Abstract
Immune checkpoint regulators such as PD‐L1 have become exciting new therapeutic targets leading to long lasting remissions in patients with advanced malignancies. However, in view of the remarkable costs and the toxicity profiles of these therapies, predictive biomarkers able to discriminate responders from non‐responders are urgently needed. In the present paper, we provide evidence that PD‐L1 is frequently expressed on metastatic cells circulating in the blood of hormone receptor‐positive, HER2‐negative breast cancer patients. We performed western blot, flow cytometry and immunocytochemical analyses to demonstrate the specificity of the PDL1 antibody used in our study and established immunoscores for PDL1 expression on single tumor cells. We then selected sixteen patients with circulating tumor cells (CTCs) using the CellSearch® system and found PD‐L1(+) CTCs in 11 patients (68.8%). The fraction of PD‐L1(+) CTCs varied from 0.2 to 100% in individual patients. This is the first report demonstrating the expression of PD‐L1 on CTCs. The established CTC/PD‐L1 assay can be used for liquid biopsy in future clinical trials for stratification and monitoring of cancer patients undergoing immune checkpoint blockade.
Keywords: Circulating tumor cells, PD-L1 expression, Breast cancer
Highlights
PD‐L1 is an immune checkpoint regulator.
PD‐L1 is frequently expressed on metastatic cells circulating in the blood of breast cancer patients.
We report an established CTC/PD‐L1 assay that can be used for liquid biopsy in cancer patients.
Detection of PD‐L1(+) CTCs may guide stratification and monitoring of cancer patients undergoing immune checkpoint blockade.
1. Introduction
The prognosis of cancer patients, even with localized primary tumors, is mainly determined by the circulation of tumor cells in the peripheral blood from the primary site to distant organs such as bone marrow, liver, lungs, or brain, and the subsequent outgrowth of a largely unknown subset of these cells into metastases in their new microenvironment (Eccles and Welch, 2007). The colonization of distant organs by disseminated tumor cells, resulting in the appearance of clinically detectable metastases, can take many years in breast cancer (Braun et al., 2005) and the mechanisms behind this “cancer dormancy” are largely unknown (Chambers et al., 2002; Goss and Chambers, 2010; Uhr and Pantel, 2011). This stringent selection process together with a potential independent genomic progression of disseminated tumor cells may explain why overt metastases can harbor unique genomic alterations differing from the bulk of the original primary tumor cells (Flatmark et al., 2002; Kang and Pantel, 2013; Riethdorf et al., 2010). Thus, the direct analysis of metastatic cells could reveal important information for systemic cancer therapy targeting metastatic disease (Alix‐Panabieres et al., 2012; Pantel et al., 2008). However, biopsy of overt metastases is an invasive procedure limited to certain locations and not easily acceptable in the clinic. Moreover, a recent work has shown that different metastatic sites harbor different genomic aberrations (Gerlinger et al., 2012) and biopsy of one or two accessible metastases may not be representative of the whole metastatic disease biology.
An alternative approach is the analysis of blood samples for circulating tumor cells (CTCs) detection and characterization as a liquid biopsy of the cancer (Alix‐Panabieres and Pantel, 2014). It can be performed frequently and might allow real‐time monitoring of cancer therapies in individual patients (Alix‐Panabieres et al., 2012). Recent reports indicated that CTCs give important complementary information on therapeutic targets and drug resistance mechanisms in carcinoma patients (Alix‐Panabieres and Pantel, 2014). CTCs isolated in the peripheral blood are a pool of cells derived from the primary tumor and different metastatic sites and may, therefore, provide a comprehensive real‐time picture of the whole tumor burden in an individual patient (Pantel and Alix‐Panabieres, 2013).
Characterization of tumor cells is of utmost importance. More specifically, PD‐L1 expressed in tumors has been recently highlighted to function as a key component of the cancer‐immunity cycle by preventing the immune system from destroying cancer cells (Butt and Mills, 2014). PD‐1 receptor (CD279) is expressed on the surface of activated T cells and its ligands, PD‐L1 (B7‐H1; CD274) and PD‐L2 (B7‐DC; CD273), are expressed on the surface of antigen‐presenting cells such as macrophages or dendritic cells. When PD‐L1 binds to PD‐1, a strong inhibitory signal is transmitted into the T cell, which induces a reduction of cytokine production and a suppression of T‐cell proliferation (Brahmer et al., 2012; Chen et al., 2012; Topalian et al., 2012): the immune system is misled by the surrounding cancer cells expressing PD‐L1 and does not destroy them.
Efforts to restore latent anti‐tumor immunity have focused on antibody‐based interventions targeting CTL antigen 4 (CTLA‐4) and programmed cell death protein 1 (PD‐1) on T lymphocytes and its principal ligand (PD‐L1) on tumor cells (Shin and Ribas, 2015). Ipilimumab, an antibody targeting CTLA‐4, appears to restore tumor immunity at the priming phase, whereas anti‐PD‐1/PD‐L1 antibodies restore immune function in the tumor microenvironment. Although ipilimumab can produce durable long‐term responses in patients with advanced melanoma, it is associated with significant immune‐related toxicities (Shin and Ribas, 2015). By contrast, antibodies targeting either PD‐1 or PD‐L1 have produced significant anti‐tumor activity with considerably less toxicity (Shin and Ribas, 2015) and are being actively investigated for the treatment of multiple cancers including lung, breast, bladder and renal cancers (Philips and Atkins, 2015). Current efforts focus on registration trials of single agents and combinations in various diseases and settings, as well as on identifying predictive biomarkers of response. Ever since the earliest reports of the effects of PD‐1 blockade, PD‐L1 expression by tumor cells has been used as potential biomarker to predict a therapeutic response. We recently showed that the level of soluble PD‐L1 present in the blood could predict patient's outcome in aggressive B‐cell lymphomas (Rossille et al., 2014). However, the predictive value of PDL1 expression measurements of primary carcinomas seems to be limited. Moreover, there is only a weak correlation in staining between the matched primary tumor and distant metastasis, suggesting that the primary tumor is not an adequate surrogate for determining PD‐L1 expression in metastatic sites (Jilaveanu et al., 2014). Tissue heterogeneity in PD‐L1 expression in both primary and metastatic sites for a given patient indicates that a single core biopsy might be not sufficient to determine tumor PD‐L1 expression (Jilaveanu et al., 2014).
CTCs are currently investigated as predictive biomarker for HER2‐targeting therapies (Bidard et al., 2013) and the same strategy could be used for immune checkpoint blockade therapies such as antibodies to PD‐L1. Jilaveanu et al. reported that distant sites have greater PD‐L1 expression than the primary tumor (Jilaveanu et al., 2014). However, there is, to date, no report on the expression of PD‐L1 on CTCs in breast cancer or other tumor entities. To investigate whether CTCs express PD‐L1, we applied the CellSearch® system to blood samples from patients with metastatic breast cancer before any treatment. Interestingly, we detected a subset of CTCs expressing PD‐L1 in the blood of patients with metastatic breast cancer using a non‐invasive liquid biopsy. This specific subset represents metastatic cells with a high potential to escape T cell‐mediated lysis and these cells are therefore the actual targets of anti‐PDL1 antibody therapy.
2. Materials and methods
2.1. Patient samples and blood collection
After informed consent was given, peripheral blood from patients with hormone receptor‐positive (HR+), HER2‐negative (HER2−) metastatic breast cancer was collected before the start of treatment (CPP number 2011‐AOI163‐38). Patients were recruited at the University Medical Centre of Montpellier (Department of Medical Oncology) and at the Montpellier Cancer Institute (ICM). Blood was drawn from the arm vein of each cancer patient in specific CellSave® tubes (Janssen, 10 mL) for CTC detection. Tubes were then conserved at room temperature and sent to the Laboratory “Detection of Rare Human Circulating Cells – LCCRH” at the University Medical Center of Montpellier where blood samples were processed immediately on the CellSearch® system. The BRISQ criteria have been listed in the Table 1 (Moore et al., 2011). This study belongs to the BMS_PD‐L1_onco clinical trial (NCT10660776), which assesses PD‐L1 as a biomarker in oncology and hematology.
Table 1.
BRISQ Checklist summarizing the lifecycle of the Biospecimen in our study. The pre‐analytical phase of the lifecycle of the biospecimen includes each stage from patient to distribution. Preanalytical variables are addressed in this BRISQ list.
| Data Elements | |
| Biospecimen type | Whole Blood |
| Disease status of patients | Metastatic breast cancer patients, Healthy control |
| Clinical diagnosis of patients | Breast cancer |
| Pathology diagnosis | HR+ Her2− Carcinoma |
| Collection mechanism | Pre‐treatment blood draw |
| Type of stabilization | CellSave Tube, Room temperature |
| Type of long‐term preservation | Fixation, Room temperature (25 °C) |
| Constitutive of preservative | CellSave Tube (Janssen patent) |
| Storage duration | Up to 96 h |
| Shipping temperature | 25 °C |
| Composition assessment & selection | Detection of at least one CTC |
HR, Hormone Receptor; CTC, Circulating Tumor Cells.
2.2. Cell culture
Mammary cancer cell lines SKBR3 (ATCC® HTB‐30™), MCF7 (ATCC® HTB‐22™), MDA‐MB‐231 (ATCC® CRMHTB‐26™) were purchased from the American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Eurobio, FR), 2 μg/ml Gentamicin and 20 UI/ml penicillin. Diffuse Large B‐Cell Lymphoma (DLBCL) cell line SU‐DHL‐4 (ATCC® CRL‐2957) cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Eurobio, FR), 2 μg/ml Gentamicin and 20UI/ml penicillin. Cells were cultured in 25 cm2 flasks at 37 °C in a humidified atmosphere containing 5% CO2.
2.3. CTC enumeration
Seven and half milliliters of blood collected in the CellSave tube were used for CTC detection using the FDA‐cleared CellSearch® system (Janssen, Beerse, Belgium).
Blood samples were processed on the CellTracks Autoprep within 72 h. The CellSearch® CXC Kit (Janssen) was used for these specific experiments. It includes a cytokeratin‐fluorescein/CD45‐allophycocyanin (APC) reagent, leaving the phycoerythrin (PE) channel of the Celltracks Analyzer II® open to incorporate a user‐defined PE‐conjugate for further characterization of the CTCs. In our project, we improved the CTC detection by using the 4th channel of the CellSearch® for PD‐L1 expression: we used the anti‐human B7‐H1/PD‐L1 PE‐conjugated monoclonal antibody (MAb, Cat N° FAB1561P, R&D Systems, Minneapolis, USA) at a final concentration of 20 μg/mL. The corresponding isotypic control (Mouse IgG1 PE, Cat N° IC002P, R&D Systems, Minneapolis, USA) was used at the same final concentration (20 μg/mL).
Briefly, fixed CTCs were first positively enriched via the epithelial cell adhesion molecule (EpCAM) expression and defined as EpCAM isolated intact cells stained positive for cytokeratins (CK8, 18, 19), positive or negative for B7H1 (PD‐L1) and negative for CD45 (the exclusion marker specific of normal hematopoietic cells): CTCs were identified as EpCAM(+)DAPI(+)CK(+)CD45(−).
As control group, we analyzed in parallel blood samples from 15 healthy controls provided by the regional centre of blood transfusion (Etablissement Français du Sang of Montpellier, France).
2.4. Flow cytometry experiments
A fixation/permeabilization kit (Beckman Coulter, Brea, USA) was used to prepare the cells before the addition of the anti‐human B7H1 Ab or Mouse IgG1 PE at a final concentration of 20 μg/mL on 0.2 million SKBR3 cells and 0.2 SU‐DHL‐4. Briefly, cells and both Abs were incubated for 60 min at room temperature in the dark. Cells were then washed with PBS supplemented with 2% fetal bovine serum, centrifuged (600 g–5 min) and the cell pellet was recovered with 400 μL PBS. Labelled cancer cells were detected by using the Cyan cytometer (Beckman Coulter) and the data analysis was performed using the Kaluza software (Beckman Coulter).
Other anti‐human B7H1 Abs have been tested: (1) clone 3.1 anti‐CD274‐PE Ab (ref IM99509, Beckman Coulter), (2) anti‐CD274‐FITC (ref 558065, Becton Dickinson), and (3) B7‐H1‐A488 (ref FAB1561G, R&D Systems). As shown by the Figure 1, a direct ELISA on different proteins showed the specificity of the anti‐human B7H1 MAb, (Cat N° FAB1561P, R&D Systems; data given by R&D systems).
Figure 1.
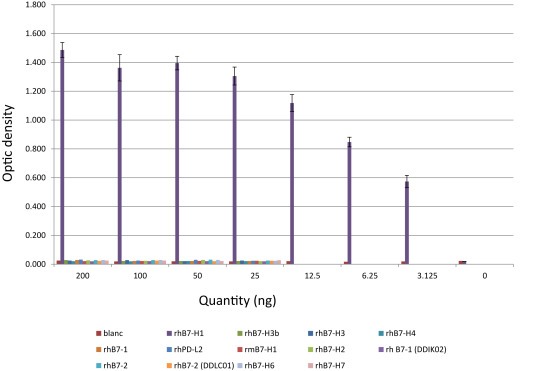
Human B7H1 Detection in direct ELISA with monoclonal anti‐human B7‐H1. Antibody: Human B7‐H1 monoclonal Ab (R&D systems). Different proteins have been analyzed: recombinant human B7‐H1, recombinant human B7‐1, recombinant human B7‐2, recombinant B7‐H2, recombinant human B7‐H3, recombinant human B7‐H3b, recombinant human B7‐H4, recombinant B7‐H6, recombinant B7‐H7, recombinant human PD‐L2 and recombinant mouse B7‐H1. The plate was coated with recombinant protein at mass/well cited in the graph. The different concentrations used were: primary Ab (0.5 μg/mL), secondary Ab (goat anti‐mouse IgG polyclonal antibody‐biotinylated at 1:10,000), enzyme (streptavidin‐AP at 1/1000); the substrate was the p‐nitrophenyl phosphate. Abbreviations: rh: recombinant human; rm: recombinant mouse.
2.5. Immunocytochemical analyses
SKBR3 cancer cells were labelled as described above using the concentration of the anti‐human B7‐H1 MAb required for the CellSearch® system (20 μg/mL) and centrifuged gently in a Cytospin™ 4 Cytocentrifuge (Thermo Scientific) on standard glass slides. A drop of the ProLong® Gold antifade reagent with DAPI (Invitrogen, Ref‐P36931) was added on the preparation to stain nuclei in blue and slides were mounted. Stained cancer cells were finally observed under a fluorescent microscope (Axio Imager.M1, Zeiss) and automatically analyzed with the software AxioVision Release 4.6.3.
2.6. Western blot analysis
The primary anti‐PD‐L1 B7‐H1/PD‐L1 MAb (IgG1 Clone #130021, Cat N° MAB1561, R&D Systems) was analyzed versus the Mouse IgG1 Isotype Control (mouse monoclonal, Clone #11711, Cat N° MAB002, R&D Systems). As a loading control, anti‐alpha‐Tubulin (11H10) antibody, from Cell Signaling Technology (Danvers, USA) was used. Protein samples from the cell lines MDA‐MB‐468, MDA‐MB‐231, BT‐20, PC‐3, LNCAP, Du145, SCC25, Cal27, MDA‐MB‐231‐SA, BC‐M1, PC‐E1 and SU‐DHL‐4 were analyzed. Cells were harvested in 9.8 M urea, 15 mM EDTA, 30 mM Tris, followed by a cell disruption step by ultrasonic treatment. Insoluble compounds were removed by centrifugation and the supernatant was collected. The protein concentration was determined using the Pierce BCA Protein Assay Kit (Pierce, Rockford, USA) using BSA as the standard. Proteins were separated by SDS‐PAGE using either the Novex XCell Sure‐Lock mini system (Invitrogen, Groningen, Netherlands) or the Protean II xi cell (Bio‐Rad, Hercules, USA). A Laemmli buffer system and 10% polyacrylamide separation gels were used. The molecular size standard was the peqGOLD protein‐marker V (Peqlab, Erlangen, Germany). The proteins were transferred to Immobilon‐PSQ membranes (Millipore GmbH, Schwalbach, Germany) using the mini VE vertical electrophoresis system equipped with tank blot transfer units (GE Healthcare, Uppsala, Sweden). The primary antibodies were applied to the membranes at 4 °C with gentle agitation over night. Protein bands were detected using the SignalFire ECL Reagent (Cell Signaling Technology) and X‐ray films (GE Healthcare). X‐ray films were digitized using the GS‐700 imaging densitometer (Bio‐Rad).
3. Results
3.1. Establishment of the B7‐H1/PD‐L1 CTC analysis
For the establishment of a novel assay detecting CTCs expressing PD‐L1, the sensitivity, specificity and the dynamic range of the PD‐L1 R&D antibody was determined by Western Blot in an array of experiments and cell lines. The PD‐L1 R&D antibody was found to detect very low levels of PD‐L1, provided only one strong signal at approx. 55 kDa and exhibited a linear increase of the signal intensity proportional to the applied protein amount. For the isotype control very weak or no signal at all was detected in the analyzed cell lines (Figure 2A). MDA‐MB‐231 served as a useful positive control for the establishment of the Western Blot. However, the extraordinary high signal intensity of the PD‐L1 R&D antibody frequently resulted in oversaturation of the signals in the Western Blots. To circumvent oversaturation of the signals in subsequent immunofluorescent assays, we therefore focused on the cell line SKBR3, because these cells show a range of levels of PD‐L1 expression, which enabled us to develop immunoscores for various degrees of PDL1 on single tumor cells.
Figure 2.
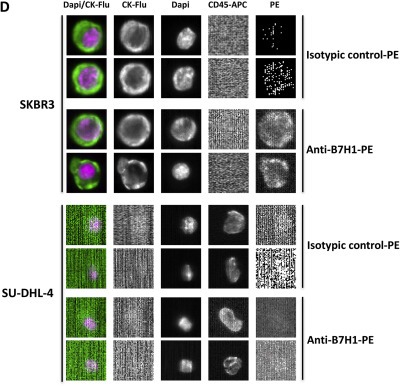
Specificity and sensitivity of the anti‐human B7H1 MAb (R&D system). A, Determination of the anti‐human B7H1 MAb recognition profile on test protein mixtures from the assigned cell lines and tumour entities by Western Blot. In parallel, the protein recognition pattern for the mouse isotype control was analyzed. For each lane 20 μg of protein was loaded. Both membranes were exposed to the same X‐ray films. To investigate the band pattern for the two antibodies, a short X‐ray film exposition time (10 s) and an overexposed X‐ray film (4 min) is shown. Alpha Tubulin served as a loading control. B, Flow cytometry analyses. SKBR3 and SU‐DHL‐4 cell lines were incubated with the anti‐human B7‐H1 MAb‐PE (in red) and the corresponding isotypic control (IgG1, in blue) – The flow cytometer used for all the analyses was the Cyan (Beckman Coulter); C, Immunocytochemical analyses. SKBR3 and SU‐DHL‐4 cells were centrifuged gently in a Cytospin™ 4 Cytocentrifuge (Thermo Scientific) on standard glass slides after being stained with the anti‐human B7‐H1‐PE MAb and the corresponding isotypic control. DAPI was added on the preparation to stain nuclei in blue. PDL1(+) tumor cells are stained in red (Fluorescent microscope Axio Imager.M1 Zeiss, software AxioVision Release 4.6.3. – magnification X20). D, CellSearch®system analyses. SKBR3 and SU‐DHL‐4 were analyzed in the CellSearch® system using either the human anti‐human B7‐H1 MAb‐PE or the corresponding isotypic control in the 4th channel (PE). All experiments were repeated 3 times and the figures show representative results.
Next, we analyzed the different breast cancer cell lines SKBR3, MCF‐7 and MDA‐MB‐231 by flow cytometry. All these cell lines were positive for PD‐L1 (data not shown). The presence of PD‐L1 on SKBR3 cells was confirmed by flow cytometry (Figure 2B) and immunofluorescent staining of cytospins (Figure 2C). The specificity of the staining was demonstrated by the fact that no signals were detected for the cell line SU‐DHL‐4 (established negative control) and for the isotype control on SKBR3 cells. Next, SKBR3 (PD‐L1(+)) and SU‐DHL‐4 (PD‐L1(−)) were analyzed for PD‐L1 using the CellSearch® system with PD‐L1 implemented in the 4th channel (Figure 2D).
We selected the anti‐human B7‐H1/PD‐L1 PE‐conjugated monoclonal antibody (MAb) from R&D. Other anti‐human B7H1 Abs have been also tested at different concentrations with the CellSearch® system: (1) clone 3.1 anti‐CD274‐PE Ab (ref IM99509, Beckman Coulter), (2) anti‐CD274‐FITC (ref 558065, Becton Dickinson), and (3) B7‐H1‐A488 (ref FAB1561G, R&D Systems). However, a concentration of only 20 μg/mL of the PE‐conjugated monoclonal antibody (MAb) from R&D was needed to get an optimal staining, whereas a concentration of 40 μg/mL was necessary for all the other antibodies (data not shown).
When we performed additional experiments with the CellSearch® system dedicated for CTC detection on pure SKBR3, we could observe single PD‐L1(+) SKBR3 tumor cells (Figure 3A) as well as clusters (Figure 3B) of PD‐L1(+) SKBR3 tumor cells. Moreover, we defined a “PD‐L1 immunoscore” with three different scores for the intensity of PD‐L1 expression by single SKBR3 tumor cells and SKBR3 tumor cell clusters: score 0 (no PD‐L1 expression), score 1 (low PD‐L1 expression) and score 2 (high PD‐L1 expression) (Figure 3).
Figure 3.
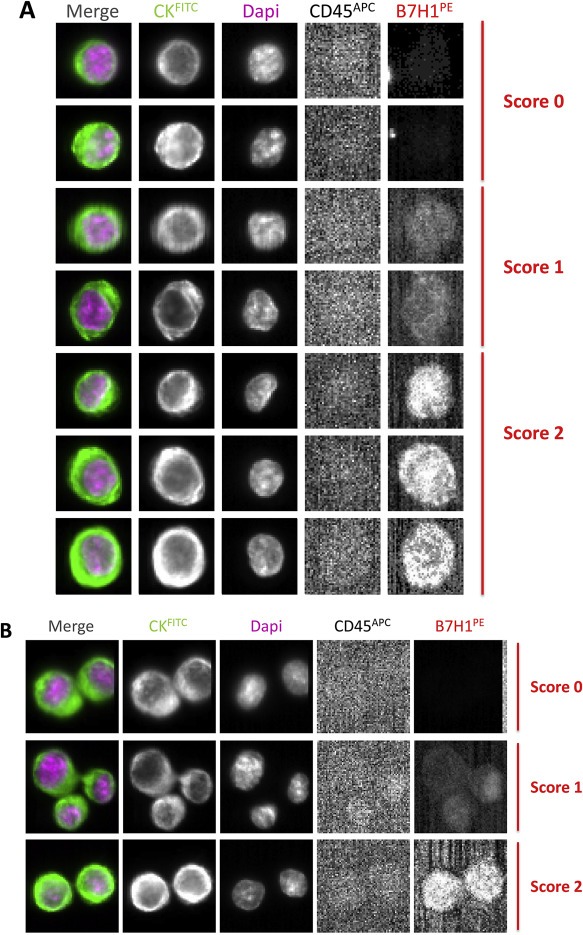
Detection of PD‐L1 expression on SKBR3 cells with the CellSearch® system. A, Single SKBR3 cells with different levels of PD‐L1 expression (B7H1PE in the 4th channel): score 0 (no PD‐L1 expression), 1 (weak PD‐L1 expression) and 2 (strong PD‐L1 expression); B, SKBR3 clusters with different PD‐L1 immunoscores.
3.2. Spiking experiments of SKBR3 in control blood
We performed spiking experiments by adding 200 SKBR3 cells in 7.5 mL of blood from healthy donors and detected the breast cancer cells using the CellSearch® system with the CellSearch® CXC Kit. These results demonstrated the feasibility to detect PD‐L1(+) tumor cells in blood samples with this technology dedicated for CTC detection. SKBR3 cells could be detected with a recovery rate of 79.5%, which is consistent with the results obtained the standard CTC kit (Riethdorf et al., 2007).
3.3. PD‐L1(+) CTC from metastatic breast cancer patients
Peripheral blood samples from HR(+), HER2(−) metastatic breast cancer patients (one male, fifteen females) were analyzed by the CellSearch® system implemented with the PD‐L1 detection by using the optimized conditions described above. For our study, we selected sixteen patients with a CTC count ≥1 using the CellSearch® system (mean, 254.5; median, 18.5; Range, 1–3146) and 13 of 16 (81.3%) patients with a CTC count ≥5 (mean, 312.5; median, 27; Range, 7–3146), the prognostic cut‐off for patients with metastatic breast cancer (Cristofanilli et al., 2004) (Table 2). Eleven patients out of 16 showed a subpopulation of CTCs expressing PD‐L1 (68.8%) scored 1 or 2 (Table 2; Figure 4A). The fraction of PD‐L1(+) CTCs varied from 0.2 to 100% (Table 2). However, this information may also depend on the number of detected CTCs. If only patients with more or equal than 10 CTCs are considered, the fraction of PD‐L1(+) CTCs varied from 0.2 to 50% (Table 2). The intensity of PD‐L1 staining varied between patients and within the same blood sample. Analyzing samples with multiple CTCs showed that six patients had CTCs with mixed PDL1 phenotypes (immunoscores 1 and 2), whereas homogeneous PDL1 immunoscores were only found in one patient (Table 2).
Table 2.
Patients characteristics, CTC detection and PD‐L1 expression on CTCs. CTCs were detected using the CellSearch® CXC Kit and stained for PD‐L1 in the 4th channel. PDL1(+) CTCs were immunoscored for PD‐L1 expression: score 1 (low PD‐L1 expression) and score 2 (high PD‐L1 expression).
| Patient ID | Age | HR Status | HER2 Status | CTCsTotal Number | PD‐L1(+) CTCs Absolute Number (%) | PD‐L1(+) CTCs Immunoscore | |
|---|---|---|---|---|---|---|---|
| 1 | 2 | ||||||
| 1 | 73 | + | – | 76 | 21 (27.6) | 10 | 11 |
| 2 | 69 | + | – | 10 | 5 (50) | 3 | 2 |
| 3 | 55 | + | – | 1 | 1 (100) | 1 | 0 |
| 4 | 69 | + | – | 10 | 5 (50) | 5 | 0 |
| 5 | 74 | + | – | 27 | 1 (3.7) | 0 | 1 |
| 6 | 60 | + | – | 7 | 3 (42.9) | 2 | 1 |
| 7 | 59 | + | – | 52 | 0 (0) | ||
| 8 | 71 | + | – | 90 | 10 (11.1) | 5 | 5 |
| 9 | 64 | + | – | 7 | 0 (0) | ||
| 10 | 71 | + | – | 3146 | 346 (11) | 106 | 240 |
| 11 | 64 | + | – | 3 | 0 (0) | ||
| 12 | 48 | + | – | 9 | 0 (0) | ||
| 13 | 56 | + | – | 518 | 1 (0.2) | 0 | 1 |
| 14 | 46 | + | – | 93 | 17 (18.3) | 10 | 7 |
| 15 | 86 | + | – | 4 | 0 (0) | ||
| 16 | 61 | + | – | 27 | 1 (3.7) | 0 | 1 |
Figure 4.
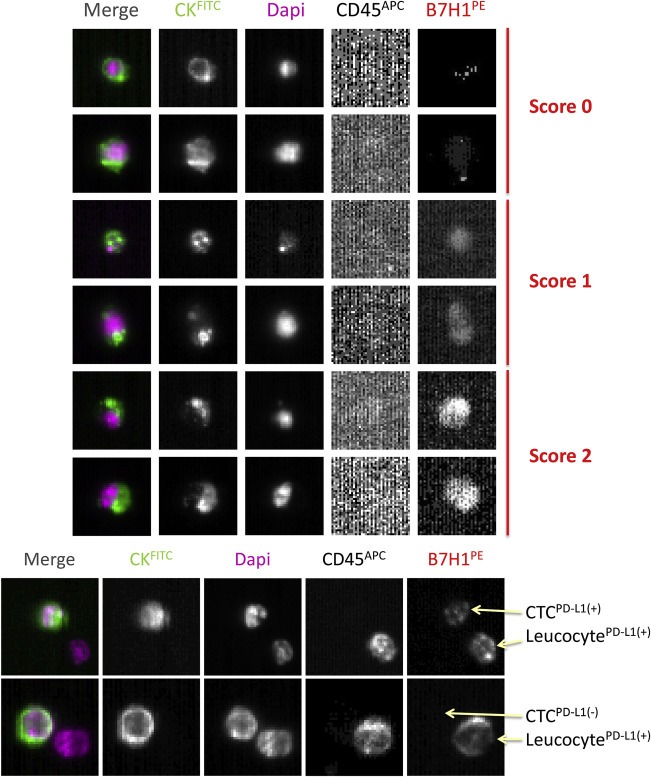
CTCs expressing various degrees of PD‐L1 detected in blood of HR(+) HER2(−) metastatic breast cancer patients with the CellSearch® system. Representative pictures of CTCs classified by scores 0, 1 and 2 for PD‐L1 expression.
As shown in the Figure 4B, we could also easily observe and distinguish PD‐L1(+) leucocytes next to CTCs positive or not for the expression of PD‐L1. Leucocytes express CD45, while CTCs are defined in the CellSearch® system as CD45(−).
As negative controls, we tested blood samples from 15 healthy controls and confirmed that none of the samples were positive for CTCs (Data not shown).
4. Discussion
In the present paper, we provide evidence that PD‐L1 is frequently expressed on metastatic cells circulating in the blood of HR(+), HER2(−) breast cancer patients. To our best knowledge, this is the first report demonstrating the expression of PD‐L1 on CTCs in any cancer entity. Detection of CTCs expressing PD‐L1 means that the patients harbor metastatic tumor cells that may have the capacity to block the immune system. These cells are potential targets for anti‐PDL1 therapies. This information is fundamentally different from the detection of shed PD‐L1 in the blood serum or plasma, which provides no information on the cellular source of PD‐L1.
The sensitivity, specificity and the dynamic range of the PD‐L1 MAb that we chose in our study was determined by Western Blot, flow cytometry and immunocytochemical analyses. We tested different mammary cancer cell lines (i.e., MDA‐MB‐231, MCF‐7 & SKBR3) and as PD‐L1 was expressed at variable degrees on SKBR3, we selected it as our positive control for the establishment of our novel assay detecting CTCs expressing PD‐L1. We could thus established immunoscores for PD‐L1 expression on single SKBR3 cells with this anti‐human B7‐H1 MAb and then validated these scores on blood samples from patients with a HR(+), HER2(−) metastatic breast cancer. This strategy has been adapted from previous reports using the CellSearch® system for immunophenotyping of CTCs (Ignatiadis et al., 2011; Riethdorf et al., 2010).
Despite the fact that we only analyzed a limited number of patients, our results clearly showed that PD‐L1 is frequently expressed on CTCs. However, the percentage of PD‐L1(+) CTCs in individual patients varied considerably. In particular, patients with a high percentage of PD‐L1(+) CTCs should be potential candidate for anti‐PD‐L1 therapy. The established CTC/PD‐L1 assay can be used in further trials for stratification and monitoring of cancer patients. It should be noted that only EpCAM(+) CTCs were analyzed, so the real number of PDL1(+) CTCs might be even higher. However, we decided to implement the PD‐L1 staining into the EpCAM‐dependent CellSearch® system because it is still the only automated technical platform cleared by the FDA‐USA with a remarkable reproducibility.
The present finding has obvious clinical implications. Immune checkpoint regulators such as PD‐L1 have become exciting new therapeutic targets leading to long lasting remissions in patients with advanced malignancies (Shin and Ribas, 2015). However, in view of the remarkable costs and the toxicity profiles (in particular during combination therapy) of these therapies, predictive biomarkers that are able to discriminate responders from non‐responders are urgently needed. The development of metastasis in breast cancer (and other solid tumors) can take many years and the metastatic cells may differ from the original cells in the operated primary lesion (Goss and Chambers, 2010). Thus, re‐staging of metastatic lesion is required to get real‐time information on the molecular characteristics of the actual metastatic cells that are the targets of antibody therapy (not the primary tumor). However, biopsies of metastatic lesions are an invasive procedure with considerable side effects depending on the site of metastasis and, moreover, different metastatic sites can have different molecular characteristics (Gerlinger et al., 2012).
As an alternative, we previously introduced a new diagnostic concept called “liquid biopsy” (Alix‐Panabieres and Pantel, 2013; Pantel and Alix‐Panabieres, 2010). Standardized automated analysis of CTCs for PD‐L1 is feasible, as demonstrated in the present investigation. This protocol can be easily expanded to other immune checkpoint molecules and other carcinoma entities. Implementing these liquid biopsy analyses into future clinical trials of checkpoint blockade might not only be useful for therapy stratification but CTC analyses performed in real‐time before and during the course of the therapy will provide, in addition, important information on the development of resistance.
Disclosure
The authors disclose no potential conflicts of interest.
Acknowledgments
The authors thank Delphine Gueroult for her excellent technical assistance for the CTC detection using the CellSearch® system.
This work was supported by the University Medical Centre of Rennes (Internal institution support in 2013), an ANR‐Roche SAS grant (TF), the FEDER and the Region Languedoc‐Roussillon (GEPETOS project, N° 38 582) (CAP), the Kastner Foundation (KP) and the European Research Council Advanced Investigator grant 269081. DISSECT (KP).
Mazel Martine, Jacot William, Pantel Klaus, Bartkowiak Kai, Topart Delphine, Cayrefourcq Laure, Rossille Delphine, Maudelonde Thierry, Fest Thierry, Alix-Panabières Catherine, (2015), Frequent expression of PD‐L1 on circulating breast cancer cells, Molecular Oncology, 9, doi: 10.1016/j.molonc.2015.05.009.
References
- Alix-Panabieres, C. , Pantel, K. , 2013. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 59, 110–118. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres, C. , Pantel, K. , 2014. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 14, 623–631. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres, C. , Schwarzenbach, H. , Pantel, K. , 2012. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 63, 199–215. [DOI] [PubMed] [Google Scholar]
- Bidard, F.C. , Fehm, T. , Ignatiadis, M. , Smerage, J.B. , Alix-Panabieres, C. , Janni, W. , Messina, C. , Paoletti, C. , Muller, V. , Hayes, D.F. , Piccart, M. , Pierga, J.Y. , 2013. Clinical application of circulating tumor cells in breast cancer: overview of the current interventional trials. Cancer Metastasis Rev. 32, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer, J.R. , Tykodi, S.S. , Chow, L.Q. , Hwu, W.J. , Topalian, S.L. , Hwu, P. , Drake, C.G. , Camacho, L.H. , Kauh, J. , Odunsi, K. , Pitot, H.C. , Hamid, O. , Bhatia, S. , Martins, R. , Eaton, K. , Chen, S. , Salay, T.M. , Alaparthy, S. , Grosso, J.F. , Korman, A.J. , Parker, S.M. , Agrawal, S. , Goldberg, S.M. , Pardoll, D.M. , Gupta, A. , Wigginton, J.M. , 2012. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, S. , Vogl, F.D. , Naume, B. , Janni, W. , Osborne, M.P. , Coombes, R.C. , Schlimok, G. , Diel, I.J. , Gerber, B. , Gebauer, G. , Pierga, J.Y. , Marth, C. , Oruzio, D. , Wiedswang, G. , Solomayer, E.F. , Kundt, G. , Strobl, B. , Fehm, T. , Wong, G.Y. , Bliss, J. , Vincent-Salomon, A. , Pantel, K. , 2005. A pooled analysis of bone marrow micrometastasis in breast cancer. N. Engl. J. Med. 353, 793–802. [DOI] [PubMed] [Google Scholar]
- Butt, A.Q. , Mills, K.H. , 2014. Immunosuppressive networks and checkpoints controlling antitumor immunity and their blockade in the development of cancer immunotherapeutics and vaccines. Oncogene. 33, 4623–4631. [DOI] [PubMed] [Google Scholar]
- Chambers, A.F. , Groom, A.C. , MacDonald, I.C. , 2002. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2, 563–572. [DOI] [PubMed] [Google Scholar]
- Chen, D.S. , Irving, B.A. , Hodi, F.S. , 2012. Molecular pathways: next-generation immunotherapy–inhibiting programmed death-ligand 1 and programmed death-1. Clin. Cancer Res. 18, 6580–6587. [DOI] [PubMed] [Google Scholar]
- Cristofanilli, M. , Budd, G.T. , Ellis, M.J. , Stopeck, A. , Matera, J. , Miller, M.C. , Reuben, J.M. , Doyle, G.V. , Allard, W.J. , Terstappen, L.W. , Hayes, D.F. , 2004. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791. [DOI] [PubMed] [Google Scholar]
- Eccles, S.A. , Welch, D.R. , 2007. Metastasis: recent discoveries and novel treatment strategies. Lancet. 369, 1742–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatmark, K. , Bjornland, K. , Johannessen, H.O. , Hegstad, E. , Rosales, R. , Harklau, L. , Solhaug, J.H. , Faye, R.S. , Soreide, O. , Fodstad, O. , 2002. Immunomagnetic detection of micrometastatic cells in bone marrow of colorectal cancer patients. Clin. Cancer Res. 8, 444–449. [PubMed] [Google Scholar]
- Gerlinger, M. , Rowan, A.J. , Horswell, S. , Larkin, J. , Endesfelder, D. , Gronroos, E. , Martinez, P. , Matthews, N. , Stewart, A. , Tarpey, P. , Varela, I. , Phillimore, B. , Begum, S. , McDonald, N.Q. , Butler, A. , Jones, D. , Raine, K. , Latimer, C. , Santos, C.R. , Nohadani, M. , Eklund, A.C. , Spencer-Dene, B. , Clark, G. , Pickering, L. , Stamp, G. , Gore, M. , Szallasi, Z. , Downward, J. , Futreal, P.A. , Swanton, C. , 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss, P.E. , Chambers, A.F. , 2010. Does tumour dormancy offer a therapeutic target?. Nat. Rev. Cancer. 10, 871–877. [DOI] [PubMed] [Google Scholar]
- Ignatiadis, M. , Rothe, F. , Chaboteaux, C. , Durbecq, V. , Rouas, G. , Criscitiello, C. , Metallo, J. , Kheddoumi, N. , Singhal, S.K. , Michiels, S. , Veys, I. , Rossari, J. , Larsimont, D. , Carly, B. , Pestrin, M. , Bessi, S. , Buxant, F. , Liebens, F. , Piccart, M. , Sotiriou, C. , 2011. HER2-positive circulating tumor cells in breast cancer. PLoS One. 6, e15624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilaveanu, L.B. , Shuch, B. , Zito, C.R. , Parisi, F. , Barr, M. , Kluger, Y. , Chen, L. , Kluger, H.M. , 2014. PD-L1 expression in Clear cell renal cell carcinoma: an analysis of Nephrectomy and sites of metastases. J. Cancer. 5, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. , Pantel, K. , 2013. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 23, 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, H.M. , Kelly, A.B. , Jewell, S.D. , McShane, L.M. , Clark, D.P. , Greenspan, R. , Hayes, D.F. , Hainaut, P. , Kim, P. , Mansfield, E. , Potapova, O. , Riegman, P. , Rubinstein, Y. , Seijo, E. , Somiari, S. , Watson, P. , Weier, H.U. , Zhu, C. , Vaught, J. , 2011. Biospecimen reporting for improved study quality (BRISQ). J. Proteome Res. 10, 3429–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel, K. , Alix-Panabieres, C. , 2010. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol. Med. 16, 398–406. [DOI] [PubMed] [Google Scholar]
- Pantel, K. , Alix-Panabieres, C. , 2013. Real-time liquid biopsy in cancer patients: fact or fiction?. Cancer Res. 73, 6384–6388. [DOI] [PubMed] [Google Scholar]
- Pantel, K. , Brakenhoff, R.H. , Brandt, B. , 2008. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer. 8, 329–340. [DOI] [PubMed] [Google Scholar]
- Philips, G.K. , Atkins, M. , 2015. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int. Immunol. 27, 39–46. [DOI] [PubMed] [Google Scholar]
- Riethdorf, S. , Fritsche, H. , Mueller, V. , Rau, T. , Schindlbeck, C. , Rack, B. , Janni, W. , Coith, C. , Beck, C. , Janicke, F. , Jackson, S. , Gornet, T. , Cristofanilli, M. , Pantel, K. , 2007. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin. Cancer Res. 13, 920–928. [DOI] [PubMed] [Google Scholar]
- Riethdorf, S. , Muller, V. , Zhang, L. , Rau, T. , Loibl, S. , Komor, M. , Roller, M. , Huober, J. , Fehm, T. , Schrader, I. , Hilfrich, J. , Holms, F. , Tesch, H. , Eidtmann, H. , Untch, M. , von Minckwitz, G. , Pantel, K. , 2010. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant geparquattro trial. Clin. Cancer Res. 16, 2634–2645. [DOI] [PubMed] [Google Scholar]
- Rossille, D. , Gressier, M. , Damotte, D. , Maucort-Boulch, D. , Pangault, C. , Semana, G. , Le Gouill, S. , Haioun, C. , Tarte, K. , Lamy, T. , Milpied, N. , Fest, T. , 2014. High level of soluble programmed cell death ligand 1 in blood impacts overall survival in aggressive diffuse large B-cell lymphoma: results from a French multicenter clinical trial. Leukemia. 28, 2367–2375. [DOI] [PubMed] [Google Scholar]
- Shin, D.S. , Ribas, A. , 2015. The evolution of checkpoint blockade as a cancer therapy: what's here, what's next?. Curr. Opin. Immunol. 33C, 23–35. [DOI] [PubMed] [Google Scholar]
- Topalian, S.L. , Drake, C.G. , Pardoll, D.M. , 2012. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr. Opin. Immunol. 24, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhr, J.W. , Pantel, K. , 2011. Controversies in clinical cancer dormancy. Proc. Natl. Acad. Sci. USA. 108, 12396–12400. [DOI] [PMC free article] [PubMed] [Google Scholar]


