Abstract
Background
Prognosis of malignant pleural mesothelioma (MPM) is poor, and predicting the outcomes of treatment is difficult. Here we investigate the potential of microRNA expression to estimate prognosis of MPM patients.
Methods
Candidate microRNAs from microarray profiling of tumor samples from 8 long (median: 53.7 months) and 8 short (median: 6.4 months) survivors following extrapleural pneumonectomy (EPP) were validated by RT‐qPCR in 48 additional EPP samples. Kaplan–Meier log ranking was used to further explore the association between microRNA expression and overall survival (OS). Binary logistic regression was used to construct a microRNA signature (miR‐Score) that was able to predict an OS of ≥20 months. Performance of the miR‐Score was evaluated by receiver operating characteristic (ROC) curve analysis and validated in a series of 43 tumor samples from patients who underwent palliative surgery [pleurectomy/decortication (P/D)].
Results
The miR‐Score, using expression data of six microRNAs (miR‐21‐5p, ‐23a‐3p, ‐30e‐5p, ‐221‐3p, ‐222‐3p, and ‐31‐5p), enabled prediction of long survival with an accuracy of 92.3% for EPP and 71.9% for palliative P/D. Hazard ratios for score‐negative patients were 4.12 (95% CI: 2.03–8.37) for EPP and 1.93 (95% CI: 1.01–3.69) for P/D. Importantly, adding the miR‐Score to a set of clinical selection criteria (histology, age, gender) increased predictive accuracy in the independent validation set from 76.3% for clinical factors only to 87.3%.
Conclusions
This study has identified a novel 6‐microRNA signature (miR‐Score) that can accurately predict prognosis of MPM patients.
Keywords: Malignant pleural mesothelioma, MicroRNAs, Prognosis, Biomarker, Survival
Highlights
MicroRNA expression in malignant pleural mesothelioma tissue is associated with prognosis.
A novel microRNA signature (miR‐Score) can predict prolonged survival.
The miR‐Score has prognostic value in malignant pleural mesothelioma patients undergoing surgery.
1. Introduction
Malignant pleural mesothelioma (MPM) is a highly aggressive cancer arising from the mesothelial lining of the thoracic cavities. With a median survival of less than 1 year and a 5‐year survival rate of less than 5% (van Meerbeeck et al., 2011), prognosis of this asbestos‐related cancer remains very poor.
According to current guidelines the majority of MPM patients will be eligible for palliative chemotherapy (van Zandwijk et al., 2013). However, there is a select group of patients that can be considered for intensive multimodality treatment including induction chemotherapy, radical removal of the diseased pleura (+/− the underlying lung) and postoperative radiotherapy. Unfortunately, prognostic criteria to select patients for intensive multimodality approaches are not available. There are prognostic scores for MPM that have been derived from data collected from phase II clinical trials in the US and Europe studying novel chemotherapy regimens (Curran et al., 1998; Herndon et al., 1998) in the previous century, but these are not sufficiently accurate to allow patient selection. More recent studies have explored the value of novel markers to aid in the selection of MPM patients but few have been validated (Bitanihirwe et al., 2014; Cedres et al., 2012; Kao et al., 2011; Opitz et al., 2008; Pass, 2012; Schramm et al., 2010).
In recent years it has become apparent that the expression of certain microRNAs in tumor cells can be closely associated with prognosis and a number of studies have profiled the microRNA content of MPM cell lines and/or MPM tumor tissue (Balatti et al., 2011; Benjamin et al., 2010; Busacca et al., 2010; Gee et al., 2010; Guled et al., 2009; Pass et al., 2010). Two studies have explored the prognostic value of specific microRNAs in MPM. One of them suggested that the expression of hsa‐miR‐17‐5p and hsa‐miR‐30c was correlated with survival in sarcomatoid tumors (Busacca et al., 2010); the second study exploring microRNA content in 129 fresh‐frozen surgical specimens concluded that elevated expression of hsa‐miR‐29c‐5p was associated with a significant survival difference (Pass et al., 2010).
With the intention to better understand the prognostic value of microRNAs in MPM we have performed a microarray profiling study using formalin‐fixed paraffin embedded (FFPE) tumor specimens from patients undergoing EPP. Twenty differentially expressed candidate microRNAs were evaluated using RT‐qPCR in a second set of EPP patients, and we were able to identify a signature of six microRNAs (miR‐Score) that allowed an accurate prediction of prolonged survival in these patients. In addition, the same signature was also prognostic in patients receiving a palliative [pleurectomy ± decortication (P/D)] surgical procedure. To our knowledge, the miR‐Score is the first multi‐microRNA signature with broad prognostic value for MPM.
2. Materials and methods
2.1. Patient samples
Waiver of consent for the use of samples in this retrospective study was granted by the Human Research Ethics Committee at Concord Repatriation General Hospital, Sydney, Australia (CH62/6/2009/078). The histopathology of all samples was independently reassessed by an expert pathologist [SK] and final diagnoses were made according to World Health Organization (WHO) criteria (Travis et al., 2004).
2.1.1. Extrapleural pneumonectomy (EPP) cohort
We studied 64 FFPE tumor tissue samples from patients who underwent EPP between October 1994 and November 2009 at Royal Prince Alfred (RPAH) or Strathfield Private Hospitals (SPH) in Sydney, Australia. These samples form part of a series of 85 consecutive patients previously used to assess the prognostic value of calretinin and neutrophil‐to‐lymphocyte ratio (Kao et al., 2011). Samples selected for the present study were those for which RNA of sufficient quality was available. Baseline characteristics of the 85 EPP patients and the subset of 64 patients used in this study are provided in Table 1.
Table 1.
Baseline characteristic of EPP and P/D cohorts.
| Variables | EPP cohort | P/D cohort | ||||
|---|---|---|---|---|---|---|
| Complete cohort (N = 85) | Patients with RNA (N = 64) | Patients without RNA (N = 21) | Complete cohort (N = 75) | Patients with RNA (N = 43) | Patients without RNA (N = 32) | |
| Median age (range) | 58 (22–74) | 62 (47–70) | 59 (41–70) | 66 (42–83) | 65 (42–79) | 66 (47–83) |
| Gender | ||||||
| Male | 68 (80%) | 49 (77%) | 19 (90%) | 59 (79%) | 34 (79%) | 25 (78%) |
| Female | 17 (20%) | 15 (23%) | 2 (10%) | 16 (21%) | 9 (21%) | 7 (22%) |
| Histological subtype | ||||||
| Epithelioid | 65 (76%) | 47 (73%) | 18 (86%) | 37 (49%) | 25 (58%) | 12 (38%) |
| Biphasic | 20 (24%) | 17 (27%) | 3 (14%) | 26 (35%) | 13 (30%) | 13 (41%) |
| Sarcomatoid | 0 (0%) | 0 (0%) | 0 (0%) | 12 (16%) | 5 (12%) | 7 (22%) |
| Pathological stagea,b | ||||||
| Complete response | 2 (2%) | 2 (10%) | N/A | |||
| I | 5 (6%) | 2 (3%) | 3 (14%) | N/A | ||
| II | 18 (21%) | 9 (14%) | 9 (43%) | N/A | ||
| III | 54 (64%) | 47 (73%) | 7 (33%) | N/A | ||
| IV | 6 (7%) | 6 (9%) | 0 (0%) | N/A | ||
| Median OS (months) | 18.86 (0.07–122.41) | 15.28 (0.07–90.48) | 26.64 (0.07–122.41) | 7.62 (0.33–224.82) | 8.64 (0.33–224.82) | 7.21 (0.56–79.74) |
Statistically different between patients with RNA and patients without RNA (defined as p < 0.05 in Mann–Whitney Test or Kaplan–Meier Analysis for OS).
Pathological stage was determined according to the American Joint Committee on Cancer Staging System (Edge et al., 2010).
Samples from 8 long (median: 53.7 months) and 8 short (median: 6.4 months) survivors were used as a discovery set (see Suppl. Table 1 for baseline characteristics), excluding patients with biphasic histology and patients who received induction therapy. The remaining 48 samples formed a training set in follow‐up RT‐qPCR studies, including patients with biphasic histology (17/48, 35.4%), those who received induction chemotherapy (13/48, 27.1%) and those who died <8 weeks after surgery (2/48, 4.2%).
2.1.2. Pleurectomy ± decortication (P/D) cohort
This cohort consisted of archival FFPE blocks from 75 consecutive patients undergoing palliative P/D at RPAH between October 1991 and October 2006 for whom tissue was available, and was previously used to study the prognostic potential of PLK1 and CDK1 (Linton et al., 2014). As part of the current study, RNA was isolated from the samples in this cohort. RNA of sufficient quality was available for 43 out of 75 patients, and used as a validation set. Baseline characteristics of the P/D patients are provided in Table 1.
2.2. RNA isolation
Prior to RNA isolation, samples were enriched for tumor content by laser‐capture microdissection (LCM). Briefly, tumor areas were marked on hematoxylin and eosin stained sections by an experienced pathologist [SK] to guide LCM. Adjacent sections were then mounted onto membrane slides for LCM using the PALM system (Zeiss, Jena, Germany), and captured tumor tissue was collected into adhesive collection tubes. Samples were deparaffinized in xylene and RNA isolated using the RNeasy FFPE Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The resulting RNA was quantified using a Nanophotometer (Implen, Munich, Germany) with readings at 260 and 280 nm. The quality of the small RNA component was assessed by capillary electrophoresis using Small RNA Chips run on an Agilent Bioanalyzer (both Agilent Technologies, Santa Clara, CA, USA) to ensure presence of an intact microRNA fraction. RNA was considered to be of sufficient quality if (i) 260/280 ratios were above 1.7; (ii) the proportion of miRNA in the sample was above 20% and (iii) the endogenous control RNU6B amplified at a quantification cycle (Cq) <33.
2.3. MicroRNA microarray analysis
MicroRNA profiling was performed on 100 ng of total RNA from each sample using the Agilent Technologies Human 8 × 15 k miRNA MicroArray Kit V3 (miRBase V12.0) as per manufacturer's instructions with hybridization to the array slides for 22 h. Hybridized array slides were stored in nitrogen gas until scanning (within 48 h) on an Agilent C MicroArray Scanner at the Ramaciotti Centre for Genomics at the University of New South Wales, Sydney. Raw array data were extracted using the Agilent Feature Extraction software (V10.5). Expression analyses were performed applying the guided workflow setting in GeneSpring 12.1 software. Data processing involved thresholding of signal values to 1, transformation into log base 2, and normalization by shift to the 90th percentile without applying baseline transformation. All expression data has been deposited in the National Centre for Biotechnology Information (NCBI) Gene Expression Omnibus (Edgar et al., 2002) under accession GEO: GSE59180.
2.4. RT‐qPCR
RT‐qPCR for selected candidates was performed using stem‐loop primers and hydrolysis probes (Life Technologies, Carlsbad, CA, USA; see Suppl. Table 2 for assay IDs) as described previously (Kirschner et al., 2012, 2013) and in the supplemental material. RNU6B was measured as endogenous control, and data were analyzed applying a variation of the 2−ΔΔCq method (Livak and Schmittgen, 2001):
-
(I)
For each sample: ΔCqx = CqmicroRNA − CqRNU6B
-
(II)
For each microRNA: ΔCqavg = mean ΔCq of all samples (in the set)
-
(III)
For each sample: 2−ΔΔCq = 2−ΔCqx−ΔCqavg = miR − Xexpr
2.5. Pathway analysis
To test the biological relevance of the prognostic microRNAs identified, we utilized pathway enrichment analysis of their target genes (identified by TargetScan 5.2) in order to determine gene modules associated with the altered microRNAs. These analyses were performed using Partek Genomics Suite 6.5 (Partek Inc, St. Louis, MO, USA). The Database for Annotation, Visualization, and Integrated Discovery [DAVID (Huang et al., 2008)] was used to identify enrichment in Gene Ontology (GO) Terms regulated by the identified microRNAs and their target genes.
2.6. Statistical analysis
Overall survival (OS) was calculated from the time of surgery to time of death or last follow‐up. Patients alive at time of analysis or lost to follow‐up were censored to the last date of follow‐up. Differences in relative expression levels on the microarray were assessed by two‐tailed independent samples t‐tests with the Benjamini–Hochberg False Discovery Rate (FDR) correction. Biological significance was determined as a difference in expression of more than ±2‐fold between groups. In the training set, the median expression of each candidate was used as cut‐off to dichotomize into low and high expression to assess the association of each microRNA with OS using the Kaplan–Meier log‐rank method. Individually significant microRNAs were entered into a multivariate Cox regression model together with the established risk factors of histological subtype, age and gender. Binary logistic regression with backward selection on likelihood ratio to determine best fit (inclusion cut‐off: P < 0.05, exclusion cut‐off: P > 0.1) was used to build a microRNA signature (on continuous variables) able to predict good prognosis (defined as OS of ≥20 months). This provides a formula to predict the probability of good prognosis [Eq. (IV)]:
| (IV) |
where P is the probability of good prognosis, X 1 − X n are the predictor variables (microRNAs), b 1 − b n, the regression coefficients, and b 0 the intercept (or constant). Predicted probabilities were entered into receiver operating characteristic (ROC) curve analysis without bootstrapping to determine the accuracy of the signature (miR‐Score) as well as the optimal cut‐off score for good prognosis. Five hundred bootstrap re‐samples were then generated and used to derive robust estimates for the average area under the curve (AUC) and Confidence Intervals (CIs). A clinical score and a combined clinical/miR‐Score were built using straightforward logistic regression without further selection, and using ≥20 months survival as dependent variable using each factor (age, gender, histotype, and miR‐Score) as binary variable. Cut‐off score‐values for dichotomization into good and bad prognosis were chosen as the score‐value associated with the highest achievable sensitivity and specificity. Forest plots of odds ratios (OR) from logistic regression were used to compare the ability of individual microRNAs (continuous variables), clinical factors and the different scores (all as categorical variables) to predict survival of ≥20 months. Unadjusted and adjusted (microarray) P‐values of ≤0.05 were considered significant. All analyses on RT‐qPCR data were performed using SPSS V21 (SPSS Inc, Chicago, Il, USA) and R v3.1.0. An overview of the study design and the use of the different samples are provided in Suppl. Figure 1.
3. Results
3.1. MicroRNA profiling identifies candidate microRNAs with differential expression between long and short survivors
Microarray‐based profiling performed on tumor samples from 8 long and 8 short survivors identified 16 microRNAs that were differentially expressed (≥2‐fold) between the two groups (Suppl. Table 3). Twelve microRNAs were present at significantly lower levels in tumors from long survivors, whereas four were present at higher levels. Of these 16 microRNAs, 6 remained significant after Benjamini–Hochberg correction (Suppl. Table 3), namely miR‐21‐5p, ‐210‐3p, ‐221‐3p, ‐27a‐3p, ‐93‐5p, and 23a‐3p, all of them present at lower levels in the group of long survivors. Candidates for validation by RT‐qPCR were selected on the basis of the following criteria: (i) significant after Benjamini–Hochberg correction; (ii) significant before Benjamini–Hochberg and fold‐difference between groups ≥2.0; (iii) classification as microRNA in miRBase v19 and (iv) availability of a hydrolysis probe. This resulted in 14 candidates for validation (Suppl. Table 3), to which we added microRNAs previously associated with prognosis [miR‐29c‐5p, ‐31‐5p (Ivanov et al., 2010; Pass et al., 2010)] or diagnosis [miR‐106a‐5p, ‐126‐3p, ‐625‐3p, ‐92a‐3p (Busacca et al., 2010; Kirschner et al., 2012; Santarelli et al., 2011)] for MPM patients. We also included miR‐23a‐3p and ‐24‐3p as these microRNAs are expressed on the same transcript as miR‐27a‐3p.
Technical validation using RT‐qPCR was performed on those samples from the discovery set (6 long and 6 short survivors) for which sufficient RNA remained following microarray studies. RT‐qPCR validation included a total of 21 microRNAs (14 array candidates + 7 linked to MPM) of which only miR‐298 was not detectable by RT‐qPCR and was subsequently excluded from further analyses. For the remaining 13 array candidates overall trends for differential expression were confirmed, with miRs‐23a‐3p, ‐30e‐5p, and ‐21‐5p reaching statistical significance (Suppl. Table 3). While some of the previously identified microRNAs showed differential expression, none reached statistical significance. Due to the relatively low number of samples in the discovery set, all selected candidates were included in the training set.
3.2. Nine candidate microRNAs are associated with prolonged survival in mesothelioma patients undergoing radical surgery (EPP)
Following measurement of microRNA levels in the training set, samples were dichotomized into low and high expression based on the median expression of each microRNA observed across all 48 samples. Categorized microRNA expression as well as established prognostic factors such as, histological subtype, age, gender and stage were entered into log‐rank regression models, and differences in survival were assessed using the Kaplan–Meier method. Only histological subtype (Figure 1A) and gender (Figure 1C), but not age (Figure 1B) or stage (Figure 1D), were associated with survival in this cohort.
Figure 1.
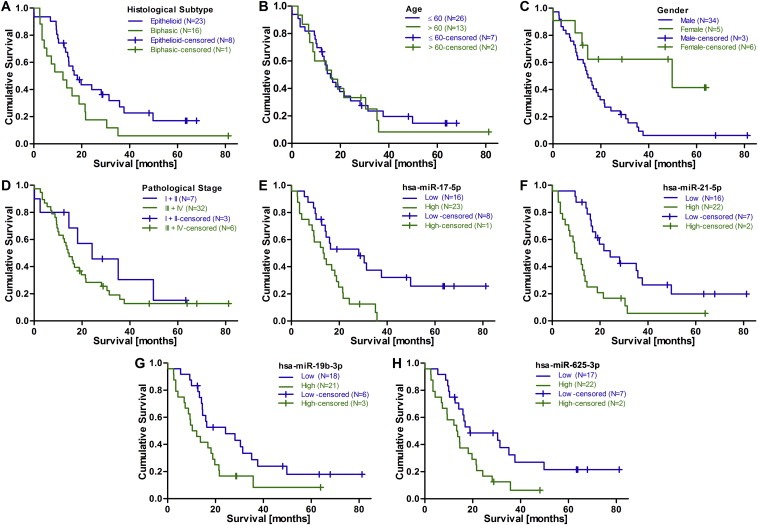
Kaplan–Meier analysis of clinical factors and microRNAs in the training set (N = 48). Kaplan–Meier analyses based on (A) tumor histology (HR for biphasic histology = 1.89, P = 0.052), (B) age (dichotomized at 60 y; HR for >60 years = 1.13, P = 0.716), (C) gender (HR for male gender = 2.96, P = 0.025), or (D) pathological stage (HR of 1.52 (P = 0.316) for stage III + IV). (E–H) For Kaplan–Meier analysis of microRNAs patients were stratified into high and low expression based on the median expression across all tumors as cut‐off. Higher tumor microRNA expression was associated with shorter survival for (E) miR‐17‐5p, (F) miR‐21‐5p, (G) miR‐19b‐3p, and (H) miR‐625‐3p.
Examining the survival curves for each microRNA revealed that lower expression of 9 of these was significantly associated with up to 14.9 months longer OS. The best candidates identified by univariate analysis were miR‐17‐5p (Figure 1E, 28.2 vs 13.3 months, HR 2.59, P = 0.006), miR‐21‐5p (Figure 1F, 24.2 vs 9.4 months, HR 2.84, P = 0.002), and miR‐19b‐3p (Figure 1G, 24.2 vs 10.3 months, HR 1.97, P = 0.039).
Using multivariate Cox regression analysis including histological subtype, gender, age (dichotomized at age 60) and the respective microRNAs, miR‐21‐5p, miR‐19b‐3p, miR‐625‐3p (Figure 1H), and miR‐106b‐5p remained significant (Table 2).
Table 2.
Summary of univariate and multivariate Cox regression analysis of microRNAs in training set (N = 48).
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | |
| hsa‐miR‐21‐5p | 2.84 | 1.48–5.45 | 0.002 | 3.35 | 1.66–6.75 | 0.001 |
| hsa‐miR‐17‐5p | 2.59 | 1.31–5.11 | 0.006 | 2.03 | 0.96–4.28 | 0.063 |
| hsa‐miR‐662 | 2.3 | 1.16–4.56 | 0.017 | 2.01 | 0.99–4.06 | 0.052 |
| hsa‐miR‐20a‐5p | 2.25 | 1.16–4.36 | 0.016 | 1.64 | 0.81–3.35 | 0.173 |
| hsa‐miR‐625‐3p | 2.16 | 1.12–4.16 | 0.022 | 2.23 | 1.12–4.41 | 0.022 |
| hsa‐miR‐27a‐3p | 2.12 | 1.11–4.06 | 0.024 | 1.85 | 0.85–3.98 | 0.113 |
| hsa‐miR‐106a‐5p | 2.11 | 1.09–1.05 | 0.027 | 1.55 | 0.77–3.10 | 0.218 |
| hsa‐miR‐210‐3p | 2.01 | 1.05–3.86 | 0.035 | 1.45 | 0.73–2.88 | 0.286 |
| hsa‐miR‐19b‐3p | 1.97 | 1.04–3.76 | 0.039 | 2.01 | 1.03–3.94 | 0.042 |
| hsa‐miR‐24‐3p | 1.77 | 0.93–3.37 | 0.081 | 1.65 | 0.85–3.20 | 0.137 |
| hsa‐miR‐23a‐3p | 1.52 | 0.80–2.86 | 0.198 | 1.24 | 0.62–2.51 | 0.546 |
| hsa‐miR‐222‐3p | 1.40 | 0.74–2.65 | 0.295 | 1.35 | 0.71–2.55 | 0.365 |
| hsa‐miR‐30e‐5p | 1.26 | 0.67–2.37 | 0.471 | 1.78 | 0.89–3.53 | 0.102 |
| hsa‐miR‐106b‐5p | 1.25 | 0.68–2.43 | 0.442 | 2.53 | 1.21–5.29 | 0.014 |
| hsa‐miR‐93‐5p | 1.24 | 0.66–2.33 | 0.504 | 1.31 | 0.69–2.49 | 0.402 |
| hsa‐miR‐221‐3p | 1.21 | 0.64–2.28 | 0.555 | 0.99 | 0.51–1.93 | 0.973 |
| hsa‐miR‐92a‐3p | 1.15 | 0.61–2.17 | 0.660 | 1.01 | 0.53–1.90 | 0.989 |
| hsa‐miR‐31‐5p | 1.06 | 0.57–2.00 | 0.849 | 0.97 | 0.50–1.90 | 0.939 |
| hsa‐miR‐29c‐5p | 0.97 | 0.52–1.82 | 0.921 | 1.33 | 0.69–2.57 | 0.392 |
| hsa‐miR‐126‐3p | 0.95 | 0.51–1.80 | 0.884 | 0.88 | 0.43–1.82 | 0.726 |
Samples were grouped into low and high expression based on the median expression of the complete set (N = 48). Hazard Ratios (HR) were estimated using univariate and multivariate Cox regression models. In the multivariate model single microRNAs were entered together with the clinical factors histological subtype, age and gender. HRs represent the estimated risk for patients with high expression of the respective microRNA. MicroRNAs significant in multivariate analysis are presented in bold.
3.3. A 6‐microRNA signature (miR‐Score) is associated with prolonged survival in mesothelioma patients undergoing EPP
While establishing an association between lower expression of certain microRNAs and prolonged survival (up to 15 months longer in the good prognosis group) is a step forward, the real question is how accurately these microRNAs can predict prolonged survival. With the median OS for the complete cohort being almost 19 months and a median OS of around 23 months being reported for multimodality treatment (van Meerbeeck et al., 2011), we chose 20 months OS as cut‐off for good prognosis. To evaluate the ability of each of the microRNAs to predict this, we entered each (as continuous variable) into binary logistic regression models with ≥20 months survival as desired outcome. None of the microRNAs, nor any of the established risk factors were significant in univariate models (Suppl. Table 4). Since combinations of predictors (signatures) can show significantly improved predictive accuracy compared to single factors we investigated the performance of multivariate binary logistic regression models, using backward selection starting with all 20 microRNAs. This approach resulted in a final model consisting of six microRNAs (miR‐21‐5p, ‐23a‐3p, ‐30e‐5p, ‐221‐3p, ‐222‐3p, and ‐31‐5p). The model formula is given by Eq. (V):
| (V) |
To assess the prognostic potential of the 6‐microRNA signature (miR‐Score), ROC curve analysis on predicted probabilities was performed, resulting in an AUC of 0.867 (95% CI: 0.76–0.96, Figure 2A), and an average AUC of 0.922 (SE = 0.053, Figure 2B) after bootstrapping 500 re‐samples. To categorize a patient into the good or poor prognosis group (according to miR‐Score) we chose the probability value with highest sensitivity and specificity, resulting in a cut‐off score of 0.44 with 82.4% sensitivity and 80.6% specificity. Kaplan–Meier analysis (Figure 2C) comparing miR‐Scores showed a highly significant difference (22.9 months) between the miR‐Score positive and negative groups. Score‐positivity was associated with an odds ratio of 19.44 (95% CI: 4.20–90.03, P = 0.0001) for survival of ≥20 months (Suppl. Table 4).
Figure 2.
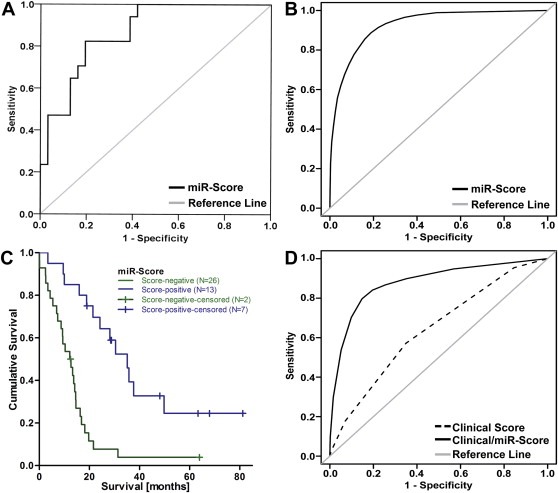
Performance of the 6‐microRNA signature (miR‐Score). (A) The predictive accuracy of the miR‐Score was assessed using ROC curve analysis. (B) The average ROC curve after bootstrapping 500 re‐samples. (C) Kaplan–Meier analysis after stratification by miR‐Score (HR for score‐negative patients = 4.12, P = 0.00009). (D) ROC curve analysis of a Clinical Score (dotted line) and a combined Clinical/miR‐Score (solid line).
To evaluate whether adding the miR‐Score to the established clinical risk factors resulted in improved predictive accuracy, we compared ROC curves obtained from straightforward logistic regression models consisting of (i) clinical factors only and (ii) clinical factors plus the miR‐Score (Figure 2D). The addition of the miR‐Score (as a binary variable) to the model increased the AUC from 0.601 (95% CI: 0.437–0.764) to 0.844 (95% CI: 0.707–0.982). Bootstrapping 500 re‐samples, and obtaining the difference between the AUC for the clinical model and the clinical/miR‐Score model revealed that including the miR‐Score increased the AUC by an average of 0.212 points (SE = 0.069). Comparing the predicted and observed outcomes for each individual case showed that addition of the miR‐Score to the prediction model resulted in re‐classification of 16 patients into the correct group (good or poor prognosis), which outweighed the incorrect re‐classification of three patients. Of the 4 cases that were classified incorrectly by every prediction model, the actual survival for 3 of them was within two months of the 20‐month cut‐off (data not shown). Investigating a possible connection between the miR‐Score and histological subtype or induction chemotherapy revealed no significant differences in score levels between the respective subgroups (data not shown).
3.4. The miR‐Score is also associated with prolonged survival in mesothelioma patients receiving palliative surgery
Independent validation of the miR‐Score was performed using tumor samples from patients undergoing palliative P/D. Univariate Kaplan–Meier analyses (Figure 3A–C) showed that there were significant differences in survival between histological subtypes (15.4 [epithelioid] vs 5.7 [biphasic] vs 3.8 months [sarcomatoid], P = 0.00001), and gender (8.6 months [male] vs 7.6 months [female], P = 0.047), but not for age (10.8 [<60] vs 6.5 months [>60], P = 0.155). Although both miR‐21‐5p (P = 0.025) and miR‐221‐3p (P = 0.017) were significantly associated with prolonged survival in univariate logistic regression (Figure 3D), the miR‐Score again outperformed single microRNAs with an OR of 9.72 for score‐positivity (95% CI: 1.70–55.75, P = 0.011, Figure 3D). ROC curve analysis of the miR‐Score then showed that in this set of patients the microRNA signature was able to predict a good prognosis with an accuracy of 71.9% (AUC 0.719, 95% CI: 0.535–0.902, Figure 3E). MiR‐Score‐positivity was associated with a survival benefit of 8.9 months (Figure 3F, 15.4 vs 6.5 months, P = 0.044). Comparing models containing (i) clinical factors only; or (ii) clinical factors and the miR‐Score, showed that the miR‐Score improved accuracy in terms of AUC, from 0.763 to 0.873 (Figure 3G) without reaching statistical significance in this cohort. Comparison of ORs for clinical prognostic factors and the miR‐Score confirmed the superior performance of the miR‐Score and the combined clinical/miR‐Score (OR for score‐positivity = 26.00; 95% CI: 2.81–240.53) over the clinical factors alone (Figure 3H). Furthermore, addition of the miR‐Score resulted in correct re‐classification of 10 patients, compared to incorrect re‐classification of only one. For the six cases that were classified wrong by each model, no common pattern could be observed.
Figure 3.
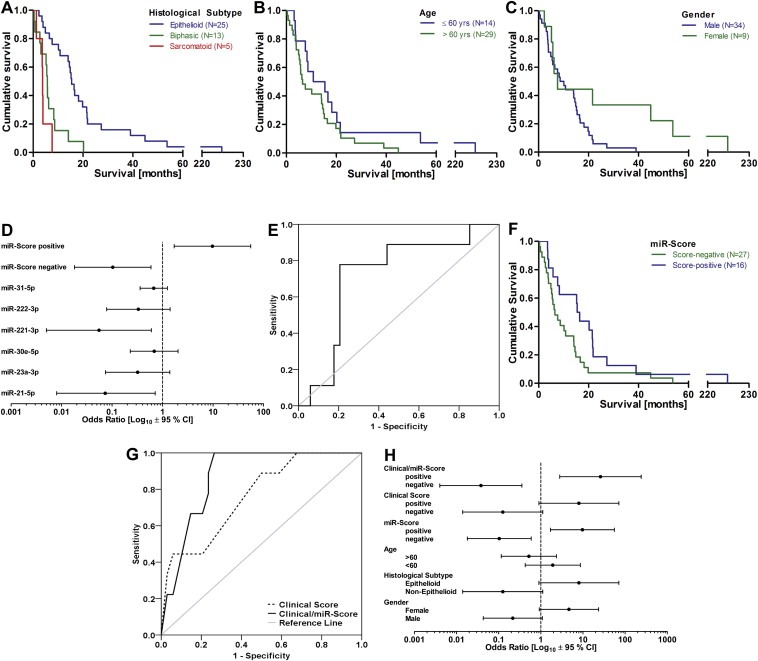
Validation of the miR‐Score in the P/D Cohort (N = 43). Kaplan–Meier analysis for the validation set based on (A) tumor histology (HRs: 3.93, P = 0.004) for biphasic and 8.55 (P = 0.002) for sarcomatoid); (B) stratification by age (HR for patients aged >60 = 1.63, P = 0.159); and (C) gender (HR for male = 2.44, P = 0.050). (D) Comparison of ORs for prediction of good prognosis. (E) ROC curve analysis of the miR‐Score. (F) Kaplan–Meier analysis after stratification based on score‐positivity (HR for miR‐Score negativity = 1.93, p = 0.047). (G) ROC curve analysis of a Clinical Score (dotted line) and a combined Clinical/miR‐Score (solid line). (H) OR comparison between scores derived from the clinical factors and the miR‐Score.
3.5. MiR‐score microRNAs target genes in pathways implicated in MPM development
Pathway enrichment analysis was performed based on the target genes of (i) the 6 microRNAs of the miR‐Score and (ii) the 11 microRNAs for which expression was found to be associated with survival. Both analyses showed significant enrichment of regulatory pathways whose dysregulation has previously been linked to MPM (de Assis et al., 2014): Wnt signaling pathway (miR‐Score: P = 7.33 × 10−5, 11‐miRs: P = 3.49 × 10−7); Hippo signaling pathway (miR‐Score: P = 6.32 × 10−4, 11‐miRs: P = 1.73 × 10−6); PI3K/Akt signaling pathway (miR‐Score: P = 9.66 × 10−3, 11‐miRs: P = 1.13 × 10−3); and pathways in cancer (miR‐Score: P = 2.88 × 10−4, 11‐miRs: P = 3.29 × 10−5). In addition, analysis based on the 11 microRNAs associated with survival also showed enrichment in the mTOR (P = 1.58 × 10−3) and Hedgehog (P = 2.22 × 10−3) signaling pathways. Functional analysis using DAVID showed enrichment of the GO terms regulation of transcription, gene expression, and translation. A summary of enriched pathways is provided in Table 3.
Table 3.
Enriched KEGG pathways for the 6 miR‐Score microRNAs and the 11 microRNAs associated with survival.
| Enriched KEGG pathway | miR‐score (6‐microRNAs) | 11 microRNAs | ||
|---|---|---|---|---|
| # of genes targeted by miRs in this pathway | Enrichment P‐value | # of genes targeted by miRs in this pathway | Enrichment P‐value | |
| Axon guidance | 44 | 1.14 × 10−5 | 66 | 2.16 × 10−6 |
| Regulation of actin cytoskeleton | 57 | 2.03 × 10−5 | 86 | 6.50 × 10−6 |
| Wnt signaling pathway | 37 | 7.33 × 10−5 | 61 | 3.49 × 10−7 |
| Endocytosis | 54 | 2.40 × 10−4 | 91 | 8.54 × 10−7 |
| Pathways in cancer | 86 | 2.88 × 10−4 | 137 | 3.29 × 10−5 |
| Glutamatergic synapse | 32 | 5.04 × 10−4 | 50 | 5.96 × 10−5 |
| Renal cell carcinoma | 25 | 6.00 × 10−4 | 38 | 1.36 × 10−4 |
| Hippo signaling pathway | 40 | 6.32 × 10−4 | 69 | 1.73 × 10−6 |
| Adherens junction | 27 | 6.54 × 10−4 | 35 | 7.10 × 10−3 |
| Focal adhesion | 51 | 6.75 × 10−4 | 80 | 1.30 × 10−4 |
| mRNA surveillance pathway | 25 | 9.81 × 10−4 | 37 | 5.48 × 10−4 |
| Tight junction | 35 | 1.67 × 10−3 | 47 | 1.60 × 10−2 |
| SNARE interactions in vesicular transport | 12 | 2.42 × 10−3 | 18 | 6.37 × 10−4 |
| Proteoglycans in cancer | 58 | 2.48 × 10−3 | 100 | 1.63 × 10−5 |
| ErbB signaling pathway | 29 | 2.89 × 10−3 | 48 | 1.35 × 10−4 |
| Phosphatidylinositol signaling system | 21 | 2.99 × 10−3 | 37 | 2.23 × 10−5 |
| Bacterial invasion of epithelial cells | 21 | 3.12 × 10−3 | 28 | 1.33 × 10−2 |
| Morphine addiction | 20 | 3.60 × 10−3 | 41 | 1.45 × 10−7 |
| Retrograde endocannabinoid signaling | 26 | 3.96 × 10−3 | 46 | 2.80 × 10−5 |
| Transcriptional misregulation in cancer | 46 | 4.20 × 10−3 | 71 | 2.65 × 10−3 |
| GABAergic synapse | 21 | 4.32 × 10−3 | 37 | 4.45 × 10−5 |
| Glycosaminoglycan biosynthesis – chondroitin sulfate/dermatan sulfate | 8 | 6.58 × 10−3 | 9 | 4.01 × 10−2 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 22 | 6.77 × 10−3 | 29 | 3.72 × 10−2 |
| Inositol phosphate metabolism | 16 | 8.63 × 10−3 | 26 | 1.14 × 10−3 |
| PI3K‐Akt signaling pathway | 72 | 9.66 × 10−3 | 119 | 1.13 × 10−3 |
The top 25 pathways from the analysis of 6 miR‐Score microRNAs are included. Enrichment p‐values and number of predicted microRNA target genes within these pathways are provided for both the 6‐microRNA and the 11‐microRNA analyses.
4. Discussion
Ideally a prognostic factor should allow the clinician to optimally tailor the treatment for an individual patient. For a malignant disease such as MPM with limited response to the few available treatment options, a personalized approach is particularly relevant. This would also ensure that radical treatment is only offered to patients who are likely to benefit. Towards this end, we have conducted a systematic investigation of microRNA expression in MPM, identifying the miR‐Score, a 6‐microRNA signature able to predict survival outcomes. To our knowledge, this is the first study in MPM to identify a microRNA signature‐based prognostic factor with the potential to predict prolonged survival in MPM patients undergoing surgery.
Although epithelioid histology, good performance status, younger age and earlier stage predict survival and are currently used to select patients for radical multimodality treatment, the outcomes are highly variable and range from a few months to more than 2 years (Cao et al., 2012, 2010, 2011, 2011). Thus it may be theorized that our inability to accurately identify patients likely to benefit from radical multimodality treatment approaches may have masked any advantages of intensive therapy in MPM.
New prognostic factors have become available (Bitanihirwe et al., 2014; Cedres et al., 2012; Kao et al., 2011; Opitz et al., 2008; Pass et al., 2010; Schramm et al., 2010) but so far they have failed to provide the accuracy needed for careful patient selection. Several recent studies have highlighted the importance of microRNA regulation in MPM biology (Gee et al., 2010; Guled et al., 2009; Ivanov et al., 2010; Kubo et al., 2011; Pass et al., 2010; Reid et al., 2013). Evaluating the prognostic potential of microRNA expression signatures in a previously reported surgical series consisting of patients undergoing EPP (Kao et al., 2011), we have identified a 6‐microRNA‐signature (miR‐Score) consisting of miR‐21‐5p, ‐23a‐3p, ‐30e‐5p, ‐221‐3p, ‐222‐3p, and ‐31‐5p, which accurately classified 92.3% of EPP patients into good and poor prognosis. Comparison of prediction models consisting of clinical prognostic factors alone or clinical factors combined with our miR‐Score showed that the addition of the miR‐Score resulted in an increase in predictive accuracy from 60.1 % to 84.4 %. The miR‐Score was also able to predict prolonged survival in a second series of samples from patients undergoing palliative surgery (P/D) with an accuracy of 71.9%, and did not seem to be affected by increased variation of histological subtype, age distribution, and performance status. Thus, the miR‐Score appears to have prognostic significance in MPM patients but still requires additional validation in a prospective study and in pre‐treatment samples.
So far, few studies have aimed to identify microRNA‐based prognostic markers for MPM. In an early cell line‐based study in which a small set of tumor tissues was analyzed, lower expression of miR‐30c and miR‐17‐5p was found to correlate with better survival in sarcomatoid mesothelioma (Busacca et al., 2010). Similarly, we found lower expression of miR‐30e‐5p [together with miR‐30c member of a microRNA family regulating TP53 expression (Li et al., 2010)] and miR‐17‐5p associated with longer survival in our series, suggesting that low expression of these microRNAs may also be associated with survival of MPM patients with other histological subtypes. In a subsequent study, miR‐29c‐5p was found to be associated with survival of patients undergoing surgery, along with two additional microRNAs (miR‐221‐3p and miR‐210‐3p) that failed to remain significant after adjustment for FDR (Pass et al., 2010). In general agreement with these results, our study identified miR‐221‐3p as part of the miR‐Score, and miR‐210‐3p as the best performing single microRNA predictor of good prognosis (P = 0.079). A formal comparison with our results is complicated by the fact that some clinical information (e.g. surgical procedure, tumor enrichment) was not reported. Nevertheless, the results reported here have identified the same microRNA families as previous studies (Busacca et al., 2010; Pass et al., 2010), confirming the importance of these in MPM biology.
MicroRNAs are important players in cellular homeostasis, and dysregulation of microRNAs has been consistently linked to the development and progression of cancer (Esquela‐Kerscher and Slack, 2006; Garzon et al., 2009; Ruan et al., 2009; Stahlhut and Slack, 2013). This may also be applicable to the prognostic microRNAs identified in the present study, and it is likely that the dysregulation of these microRNAs has contributed to tumor progression. The microRNA signature is best described as a combination of microRNAs with complementary prognostic value. Therefore it is not surprising that the signature includes microRNAs which did not exhibit convincing correlation with survival in univariate analysis. Conversely, despite strong correlations between survival and expression miR‐17‐5p was not included in the miR‐Score as the information contained in its expression levels was similar to that of miR‐21‐5p. Nevertheless, microRNAs differentially expressed in long and short survivors but not part of the signature are likely to contribute to the progression of the disease. This is reflected by the fact that many of the microRNAs identified in this study have been linked to development (Esquela‐Kerscher and Slack, 2006; Garzon et al., 2009; Ruan et al., 2009; Stahlhut and Slack, 2013) and prognosis (Kneitz et al., 2014; Menendez et al., 2013; Nair et al., 2012; Qu et al., 2014) of other cancers. For example miR‐21‐5p, miR‐221‐3p and members of the miR‐17∼92 cluster (miR‐17‐5p, miR‐19b‐3p, and miR‐20a‐5p) have been shown to regulate PTEN protein expression, and modulate the PI3K/Akt signaling axis (Concepcion et al., 2012; Pan et al., 2010). Loss of PTEN expression has been linked to poor prognosis in MPM patients (Bitanihirwe et al., 2014; Opitz et al., 2008; Schramm et al., 2010), and a recent study showed that loss of PTEN expression during chemotherapy was associated with worse survival outcomes (Bitanihirwe et al., 2014). As the aforementioned microRNAs target PTEN, and higher expression of these microRNAs was found to be associated with shorter survival, it is plausible to speculate that loss of PTEN is a result of increased microRNA expression, providing a direct link between them and the PI3K/Akt pathway. This is further supported by the fact that target genes within the PI3K/Akt signaling were significantly over‐represented in our pathway enrichment analysis, with 119 genes within the pathway being targeted by the microRNAs associated with survival in this study.
A microRNA with a previously reported link to MPM is miR‐31‐5p (Ivanov et al., 2010). Loss of MIR31 via the common deletion of the locus at 9p21.3 in MPM was shown to have pro‐tumorigenic effects, and re‐introduction of this microRNA inhibited proliferation and invasion in vitro (Ivanov et al., 2010). Consistent with these data, we have identified expression of miR‐31‐5p to be predictive of prognosis. In addition, expression levels of other microRNAs identified in this study are associated with prognosis in other cancers (Esquela‐Kerscher and Slack, 2006; Garzon et al., 2009; Ruan et al., 2009; Stahlhut and Slack, 2013). That similar microRNAs are linked to prognosis for a variety of cancer types suggests that their dysregulation is a common ‘cancer phenotype’ which leads to progression of the disease through disruption of cell homeostasis. Our pathway analyses show that the target genes of these microRNAs are involved in pathways previously linked to MPM in the literature, such as the Wnt and Hippo signaling networks (de Assis et al., 2014; Sekido, 2011). This suggests the possibility for these microRNAs to serve as therapeutic targets for MPM using, for example, the targeted delivery strategy we have recently shown to be effective in restoring levels of miR‐16 in an MPM tumor model (Reid et al., 2013).
Our results suggest that the miR‐Score has promise as a prognostic factor. Nevertheless, there are some limitations to the present study. All of the prognostic calculations have been done in retrospect from relatively small sample numbers and sufficient RNA could not be isolated from all samples. However, comparison of baseline characteristics of patients with and without RNA showed that except for the distribution of pathological staging in the EPP cohort, there were no major differences between the respective groups. The discrepancy in pathological staging did not affect the overall analysis as we deliberately excluded this clinical factor from analysis, because this information will only be available after surgery. That both cohorts are from a single institution with most surgeries (all EPPs and 71% of P/Ds) being performed by a single surgeon could be considered as a confounding factor, however, it has been suggested that in the case of EPP in particular surgery should only be performed by experienced hands in a high volume centre (Burt et al., 2014; Cao et al., 2010). Although the lack of a validation cohort receiving the same treatment as the training cohort, namely EPP, could be considered a potential weakness of our study, the fact that our miR‐Score can also predict prognosis in patients undergoing P/D, suggests that the miR‐Score has general prognostic value.
As for any microRNA‐based test, translation of our miR‐Score into a clinically useful tool to be applied on a patient‐by‐patient basis presents minor technical challenges. The miR‐Score was developed based on a comparison of individual patients relative to all patients in the series. However, if the distribution of patients with good and poor prognosis in independent cohorts does not vary from that observed in the present study, the patient series described here could be used as universal reference for any patient investigated in the future [consistent with the approach used in the miRView tests developed by Rosetta Genomics (Gilad et al., 2012; Meiri et al., 2012; Spector et al., 2013)]. Alternatively, absolute quantification of microRNA levels could be used in a clinical setting. For such an approach digital PCR would be more suitable than RT‐qPCR as this technology provides higher accuracy in absolute measurements (Hindson et al., 2013). Despite these caveats, the ability of our miR‐Score to accurately predict outcome both in patients treated with EPP/multimodality treatment and in those treated with palliative intent warrants further validation.
In summary, this study has identified a novel 6‐microRNA signature, named the miR‐Score, which has an accuracy of 92.3% in predicting survival of ≥20 months following EPP and 71.9% following palliative P/D. Addition of the miR‐Score to currently used clinical factors resulted in models with increased predictive accuracy compared to the clinical factors alone for both patient groups. Thus the miR‐Score addresses the unmet need for reliable markers to accurately predict prolonged survival of MPM patients, and is a novel tool ready to be tested in a prospective fashion.
Funding
This work was supported by a grant from the Dust Diseases Board New South Wales to MBK and GR, and a Translational Program Grant (11/TPG/3‐06) from the Cancer Institute New South Wales to NvZ and GR. MBK was supported by the ‘Swift Family Bequest and Mr Jim Tully Fellowship’, and AL by a Fellowship from the Biaggio Signorelli Foundation. The funding sources had no role in the design and conduct of the study or in the preparation of the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Suppl. Figure 1 Flow‐chart of study design. Black arrows indicate consecutive steps within a sample set, red arrows indicate move from one sample set to the next set.
Acknowledgments
We thank our colleagues, in particular James Edelman, for helpful discussions and critical evaluation of the manuscript.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2014.11.007.
Kirschner Michaela B., Cheng Yuen Yee, Armstrong Nicola J., Lin Ruby C.Y., Kao Steven C., Linton Anthony, Klebe Sonja, McCaughan Brian C., van Zandwijk Nico, Reid Glen, (2015), MiR‐Score: A novel 6‐microRNA signature that predicts survival outcomes in patients with malignant pleural mesothelioma, Molecular Oncology, 9, doi: 10.1016/j.molonc.2014.11.007.
Contributor Information
Michaela B. Kirschner, Email: michaela.kirschner@sydney.edu.au, Email: mkirschner@gmx.com
Yuen Yee Cheng, Email: yuen.cheng@sydney.edu.au.
Nicola J. Armstrong, Email: nicola.armstrong@sydney.edu.au
Ruby C.Y. Lin, Email: ruby.lin@sydney.edu.au
Steven C. Kao, Email: Steven.Kao@lh.org.au
Anthony Linton, Email: anthony.linton@sydney.ed.au.
Sonja Klebe, Email: Sonja.Klebe@health.sa.gov.au.
Brian C. McCaughan, Email: brianmccaughan@gmail.com
Nico van Zandwijk, Email: nico.vanzandwijk@sydney.edu.au.
Glen Reid, Email: glen.reid@sydney.edu.au.
References
- Balatti, V. , Maniero, S. , Ferracin, M. , Veronese, A. , Negrini, M. , Ferrocci, G. , Martini, F. , Tognon, M.G. , 2011. MicroRNAs dysregulation in human malignant pleural mesothelioma. J. Thorac. Oncol. 6, 844–851. [DOI] [PubMed] [Google Scholar]
- Benjamin, H. , Lebanony, D. , Rosenwald, S. , Cohen, L. , Gibori, H. , Barabash, N. , Ashkenazi, K. , Goren, E. , Meiri, E. , Morgenstern, S. , Perelman, M. , Barshack, I. , Goren, Y. , Edmonston, T.B. , Chajut, A. , Aharonov, R. , Bentwich, Z. , Rosenfeld, N. , Cohen, D. , 2010. A diagnostic assay based on microRNA expression accurately identifies malignant pleural mesothelioma. J. Mol. Diagn. 12, 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitanihirwe, B.K. , Meerang, M. , Friess, M. , Soltermann, A. , Frischknecht, L. , Thies, S. , Felley-Bosco, E. , Tsao, M.S. , Allo, G. , de Perrot, M. , Seifert, B. , Moch, H. , Stahel, R. , Weder, W. , Opitz, I. , 2014. PI3K/mTOR signaling in mesothelioma patients treated with induction chemotherapy followed by extrapleural pneumonectomy. J. Thorac. Oncol. 9, 239–247. [DOI] [PubMed] [Google Scholar]
- Burt, B.M. , Cameron, R.B. , Mollberg, N.M. , Kosinski, A.S. , Schipper, P.H. , Shrager, J.B. , Vigneswaran, W.T. , 2014. Malignant pleural mesothelioma and the Society of Thoracic Surgeons Database: an analysis of surgical morbidity and mortality. J. Thorac. Cardiovasc. Surg. 148, 30–35. [DOI] [PubMed] [Google Scholar]
- Busacca, S. , Germano, S. , De Cecco, L. , Rinaldi, M. , Comoglio, F. , Favero, F. , Murer, B. , Mutti, L. , Pierotti, M. , Gaudino, G. , 2010. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am. J. Respir. Cell Mol. Biol. 42, 312–319. [DOI] [PubMed] [Google Scholar]
- Cao, C. , Tian, D. , Manganas, C. , Matthews, P. , Yan, T.D. , 2012. Systematic review of trimodality therapy for patients with malignant pleural mesothelioma. Ann. Cardiothorac. Surg. 1, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, C.Q. , Yan, T.D. , Bannon, P.G. , McCaughan, B.C. , 2010. A systematic review of extrapleural pneumonectomy for malignant pleural mesothelioma. J. Thorac. Oncol. 5, 1692–1703. [DOI] [PubMed] [Google Scholar]
- Cedres, S. , Montero, M.A. , Martinez, P. , Martinez, A. , Rodriguez-Freixinos, V. , Torrejon, D. , Gabaldon, A. , Salcedo, M. , Ramon, Y.C.S. , Felip, E. , 2012. Exploratory analysis of activation of PTEN-PI3K pathway and downstream proteins in malignant pleural mesothelioma (MPM). Lung Cancer. 77, 192–198. [DOI] [PubMed] [Google Scholar]
- Concepcion, C.P. , Bonetti, C. , Ventura, A. , 2012. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 18, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, D. , Sahmoud, T. , Therasse, P. , van Meerbeeck, J. , Postmus, P.E. , Giaccone, G. , 1998. Prognostic factors in patients with pleural mesothelioma: the European Organization for Research and Treatment of Cancer experience. J. Clin. Oncol. 16, 145–152. [DOI] [PubMed] [Google Scholar]
- de Assis, L.V. , Locatelli, J. , Isoldi, M.C. , 2014. The role of key genes and pathways involved in the tumorigenesis of Malignant Mesothelioma. Biochim. Biophys. Acta. 1845, 232–247. [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. , Lash, A.E. , 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge, S. , Byrd, D.R. , Comptom, C.C. , Fritz, A.G. , Greene, F.L. , Trotti, A. , 2010. AJCC Cancer Staging Manual seventh ed. Springer; New York: [Google Scholar]
- Esquela-Kerscher, A. , Slack, F.J. , 2006. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 6, 259–269. [DOI] [PubMed] [Google Scholar]
- Garzon, R. , Calin, G.A. , Croce, C.M. , 2009. MicroRNAs in cancer. Annu. Rev. Med. 60, 167–179. [DOI] [PubMed] [Google Scholar]
- Gee, G.V. , Koestler, D.C. , Christensen, B.C. , Sugarbaker, D.J. , Ugolini, D. , Ivaldi, G.P. , Resnick, M.B. , Houseman, E.A. , Kelsey, K.T. , Marsit, C.J. , 2010. Downregulated microRNAs in the differential diagnosis of malignant pleural mesothelioma. Int. J. Cancer. 127, 2859–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, S. , Lithwick-Yanai, G. , Barshack, I. , Benjamin, S. , Krivitsky, I. , Edmonston, T.B. , Bibbo, M. , Thurm, C. , Horowitz, L. , Huang, Y. , Feinmesser, M. , Hou, J.S. , St Cyr, B. , Burnstein, I. , Gibori, H. , Dromi, N. , Sanden, M. , Kushnir, M. , Aharonov, R. , 2012. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J. Mol. Diagn. 14, 510–517. [DOI] [PubMed] [Google Scholar]
- Guled, M. , Lahti, L. , Lindholm, P.M. , Salmenkivi, K. , Bagwan, I. , Nicholson, A.G. , Knuutila, S. , 2009. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer. 48, 615–623. [DOI] [PubMed] [Google Scholar]
- Herndon, J.E. , Green, M.R. , Chahinian, A.P. , Corson, J.M. , Suzuki, Y. , Vogelzang, N.J. , 1998. Factors predictive of survival among 337 patients with mesothelioma treated between 1984 and 1994 by the Cancer and Leukemia Group B. Chest. 113, 723–731. [DOI] [PubMed] [Google Scholar]
- Hindson, C.M. , Chevillet, J.R. , Briggs, H.A. , Gallichotte, E.N. , Ruf, I.K. , Hindson, B.J. , Vessella, R.L. , Tewari, M. , 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 10, 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.W. , Sherman, B.T. , Lempicki, R.A. , 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ivanov, S.V. , Goparaju, C.M. , Lopez, P. , Zavadil, J. , Toren-Haritan, G. , Rosenwald, S. , Hoshen, M. , Chajut, A. , Cohen, D. , Pass, H.I. , 2010. Pro-tumorigenic effects of miR-31 loss in mesothelioma. J. Biol. Chem. 285, 22809–22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, S.C. , Klebe, S. , Henderson, D.W. , Reid, G. , Chatfield, M. , Armstrong, N.J. , Yan, T.D. , Vardy, J. , Clarke, S. , van Zandwijk, N. , McCaughan, B. , 2011. Low calretinin expression and high neutrophil-to-lymphocyte ratio are poor prognostic factors in patients with malignant mesothelioma undergoing extrapleural pneumonectomy. J. Thorac. Oncol. 6, 1923–1929. [DOI] [PubMed] [Google Scholar]
- Kirschner, M.B. , Cheng, Y.Y. , Badrian, B. , Kao, S.C. , Creaney, J. , Edelman, J.J. , Armstrong, N.J. , Vallely, M.P. , Musk, A.W. , Robinson, B.W. , McCaughan, B.C. , Klebe, S. , Mutsaers, S.E. , van Zandwijk, N. , Reid, G. , 2012. Increased circulating miR-625-3p: a potential biomarker for patients with malignant pleural mesothelioma. J. Thorac. Oncol. 7, 1184–1191. [DOI] [PubMed] [Google Scholar]
- Kirschner, M.B. , Edelman, J.J. , Kao, S.C. , Vallely, M.P. , van Zandwijk, N. , Reid, G. , 2013. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 4, 94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz, B. , Krebs, M. , Kalogirou, C. , Schubert, M. , Joniau, S. , van Poppel, H. , Lerut, E. , Kneitz, S. , Scholz, C.J. , Ströbel, P. , Gessler, M. , Riedmiller, H. , Spahn, M. , 2014. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate Cancer cells by inhibiting IRF2 and SOCS3. Cancer Res. 74, 2591–2603. [DOI] [PubMed] [Google Scholar]
- Kubo, T. , Toyooka, S. , Tsukuda, K. , Sakaguchi, M. , Fukazawa, T. , Soh, J. , Asano, H. , Ueno, T. , Muraoka, T. , Yamamoto, H. , Nasu, Y. , Kishimoto, T. , Pass, H.I. , Matsui, H. , Huh, N.H. , Miyoshi, S. , 2011. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin. Cancer Res. 17, 4965–4974. [DOI] [PubMed] [Google Scholar]
- Li, J. , Donath, S. , Li, Y. , Qin, D. , Prabhakar, B.S. , Li, P. , 2010. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 6, e1000795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton, A. , Cheng, Y.Y. , Griggs, K. , Kirschner, M.B. , Gattani, S. , Srikaran, S. , Chuan-Hao Kao, S. , McCaughan, B.C. , Klebe, S. , van Zandwijk, N. , Reid, G. , 2014. An RNAi-based screen reveals PLK1, CDK1 and NDC80 as potential therapeutic targets in malignant pleural mesothelioma. Br. J. Cancer. 110, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. , Schmittgen, T.D. , 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Meiri, E. , Mueller, W.C. , Rosenwald, S. , Zepeniuk, M. , Klinke, E. , Edmonston, T.B. , Werner, M. , Lass, U. , Barshack, I. , Feinmesser, M. , Huszar, M. , Fogt, F. , Ashkenazi, K. , Sanden, M. , Goren, E. , Dromi, N. , Zion, O. , Burnstein, I. , Chajut, A. , Spector, Y. , Aharonov, R. , 2012. A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist. 17, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez, P. , Villarejo, P. , Padilla, D. , Menendez, J.M. , Rodriguez-Montes, J.A. , 2013. Implications of the histological determination of microRNAs in the screening, diagnosis and prognosis of colorectal cancer. J. Surg. Oncol. 108, 70–73. [DOI] [PubMed] [Google Scholar]
- Nair, V.S. , Maeda, L.S. , Ioannidis, J.P. , 2012. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J. Natl. Cancer Inst. 104, 528–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz, I. , Soltermann, A. , Abaecherli, M. , Hinterberger, M. , Probst-Hensch, N. , Stahel, R. , Moch, H. , Weder, W. , 2008. PTEN expression is a strong predictor of survival in mesothelioma patients. Eur. J. Cardiothorac. Surg. 33, 502–506. [DOI] [PubMed] [Google Scholar]
- Pan, X. , Wang, Z.X. , Wang, R. , 2010. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol. Ther. 10, 1224–1232. [DOI] [PubMed] [Google Scholar]
- Pass, H.I. , 2012. Biomarkers and prognostic factors for mesothelioma. Ann. Cardiothorac. Surg. 1, 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pass, H.I. , Goparaju, C. , Ivanov, S. , Donington, J. , Carbone, M. , Hoshen, M. , Cohen, D. , Chajut, A. , Rosenwald, S. , Dan, H. , Benjamin, S. , Aharonov, R. , 2010. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 70, 1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, A. , Du, L. , Yang, Y. , Liu, H. , Li, J. , Wang, L. , Liu, Y. , Dong, Z. , Zhang, X. , Jiang, X. , Wang, H. , Li, Z. , Zheng, G. , Wang, C. , 2014. Hypoxia-inducible MiR-210 is an independent prognostic factor and contributes to metastasis in colorectal cancer. PLoS One. 9, e90952 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Reid, G. , Pel, M.E. , Kirschner, M.B. , Cheng, Y.Y. , Mugridge, N. , Weiss, J. , Williams, M. , Wright, C. , Edelman, J.J. , Vallely, M.P. , McCaughan, B.C. , Klebe, S. , Brahmbhatt, H. , Macdiarmid, J.A. , van Zandwijk, N. , 2013. Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. 24, 3128–3135. [DOI] [PubMed] [Google Scholar]
- Ruan, K. , Fang, X. , Ouyang, G. , 2009. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 285, 116–126. [DOI] [PubMed] [Google Scholar]
- Santarelli, L. , Strafella, E. , Staffolani, S. , Amati, M. , Emanuelli, M. , Sartini, D. , Pozzi, V. , Carbonari, D. , Bracci, M. , Pignotti, E. , Mazzanti, P. , Sabbatini, A. , Ranaldi, R. , Gasparini, S. , Neuzil, J. , Tomasetti, M. , 2011. Association of MiR-126 with soluble mesothelin-related peptides, a marker for malignant mesothelioma. PLoS One. 6, e18232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, A. , Opitz, I. , Thies, S. , Seifert, B. , Moch, H. , Weder, W. , Soltermann, A. , 2010. Prognostic significance of epithelial-mesenchymal transition in malignant pleural mesothelioma. Eur. J. Cardiothorac. Surg. 37, 566–572. [DOI] [PubMed] [Google Scholar]
- Sekido, Y. , 2011. Inactivation of Merlin in malignant mesothelioma cells and the Hippo signaling cascade dysregulation. Pathol. Int. 61, 331–344. [DOI] [PubMed] [Google Scholar]
- Spector, Y. , Fridman, E. , Rosenwald, S. , Zilber, S. , Huang, Y. , Barshack, I. , Zion, O. , Mitchell, H. , Sanden, M. , Meiri, E. , 2013. Development and validation of a microRNA-based diagnostic assay for classification of renal cell carcinomas. Mol. Oncol. 7, 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut, C. , Slack, F.J. , 2013. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 5, 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis, W.D. , Brambilla, E. , Müller-Hermelink, H.K. , Harris, C.C. , 2004. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart IACR Press; Lyon: [Google Scholar]
- Treasure, T. , Lang-Lazdunski, L. , Waller, D. , Bliss, J.M. , Tan, C. , Entwisle, J. , Snee, M. , O'Brien, M. , Thomas, G. , Senan, S. , O'Byrne, K. , Kilburn, L.S. , Spicer, J. , Landau, D. , Edwards, J. , Coombes, G. , Darlison, L. , Peto, J. , 2011. Extra-pleural pneumonectomy versus no extra-pleural pneumonectomy for patients with malignant pleural mesothelioma: clinical outcomes of the Mesothelioma and Radical Surgery (MARS) randomised feasibility study. Lancet Oncol. 12, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meerbeeck, J.P. , Scherpereel, A. , Surmont, V.F. , Baas, P. , 2011. Malignant pleural mesothelioma: the standard of care and challenges for future management. Crit. Rev. Oncol. Hematol. 78, 92–111. [DOI] [PubMed] [Google Scholar]
- van Zandwijk, N. , Clarke, C. , Henderson, D. , Musk, A.W. , Fong, K. , Nowak, A. , Loneragan, R. , McCaughan, B. , Boyer, M. , Feigen, M. , Currow, D. , Schofield, P. , Nick Pavlakis, B.I. , McLean, J. , Marshall, H. , Leong, S. , Keena, V. , Penman, A. , 2013. Guidelines for the diagnosis and treatment of malignant pleural mesothelioma. J. Thorac. Dis. 5, E254–E307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Suppl. Figure 1 Flow‐chart of study design. Black arrows indicate consecutive steps within a sample set, red arrows indicate move from one sample set to the next set.


