Abstract
Triple negative (TN) breast cancers make up some 15% of all breast cancers. Approximately 10–15% are mutant for the tumor suppressor, BRCA1. BRCA1 is required for homologous recombination‐mediated DNA repair and deficiency results in genomic instability. BRCA1‐mutated tumors have a specific pattern of genomic copy number aberrations that can be used to classify tumors as BRCA1‐like or non‐BRCA1‐like. BRCA1 mutation, promoter methylation, BRCA1‐like status and genome‐wide expression data was determined for 112 TN breast cancer samples with long‐term follow‐up. Mutation status for 21 known DNA repair genes and PIK3CA was assessed. Gene expression and mutation frequency in BRCA1‐like and non‐BRCA1‐like tumors were compared. Multivariate survival analysis was performed using the Cox proportional hazards model. BRCA1 germline mutation was identified in 10% of patients and 15% of tumors were BRCA1 promoter methylated. Fifty‐five percent of tumors classified as BRCA1‐like. The functions of genes significantly up‐regulated in BRCA1‐like tumors included cell cycle and DNA recombination and repair. TP53 was found to be frequently mutated in BRCA1‐like (P < 0.05), while PIK3CA was frequently mutated in non‐BRCA1‐like tumors (P < 0.05). A significant association with worse prognosis was evident for patients with BRCA1‐like tumors (adjusted HR = 3.32, 95% CI = 1.30–8.48, P = 0.01). TN tumors can be further divided into two major subgroups, BRCA1‐like and non‐BRCA1‐like with different mutation and expression patterns and prognoses. Based on these molecular patterns, subgroups may be more sensitive to specific targeted agents such as PI3K or PARP inhibitors.
Keywords: Breast cancer, Triple negative breast cancer, BRCA1, Genomic instability, Targeted therapy
Highlights
Triple negative breast cancers subdivide into 2 groups: BRCA1‐like, non‐BRCA1‐like.
In a retrospective analysis, patients with BRCA1‐like tumors have a worse outcome.
Mutation and gene expression patterns reveal potential therapy targets in subgroups.
Abbreviations
- TN
triple negative
- ER
estrogen receptor
- PR
progesterone receptor
- HER2
human epidermal growth factor receptor
- CNA
copy number aberration
- HR
hazard ratio
- DRFS
distant recurrence-free survival
1. Introduction
The heterogeneous nature of breast cancer, both at the histological and molecular levels, has been well documented and this information is routinely used to guide treatment decisions (Curtis et al., 2012; Dvinge et al., 2013; Perou et al., 2000; Sørlie et al., 2001; van de Vijver et al., 2002). Despite the reduced incidence of death from breast cancer overall in the last two decades in the industrialized world, certain subtypes remain difficult to treat due to limited treatment options (Hudis and Gianni, 2011). One such subtype which makes up around 12–17% of all breast cancers, triple negative (TN) breast cancer, is characterized by low or lack of expression of estrogen (ER) and progesterone (PR) receptors and lack of human epidermal growth factor receptor 2 (HER2) over‐expression (Criscitiello et al., 2012; Foulkes et al., 2010). Other than conventional chemotherapy few treatment options are currently available for these patients (Linn and Van 't Veer, 2009). Multiple studies have shown poor recurrence‐free and overall survival for patients with TN breast cancer, which tends to be aggressive and metastasize early, independent of other known breast cancer prognostic factors such as tumor size, grade and number of positive lymph nodes (Hudis and Gianni, 2011).
Depending on the ethnic background and age of the investigated cohort, around 10–15% TN breast cancers are mutant for the tumor suppressor, BRCA1 (Foulkes et al., 2003). BRCA1‐associated breast cancer displays a high frequency of TP53 mutations (Manié et al., 2009). Furthermore, BRCA1‐mutant breast cancers are commonly high‐grade and most frequently classified as basal‐like breast cancers – those that display basal cellular markers, such as cytokeratin5/6 (Foulkes et al., 2003). The categories of basal‐like and TN do not completely overlap, although, it has been previously reported that a substantial proportion of BRCA1‐mutant tumors are TN, basal‐like or both (Linn and Van 't Veer, 2009).
DNA double‐strand‐breaks (DSBs), most frequently caused by UV light and metabolic processes are repaired by several mechanisms. Homologous recombination (HR) repair – is the cell's most error‐free mechanism (Bouwman and Jonkers, 2012; Moynahan et al., 1999). Cells without functional BRCA1, most often through mutation and loss‐of‐heterozygosity or promoter methylation, are deficient in HR. These cells utilize an alternate mechanism to repair DSBs known to be highly error‐prone, called non‐homologous‐end‐joining (NHEJ), which results in genomic instability (Turner et al., 2004; Wang et al., 2001). Thus, BRCA1‐mutant tumors have numerous copy number aberrations (CNAs). Importantly, these tumors display a very characteristic pattern of gains and losses of genomic DNA. This specific pattern of CNAs was used to develop classifiers to identify tumors with the same pattern and to identify some sporadic, non‐BRCA1‐mutant tumors that have the same pattern of CNAs as BRCA1‐mutated tumors. This group with the characteristic CNAs pattern, larger than the BRCA1‐mutant tumors alone, is referred to as BRCA1‐like (Lips et al., 2011; Schouten et al., 2013; Vollebergh et al., 2011; Wessels et al., 2002). These tests assign copy number profiles to BRCA1‐like or non‐BRCA1‐like status based on the copy number pattern alone and can be implemented on all types of copy number data such as array data, next generation sequencing data and MLPA (multiplex ligation‐dependent probe amplification) data. The BRCA1‐like category, which is based on only copy number pattern frequently includes tumors with a BRCA1 mutation (as the classifiers were trained on these samples) and tumors with BRCA1 promoter methylation. However, a large number of tumors identified by these classifiers lack an apparent defect in BRCA1 itself. A number of these classifiers are capable of predicting benefit from specific therapies regardless of mutation status, particularly those utilizing DSB‐inducing agents, such as bifunctional alkylators and intensified platinum‐based chemotherapy (Lips et al., 2011; Schouten et al., 2015; Vollebergh et al., 2011).
The mechanisms underlying genomic instability in TN breast cancer are complex and although these tumors are frequently BRCA1‐associated, it remains unclear what role BRCA1 deficiency may play in the process (Bouwman and Jonkers, 2012; Turner et al., 2004). In this study, we first aimed to characterize our TN cohort with respect to BRCA1 mutation, promoter methylation and BRCA1‐like status. To gain insight into the mechanism that results in genomic instability in TN BRCA1‐like tumors, we sought to identify genes and their functions that are differentially expressed between BRCA1‐like and non‐BRCA1‐like tumors. In addition, we compared the mutation frequency of non‐BRCA1‐like and BRCA1‐like tumors in a subset of DNA repair genes and PIK3CA, the second most frequently mutated gene in breast cancer besides TP53 (Cancer Genome Atlas Network, 2012). Finally, we retrospectively assessed outcome of BRCA1‐like patients in comparison to non‐BRCA1‐like patients.
2. Methods
2.1. Patient selection and characteristics
The FP7 European Union funded RATHER Project (Rational Therapy for Breast Cancer) is a collaborative effort that aims to integrate gene expression profiling, copy number aberrations, kinome variation and kinase activation status in an effort to identify new targets for therapy of difficult‐to‐treat breast cancer subtypes, including TN breast cancer (www.ratherproject.com). The RATHER Project retrospectively identified 112 TN breast cancer patients in total: 64 from Netherlands Cancer Institute (NKI), Amsterdam, the Netherlands and 48 from Addenbrooke's Hospital, Cambridge, UK.
Potential TN primary tumors were selected from tumor registration and existing study databases at both sites (biobank). The primary inclusion criterion for this RATHER TN cohort was availability of sufficient frozen tissue for DNA, RNA and protein isolation for all RATHER project assays in the tissue bank. As a result the cohort may be skewed towards larger tumors. Neoadjuvant treated patients were an exclusion criterion when establishing the cohort. In addition, we enriched for patients diagnosed before 1999, since in that era only node‐positive patients received adjuvant systemic therapy. Premenopausal node‐positive patients would receive adjuvant chemotherapy while postmenopausal node‐positive patients would receive adjuvant endocrine therapy. Only from 2000 onwards was the estrogen‐receptor status taken into account when prescribing adjuvant endocrine therapy. These historical treatments mean 23/112 patients were treated with endocrine‐therapy despite having TN disease. The criteria allowed us to study a cohort of mainly adjuvant systemic therapy‐naïve patients with long‐term follow‐up. We collected database information on ER, PR and HER2 status, tumor size and grade, number of tumor positive lymph nodes, surgery and treatment information, age at diagnosis, diagnosis date as well as follow‐up data.
Frozen tumors with 30% or greater tumor content (based on average score before and after sectioning, 2 × 8 μm serial sections, hematoxylin and eosin‐stained (JJFM)) were used for further analyses. Formalin fixed paraffin‐embedded (FFPE) material was used to construct tissue microarrays (TMAs) for expert pathological review (KJ), which included ER, PR and HER2 status, grade and tumor percentage determination. Samples were defined as ER‐ or PR‐positive when 10% or more of tumor cells stained positive with immunohistochemical staining. HER2 samples with intensity ≥2 were considered positive and confirmed when possible using TargetPrint® (Agendia BV, Amsterdam, Netherlands) (Roepman et al., 2009). Samples with missing ER, PR or HER2 status upon review were included, as diagnostic information was originally indicative of TN status. The local medical ethical authorities of both centers approved of the collection protocols.
2.2. DNA/RNA isolation
All samples were processed following one standard operating protocol to isolate high quality nucleic acids. Each frozen tumor was serially sectioned for DNA and RNA isolation (30 × 30 μm serial sections for both). DNA was isolated using the DNeasy kit for purification of total DNA for animal tissues using two spin columns (Qiagen). On each column samples were eluted twice with 100 μl volumes of buffer AE for a final volume of 400 μl. For RNA extraction, depending on the size of the tumor sample, 20–30 sections of 30 μm were used for RNA extraction. Sections were homogenized in Qiazol (Qiagen) using a tissue lyser (Qiagen) and total RNA was isolated with Qiazol according the manufacturer's instructions. The RNA was further purified using the RNeasy Mini Kit (Qiagen).
2.3. Microarray hybridization and analysis
The RNA quality was assessed by a 2100 Bioanalyzer (Agilent Technologies) and samples with RIN above 5 were selected for further analysis. RNA was amplified, labeled and hybridized to the Agendia custom‐designed whole genome microarrays (Agilent Technologies) and raw fluorescence intensities were quantified using Feature Extraction software (Agilent Technologies) according to the manufacturer's protocols. The microarray expression dataset was imported into R/Bioconductor software (R version 3.0.2, www.bioconductor.org) for pre‐processing.
Feature signal intensities were processed and extracted according to the ‘limma’ Bioconductor R package (Bolstad et al., 2003) with background subtraction using an offset of 10. All probe intensities <1 were set as missing values. The log2‐transformed probe intensities were quantile normalized using ‘limma’. A principal component analysis showed a batch effect for biobank, which was adjusted for using ComBat (Johnson et al., 2007). Missing values were imputed by 10‐nearest neighbor imputation. Genes with multiple probes were summarized by the first principal component of a correlating subset.
Differential analysis, clustering and visualization of the data was performed with R (version 3.1.2) using the ‘heatmap.3’ package standard settings. Differential expression between classes was assessed using ANOVA in R with the significant genes selected univariately with FDR <0.001 and a fold change >1.
2.4. Capture library and next‐generation sequencing and analysis
For each sample, Illumina TruSeq indexed libraries were constructed according to manufacturer's instructions (Illumina) before enrichment by capture with a biotinylated RNA probe set targeting the human kinome and a range of cancer related genes (Agilent Technologies). We sequenced 10–12 samples on a single Illumina HiSeq lane to generate 50, 51 or 60 bp paired‐end reads. Raw sequence data were aligned using Burrows–Wheeler Aligner (BWA) to the human genome (Ensembl 37). Single nucleotide variants and indels were called using SAMTools on unique paired aligned data. Matched normal germline DNA was unavailable for most samples so we used dbSNP and variant data from the Exome Variant Server to remove potential germline variants. We further focused on variants predicted to alter protein coding sequence or splicing of genes according to Ensembl VariantEffectPredictor and not identified in a pool of 80 normal DNAs taken from various tissues. All variants found in the COSMIC database were retained. In addition, we retained any BRCA1 or BRCA2 variants that were clinically relevant according to the Breast Cancer Information Core database (BIC, http://research.nhgri.nih.gov/bic/). Following our filtering steps, samples with variants were termed BRCA1/2‐mutant. For all other genes, the same criteria were applied except the BIC database step and the remaining variants were termed mutant. BRCA1 mutations were validated when possible by germ‐line sequencing using the Nextera Custom Enrichment kit (Illumina) on matched normal DNA according to manufacturer's instructions, traditional capillary sequencing, or small PCR amplicon pooling targeting the variant using Illumina TruSeq indexing.
2.5. BRCA1‐like classification
We used the MLPA (multiplex ligation‐dependent probe amplification) method to determine the BRCA1‐like status of the tumor DNAs. The assay was performed, fragments analyzed and data normalized according to the manufacturer's protocol using the SALSA MLPA P376 BRCA1ness probemix (MRC‐Holland). Class prediction (BRCA1‐like/non‐BRCA1‐like) was carried out on the normalized data according to published instructions (Lips et al., 2011).
2.6. BRCA1 promoter methylation
Semi‐quantitative BRCA1 promoter methylation was determined using the MS‐MLPA (methylation‐specific‐MLPA) method. This assay combines copy number detection with methylation‐specific enzymatic restriction. The assay was performed, fragments analyzed, data normalized and a cutoff of 20% was used to call a sample methylated according to manufacturer's protocols using the SALSA MLPA ME001 Tumour suppressor probemix 1 (MRC‐Holland).
2.7. Statistical analysis
Patient characteristics were compared between BRCA1‐like and non‐BRCA1‐like classes and statistical significance was examined by a Wilcoxon rank‐sum test or Pearson's chi‐squared test using R version 3.1.2. Survival analysis was conducted in R using the ‘survival’ package to employ the Cox proportional hazards model. We observed patients from date of diagnosis until 2012 for distant recurrence‐free survival and censored data in accordance with the STEEP (Standard Efficacy Endpoint) system (Hudis et al., 2007). We used only follow‐up data up to 10 years. An event includes distant recurrence, death from breast cancer, death from non‐breast cancer cause and death from unknown cause. Co‐variates in the Cox proportional model included BRCA1‐like status, patient age at diagnosis, treatment (radiotherapy/hormonal/chemotherapy), tumor size, grade and number of tumor positive lymph nodes. We included stratification of all Cox models for biobank (NKI/Addenbrooke's Hospital). To assess the accuracy of the model we included a test for the Proportional Hazards (PH) assumptions using cox.zph in the ‘survival’ package.
The ‘Mutascape’ R package (manuscript in preparation) was employed to test differences in gene mutation frequency for two analyses: 1) within classes and 2) between classes (BRCA1‐like and non‐BRCA1‐like). Within classes a binomial test was employed to determine if the number of mutations in a gene was greater than expected by chance. Given the total number of mutations in the dataset and the size of the gene, probabilities of occurrence were computed, which can be interpreted as the probability of a gene's mutation frequency being random (modeled by the null binomial distribution) or not. A multiple testing correction was applied to the p‐values with the Benjamini–Hochberg method. The distribution of mutation location was also determined and visualized in a bubble plot to examine mutation recurrence as well as frequency within genes. For genes identified as significantly mutated we used the Fisher's exact test to compare the distribution between the BRCA1‐like and non‐BRCA1‐like groups. The p‐values were adjusted as above.
3. Results
112 TN samples were available for further analysis based on inclusion criteria, central pathological review of TN immunohistochemical status and BRCA1‐like status data availability. To characterize the TN cohort with respect to BRCA1 deficiency and characteristic genomic instability, we assessed the samples for BRCA1 mutation, promoter methylation and BRCA1‐like status. We identified 62 of 112 as BRCA1‐like tumors, 10 of 104 as BRCA1 mutated tumors (8 with missing data) and 14 of 94 (18 with missing data) as BRCA1 promoter methylated tumors (Figure 1). ‘Missing data’ indicates a failed experiment. We found BRCA1 germline mutation and BRCA1 promoter methylation overlap with BRCA1‐like status in 70% (7/10) and 79% (11/14) of the samples, respectively. Patient characteristics and association with BRCA1‐like status are found in Table 1.
Figure 1.
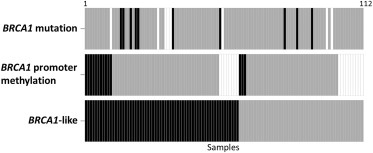
Heatmap indicating data collected for all 112 TN samples. Each box represents one sample, with color indicating type of data for that sample in each row: positive (black), negative (gray) and no data due to failed experiment (white). BRCA1 mutation, promoter methylation and ‐like status data were obtained for 104, 98 and 112 samples, respectively.
Table 1.
Patient characteristics.
| Variable | Non‐BRCA1‐like N = 50No. (%) | BRCA1‐like N = 62No. (%) | P |
|---|---|---|---|
| Year of diagnosis | 0.69a | ||
| Mean | 2001 | 2001 | |
| Range | 1989–2009 | 1988–2009 | |
| Age at diagnosis | 0.02a | ||
| Mean | 59 | 52 | |
| Range | 31–88 | 26–78 | |
| Time to last followup (days) | 0.10a | ||
| Mean | 1973 | 2024 | |
| Range | 0–5267 | 242–5634 | |
| Tumor size (cm) | 0.42b | ||
| 0–1 | 5 (10.0) | 2 (3.2) | |
| >1–2 | 15 (30.0) | 21 (33.9) | |
| >2–5 | 22 (44.0) | 34 (55.0) | |
| >5 | 3 (6.0) | 3 (4.8) | |
| Mean | 2.7 | 2.8 | |
| Number positive lymph nodes | 0.51b | ||
| 0 | 29 (58.0) | 42 (67.7) | |
| 1–3 | 12 (24.0) | 12 (19.4) | |
| 4–9 | 3 (6.0) | 1 (1.6) | |
| ≥10 | 3 (6.0) | 5 (8.1) | |
| Mean | 2.0 | 1.6 | |
| Tumor histological grade | 0.20b | ||
| 1 | 0 (0.0) | 0 (0.0) | |
| 2 | 7 (14.0) | 4 (6.5) | |
| 3 | 33 (66.0) | 53 (85.5) | |
| Mean | 2.8 | 2.9 | |
| Adj. chemotherapy | 0.01b | ||
| No treatment | 32 (64.0) | 23 (37.1) | |
| Treatment | 18 (36.0) | 39 (63.0) | |
| Biobank | 0.02b | ||
| Addenbrooke's Hosp. | 28 (56.0) | 20 (32.3) | |
| NKI | 22 (44.0) | 42 (67.7) |
Abbreviations: Adj., adjuvant; Hosp., hospital; NKI, Netherlands Cancer Institute.
Wilcoxon‐rank‐sum‐test.
Pearson's chi‐squared test.
To better understand the mechanisms that can result in genomic instability, we analyzed gene expression data of the TN samples in combination with BRCA1 mutation/promoter methylation and BRCA1‐like status. We aimed to explore the gene expression data for association with BRCA1‐like status (top variable genes with a fold change >1, N = 3569). Figure 2A shows the unsupervised clustering of the 279 most significantly differentially expressed genes between BRCA1‐like and non‐BRCA1‐like samples (ANOVA, FDR < 0.001, fold‐change >1). We also classified our samples according to known gene expression TN subgroups from the Lehmann group with the TNBCType tool (http://cbc.mc.vanderbilt.edu/tnbc/) using the top variable genes with a fold change >1 (N = 3569). Significantly different genes showed no specific association with TNBCType classifications (Chen et al., 2012) (Supplementary Figure 1).
Figure 2.
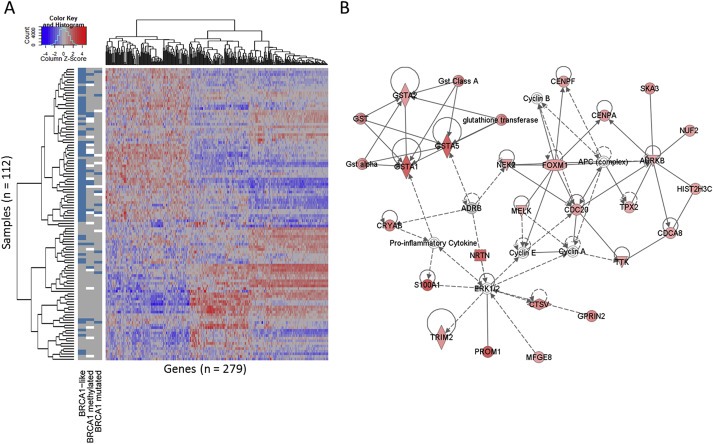
A, Unsupervised clustering of 279 top variable genes most differentially expressed between BRCA1‐like and non‐BRCA1‐like status (ANOVA, FDR <0.001, fold‐change >1) in 112 TN breast tumors. Scaled expression value is denoted as the column Z‐score and plotted in red–blue color scale. Red indicates high expression and blue low expression. Information Columns 1, 2 and 3 depict BRCA1‐like status, BRCA1 promoter methylation status and BRCA1 mutation status, respectively. For all sample columns, assay positive is indicated by blue, negative by gray and no data due to failed experiment by white. B, Network analysis of up‐regulated differentially expressed genes, indicating level of up‐regulation in BRCA1‐like compared with non‐BRCA1‐like (red shading), direct relationships (solid lines), and indirect relationships (interrupted lines).
Ingenuity Pathway Analysis (IPA, Ingenuity) was used to identify key biological processes regulated by the differentially expressed genes. The most down‐regulated genes in BRCA1‐like tumors are related to cellular maintenance and proliferation and development of lymphocytes (Supplementary File 1). The most up‐regulated genes were enriched for cell cycle and DNA replication, associated with recombination and repair, within a network centered on FOXM1 (Figure 2B). FOXM1 gene expression was significantly up‐regulated in BRCA1‐like samples (P < 0.001) along with CDK4 and CDK6 (P < 0.001 and P = 0.03, respectively). Although MYC has been found to be amplified in BRCA1 germline mutated tumors (Adem et al., 2004; Grushko et al., 2004) with associated over‐expression (Blancato et al., 2004) we did not observe significant increased MYC gene expression in BRCA1‐like versus non‐BRCA1‐like tumors (P = 0.78). BRCA1‐mutant versus wild type (BRCA1‐like removed) showed no significant difference in MYC gene expression (P = 0.41).
Using next‐generation sequencing, we analyzed the exons of 21 genes known to be involved in DNA repair for mutations as well as PIK3CA (Supplementary File 1). Significantly mutated genes were identified taking their genomic size into account (see Methods). In both classes, TP53 was significantly more frequently mutated than expected by chance, with non‐BRCA1‐like tumors mutated at 50% and BRCA1‐like at 84% (adjusted P < 0.001 and adjusted P < 0.001, respectively) (Figure 3A and B). Only TP53 was significantly differentially mutated between classes with BRCA1‐like tumors more frequently mutated than non‐BRCA1‐like tumors (adjusted P = 0.002). Additionally, we investigated the type of TP53 mutations (non‐truncating or truncating) identified in both classes and found a trend indicating more truncating mutations in BRCA1‐like tumors than in non‐BRCA1‐like tumors (Figure 3B, Supplementary File 1). Interestingly, we only observed a high frequency of PIK3CA mutations in the non‐BRCA1‐like tumors (21%; adjusted P = <0.001). There was no evidence of significant associations with PIK3CA hotspot mutations between the two classes although numbers were very small (Supplementary File 1).
Figure 3.
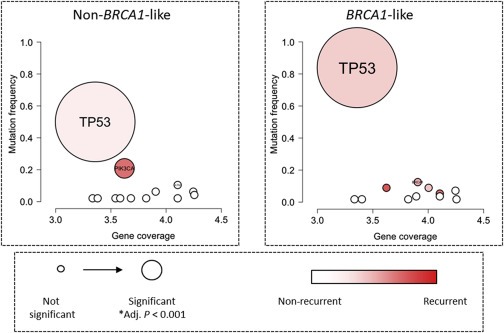
Bubble plot of mutational analysis of 21 DNA repair genes and PIK3CA. Panels A and B depict analysis within non‐BRCA1‐like (n = 48 samples) and BRCA1‐like classes (n = 56 samples), respectively. Each mutated gene is represented as a bubble positioned according to its size on the x‐axis (Gene coverage = log basepair) and its mutation frequency within the group on the y‐axis. Bubble size indicates the statistical significance and color represents the type of mutation pattern, recurrent or non‐recurrent (genes in red tend to have mutations at recurrent positions, e.g. ‘hotspots’, while genes in white tend to have mutations at unique positions in the various samples). Genes are listed in Supplementary File 1. *Adj. P indicates the Benjamini–Hochberg adjusted p‐value.
We observed 33 events in 112 patients. Distant recurrence‐free survival of the cohort was visualized with respect to BRCA1‐like status (univariate analysis stratified for biobank) using the Kaplan–Meier method, which indicated a trend in association with worse outcome for BRCA1‐like patients (adjusted log rank, P = 0.08) (Figure 4). We calculated the adjusted hazard ratios in multivariate and found patients with a BRCA1‐like tumor had a significantly worse prognosis than patients with a non‐BRCA1‐like tumor (HR = 3.32, 95% CI = 1.30–8.48, P = 0.01) (Table 2). The proportional hazards assumption test was not significant (P = 0.13) in the global model indicating the accuracy of the constructed Cox model for the dataset. The significant prognostic findings were also true for breast cancer specific survival and recurrence‐free survival. The test for interaction between chemotherapy and BRCA1‐like status was not significant (P = 0.75, Supplementary Table 1). There are no patients for whom treatment data were not available. Because of the significant difference in TP53 mutation frequency between classes, we also calculated adjusted hazard ratios in a multivariate model for TP53 mutation status. We observed no significant association with prognosis for TP53 mutation status (HR = 1.39, 95% CI = 0.55–3.53, P = 0.49).
Figure 4.
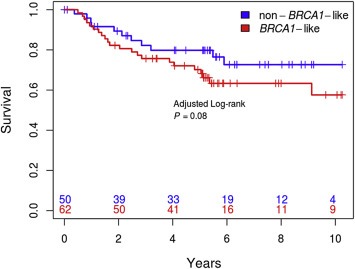
Survival analysis. Distant recurrence‐free survival (univariate) of the cohort with respect to BRCA1‐like status using the Kaplan–Meier method (adjusted log‐rank, P = 0.08). The blue line indicates non‐BRCA1‐like patients and the red line indicates BRCA1‐like patients. Patients at risk are shown on the x‐axis in corresponding colors.
Table 2.
Cox proportional hazards model for DRFS in TN patients (N = 112).
| Variable | HR | 95% CI | P |
|---|---|---|---|
| BRCA1‐like (BRCA1‐like vs non‐BRCA1‐like) | 3.32 | 1.30–8.48 | 0.01 |
| Tumor size (cm) | 1.41 | 1.02–1.95 | 0.04 |
| Number of positive lymph nodes | 1.11 | 1.01–1.21 | 0.03 |
| Tumor histological grade | 0.88 | 0.23–3.34 | 0.85 |
| Hormonal treatment (true vs false) | 0.89 | 0.31–2.55 | 0.83 |
| Adj. chemotherapy treatment (true vs false) | 0.43 | 0.17–1.12 | 0.09 |
| Radiotherapy treatment (true vs false) | 0.59 | 0.23–1.52 | 0.27 |
| Age at diagnosis | 0.98 | 0.94–1.01 | 0.19 |
Abbreviations: DRFS, distant recurrence‐free survival; TN, triple negative; HR, hazard ratio; Adj., adjuvant; CI, confidence interval.
4. Discussion
The underlying mechanisms responsible for genomic instability are complex and although BRCA1 deficiency and genomic instability are frequently associated, the exact role of BRCA1 in the process remains elusive. To further explore the role of BRCA1 and characteristic genomic instability in TN tumors, we examined differences in mutation and gene expression patterns between BRCA1‐like and non‐BRCA1‐like tumors combined with BRCA1 mutation and promoter methylation data. The percentages of BRCA1 mutation, promoter methylation and BRCA1‐like status indicate the group is representative of the larger TN population. In addition, although our selection criteria may have skewed the cohort toward larger tumors, we did not observe evidence for this when comparing tumor size classes with a similar previously published cohort (n = 180, Fisher's exact test, P = 0.69) (Dent et al., 2007) (data not shown). We did find tumors of grade III were more frequent in the RATHER cohort compared to the Dent cohort (Fisher's exact test, P < 0.001) (data not shown). As has been reported previously, we found overlap of BRCA1‐like status with most BRCA1 germline mutation and promoter methylation cases. Consistent with other reports, not all BRCA1 mutations are co‐detected with the BRCA1‐like assay. The relative ability to detect true signal between mutation detection methods and the BRCA1‐like status assay may be a contributing factor in these apparent discrepant samples. It is also possible that a BRCA1 germline mutation carrier may develop a non‐BRCA1‐like (sporadic) tumor. Our definition of mutation status is however imperfect as we have no supporting functional data to determine the impact of the putative mutation on the protein function.
Both classes, BRCA1‐like and non‐BRCA1‐like alone, had a significantly higher frequency of TP53 mutation than expected by chance. While TN tumors are known to be enriched for TP53 mutations and are frequently associated with BRCA1 associated breast cancer (Manié et al., 2009), we observed TP53 was more frequently mutated in the BRCA1‐like than the non‐BRCA1‐like class. Although TP53 has been reported to be mutated at around 80–90% in basal‐like breast cancers (Cancer Genome Atlas Network, 2012; Manié et al., 2009), this study and another using similar sequencing technology have found a slightly lower frequency in TN breast cancer (54 and 68%, respectively) (Shah et al., 2012). This difference may be reflective of the difference between basal‐like and TN disease or because many of the reports of TP53 mutation and basal‐like breast cancer have been carried out using capillary sequencing technology. This technology is likely to have more precision in mutation detection in low complexity regions and for insertions/deletions, but is economically unfeasible for large sequencing projects. The prognostic value in breast cancer of mutations in TP53 has been found to be specific to ER‐positive disease (Silwal‐Pandit et al., 2014). However, the predictive capacity of TP53 mutations in breast cancer for various therapies has not been thoroughly examined. In recent findings a combination of Chk1 inhibition with irinotecan, a DNA‐damage inducing agent has shown promise in TN xenograft experiments in mice (Ma et al., 2012). BRCA1‐like tumors may be more susceptible to such treatments due to their high frequency of TP53 mutation.
It has been shown that TN breast cancer can be further subdivided into different molecular subtypes based on mRNA expression (Chen et al., 2012; Teschendorff et al., 2007; Waddell et al., 2010) and that these subtypes differ in their response to therapy (Lehmann et al., 2011; Masuda et al., 2013). We added the Lehmann TNBCType classifications to the heatmap depicting the most differentially expressed genes based on BRCA1‐like status and found both ‘basal‐like 1’ and ‘mesenchymal’ categories are most associated with BRCA1‐like status (Pearson's chi squared test, P < 0.001). This indicates that BRCA1‐like based gene expression patterns, which identify a different group of patients are novel in TN breast cancer (Supplementary Figure 1). Through differential gene expression analysis, we have identified BRCA1‐like TNs may be susceptible to therapies targeting DNA repair and cell cycle pathways. The up‐regulated genes in BRCA1‐like tumors formed a network centered on the FOXM1 gene, which is a key regulator of cell cycle progression and DNA damage repair and has been found to be over‐expressed in most human cancers (Alvarez‐Fernández and Medema, 2013). This may explain the BRCA1‐like status of these samples regardless of their BRCA1 mutation/promoter methylation status as aberrant FOXM1 can lead to re‐entry into the cell cycle after DNA damage induced arrest rather than apoptosis (Alvarez‐Fernández et al., 2010). Furthermore, breast cancer cell lines with FOXM1 over‐expression are linked to acquired resistance to specific chemotherapeutics; these cell lines can be re‐sensitized to these treatments when FOXM1 is depleted, potentially explaining the poor prognosis of BRCA1‐like patients in our study (Kwok et al., 2010). Targeting FOXM1 through CDK4/6 inhibitors has shown promising results in melanoma cell lines (Anders et al., 2011) and FOXM1 suppression increases sensitivity to certain DNA damaging agents in tumor cell lines (Kwok et al., 2010; Zhang et al., 2012).
Our finding that FOXM1, CDK4 and CDK6 are highly expressed in BRCA1‐like tumors is of particular interest in light of recent reports that CDK4/6 inhibitors are effective in various breast tumors (Dean et al., 2012; Finn et al., 2009). BRCA1‐mutated breast tumors, which are deficient in HR‐mediated DNA double‐strand‐break‐repair are known to respond to PARP inhibitors, such as olaparib (Gelmon et al., 2011; Kaufman et al., 2014; Tutt et al., 2010). Inhibition of FOXM1 may sensitize cells to PARP inhibition allowing for effective combination treatments. In addition, previous studies have shown that BRCA1‐like breast cancer patients benefit substantially from intensified alkylating chemotherapy in comparison to those treated with conventional chemotherapy (Lips et al., 2011; Schouten et al., 2015; Vollebergh et al., 2011).
Multivariate survival analysis is routinely employed in retrospective studies to determine the independent prognostic factors, which reduce survival time. The Cox proportional hazards model was used to estimate the hazard for each co‐variable including all potentially confounding co‐variables in the model. Using this analysis we identified BRCA1‐like status to be an independent prognostic factor with BRCA1‐like patients associated with a worse prognosis. In the multivariate Cox model, although it is not significant, there is an indication that chemotherapy treatment may be an influencing factor on prognosis when stratifying patients for BRCA1‐like status (Table 2). To rule out that the prognostic association is influenced by a differential treatment effect, we employed a test for interaction with chemotherapy and BRCA1‐like status. This test was not significant (P = 0.75) indicating the differential prognosis is not influenced by chemotherapy treatment (Supplemental Table 1). In addition we tested association of biobank with chemotherapy and found no significant association (P = 0.52). These findings suggest BRCA1‐like status is a prognostic marker in TN breast cancer and is not influenced by treatment effect in this cohort. It is important to note, we have only examined the prognostic capacity of the biomarker, BRCA1‐like status in our series and are unable to determine its power to predict derived benefit from specific treatments such as platinum salts as we do not have access to those data and the setting is not amenable to those analyses. It remains that specific treatments which may be present in our series may mask the BRCA1‐like prognostic effect. To examine this, we employed multivariate survival analysis on only the non‐adjuvant‐systemically treated patients in the series (no chemo‐/hormonal therapy). We observe in this subgroup the same trend of BRCA1‐like status associated with a worse prognosis (HR = 6.75, 95% CI = 0.85–53.72, P = 0.07) lending further support that BRCA1‐like status is an independent prognostic factor in this series (Supplementary Figure 2 and Supplementary Table 2).
Both these and previous findings identified that a portion of BRCA1‐like tumors are not BRCA1 mutated/promoter methylated (Lips et al., 2013; Vollebergh et al., 2011), indicating that alterations in another gene or genes involved in DNA‐repair besides BRCA1 may be associated with the characteristic genomic instability of BRCA1‐like tumors. Based on previous reports, the BRCA1‐like group is likely to be susceptible to specific treatments aimed at DNA damage and cell cycle pathways, such as PARP, Chk1 or CDK4/6‐FOXM1 inhibitors and/or intensified alkylating agents or platinum compounds, regardless of BRCA1 mutation or promoter methylation status. Prospective clinical trials should deliver final proof for these assumptions. Recently we found evidence that BRCA1‐like breast cancer patients derived more benefit from neoadjuvant carboplatin/veliparib added to a standard regimen of doxorubicin‐cyclophosphamide, followed by paclitaxel, than non‐BRCA1‐like patients in the I‐SPY 2 trial (Glas et al., 2014). The biomarker × treatment interaction odds ratio of achieving a pathological complete remission was 9.3 (P = 0.02) with carboplatin/veliparib added to standard chemotherapy, when compared to standard chemotherapy alone, according to BRCA1‐like status (Glas et al., 2014). These interesting data need confirmation in a second (neo)adjuvant trial where the addition of DNA damaging agents to standard chemotherapy has been studied. Recently Tutt et al. presented very interesting response and progression‐free survival data regarding first line treatment of M1 TNBC patients randomized between docetaxel or carboplatin. Only BRCA1 mutation status interacted significantly with treatment, while another homologous recombination deficiency test (Myriad HRD Assay, Myriad Genetics) did not (Tutt et al., 2014) indicating some BRCA‐associated tests are not capable of predicting response in the metastatic setting. We have recently initiated a prospective randomized‐controlled trial with a 2 × 2 factorial design of paclitaxel ± bevacizumab versus carboplatin‐cyclophosphamide ± bevacizumab in first line metastatic TNBC patients (NCT01898117). One of the primary endpoints of this trial is to validate the BRCA1‐like status as a biomarker for alkylating chemotherapy benefit.
PIK3CA is a frequently mutated gene in breast cancer, although most often associated with luminal‐type tumors rather than basal‐like or TN tumors (Cancer Genome Atlas Network, 2012). Patients with tumors mutated in PIK3CA may benefit from inhibitors of the PI3K/AKT/mTOR pathway. We found PIK3CA mutation frequency significantly higher than expected within the non‐BRCA1‐like class alone with a frequency of 21%. This is roughly 2 times higher than previous reports of basal‐like tumors (Cancer Genome Atlas Network, 2012) suggesting non‐BRCA1‐like classification enriches for patients who may benefit from PI3K inhibitors. To substantiate this, a small trial with a Bayesian design would be helpful. It has been reported that not all hotspot mutations confer pathway activation as measured by downstream‐activated proteins (Beelen et al., 2014). For this reason, downstream‐activated proteins should be included in any trials assessing the inhibition of PI3K pathway in PIK3CA mutated patients. Patients with TN tumors are currently unlikely to be tested for PIK3CA mutations or pathway activation, however, our findings indicate a portion of these patients may benefit from PI3K/AKT/mTOR pathway inhibition. Importantly, the non‐BRCA1‐like subgroup makes up ±45% of all TNs.
In summary, our data indicate one sizeable subgroup (around 9.5%) may be more susceptible to PI3K/AKT/mTOR inhibitors which are currently available in clinical practice but are not routinely administered to TN patients. In addition, we have indicated a second subgroup which is more likely to respond to DNA damaging agents and putatively also to CDK4/6 inhibitors based on the gene expression pattern and high frequency of TP53 mutations. In conclusion, while TN breast cancer currently has few treatment options, using mutation and gene expression analysis to molecularly characterized these tumors we have identified relatively large subgroups within the subtype that may benefit from specific tailored treatments, potentially impacting a substantial portion of TN breast cancers in total.
Disclosure of potential conflicts of interest
SCL is an advisory board member for Cergentis, Novartis, Roche, and Sanofi. SCL received research support funding from Amgen, AstraZeneca, Roche, and Sanofi. SCL is named inventor on a patent application for the BRCA1‐like classifier. SLC, TMS, IMS and JP are all named co‐inventors on a patent application for a BRCAness gene expression classifier. All other authors have no competing interests.
Authors' contributions
TMS, JP and SCL lead the analysis. TMS wrote the manuscript with contributions from all authors. IM, MM, AB, PCS, S‐FC, BP, MAG, TB, RJCK, JJFM, KJ, RK, LW, RB, IMS, CC and SCL contributed to acquisition of data, data collection and analysis. JJFM and KJ provided pathological expertise. TMS, JP and SCL conceived of and designed the study. SCL and IMS provided project supervision.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary Figure 1 Hierarchical clustering of 279 top variable genes most differentially expressed between BRCA1‐like and non‐BRCA1‐like status (ANOVA, FDR < 0.001, fold‐change > 1) in 125 TN breast tumors. Scaled expression value is denoted as the column Z‐score and plotted in red–blue color scale. Red indicates high expression and blue low expression. Information Columns 1, 2 and 3 depict BRCA1‐like status, BRCA1 promoter methylation status and BRCA1 mutation status, respectively. For all sample columns, assay positive is indicated by blue, negative by gray and no data due to failed assay in white. Column 4 depicts Lehmann TNBCTypes basal‐like 1 (BL1), basal‐like 2 (BL2), immunomodulatory (IM), luminal androgen receptor (LAR), mesenchymal (M), mesenchymal stem‐like (MSL), and unidentified (U) as teal, yellow, purple, salmon, light blue, orange and pink, respectively. No data due to failed Lehmann TNBCType assay indicated in lime green.
Supplementary Figure 2 Survival analysis. Distant recurrence‐free survival (univariate) of the non‐adjuvant‐systemic therapy treated cohort with respect to BRCA1‐like status using the Kaplan–Meier method (adjusted log‐rank, P = 0.08). The blue line indicates non‐BRCA1‐like patients and the red line indicates BRCA1‐like patients. Patients at risk are shown on the x‐axis in corresponding colors.
Acknowledgments
We thank all patients who contributed to this study. The authors would like to acknowledge all the members of the RATHER consortium for their helpful discussions (http://www.ratherproject.com/) and the European Union Seventh Framework Programme (FP7) which supported this work, project number 258967. The authors would also like to acknowledge the effort and support of the Netherlands Cancer Institute (NKI) Genomics Core Facility and the Netherlands Cancer Institute‐Antoni van Leeuwenhoek Ziekenhuis (NKI‐AVL) Core Facility Molecular Pathology & Biobanking (CFMPB) for supplying NKI‐AVL Biobank material and/or lab support.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.04.011.
Severson Tesa M., Peeters Justine, Majewski Ian, Michaut Magali, Bosma Astrid, Schouten Philip C., Chin Suet-Feung, Pereira Bernard, Goldgraben Mae A., Bismeijer Tycho, Kluin Roelof J.C., Muris Jettie J.F., Jirström Karin, Kerkhoven Ron M., Wessels Lodewyk, Caldas Carlos, Bernards René, Simon Iris M., Linn Sabine, (2015), BRCA1‐like signature in triple negative breast cancer: Molecular and clinical characterization reveals subgroups with therapeutic potential, Molecular Oncology, 9, doi: 10.1016/j.molonc.2015.04.011.
References
- Adem, C. , Soderberg, C.L. , Hafner, K. , Reynolds, C. , Slezak, J.M. , Sinclair, C.S. , Sellers, T.A. , Schaid, D.J. , Couch, F. , Hartmann, L.C. , 2004. ERBB2, TBX2, RPS6KB1, and MYC alterations in breast tissues of BRCA1 and BRCA2 mutation carriers. Genes Chromosomes Cancer. 41, 1–11. [DOI] [PubMed] [Google Scholar]
- Alvarez-Fernández, M. , Medema, R.H. , 2013. Novel functions of FoxM1: from molecular mechanisms to cancer therapy. Front. Oncol. 3, 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Fernández, M. , Halim, V.A. , Krenning, L. , Aprelia, M. , Mohammed, S. , Heck, A.J. , Medema, R.H. , 2010. Recovery from a DNA-damage-induced G2 arrest requires Cdk-dependent activation of FoxM1. EMBO Rep. 11, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, L. , Ke, N. , Hydbring, P. , Choi, Y.J. , Widlund, H.R. , Chick, J.M. , Zhai, H. , Vidal, M. , Gygi, S.P. , Braun, P. , 2011. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 20, 620–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelen, K. , Opdam, M. , Severson, T.M. , Koornstra, R.H. , Vincent, A.D. , Wesseling, J. , Muris, J.J. , Berns, E.M. , Vermorken, J.B. , van Diest, P.J. , 2014. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res. BCR. 16, R13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancato, J. , Singh, B. , Liu, A. , Liao, D.J. , Dickson, R.B. , 2004. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br. J. Cancer. 90, 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad, B.M. , Irizarry, R.A. , Astrand, M. , Speed, T.P. , 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinforma. Oxf. Engl. 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Bouwman, P. , Jonkers, J. , 2012. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer. 12, 587–598. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network, 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Li, J. , Gray, W.H. , Lehmann, B.D. , Bauer, J.A. , Shyr, Y. , Pietenpol, J.A. , 2012. TNBCtype: a subtyping tool for triple-negative breast cancer. Cancer Inform. 11, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello, C. , Azim, H.A. , Schouten, P.C. , Linn, S.C. , Sotiriou, C. , 2012. Understanding the biology of triple-negative breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO. 23, (Suppl. 6) vi13–vi18. [DOI] [PubMed] [Google Scholar]
- Curtis, C. , Shah, S.P. , Chin, S.-F. , Turashvili, G. , Rueda, O.M. , Dunning, M.J. , Speed, D. , Lynch, A.G. , Samarajiwa, S. , Yuan, Y. , 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, J.L. , McClendon, A.K. , Knudsen, E.S. , 2012. Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J. Biol. Chem. 287, 29075–29087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, R. , Trudeau, M. , Pritchard, K.I. , Hanna, W.M. , Kahn, H.K. , Sawka, C.A. , Lickley, L.A. , Rawlinson, E. , Sun, P. , Narod, S.A. , 2007. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 13, 4429–4434. [DOI] [PubMed] [Google Scholar]
- Dvinge, H. , Git, A. , Gräf, S. , Salmon-Divon, M. , Curtis, C. , Sottoriva, A. , Zhao, Y. , Hirst, M. , Armisen, J. , Miska, E.A. , 2013. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 497, 378–382. [DOI] [PubMed] [Google Scholar]
- Finn, R.S. , Dering, J. , Conklin, D. , Kalous, O. , Cohen, D.J. , Desai, A.J. , Ginther, C. , Atefi, M. , Chen, I. , Fowst, C. , 2009. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. BCR. 11, R77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes, W.D. , Stefansson, I.M. , Chappuis, P.O. , Bégin, L.R. , Goffin, J.R. , Wong, N. , Trudel, M. , Akslen, L.A. , 2003. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 95, 1482–1485. [DOI] [PubMed] [Google Scholar]
- Foulkes, W.D. , Smith, I.E. , Reis-Filho, J.S. , 2010. Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948. [DOI] [PubMed] [Google Scholar]
- Gelmon, K.A. , Tischkowitz, M. , Mackay, H. , Swenerton, K. , Robidoux, A. , Tonkin, K. , Hirte, H. , Huntsman, D. , Clemons, M. , Gilks, B. , 2011. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 12, 852–861. [DOI] [PubMed] [Google Scholar]
- Glas, A. , Peeters, J. , Yau, C. , Wolf, D. , Sanil, A. , Li, Y. , Severson, T. , Linn, S. , I-SPY 2 Trial Investigators, Buxton, M. , 2014. Evaluation of a BRCAness Signature as a Predictive Biomarker of Response to Veliparib/carboplatin Plus Standard Neoadjuvant Therapy in High-risk Breast Cancer: Results from the I-SPY 2 TRIAL. (Barcelona, Spain) [Google Scholar]
- Grushko, T.A. , Dignam, J.J. , Das, S. , Blackwood, A.M. , Perou, C.M. , Ridderstråle, K.K. , Anderson, K.N. , Wei, M.-J. , Adams, A.J. , Hagos, F.G. , 2004. MYC is amplified in BRCA1-associated breast cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 10, 499–507. [DOI] [PubMed] [Google Scholar]
- Hudis, C.A. , Gianni, L. , 2011. Triple-negative breast cancer: an unmet medical need. Oncologist. 16, (Suppl. 1) 1–11. [DOI] [PubMed] [Google Scholar]
- Hudis, C.A. , Barlow, W.E. , Costantino, J.P. , Gray, R.J. , Pritchard, K.I. , Chapman, J.-A.W. , Sparano, J.A. , Hunsberger, S. , Enos, R.A. , Gelber, R.D. , 2007. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 25, 2127–2132. [DOI] [PubMed] [Google Scholar]
- Johnson, W.E. , Li, C. , Rabinovic, A. , 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat. Oxf. Engl. 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Kaufman, B. , Shapira-Frommer, R. , Schmutzler, R.K. , Audeh, M.W. , Friedlander, M. , Balmaña, J. , Mitchell, G. , Fried, G. , Stemmer, S.M. , Hubert, A. , 2014. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 33, (3) 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, J.M.-M. , Peck, B. , Monteiro, L.J. , Schwenen, H.D.C. , Millour, J. , Coombes, R.C. , Myatt, S.S. , Lam, E.W.-F. , 2010. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol. Cancer Res. MCR. 8, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, B.D. , Bauer, J.A. , Chen, X. , Sanders, M.E. , Chakravarthy, A.B. , Shyr, Y. , Pietenpol, J.A. , 2011. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn, S.C. , Van 't Veer, L.J. , 2009. Clinical relevance of the triple-negative breast cancer concept: genetic basis and clinical utility of the concept. Eur. J. Cancer Oxf. Engl. 45, (Suppl. 1) 11–26. [DOI] [PubMed] [Google Scholar]
- Lips, E.H. , Laddach, N. , Savola, S.P. , Vollebergh, M.A. , Oonk, A.M.M. , Imholz, A.L.T. , Wessels, L.F.A. , Wesseling, J. , Nederlof, P.M. , Rodenhuis, S. , 2011. Quantitative copy number analysis by Multiplex Ligation-dependent Probe Amplification (MLPA) of BRCA1-associated breast cancer regions identifies BRCAness. Breast Cancer Res. BCR. 13, R107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips, E.H. , Mulder, L. , Oonk, A. , van der Kolk, L.E. , Hogervorst, F.B.L. , Imholz, A.L.T. , Wesseling, J. , Rodenhuis, S. , Nederlof, P.M. , 2013. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br. J. Cancer. 108, 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C.X. , Cai, S. , Li, S. , Ryan, C.E. , Guo, Z. , Schaiff, W.T. , Lin, L. , Hoog, J. , Goiffon, R.J. , Prat, A. , 2012. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J. Clin. Invest. 122, 1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manié, E. , Vincent-Salomon, A. , Lehmann-Che, J. , Pierron, G. , Turpin, E. , Warcoin, M. , Gruel, N. , Lebigot, I. , Sastre-Garau, X. , Lidereau, R. , 2009. High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res. 69, 663–671. [DOI] [PubMed] [Google Scholar]
- Masuda, H. , Baggerly, K.A. , Wang, Y. , Zhang, Y. , Gonzalez-Angulo, A.M. , Meric-Bernstam, F. , Valero, V. , Lehmann, B.D. , Pietenpol, J.A. , Hortobagyi, G.N. , 2013. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 19, 5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan, M.E. , Chiu, J.W. , Koller, B.H. , Jasin, M. , 1999. Brca1 controls homology-directed DNA repair. Mol. Cell. 4, 511–518. [DOI] [PubMed] [Google Scholar]
- Perou, C.M. , Sørlie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Rees, C.A. , Pollack, J.R. , Ross, D.T. , Johnsen, H. , Akslen, L.A. , 2000. Molecular portraits of human breast tumours. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Roepman, P. , Horlings, H.M. , Krijgsman, O. , Kok, M. , Bueno-de-Mesquita, J.M. , Bender, R. , Linn, S.C. , Glas, A.M. , van de Vijver, M.J. , 2009. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 15, 7003–7011. [DOI] [PubMed] [Google Scholar]
- Schouten, P.C. , van Dyk, E. , Braaf, L.M. , Mulder, L. , Lips, E.H. , de Ronde, J.J. , Holtman, L. , Wesseling, J. , Hauptmann, M. , Wessels, L.F.A. , 2013. Platform comparisons for identification of breast cancers with a BRCA-like copy number profile. Breast Cancer Res. Treat. 139, 317–327. [DOI] [PubMed] [Google Scholar]
- Schouten, P.C. , Marmé, F. , Aulmann, S. , Sinn, H.-P. , van Essen, H.F. , Ylstra, B. , Hauptmann, M. , Schneeweiss, A. , Linn, S.C. , 2015. Breast cancers with a BRCA1-like DNA copy number profile recur less often than expected after high-dose alkylating chemotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 21, 763–770. [DOI] [PubMed] [Google Scholar]
- Shah, S.P. , Roth, A. , Goya, R. , Oloumi, A. , Ha, G. , Zhao, Y. , Turashvili, G. , Ding, J. , Tse, K. , Haffari, G. , 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 486, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silwal-Pandit, L. , Vollan, H.K.M. , Chin, S.-F. , Rueda, O.M. , McKinney, S. , Osako, T. , Quigley, D.A. , Kristensen, V.N. , Aparicio, S. , Børresen-Dale, A.-L. , 2014. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 20, 3569–3580. [DOI] [PubMed] [Google Scholar]
- Sørlie, T. , Perou, C.M. , Tibshirani, R. , Aas, T. , Geisler, S. , Johnsen, H. , Hastie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff, A.E. , Miremadi, A. , Pinder, S.E. , Ellis, I.O. , Caldas, C. , 2007. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 8, R157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, N. , Tutt, A. , Ashworth, A. , 2004. Hallmarks of “BRCAness” in sporadic cancers. Nat. Rev. Cancer. 4, 814–819. [DOI] [PubMed] [Google Scholar]
- Tutt, A. , Robson, M. , Garber, J.E. , Domchek, S.M. , Audeh, M.W. , Weitzel, J.N. , Friedlander, M. , Arun, B. , Loman, N. , Schmutzler, R.K. , 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 376, 235–244. [DOI] [PubMed] [Google Scholar]
- Tutt, A. , Ellis, P. , Kilburn, L. , Gilett, C. , Pinder, S. , Abraham, J. , Barrett, S. , Barrett-Lee, P. , Chan, S. , Cheang, M. , 2014. [S3-01] The TNT trial: a randomized phase III trial of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012). San Antonio Breast Cancer Conference 2014, (San Antonio, TX, USA). [Google Scholar]
- Van de Vijver, M.J. , He, Y.D. , van’t Veer, L.J. , Dai, H. , Hart, A.A.M. , Voskuil, D.W. , Schreiber, G.J. , Peterse, J.L. , Roberts, C. , Marton, M.J. , 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009. [DOI] [PubMed] [Google Scholar]
- Vollebergh, M.A. , Lips, E.H. , Nederlof, P.M. , Wessels, L.F.A. , Schmidt, M.K. , van Beers, E.H. , Cornelissen, S. , Holtkamp, M. , Froklage, F.E. , de Vries, E.G.E. , 2011. An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO. 22, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell, N. , Arnold, J. , Cocciardi, S. , da Silva, L. , Marsh, A. , Riley, J. , Johnstone, C.N. , Orloff, M. , Assie, G. , Eng, C. , 2010. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res. Treat. 123, 661–677. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zeng, Z.C. , Bui, T.A. , DiBiase, S.J. , Qin, W. , Xia, F. , Powell, S.N. , Iliakis, G. , 2001. Nonhomologous end-joining of ionizing radiation-induced DNA double-stranded breaks in human tumor cells deficient in BRCA1 or BRCA2. Cancer Res. 61, 270–277. [PubMed] [Google Scholar]
- Wessels, L.F.A. , van Welsem, T. , Hart, A.A.M. , van't Veer, L.J. , Reinders, M.J.T. , Nederlof, P.M. , 2002. Molecular classification of breast carcinomas by comparative genomic hybridization: a specific somatic genetic profile for BRCA1 tumors. Cancer Res. 62, 7110–7117. [PubMed] [Google Scholar]
- Zhang, N. , Wu, X. , Yang, L. , Xiao, F. , Zhang, H. , Zhou, A. , Huang, Z. , Huang, S. , 2012. FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene Rad51. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 18, 5961–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary Figure 1 Hierarchical clustering of 279 top variable genes most differentially expressed between BRCA1‐like and non‐BRCA1‐like status (ANOVA, FDR < 0.001, fold‐change > 1) in 125 TN breast tumors. Scaled expression value is denoted as the column Z‐score and plotted in red–blue color scale. Red indicates high expression and blue low expression. Information Columns 1, 2 and 3 depict BRCA1‐like status, BRCA1 promoter methylation status and BRCA1 mutation status, respectively. For all sample columns, assay positive is indicated by blue, negative by gray and no data due to failed assay in white. Column 4 depicts Lehmann TNBCTypes basal‐like 1 (BL1), basal‐like 2 (BL2), immunomodulatory (IM), luminal androgen receptor (LAR), mesenchymal (M), mesenchymal stem‐like (MSL), and unidentified (U) as teal, yellow, purple, salmon, light blue, orange and pink, respectively. No data due to failed Lehmann TNBCType assay indicated in lime green.
Supplementary Figure 2 Survival analysis. Distant recurrence‐free survival (univariate) of the non‐adjuvant‐systemic therapy treated cohort with respect to BRCA1‐like status using the Kaplan–Meier method (adjusted log‐rank, P = 0.08). The blue line indicates non‐BRCA1‐like patients and the red line indicates BRCA1‐like patients. Patients at risk are shown on the x‐axis in corresponding colors.


