Abstract
Fibroepithelial tumors (FTs) of the breast are a heterogeneous group of lesions ranging from fibroadenomas (FAD) to phyllodes tumors (PT) (benign, borderline, malignant). Further understanding of their molecular features and classification might be of clinical value. In this study, we analysed the expression of 105 breast cancer‐related genes, including the 50 genes of the PAM50 intrinsic subtype predictor and 12 genes of the Claudin‐low subtype predictor, in a panel of 75 FTs (34 FADs, 5 juvenile FADs, 20 benign PTs, 5 borderline PTs and 11 malignant PTs) with clinical follow‐up. In addition, we compared the expression profiles of FTs with those of 14 normal breast tissues and 49 primary invasive ductal carcinomas (IDCs). Our results revealed that the levels of expression of all breast cancer‐related genes can discriminate the various groups of FTs, together with normal breast tissues and IDCs (False Discovery Rate < 5%). Among FTs, the levels expression of proliferation‐related genes (e.g. CCNB1 and MKI67) and mesenchymal/epithelial‐related (e.g. CLDN3 and EPCAM) genes were found to be most discriminative. As expected, FADs showed the highest and lowest expression of epithelial‐ and proliferation‐related genes, respectively, whereas malignant PTs showed the opposite expression pattern. Interestingly, the overall profile of benign PTs was found more similar to FADs and normal breast tissues than the rest of tumours, including juvenile FADs. Within the dataset of IDCs and normal breast tissues, the vast majority of FADs, juvenile FADs, benign PTs and borderline PTs were identified as Normal‐like by intrinsic breast cancer subtyping, whereas 7 (63.6%) and 3 (27.3%) malignant PTs were identified as Claudin‐low and Basal‐like, respectively. Finally, we observed that the previously described PAM50 risk of relapse prognostic score better predicted outcome in FTs than the morphological classification, even within PTs‐only. Our results suggest that classification of FTs using gene expression‐based data is feasible and might provide clinically useful biological and prognostic information.
Keywords: Fibroepithelial, Fibroadenoma, Juvenile fibroadenoma, Phyllodes tumours, Gene expression, Intrinsic subtypes and claudin-low subtype
Highlights
The levels expression of proliferation‐ and mesenchymal/epithelial‐related genes were found to be the most discriminative.
The overall profile of benign phyllodes was very similar to fibroadenomas.
The vast majority of fibroepithelial tumors (FTs) were identified as Normal‐like by the PAM50 and Claudin‐low predictors.
The PAM50 risk of relapse prognostic score better predicted outcome in FTs than the morphological classification.
Classification of FTs using gene expression‐based data provides clinically useful information.
1. Introduction
Fibroepithelial tumours (FT) of the breast represent a heterogeneous group of biphasic neoplasms, composed of both epithelial and stromal components, that account for about 0.5–1 % of all breast tumours (Fattaneh, 2003; Reinfuss et al., 1996). To date, 3 main groups of FTs of the breast have been identified based on morphology: fibroadenoma (FAD), juvenile FAD and phyllodes tumour (PT). PTs are further subclassified into benign, borderline or malignant categories on the basis of a series of histological features such as stromal cellularity, nuclear atypia and mitotic activity (Contarini et al., 1982). However, reliable classification of FTs based on morphology remains challenging (Contarini et al., 1982; Hart et al., 1988; Niezabitowski et al., 2001; Yonemori et al., 2006).
From a clinical perspective, FADs may be safely followed without further investigation or treated with simple enucleation, whereas PTs are usually treated with mastectomy or wide excision with adequate margins. Although surgical resection is sufficient to cure the vast majority of PTs, PTs can recur locally and/or undergo metastatic spread. Indeed, local recurrence rate of PTs is 10%–18% with negative and positive resection margins, respectively, and 9–27% of malignant PTs metastasize to distant organs (Barrio et al., 2007; Kracht et al., 1998; Lester and Stout, 1954; Lindquist et al., 1982). However, reports of benign and borderline PTs metastasizing also exist (Kracht et al., 1998; Lester and Stout, 1954; Lindquist et al., 1982). Thus, there is a need for an accurate diagnosis and management of FTs of the breast (Jones et al., 2008a; Tan and Ellis, 2013).
Similar to FTs, invasive breast carcinoma is a heterogeneous disease with respect to molecular alterations, cellular compositions and clinical outcomes. Over the last decade, studies based on gene expression analysis have identified and extensively studied 5 major classes of breast cancer (Luminal A, Luminal B, HER2‐enriched, Basal‐like and Claudin‐low) (Perou et al., 2000; Prat and Perou, 2011; TCGA, 2012a). Known as the “intrinsic subtypes of breast cancer”, these groups of tumours have revealed critical differences in incidence, survival, dissemination sites and response to treatment (Parker et al., 2009; Prat et al., 2014a; TCGA, 2012b). To date, it is unknown how the FTs are classified according to the biology of the intrinsic breast cancer subtypes.
Among the different intrinsic subtypes, the Claudin‐low shows a stromal‐like phenotype characterized by the low expression of many tight junction‐related genes such as claudins −3, −4 and −7 and E‐cadherin and high expression of mesenchymal‐related genes such as vimentin or ZEB1 (Prat et al., 2013, 2010). Clinically, these tumours are usually aggressive and have a poor outcome. Interestingly, metaplastic breast cancer, which resembles many phenotypic features of malignant PTs, usually belongs to the Claudin‐low and Basal‐like intrinsic subtype (Prat et al., 2010). Both Claudin‐low and metaplastic breast cancer have previously show high enrichment for cancer stem cell‐related biological processes (Hennessy et al., 2009).
In this study, we analysed the expression of 105 breast cancer‐related genes, including the genes that define the intrinsic subtypes of breast cancer, in a panel of FTs with clinical follow‐up. In addition, we compared the expression profiles of FTs with those of normal breast tissues and invasive breast carcinomas.
2. Materials and methods
2.1. Patient samples
This is a retrospective and exploratory study. From 1998 to 2013, we identified all consecutive patients (n = 41) diagnosed of juvenile FAD, benign PTs, borderline PTs or malignant PTs who had undergone local treatment at the breast surgery unit of the Vall d'Hebron University Hospital. In addition, we randomly selected 34 surgically resected FADs from our records from the same period of time for a total of 75 FTs. All FADs were enucleated and PTs were resected with free margins. All FTs were classified by V.P. according to the 2012 World Health Organization (WHO) guidelines (Lakhani et al., 2012). Clinical reports and follow‐up data were available for 64 patients. Moreover, we included an in‐house FFPE‐based dataset of 49 primary invasive ductal carcinomas and 14 normal breast tissue obtained from reduction mammoplasties. The project was approved by the ethics committee of our institution.
2.2. Immunohistochemistry
Immunohistochemical Ki‐67 staining of 5 representative FTs was performed in sections from paraffin‐embedded tissue blocks with the avidin‐biotin‐peroxidase technique. Five micrometer‐thick sections were cut from the tissue specimens and placed on poly‐l‐lysine‐coated glass slides. Sections were deparaffinized with xylene and rehydrated in graded alcohol. Endogenous peroxidase was blocked by immersing the sections in 0.1% hydrogen peroxidase in absolute methanol for 20 min. For antigen retrieval, tissue sections were heated in a pressure cooker in 10 mM citric acid monohydrate, pH 6.0 for 5 min, and then incubated with the primary antibody at room temperature. The primary antibody used was CONFIRM anti‐Ki‐67 (30–9) (Ventana Medical Systems, Tucson, AZ). Immunohistochemistry was performed with the Ventana BenchMark XT slide processing system and the iView detection kit (Ventana Medical Systems, Tucson, AZ). All slides were counterstained with hematoxylin, dehydrated, and mounted. Negative controls were performed by omitting the primary antibody.
2.3. Gene expression analysis
A section of the formalin‐fixed paraffin‐embedded (FFPE) breast tissue was first examined with a hematoxylin and eosin staining to confirm the diagnosis and determine the tumour area. For RNA purification (Roche® High Pure FFPET RNA isolation kit), 3 10 μm FFPE slides were cut for each tumour, and macrodissection was performed, when needed, to avoid normal breast contamination. A minimum of ∼100 ng of total RNA was used to measure the expression of 105 breast cancer‐related genes and 5 house‐keeping genes using the nCounter platform (Nanostring Technologies, Seattle, WA, US) (Geiss et al., 2008). Data was log base 2 transformed and normalized using 5 house‐keeping genes (ACTB, MRPL19, PSMC4, RPLP0 and SF3A1). Raw gene expression data has been deposited in Gene Expression Omnibus (GSE55224).
2.4. Genes and gene signatures
The list of 105 genes includes genes from the following 3 signatures: PAM50 intrinsic subtype predictor (n = 50) (Parker et al., 2009), Claudin‐low subtype predictor (n = 43) (Prat et al., 2010), 13‐VEGF/Hypoxia signature (n = 13) (Hu et al., 2009). In addition, we included 8 individual genes that have been found to play an important role in breast cancer (i.e. CD24 (Prat et al., 2010), CRYAB (Moyano et al., 2006), ERBB4 (Sundvall et al., 2008), PIK3CA(TCGA, 2012b), PTEN (TCGA, 2012b), RAD17 (Weigman et al., 2012), RAD50 (Weigman et al., 2012) and RB1 (TCGA, 2012b)).
2.5. Breast cancer intrinsic subtyping
All tumors were assigned to an intrinsic molecular subtype of breast cancer (Luminal A, Luminal B, HER2‐enriched, Basal‐like and Claudin‐low) and the Normal‐like group using the previously reported PAM50 subtype and the Claudin‐low subtype predictors (Nielsen et al., 2010; Parker et al., 2009; Prat et al., 2010). In addition, we also evaluated the previously reported PAM50‐based Risk of Relapse (ROR) score, as a continuous variable and as group categories using the previously reported cutoffs (Nielsen et al., 2010). ROR was trained to predict distant relapse‐free survival in a node‐negative breast cancer patient dataset representative of all subtypes and no patient received adjuvant systemic therapy (Nielsen et al., 2010; Parker et al., 2009; Prat et al., 2010).
2.6. Statistical analysis
To identify genes whose expression is significantly different between ≥2 groups, we used either a two class unpaired Analysis of Microarrays (SAM) or a multiclass SAM (Hennessy et al., 2009). Average linkage hierarchical clustering was performed using Cluster v3.0 (Eisen et al., 1998), and heatmaps were displayed using Java Treeview v1.1.4r2. Biologic analysis of gene lists was performed with DAVID annotation tool (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003).
Survival functions to relapse‐free survival (RFS) were from the Kaplan–Meier product‐limit estimator with tests of differences by the log‐rank test. RFS was defined as the period of time from surgery to either the last follow‐up date or the date of the first local or distant relapse of the disease. Cox proportional hazard models adjusted for standard clinical‐pathological variables were used to test the associations with RFS of each variable. All statistical computations were carried out in R v2.15.1 (http://cran.r‐project.org). All statistical tests were two sided, and the statistical significance level was set to less than 0.05.
3. Results
3.1. Characteristics of the patient population with FTs
The main clinical‐pathological characteristics of the FTs evaluated in this study are shown in Table 1. Of 75 FTs analysed, 34 (45.3%) were FADs, 5 (6.7%) were juvenile FADs, 20 (26.7%) were benign PTs, 5 (6.7%) were borderline PTs and 11 were malignant PTs (14.7%). Of note, all cases underwent surgical removal, including FADs. In addition, we included 49 cases of invasive breast carcinoma and 14 normal breast tissues from reduction mammoplasties. The distribution of the intrinsic subtypes, based on the PAM50 predictor, within the 49 invasive breast carcinomas was the following: 34.7% Luminal A, 28.6% Basal‐like, 18.4% HER2‐enriched, 16.3% Luminal B and 2.0% normal breast‐like. As expected, all normal breast tissues were identified as normal breast‐like by PAM50.
Table 1.
Characteristics of the patient population.
| FAD | Juvenile FAD | Benign PT | Borderline PT | Malignant PT | Total | |
|---|---|---|---|---|---|---|
| N | 34 | 5 | 20 | 5 | 11 | 75 |
| Age (years) | 34 (13–65) | 17 (14–20) | 38.6 (21–74) | 59 (29–85) | 51 (32–77) | 39 (13–85) |
| mean, (range) | ||||||
| Size (mm) | 32.79 (15–70) | 36.5 (26–35) | 36.6 (16–80) | 93 (30–180) | 63.91 (20–160) | 42.75 (15–180) |
| mean, (range) | ||||||
| Type of Surgery | ||||||
| Enucleation | 34 | 0 | 0 | 0 | 0 | 34 |
| Lumpectomy | 0 | 5 | 5 | 4 | 4 | 18 |
| Mastectomy | 0 | 0 | 0 | 1 | 5 | 6 |
| Adjuvant Therapy | ||||||
| Adjuvant chemotherapy | 0 | 0 | 0 | 0 | 2 | 2 |
| Radiotherapy | 0 | 0 | 0 | 0 | 5 | 5 |
3.2. Gene expression differences among groups of FTs
To examine the gene expression differences among the 5 groups of FTs (i.e. FADs, juvenile FAD, benign PTs, borderline PTs and malignant PTs), normal breast tissues and invasive ductal carcinomas, we analysed the expression of 105 breast cancer‐related genes and identified the genes whose expression is significantly different across the 7 groups (multi‐class SAM; False Discovery Rate < 5%). The results revealed that the expression of all 105 genes was discriminative (Supplemental Data). Similar results were obtained when normal breast tissues and invasive breast carcinomas were excluded (Supplemental Data).
To visualize the differences in expression levels of the 105 breast cancer‐related genes across the 7 groups, we performed a hierarchical clustering analysis using the SAM scores of each 105 gene in each group (Figure 1). The heatmap revealed 4 main gene clusters (A, B, C and D) dominating the entire clustering. In addition, interesting relationships of the 105‐gene profile among the 5 groups of FTs were observed. On one hand, the profiles of FADs and benign PTs were found to be more similar to each other than to any of the profiles of the other groups of FTs. On the other hand, the profile of juvenile FADs was found to be more similar to the profiles of borderline and malignant PTs than to the profile of FADs.
Figure 1.
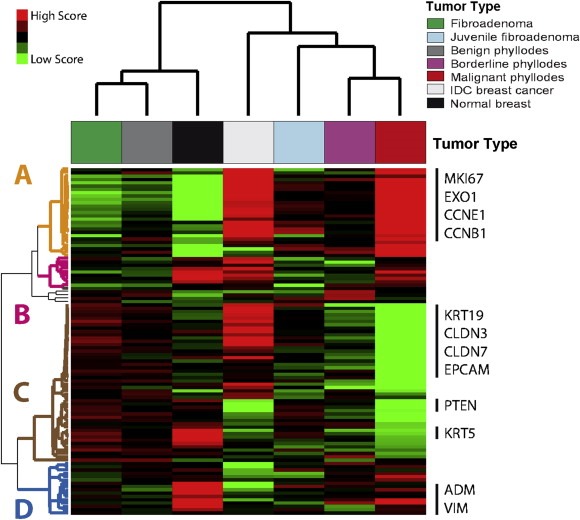
Group‐specific gene expression profiles. Each coloured square represents the relative mean gene score for each subtype, with highest expression shown in red, average expression in black, and lowest expression in green. This gene list was obtained by performing a 7‐class significance analysis of microarrays (False Discovery Rate < 5%). On the right, selected genes symbols of several gene clusters are shown.
3.3. Gene cluster A: proliferation‐related
Cluster A (n = 27 genes) was found enriched for genes enriched for cell cycle‐related (e.g. MKI67, CDC20 and CCNB1) and chromosome segregation‐related (e.g. CENPF, BIRC5 and NDC80) biological processes. This cluster was found highly expressed in malignant PTs and invasive breast carcinomas (especially in the Basal‐like subtype), and lowly expressed in normal breast tissue and FADs followed by benign PTs (Figure 1 and Figure 2 A–B). Similar results are observed with Ki‐67 IHC staining (Figure 3).
Figure 3.
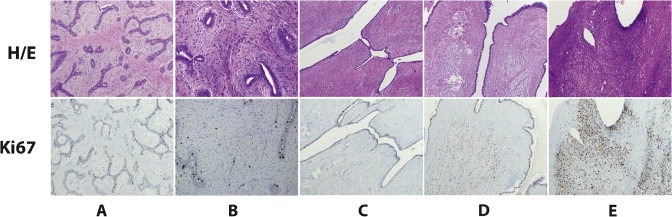
Ki‐67 IHC staining in 5 representative cases of FTs. (A) FAD; (B) Juvenile FAD; (C) Benign PT; (D) Borderline PT; (E) Malignant PT.
3.4. Gene cluster B: hypoxia‐related
Gene cluster B (n = 10 genes) was not found enriched for any gene ontology biological process. However, 60% of the genes (i.e. PNP, GAL, FLVCR2, NDRG1, FABP5 and DDIT4) were found to be part of a previously reported breast cancer 13‐VEGF hypoxia signature (Hu et al., 2009). Among FTs, gene cluster B, as well as the complete 13‐VEGF signature, was found highly expressed in malignant PTs (Figure 2C).
Figure 2.
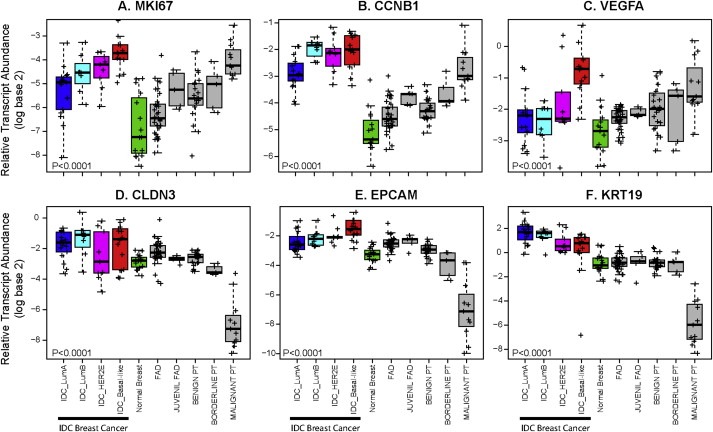
Box‐and‐whisker plots for expression of 6 selected breast cancer‐related genes across invasive breast carcinomas, normal breast tissues and FTs. P‐values were calculated by comparing mean values across all groups. IDC, invasive ductal carcinoma.
3.5. Gene cluster C: epithelial‐ and luminal‐related
Gene cluster C (n = 47 genes) was found enriched for genes enriched for ectoderm/epidermis development (e.g. KRT5, KRT14 and KRT17), cell adhesion (e.g. CLDN3, CLDN4, CLDN7 and CD24), mammary gland development (e.g. MET, PGR, ERBB3 and ERBB4), response to oestrogen stimulus (e.g. GATA3, ESR1, KRT19 and BCL2) biological processes (Figure 1 and Figure 2D–F). Among FTs, gene cluster C was found more expressed in FADs compared (in order of expression) to benign PTs, juvenile FADs, borderline PTs and malignant PTs.
3.6. Gene cluster D: apoptosis‐ and angiogenesis‐related
Gene cluster D (n = 16 genes) was found enriched for genes enriched for negative regulation of apoptosis (e.g. ANGPTL4, EGFR and PIK3CA), blood vessel morphogenesis (e.g. ANGPTL4, CAV1 and FOXC1) and negative regulation of cell cycle (e.g. RB1 and FOXC1) biological processes. This cluster was found highly expressed in normal breast tissues, followed by malignant PTs, and lowly expressed in invasive breast carcinomas.
3.7. PAM50‐and claudin‐low‐based classifications of FTs
To determine which breast cancer intrinsic subtype each group of FT best resemble, we applied the PAM50 predictor, with or without the Claudin‐low predictor, on the 75 FT‐sample set (Table 2). Regardless of the Claudin‐low predictions, 82.7% of FTs were identified as normal breast‐like and 2.7% as Luminal A (1 FAD and 1 benign PT). Interestingly, no FAD, benign PT or borderline PT was identified as Basal‐like or Claudin‐low, whereas most malignant PTs were identified as Basal‐like or Claudin‐low. Interestingly, 1 juvenile FAD out of 5 was identified as Basal‐like.
Table 2.
PAM50, Claudin‐low and ROR of FTs breast tumors.
| Group | FAD | Juvenile FAD | Benign PT | Borderline PT | Malignant PT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | ||
| PAM50 | Basal‐like | 0 | 0% | 1 | 20% | 0 | 0% | 0 | 0% | 10 | 91% |
| Luminal A | 1 | 4% | 0 | 0% | 1 | 5% | 0 | 0% | 0 | 0% | |
| Normal | 25 | 96% | 4 | 80% | 18 | 95% | 5 | 100% | 1 | 9% | |
| Total | 26 | – | 5 | – | 19 | – | 5 | – | 11 | – | |
| PAM50 + Claudin‐low | Basal‐like | 0 | 0% | 1 | 20% | 0 | 0% | 0 | 0% | 3 | 27% |
| Claudin‐low | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 7 | 64% | |
| Luminal A | 1 | 4% | 0 | 0% | 1 | 5% | 0 | 0% | 0 | 0% | |
| Normal | 25 | 96% | 4 | 80% | 18 | 95% | 5 | 100% | 1 | 9% | |
| Total | 26 | – | 5 | – | 19 | – | 5 | – | 11 | – | |
| PAM50‐ROR | High | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 8 | 73% |
| Med | 2 | 8% | 2 | 40% | 1 | 5% | 2 | 40% | 3 | 27% | |
| Low | 24 | 92% | 3 | 60% | 18 | 95% | 3 | 60% | 0 | 0% | |
| Total | 26 | – | 5 | – | 19 | – | 5 | – | 11 | – | |
To visualize these results, we clustered all FTs, normal breast tissues and invasive breast carcinomas using the PAM50 genes (Figure 4). The results revealed that the PAM50 expression profile of most FADs and benign PTs are undistinguishable from normal breast tissues. On the other hand, most malignant PTs have very similar gene expression profiles as Basal‐like/Claudin‐low breast carcinomas.
Figure 4.
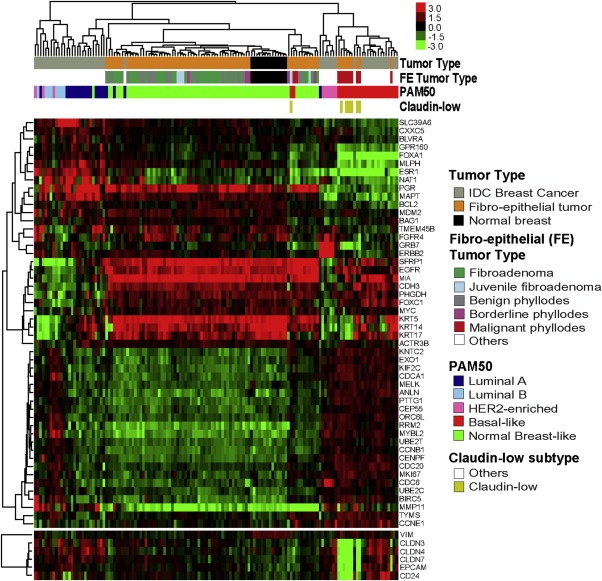
Hierarchical clustering of 124 tumours and 14 normal breast tissues using the PAM50 genes. Each coloured square represents the relative mean transcript abundance (in log2 space) for each sample, with highest expression shown in red, median expression in black, and lowest expression in green. Tumour type, fibro‐epithelial (FE) tumour type, PAM50 and Claudin‐low subtype calls are identified below the array tree. Below the heatmap, the expression of selected genes related to the Claudin‐low subtype is shown.
3.8. Predicting risk of relapse among FTs
Of the 64 patients with an FT and clinical follow‐up, 7 cases (10.9%) presented a recurrence. No patients with a FAD or juvenile FAD presented a recurrence, and the 2 distant recurrences occurred in patients with a malignant PT. Overall, the local and distant recurrence rate in benign PTs, borderline PTs and malignant PTs was 5.9% (1/17), 40.0% (2/5) and 36.4% (4/11), respectively (P = 0.069, Chi‐square test). The 5‐year RFS of benign PTs, borderline PTs and malignant PTs was 92.9% (95% Confidence Interval [CI] 80.3–100.0), 80.0% (95% CI 51.6–100.0) and 56.8% (95% CI 32.2–100.0), respectively. Despite these differences, the type of PT was not found significantly associated with RFS (Figure 5 and Table 3) with a hazard ratio (HR) between the malignant PT vs. benign PT of 8.83 (0.98–79.50 95% Confidence interval [CI], P = 0.052), although a clear tendency was noted.
Figure 5.
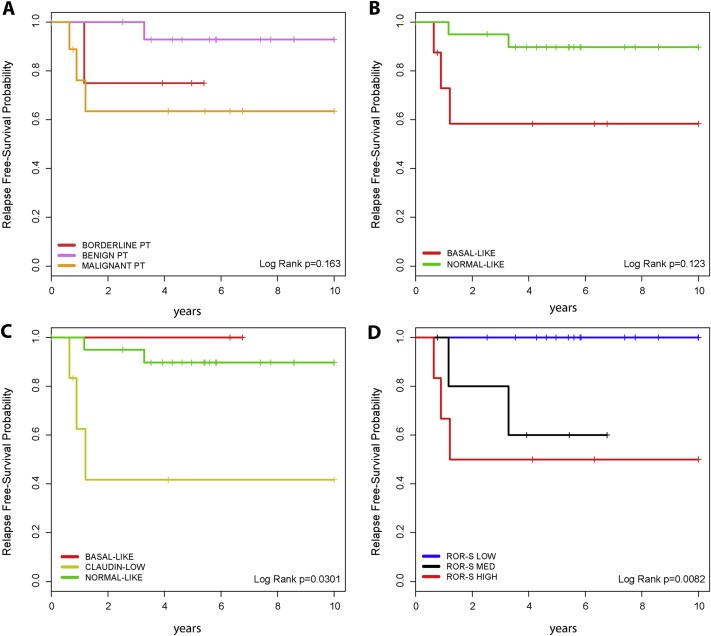
Kaplan–Meier relapse‐free survival analysis within breast PTs. (A) Based on morphological classification; (B) Based on Basal‐like versus Normal‐like classification; (C) Based on Basal‐like, Claudin‐low and Normal‐like classification; (D) Based on ROR‐S group.
Table 3.
Univariate Cox model 10‐year relapse‐free survival analyses in patients with PTs.
| Variables | HR | Lower 95% | Upper 95% | P‐value |
|---|---|---|---|---|
| Age at diagnosis (cont. variable) | 1.029 | 0.989 | 1.070 | 0.158 |
| Tumor size (cont. variable) | 0.982 | 0.951 | 1.013 | 0.259 |
| Radiotherapy (yes vs. no) | 1.480 | 0.176 | 12.460 | 0.718 |
| Histological Type (Benign Phyllodes as reference) | 1.0 | – | – | – |
| Borderline phyllodes | 6.839 | 0.618 | 75.680 | 0.117 |
| Malignant phyllodes | 8.833 | 0.981 | 79.500 | 0.052 |
| 13‐gene VEGF signature (continuous variable) | 0.746 | 0.263 | 2.115 | 0.582 |
| Claudin‐low (continuous variable) | 2.350 | 1.197 | 4.614 | 0.013 |
| ROR‐S (continuous variable) | 1.039 | 1.010 | 1.070 | 0.009 |
| ROR‐S Groups(ROR‐S Low as reference) | 1.0 | – | – | – |
| ROR‐S interm | 7.784 | 0.702 | 86.360 | 0.095 |
| ROR‐S high | 15.678 | 1.729 | 142.190 | 0.014 |
| PAM50 (Basal‐like vs. Normal‐like) | 4.811 | 1.064 | 21.750 | 0.041 |
| PAM50+Claudin‐low (Normal‐like as reference) | 1.0 | – | – | – |
| Basal‐like | 2.668 | 0.276 | 25.760 | 0.396 |
| Claudin‐low | 6.678 | 1.309 | 34.070 | 0.022 |
To evaluate if gene expression‐based data can predict RFS, we tested the prognostic value of the 13‐VEGF/Hypoxia signature, the Claudin‐low signature, intrinsic subtyping with and without Claudin‐low predictions and the PAM50‐based ROR score (Table 3). The results revealed that all signatures predict outcome either as group categories or as a continuous variable, except for the 13‐VEGF/Hypoxia signature (P = 0.582). Of note, the ROR risk groups, whose cut‐offs were determined in a pure prognostic breast cancer dataset (Nielsen et al., 2010; Parker et al., 2009; Prat et al., 2010), predicted outcome better than the morphological classification of PTs (Figure 5). The HR between the ROR‐high vs. ROR‐low groups was 15.68 (1.73–142.19 95% CI, P = 0.014).
4. Discussion
In this study, we used gene expression data to classify and better understand the underlying biology of breast FTs. Our results revealed that the levels of expression of genes involved in important breast cancer‐related biological processes such as cell proliferation, hypoxia or epithelial differentiation can also discriminate the various groups of FTs. In addition, we showed that gene expression‐based classifications that are prognostic in breast cancer can also predict the clinical behaviour of breast FTs. Overall, our results should help improve our understanding of the biologic heterogeneity of breast FTs and improve their clinical management.
Two previous studies have started to dissect the underlying genetics of breast FTs (Jones et al., 2008b; Wang et al., 2006). Wang and colleagues (Wang et al., 2006) studied genome wide loss of heterozygosity (LOH) in 11 PTs and 22 FADs. The authors showed that LOH is frequent and sometimes extensive in PTs, but is rarely seen in FADs. Interestingly, although no LOH marker identified the majority of these lesions, a subset of 4 LOH regions (i.e. 7p12, 3p24, 10p12 and 9p21) occurred in multiple cases of PTs and was not found in FADs. At the same time, Jones and colleagues (Jones et al., 2008b) performed array‐CGH analyses in 126 PTs (37 benign, 41 borderline, 48 malignant). The results revealed clear chromosomal copy‐number changes in borderline and malignant PTs, such as deletion of 9p21 that involved p16INK4a, supporting the division of malignant and borderline PTs into two separate groups, one comprising almost all malignant lesions and the other, benign and borderline tumours. Following this work, Jones and colleagues (Jones et al., 2008a) studied gene expression‐based data of 23 PTs and identified 4 genes (i.e. PAX3, SIX1, TGFB2 and HMGA2) that might be important in the transition from the benign to borderline/malignant phenotype.
In the era of personalized medicine, new tools are needed that provide clinically useful prognostic and predictive information for patients with lesions in the breast (Campbell et al., 2014). However, few studies have tried to identify biomarkers that predict outcome in breast FTs. For example, Tan et al. have proposed a nomogram based on 3 histological criteria (i.e. cell atypia, mitoses and overgrowth and surgical margins) that can be used to calculate RFS of an individual diagnosed of a PT (Tan et al., 2012). Moreover, the same group identified Six1 and Pax3 IHC‐based expression to correlate with poorer clinical outcome (Tan et al., 2014). Finally, Yonemori and colleagues (Yonemori et al., 2006) examined the IHC expression of the epidermal growth factor receptor (EGFR), HER2/neu, CD117/c‐kit, p53, and Ki‐67 in 41 PTs, and looked for associations with survival. The authors found that p53 expression and the Ki‐67 index, but not the expression of EGFR, were significantly correlated with the RFS and overall survival.
Our results suggest that gene expression‐based classifications of FTs could be clinically useful. On one hand, since reliable classification of breast FTs based on morphology is challenging (Contarini et al., 1982; Hart et al., 1988; Niezabitowski et al., 2001; Yonemori et al., 2006), a standardized gene expression‐based predictor could provide an objective and reproducible diagnosis similar to the PAM50 subtype predictor (Nielsen et al., 2014, 2013, 2013, 2012, 2014). This would be especially useful within PTs, where current WHO classification (Lakhani et al., 2012) into benign, borderline, and malignant categories is based on multiple and subjective clinical‐pathological criteria (i.e. degree of stromal cellular atypia, mitotic activity per 10 high power fields, degree of stromal overgrowth, tumor necrosis and margin appearance). On the other hand, gene expression‐based classifications could be used to identify, beyond the WHO classification, those patients with FTs with either an outstanding or a very bad prognosis. In our dataset, 7 of 35 PTs (20%) were re‐classified by PAM50 ROR to other risk categories (0.654 concordance Cohen's kappa score) with 3 borderline cases being classified as having a low‐risk, and 4 benign or malignant PTs being re‐classified as having an intermediate risk. In addition, 4 of 11 (36.4%) malignant PTs were not identified as being Claudin‐low by gene expression and showed a better outcome than Claudin‐low malignant PTs. Based on all of these data, we propose to further develop and validate a predictor of breast FTs in a large series of well‐characterized breast FTs with follow‐up data.
Our study has several caveats that need attention. First, although our dataset has a total of 75 FTs, which is in line with the numbers of previous reports, some of our groups, such as juvenile FAD or borderline PTs, have each a limited number of samples (n = 5). Thus, further studies are needed to better characterize these two groups of tumours. Second, our survival outcome association analyses need to be taken with caution due to the limited number of events of our dataset. Finally, we did not explore the expression of all genes in the genome but rather focused on the expression of 105 breast cancer‐related genes. Thus, it is likely that larger discovery efforts will identify new biological processes that are breast FTs‐specific and that might better predict outcome.
To conclude, classification of FTs using gene expression is feasible and might provide clinically useful biologic and prognostic information. Further studies are needed to develop and validate a molecular predictor of FTs of the breast.
Statement of author contributions
MV and AP contributed to experimental design. MV, VP, PG, JC, IR and AP were responsible for performing experiments and contributed to data analysis. All authors contributed to manuscript preparation.
Conflict of interest
Uncompensated advisory role of A.P. for Nanostring Technologies.
Supporting information
The following is the supplementary data related to this article:
Results of all Significance Analyses of Microarrays (SAM) performed in the study.
Fig. S1 Kaplan–Meier relapse‐free survival analysis within Fibroepithelial tumors (FTs). (A) Based on morphological classification; (B) Based on Basal‐like versus Normal‐like classification; (C) Based on Basal‐like, Claudin‐low and Normal‐like classification; (D) Based on ROR‐S group.
Acknowledgments
This work was supported by funds from the Banco Bilbao Vizcaya Argentaria (BBVA) Foundation (to AP).
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.01.003.
Vidal Maria, Peg Vicente, Galván Patricia, Tres Alejandro, Cortés Javier, Ramón y Cajal Santiago, Rubio Isabel T., Prat Aleix, (2015), Gene expression-based classifications of fibroadenomas and phyllodes tumours of the breast, Molecular Oncology, 9, doi: 10.1016/j.molonc.2015.01.003.
References
- Barrio, A. , Clark, B. , Goldberg, J. , Hoque, L. , Bernik, S. , Flynn, L. , Susnik, B. , Giri, D. , Polo, K. , Patil, S. , Van Zee, K. , 2007. Clinicopathologic features and long-term outcomes of 293 phyllodes tumors of the breast. Ann. Surg. Oncol.. 14, 2961–2970. [DOI] [PubMed] [Google Scholar]
- Campbell, R.M. , Anderson, B.D. , Brooks, N. , Brooks, H. , Chan, E.M. , De Dios, A. , Gilmour, R. , Graff, J.R. , Jambrina, E. , Mader, M. , McCann, D. , Na, S. , Parsons, S.H. , Pratt, S.E. , Shih, C. , Stancato, L.F. , Starling, J.J. , Tate, C. , Velasco, J.A. , Wang, Y.Y. , Ye, X.S. , 2014. Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with anti-tumor activity. Mol. Cancer Ther.. 13, (2) 364–374. [DOI] [PubMed] [Google Scholar]
- Contarini, O. , Urdaneta, L.F. , Hagan, W. , Stephenson, S.E. , 1982. Cystosarcoma phylloides of the breast: a new therapeutic proposal. Am. Surg.. 48, 157–166. [PubMed] [Google Scholar]
- Dennis, G. , Sherman, B.T. , Hosack, D.A. , Yang, J. , Gao, W. , Lane, H.C. , Lempicki, R.A. , 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol.. 4, R60 [PubMed] [Google Scholar]
- Eisen, M.B. , Spellman, P.T. , Brown, P.O. , Botstein, D. , 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci.. 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattaneh, A. , Devilee, P. , 2003. WHO Classification of Tumors: Tumors of the Breast and Female Genital Organs IARC; Lyon: [Google Scholar]
- Geiss, G.K. , Bumgarner, R.E. , Birditt, B. , Dahl, T. , Dowidar, N. , Dunaway, D.L. , Fell, H.P. , Ferree, S. , George, R.D. , Grogan, T. , James, J.J. , Maysuria, M. , Mitton, J.D. , Oliveri, P. , Osborn, J.L. , Peng, T. , Ratcliffe, A.L. , Webster, P.J. , Davidson, E.H. , Hood, L. , Dimitrov, K. , 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotech.. 26, 317–325. [DOI] [PubMed] [Google Scholar]
- Hart, J. , Layfield, L.J. , Trumbull, W.E. , Brayton, D. , Barker, W.F. , Giuliano, A.E. , 1988. Practical aspects in the diagnosis and management of cystosarcoma phyllodes. Arch. Surg.. 123, 1079–1083. [DOI] [PubMed] [Google Scholar]
- Hennessy, B.T. , Gonzalez-Angulo, A.M. , Stemke-Hale, K. , Gilcrease, M.Z. , Krishnamurthy, S. , Lee, J.S. , Fridlyand, J. , Sahin, A. , Agarwal, R. , Joy, C. , Liu, W. , Stivers, D. , Baggerly, K. , Carey, M. , Lluch, A. , Monteagudo, C. , He, X. , Weigman, V. , Fan, C. , Palazzo, J. , Hortobagyi, G.N. , Nolden, L.K. , Wang, N.J. , Valero, V. , Gray, J.W. , Perou, C.M. , Mills, G.B. , 2009. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res.. 69, 4116–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Fan, C. , Livasy, C. , He, X. , Oh, D. , Ewend, M. , Carey, L. , Subramanian, S. , West, R. , Ikpatt, F. , 2009. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med.. 7, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M. , Mitter, R. , Poulsom, R. , Gillett, C. , Hanby, A.M. , Tomlinson, I.P. , Sawyer, E.J. , 2008. mRNA expression profiling of phyllodes tumours of the breast: identification of genes important in the development of borderline and malignant phyllodes tumours. J. Pathol.. 216, 408–417. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. , Mitter, R. , Springall, R. , Graham, T. , Winter, E. , Gillett, C. , Hanby, A.M. , Tomlinson, I.P. , Sawyer, E.J. , 2008. A comprehensive genetic profile of phyllodes tumours of the breast detects important mutations, intra-tumoral genetic heterogeneity and new genetic changes on recurrence. J. Pathol.. 214, 533–544. [DOI] [PubMed] [Google Scholar]
- Kracht, J. , Sapino, A. , Bussolati, G. , 1998. Malignant phyllodes tumor of breast with lung metastases mimicking the primary. Am. J. Surg. Pathol.. 22, 1284–1290. [DOI] [PubMed] [Google Scholar]
- Lakhani, Sunil R. , Ellis, I.O. , Schnitt, Stuart JH. , Tan, Puay Hoon , van de Vijver, Marc J. , 2012. WHO Classification of Tumours of the Breast. [Google Scholar]
- Lester, J. , Stout, A.P. , 1954. Cystosarcoma phyllodes. Cancer. 7, 335–353. [DOI] [PubMed] [Google Scholar]
- Lindquist, K.D. , van Heerden, J.A. , Weiland, L.H. , Martin, J.K. , 1982. Recurrent and metastatic cystosarcoma phyllodes. Am. J. Surg.. 144, 341–343. [DOI] [PubMed] [Google Scholar]
- Moyano, J.V. , Evans, J.R. , Chen, F. , Lu, M. , Werner, M.E. , Yehiely, F. , Diaz, L.K. , Turbin, D. , Karaca, G. , Wiley, E. , Nielsen, T.O. , Perou, C.M. , Cryns, V.L. , 2006. αB-Crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J. Clin. Invest.. 116, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, T. , Wallden, B. , Schaper, C. , Ferree, S. , Liu, S. , Gao, D. , Barry, G. , Dowidar, N. , Maysuria, M. , Storhoff, J. , 2014. Analytical validation of the PAM50-based prosigna breast cancer prognostic gene signature assay and ncounter analysis system using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 14, 177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, T.O. , Parker, J.S. , Leung, S. , Voduc, D. , Ebbert, M. , Vickery, T. , Davies, S.R. , Snider, J. , Stijleman, I.J. , Reed, J. , Cheang, M.C.U. , Mardis, E.R. , Perou, C.M. , Bernard, P.S. , Ellis, M.J. , 2010. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast Cancer. Clin. Cancer Res.. 16, 5222–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niezabitowski, A. , Lackowska, B. , Rys, J. , Kruczak, A. , Kowalska, T. , Mitus, J. , Reinfuss, M. , Markiewicz, D. , 2001. Prognostic evaluation of proliferative activity and DNA content in the phyllodes tumor of the breast: immunohistochemical and flow cytometric study of 118 cases. Breast Cancer Res. Treat. 65, 77–85. [DOI] [PubMed] [Google Scholar]
- Parker, J.S. , Mullins, M. , Cheang, M.C.U. , Leung, S. , Voduc, D. , Vickery, T. , Davies, S. , Fauron, C. , He, X. , Hu, Z. , Quackenbush, J.F. , Stijleman, I.J. , Palazzo, J. , Marron, J.S. , Nobel, A.B. , Mardis, E. , Nielsen, T.O. , Ellis, M.J. , Perou, C.M. , Bernard, P.S. , 2009. Supervised risk predictor of breast Cancer based on intrinsic subtypes. J. Clin. Oncol.. 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Sorlie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Rees, C.A. , Pollack, J.R. , Ross, D.T. , Johnsen, H. , Akslen, L.A. , 2000. Molecular portraits of human breast tumours. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Prat, A. , Adamo, B. , Cheang, M.C.U. , Anders, C.K. , Carey, L.A. , Perou, C.M. , 2013. molecular characterization of basal-like and non-basal-like triple-negative breast cancer. The Oncologist. 18, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Carey, L.A. , Adamo, B. , Vidal, M. , Tabernero, J. , Cortés, J. , Parker, J.S. , Perou, C.M. , Baselga, J. , 2014. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast Cancer. J. Natl. Cancer Inst.. 106, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Cheang, M.C.U. , Martín, M. , Parker, J.S. , Carrasco, E. , Caballero, R. , Tyldesley, S. , Gelmon, K. , Bernard, P.S. , Nielsen, T.O. , Perou, C.M. , 2013. Prognostic significance of progesterone receptor–positive tumor cells within immunohistochemically defined luminal a breast Cancer. J. Clin. Oncol.. 31, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Ellis, M.J. , Perou, C.M. , 2012. Practical implications of gene-expression-based assays for breast oncologists. Nat. Rev. Clin. Oncol.. 9, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Karginova, O. , Parker, J. , Fan, C. , He, X. , Bixby, L. , Harrell, J.C. , Roman, E. , Adamo, B. , Troester, M. , Perou, C. , 2013. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat.. 142, 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Lluch, A. , Albanell, J. , Barry, W.T. , Fan, C. , Chacon, J.I. , Parker, J.S. , Calvo, L. , Plazaola, A. , Arcusa, A. , Segui-Palmer, M.A. , Burgues, O. , Ribelles, N. , Rodriguez-Lescure, A. , Guerrero, A. , Ruiz-Borrego, M. , Munarriz, B. , Lopez, J.A. , Adamo, B. , Cheang, M.C.U. , Li, Y. , Hu, Z. , Gulley, M.L. , Vidal, M.J. , Pitcher, B.N. , Liu, M.C. , Citron, M.L. , Ellis, M.J. , Mardis, E. , Vickery, T. , Hudis, C.A. , Winer, E.P. , Carey, L.A. , Caballero, R. , Carrasco, E. , Martin, M. , Perou, C.M. , Alba, E. , 2014. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br. J. Cancer. 111, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Parker, J. , Karginova, O. , Fan, C. , Livasy, C. , Herschkowitz, J. , He, X. , Perou, C. , 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast Cancer. Breast Cancer Res.. 12, R68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat, A. , Perou, C.M. , 2011. Deconstructing the molecular portraits of breast cancer. Mol. Oncol.. 5, 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinfuss, M. , Mitus, J. , Duda, K. , Stelmach, A. , Rys, J. , Smolak, K. , 1996. The treatment and prognosis of patients with phyllodes tumor of the breast: an analysis of 170 cases. Cancer. 77, 910–916. [DOI] [PubMed] [Google Scholar]
- Sundvall, M. , Iljin, K. , Kilpinen, S. , Sara, H. , Kallioniemi, O.-P. , Elenius, K. , 2008. Role of ErbB4 in breast Cancer. J. Mammary Gland Biol. Neoplasia. 13, 259–268. [DOI] [PubMed] [Google Scholar]
- Tan, P.H. , Ellis, I.O. , 2013. Myoepithelial and epithelial-myoepithelial, mesenchymal and fibroepithelial breast lesions: updates from the WHO Classification of Tumours of the Breast 2012. J. Clin. Pathol.. 66, 465–470. [DOI] [PubMed] [Google Scholar]
- Tan, P.H. , Thike, A.A. , Tan, W.J. , Thu, M.M. , Busmanis, I. , Li, H. , Chay, W.Y. , Tan, M.H. , 2012. Predicting clinical behaviour of breast phyllodes tumours: a nomogram based on histological criteria and surgical margins. J. Clin. Pathol.. 65, 69–76. [DOI] [PubMed] [Google Scholar]
- Tan, W.J. , Thike, A.A. , Bay, B.H. , Tan, P.H. , 2014. Immunohistochemical expression of homeoproteins Six1 and Pax3 in breast phyllodes tumours correlates with histological grade and clinical outcome. Histopathology. 64, 807–817. [DOI] [PubMed] [Google Scholar]
- TCGA, 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.C. , Buraimoh, A. , Iglehart, J.D. , Richardson, A.L. , 2006. Genome-wide analysis for loss of heterozygosity in primary and recurrent phyllodes tumor and fibroadenoma of breast using single nucleotide polymorphism arrays. Breast Cancer Res. Treat. 97, 301–309. [DOI] [PubMed] [Google Scholar]
- Weigman, V. , Chao, H.-H. , Shabalin, A. , He, X. , Parker, J. , Nordgard, S. , Grushko, T. , Huo, D. , Nwachukwu, C. , Nobel, A. , Kristensen, V. , Børresen-Dale, A.-L. , Olopade, O. , Perou, C. , 2012. Basal-like breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res. Treat.. 133, 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemori, K. , Hasegawa, T. , Shimizu, C. , Shibata, T. , Matsumoto, K. , Kouno, T. , Ando, M. , Katsumata, N. , Fujiwara, Y. , 2006. Correlation of p53 and MIB-1 expression with both the systemic recurrence and survival in cases of phyllodes tumors of the breast. Pathol. Res. Pract.. 202, 705–712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Results of all Significance Analyses of Microarrays (SAM) performed in the study.
Fig. S1 Kaplan–Meier relapse‐free survival analysis within Fibroepithelial tumors (FTs). (A) Based on morphological classification; (B) Based on Basal‐like versus Normal‐like classification; (C) Based on Basal‐like, Claudin‐low and Normal‐like classification; (D) Based on ROR‐S group.


