Scholar]
- Stupp, R. , Hegi, M.E. , Gilbert, M.R. , Chakravarti, A. , 2007. Chemoradiotherapy in malignant glioma: standard of care and future directions. J. Clin. Oncol. 25, 4127–4136. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Shi, Z.M. , Jiang, C.F. , Liu, X. , Chen, Q.D. , Qian, X. , Li, D.M. , Ge, X. , Wang, X.F. , Liu, L.Z. , You, Y.P. , Liu, N. , Jiang, B.H. , 2014. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 5, 5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Southard, R.C. , Allred, C.D. , Talbert, D.R. , Wilson, M.E. , Kilgore, M.W. , 2008. MAZ drives tumor-specific expression of PPAR gamma 1 in breast cancer cells. Breast Cancer Res. Treat. 111, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B. , Song, Y. , Zhang, Y. , Hu, M. , 2013. microRNA-449a functions as a tumor-suppressor in gastric adenocarcinoma by targeting Bcl-2. Oncol. Lett. 6, 1713–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y. , Xue, Y. , Ma, J. , Shang, C. , Wang, P. , Liu, L. , Liu, W. , Li, Z. , Qu, S. , Li, Z. , Liu, Y. , 2014. MiR-330-Mediated regulation of SH3GL2 expression enhances malignant behaviors of glioblastoma stem cells by activating ERK and PI3K/AKT signaling pathways. PLoS One. 9, e95060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, W. , Xue, J. , Zhang, Q. , Li, F. , Zhang, W. , Chen, H. , Huang, Y. , Zheng, F. , 2014. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol. Rep. 32, 1193–1199. [DOI] [PubMed] [Google Scholar]
- Zaytseva, Y.Y. , Wang, X. , Southard, R.C. , Wallis, N.K. , Kilgore, M.W. , 2008. Down-regulation of PPARgamma1 suppresses cell growth and induces apoptosis in MCF-7 breast cancer cells. Mol. Cancer. 7, 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Ruvolo, V.R. , Gao, C. , Zhou, L. , Bornmann, W. , Tsao, T. , Schober, W.D. , Smith, P. , Guichard, S. , Konopleva, M. , Andreeff, M. , 2014. Evaluation of apoptosis induction by concomitant inhibition of MEK, mTOR, and Bcl-2 in human acute myeloid leukemia cells. Mol. Cancer Ther. 13, 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.B. , Zhang, H. , Damelin, M. , Geles, K.G. , Grindley, J.C. , Dirks, P.B. , 2009. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat. Rev. Drug Discov. 8, 806–823. [DOI] [PubMed] [Google Scholar]


