Abstract
Background
Development of an effective non-viral vaccine against hepatitis C virus infection is of a great importance. Gelatin nanoparticles (Gel.NPs) have an attention and promising approach as a viable carrier for delivery of vaccine, gene, drug and other biomolecules in the body.
Aim of work
The present study aimed to develop stable Gel.NPs conjugated with nonstructural protein 2 (NS2) gene of Hepatitis C Virus genotype 4a (HCV4a) as a safe and an efficient vaccine delivery system.
Methods and results
Gel.NPs were synthesized and characterized (size: 150±2 nm and zeta potential +17.6 mv). NS2 gene was successfully cloned and expressed into E. coli M15 using pQE-30 vector. Antigenicity of the recombinant NS2 protein was confirmed by Western blotting to verify the efficiency of NS2 as a possible vaccine. Then NS2 gene was conjugated to gelatin nanoparticles and a successful conjugation was confirmed by labeling and imaging using Confocal Laser Scanning Microscope (CLSM). Interestingly, the transformation of the conjugated NS2/Gel.NPs complex into E. coli DH5-α was 50% more efficient than transformation with the gene alone. In addition, conjugated NS2/Gel.NPs with ratio 1:100 (w/w) showed higher transformation efficiency into E. coli DH5-α than the other ratios (1:50 and 2:50).
Conclusion
Gel.NPs effectively enhanced the gene delivery in bacterial cells without affecting the structure of NS2 gene and could be used as a safe, easy, rapid, cost-effective and non-viral vaccine delivery system for HCV.
Introduction
Egypt has the highest prevalence of Hepatitis C Virus (HCV) infection in the world, making it one of the major public health challenges facing the country. Approximately 14.7% (about 10 million) of the Egyptian population are anti-HCV positive, mainly genotype 4a [1,2]. Only 20–30% of HCV patients are able to clear the virus spontaneously [3], while the remaining 70–80% develop chronic infection and have a risk to develop cirrhosis and hepatocellular carcinoma (HCC) [4,5].
Approved combination therapy of Poly Ethylene Glycated (PEGylated) Interferon and Ribavirin is expensive and causes many side effects including anemia and decrease of the neutrophils count [6,7]. Moreover, only about 55% of patients respond to the treatment and can successfully clear the virus depending on virological, immunological and genetic factors [8,9]. New regimens of Sofosbuvir provide HCV–g4 high cure rates by inhibiting HCV polymerase but are still the highest cost treatment [10]. As prevention is better than treatment, and to avoid the expected mortality from HCV-related cirrhosis or HCC in the next decade; an effective vaccine against HCV should be developed [11]. For this purpose; nonstructural (NS2) protease domain, the most genetically conserved viral antigen among HCV genotypes, has been used for the induction of cellular immunity in animal models in different vaccine studies [12], this might help in the design of a vaccine against the predominant genotype 4a.
Non-structural protein 2 (NS2) is an integral membrane protein (23-kDa of 217 amino acids) [13]. The N-terminus of NS2 (residues 1–94) forms three transmembrane domains, while the C-terminus (residues 94–217) exists in the cytoplasm [14]. NS2 is suggested to play a vital role in HCV assembly and replication through its interaction with structural (E1 and E2) and non-structural (p7, NS3-4A, and NS5A) proteins [15,16]. NS2 is an essential key enzyme for the viral life cycle, making it an excellent candidate molecule for the development of antiviral therapies and vaccines against HCV infection [17]. The safest and most efficient method of gene delivery into the human cell is still a point of interest in biotechnological research. Direct administration of naked DNA is the easiest route but limited only to rigid tumors [18] and muscle tissue [19]. In addition, naked DNA can be rapidly degraded by blood enzymes and repulsed with the negatively charged cell membrane that consequently inhibits efficient delivery [20]. Viral carriers are prohibited by Food and Drug Administration (FDA) because of their high risk of toxicity, inflammatory and immune responses [21,22]. Recently, biocompatible nanomaterials are promising carriers with low toxicity and well-controlled gene delivery [23]. Gelatin nanoparticles (Gel.NPs) have significant uses in biomedical and pharmaceutical research. Gelatin is nontoxic, biodegradable, bioactive, inexpensive, non-immunogenic and biocompatible to human tissues. Gel.NPs are very efficient in vaccine, gene or drug delivery in the body [24]. Properties of Gel.NPs such as size, swelling behavior, and thermal properties; depend on the crosslinking degree between cationic and anionic groups that could be controlled during preparation. Gel.NPs can be prepared by the desolvation/coacervation or the emulsion methods [24,25].
The present study aimed to investigate NS2 antigenicity in order to develop a safe, rapid, easy, non-immunogenic, more effective and efficient HCV vaccine using Gel.NPs as a delivery system. Gel.NPs were prepared with optimum particle size and high positive zeta charge. NS2 gene was amplified from the Egyptian HCV genotype 4a (isolate ED43), then successfully cloned and expressed in E.coli using pQE-30 vector. The antigenicity of the recombinant NS2 protein was confirmed when reacted with sera from patients with HCV. NS2 and Gel.NPs were conjugated and the NS2/Gel.NPs conjugate showed higher efficiency in bacterial transformation than NS2 gene alone.
Materials and methods
Materials
Gelatine type B (gel strength ~300 g Bloom, Sigma-Aldrich, US), Glutaraldehyde (25%, Sigma-Aldrich, US), Acetone (99.9%, Sigma-Aldrich, US), E.coli M15 (Qiagen Inc., Valencia, USA). LB broth Miller (Luria-Bertani, Amresco,US), Ampicillin (Sigma-Aldrich, US), Kanamycin (Sigma-Aldrich, US), Isopropyl b-D-thiogalactoside (IPTG) (≥99% (TLC), ≤0.1% Dioxane, Sigma-Aldrich, US), PBS (Sigma, US), PMSF (Sigma, Ltd. Dorset, England), 8 M urea (Sigma, US), 20 mM β-mercaptoethanol (β-ME) (Sigma-Aldrich,US) Ni2+/nitrilotriacetate (NTA)–agarose (Qiagen Inc., Valencia, USA), Bovine serum albumin (Sigma, US), Tris-buffered saline (Sigma, US), Horseradish peroxidase (HRP)-labeled Protein A (Sigma, Ltd. Dorset, England), ECL Plus™ Western Blotting Reagents from GE Healthcare (formerly Amersham Biosciences), Rhodamine red dye (PowerPlex® 16 BIO System, Promega) and Quantifluor (QuantiFluor® dsDNA System, Promega), High Pure PCR Purification Kit (Biobasic Inc., Ontario, Canada), QIAexpressionest kit (Qiagen Inc., Valencia, USA), Proteins and Molecular weight standards (Sigma-Aldrich, US), Nitrocellulose membranes (Wattman, US), QIAGEN EZ Competent Cells (Qiagen Inc., Valencia, USA), QIAprep SpinMiniprep kit (Qiagen Inc., Valencia, USA), SphI and HindIII (Roche), QIAGEN® PCR Cloning kit (Qiagen Inc., Valencia, USA), GoTaq® Green Master Mix (Promega).
Methods
Preparation and characterization of gelatine nanoparticles (Gel.NPs)
Gel.NPs were synthesized by the two step desolvation method, described by Coester and colleagues, [26] with some modifications in the temperature, pH, concentration of glutaraldehyde and the type of desolvating agent “S1 Table”. Briefly, one gram of gelatin type B was dissolved in 20 ml distilled water with gentle heating. For the first desolvation step; 20 ml acetone was added; gelatin fractions were precipitated after 15 min and the supernatant was discarded. To dissolve the precipitate; 20 ml water was added under gentle heating at pH 3 that was adjusted using 0.1 N hydrochloric acid (HCL). In situ, Gel.NPs were formed during the second desolvation step by dropwise addition of 80 ml acetone while stirring (500 rpm). After 10 min, 100 μl of 25% glutaraldehyde were added to crosslink the nanoparticles under stirring for 12 h. Gel.NPs were purified by three-fold centrifugation (16000 g for 20 min) in 30% acetone. Purified Gel.NPs were dried, suspended in a highly purified water (conductivity < 0.04 μs/cm) and stored at 4–8°C.
Physico-chemical properties of Gel.NPs were characterized using high-resolution transmission electron microscope (HR-TEM, FEI, Tecnia G20, Eindhoven, and Netherland), X-ray Diffraction (XRD, PanAnalytical, X’pert Pro, Almelo, Netherland) and particle size analyzer (Zeta sizer a nano series s’, Malvern, Worcestershire, UK) “S1–S3 Figs”.
Amplification and cloning of NS2 gene
NS2 gene was amplified from the recombinant HCV genotype 4a (isolate ED43) genome (GenBank accession number: Y11604) (kindly provided by Dr. Richard Eliott lab, Institute of Virology, University of Glasgow, Church Street, Glasgow). The amplification of NS2 gene (650 bp, extends from 2707 to 3357 bp of the genome) was performed as mentioned in the manufacturer’s manual of the PCR master mix. SphI and HindIII restrictions sites (found to have zero cuts using the online program NEB cutter version 2.0 (nc2.neb.com/NEBcutter2) have been added at the 5’end of the forward and reverse primers respectively, and their sequences were as follows: 5' GCATGCTACGACCAGGAAGTGGCAGG –3', and 5'- AAGCTTAAGGAGTCTCCACCCCTTTGA -3'. PCR program consisted of 30 cycles with initial denaturation 95°c for 5 min, denaturation 95°c for 30 sec, annealing 55°c for 45 sec, extension 72°c for one min, and final extension72°c for 10 min. The NS2 amplicon was purified from gel using High Pure PCR Purification Kit (Biobasic Inc., Ontario, Canada) [27] as per manufacturer’s instructions, double digested with SphI and HindIII and repurified Then, NS2 was cloned using QIAGEN® PCR Cloning kit and the steps were performed as mentioned in the manufacturer’s manual, with the only modification in using pQE-30 cloning and expression vector instead of pDrive cloning vector provided with the kit in order to perform both cloning and expression with the same construct. Recombinant NS2 concentration and purity were measured using UV spectrophotometer (Q5000, Quawell, USA)
Expression of recombinant NS2 and antigenicity
The pQE-30 vector contains the sequence for the expression of N-terminally 6xHis-tagged proteins, and it also provides the multiple cloning sites in the first reading frame.
After cloning; Recombinant NS2 was transformed into M15 competent cells using QIAexpressionest kit (Qiagen Inc., Valencia, USA). Bacteria were grown overnight at 37°C and the recombinant hexahistidine-tagged NS2 protein was purified under denaturing conditions on Ni2+-nitrilotriacetate (NTA)–agarose (Qiagen Inc., Valencia, USA) according to the manufacturer’s instructions. NS2 recombinant protein was detected by electrophoresing denaturing SDS-PAGE acrylamide gel which was then stained with coomassie blue.
The antigenicity of NS2 protein was then investigated by Western blotting. Briefly, NS2 protein was electrophoresed in 12% SDS-PAGE acrylamide gels and transferred onto nitrocellulose membranes (Wattman, US). Membranes were then blocked with 1% bovine serum albumin in Tris-buffered saline for 2 h at room temperature with shaking. Then, the membranes were incubated with anti-HCV human sera of Egyptian blood donors infected with hepatitis C, washed, incubated with horseradish peroxidase (HRP)-conjugated second antibody for 1 h at 37°C and the membrane was developed using the enhanced chemiluminescent (ECL®) kit following the manufacturer’s instructions. The same steps were repeated using sera from healthy Egyptian donors to ensure the specificity of the recognized protein.
Conjugation of the recombinant NS2 gene with Gel.NPs
Gel.NPs were labeled with Rhodamine red (0.001g of Gel.NPs dissolved in 400μl dH2O with 2μl Rhodamine red dye). The recombinant NS2 gene was labeled with fluorescein dye (1μl NS2 gene + 199μl Quantifluor), then excited with the corresponding laser lines, 633 nm for Rhodamine red dye (red color) and 514 nm for Quantifluor dye (green color).
Recombinant NS2 concentration was estimated to be 50 ng/μl by UV spectrophotometer and the final concentration of Gel.NPS after labelling was 2500 ng/μl. To prepare recombinant NS2/Gel.NPs conjugates; different ratios have been tried but the ratios 1:50, 2:50 and 1:100 (w/w) of NS2 gene and the labeled Gel.NPs respectively have been used in further experiments as they have resulted in successful and consistent transformation. For this purpose; labeled NS2 gene and Gel.NPs were co-incubated over one day on 500 rpm shaker to form NS2/Gel.NPs conjugate by physical conjugation. To estimate the amount of recombinant NS2 bound to Gel.NPs, NS2/Gel.NPs conjugate was precipitated by three-fold centrifugation at 16000 g for 20 min (till the purity of DNA in supernatant reached 1.8–2), the amount of recombinant NS2 was measured in the supernatant by UV spectrophotometer (Q5000, Quawell, USA), then the amount of recombinant NS2 conjugated to Gel.NPs was estimated as previously described (Singh and Mishra, 2014). To get rid of the excess unbound NS2 gene, the supernatant was discarded and the conjugated NS2/Gel.NPs were resuspended in deionized water.
Confocal Laser Scanning Microscope (CLMS) imaging was utilized to validate the conjugation process between recombinant NS2 gene and Gel.NPs.
Transformation of NS2/Gel.NPs into DH5-α bacteria
Transformations of the labeled Gel.NPs, NS2 gene, and the NS2 /Gel.NPs conjugate into DH5-α bacteria were assessed using four replicates for each sample. CLSM technique was used to study the efficiency of Gel.NPs to deliver NS2 gene into DH5-α bacteria compared to NS2 gene alone; one colony from the transformed DH5-α bacteria was picked from each sample, fixed on the slide by 20% glutaraldehyde and excited using the suitable laser as described in Table 1. For quantitative analysis, bacterial replicates were counted using ImageJ software (National Institutes of Health, Bethesda, MA, USA).
Table 1. Different laser wavelengths used to detect prepared samples.
| Sample | Type of excited laser | |
|---|---|---|
| labeled Gel.NPs | 633nm | |
| labeled recombinant NS2 gene | 405nm for gene that absorb at UV region | 514nm for Quantifluor dye labeling gene |
| Conjugate of labeled (Gel.NPs+ recombinant NS2 gene) | 633nm,405nm and 514nm | |
Finally, in order to confirm that Gel.NPs didn’t affect the recombinant NS2 gene structure, minipreps of the transformed bacteria with the three tested concentrations were prepared using QIAprep SpinMiniprep kit (Qiagen Inc., Valencia, USA) following the manufacturer instructions, in which a single colony from each plate was picked, inoculated into 5 ml of LB containing 5μl ampicillin (LBAmp broth) and grown overnight at 37°C.
Results
Characterization of gelatin nanoparticles
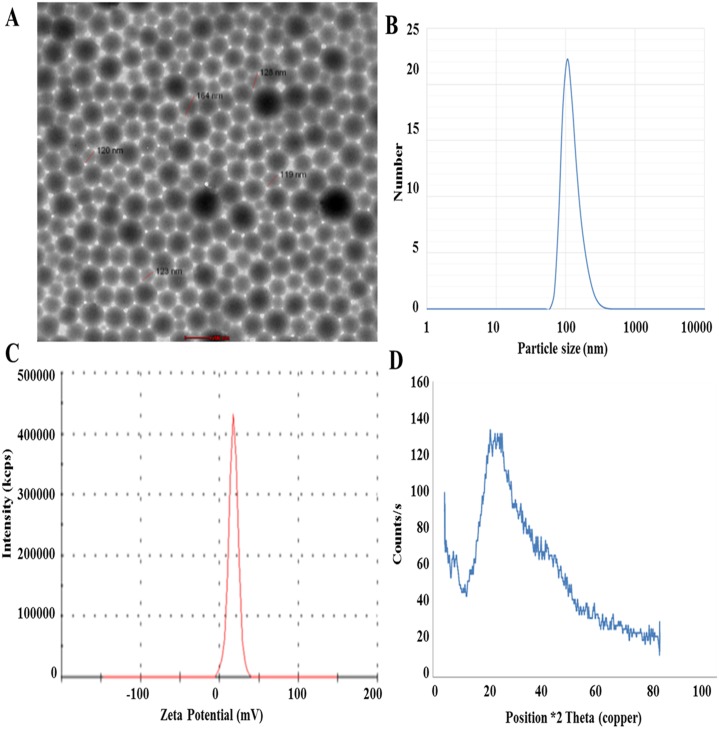
Morphological characterization of Gel.NPs showed nearly perfect unitized and well dispersed spherical particles with an average size of 150.0 ± 2.0 nm, using high-resolution TEM. These observations were supported by particle size analysis done by Dynamic Light Scattering (DLS) technique (Fig 1A). The particle size was measured showing to have an average size of 150 nm with low Polydispersity Index (PDI) 0.09 which indicates its narrow size distribution (Fig 1B). Zeta-Potential (surface charge) of the prepared Gel.NPs was improved to be more electro-positive (+17.6 mV) (Fig 1C). Phase analysis was performed using X-Ray Diffraction technique (XRD) (Fig 1D). XRD pattern showed a characteristic broad peak at angle θ = 20° indicating that the prepared Gel.NPs have an amorphous structure.
Fig 1. Characterization of Gel.NPs.
A. HR-TEM image of the prepared Gel.NPs resuspended in water and adsorbed onto solid support and stained with phosphotangestic acid which shows that particles have well dispersed spherical shape with average particles. B. Graph showing particle size distribution by number for the synthesized Gel.NPs that was obtained by particle size analyzer. As shown from the observed peak the size of Gel.NPs is 150±2 nm. C. Graph showing the Zeta potential for synthesized Gel.NPs that was determined by measuring the electrophoretic mobility of the Gel.NPs using a Malvern zeta sizer. As shown the observed Zeta potential is +17.6 mV. D. XRD of prepared GNPs.
Cloning and Expression of the NS2 gene
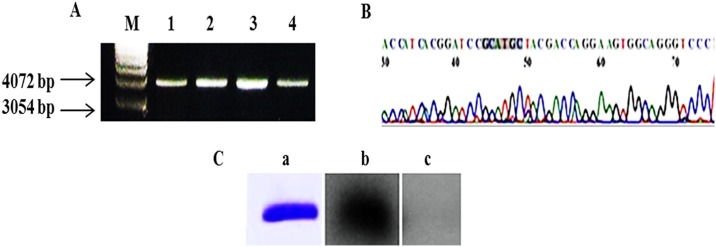
NS2 gene was successfully amplified, cloned and expressed into protein and the antigenicity of NS2 protein was confirmed by Western blotting in which a band of ~23 kDa corresponding to the expressed NS2 protein was recognized by anti-HCV antibodies present in the sera of Egyptian patients infected with HCV, while healthy donors showed no specific recognition of the NS2 protein (Fig 2).
Fig 2. Cloning and expression of NS2 gene in pQE30.
A. Agarose gel electrophoresis for single digestion of the miniprep of the recombinant NS2 samples with SphI. Lane M: 1 kb DNA marker, Lane 1–4: DNA resulted from minipreps of four different clones digested with SphI showing a band of ~ 4 kb indicating NS2 insert (650 bp) within the pQE-30 vector (3.4 kb). B. Representative DNA sequence analysis of the NS2 insert into pQE-30 vector using ABI PRISM model 310 DNA automated sequencer. Bases 1–50 are pQE-30 vector sequence, bases from 50 to the end are partial NS2 sequene. Shadowed sequence is the restriction site for SphI. C. a: SDS-PAGE of M15 bacterial lysate resulted from expression of the NS2 protein Lane M: prestained wide range MW marker. Lane 1: expressed NS2 protein after purification. b: Western blot of the purified NS2 protein recognized by sera of Egyptian patients infected with HCV. c: Western blot for healthy donors showed no specific recognition of the NS2 protein.
Conjugation of recombinant NS2 with Gel.NPs
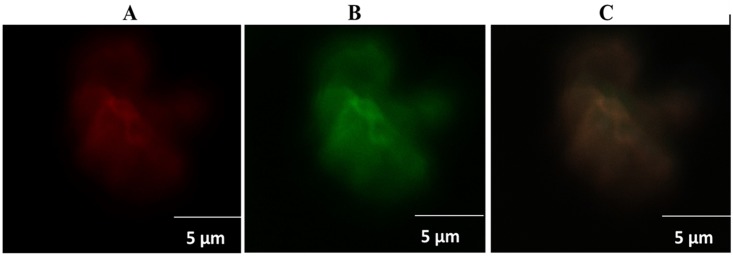
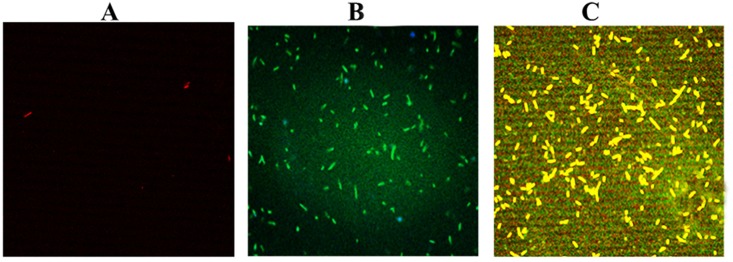
It was very important to estimate the amount of the recombinant NS2 gene that gets conjugated to Gel.NPs and to confirm the conjugation between NS2 gene and Gel.NPs. We found that NS2/Gel.NPs with ratio of 1:100 have shown the highest conjugation percentage as 62.6% of the recombinant NS2 gene were conjugated to Gel.NPs, while 43%, 44.2% of the recombinant NS2 gene in NS2/Gel.NPs with ratios of 1:50, 2:50 respectively were successfully conjugated “S2 Table”. Then, confirmation of conjugation was carried out using CLMS technique. Excitation of Gel.NPs labeled with Rhodamine red at 633 nm showed highly intense red aggregations of gelatin nanoparticles, (Fig 3A). NS2 gene labeled with Quantifluor was exited at 514 nm laser line showed green aggregations of NS2 gene, (Fig 3B). While simultaneous excitation of NS2/Gel.NPs conjugate with both laser lines (633 nm and 514 nm) showed yellowish-orange clusters as a result of the crossing over between the two laser dyes Rhodamine (red) and Quantifluor (green), corresponding to the conjugation between the Gel.NPs and the NS2 gene and confirm the generation of the new NS2/Gel.NPs conjugate that can be used as a simple and direct gene delivery system, (Fig 3C).
Fig 3. Confocal Laser Scanning Microscope (CLSM) imaging.
A: Gel.NPs labeled with for Rhodamine red dye excited at 633 nm. B: NS2 gene labeled with Quantifluor exited at 514 nm. C: labeled NS2 /Gel.NPs conjugate excited simultaneously at both 633 nm and 514 nm.
Gelatin nanoparticles enhanced NS2 transformation into E. coli
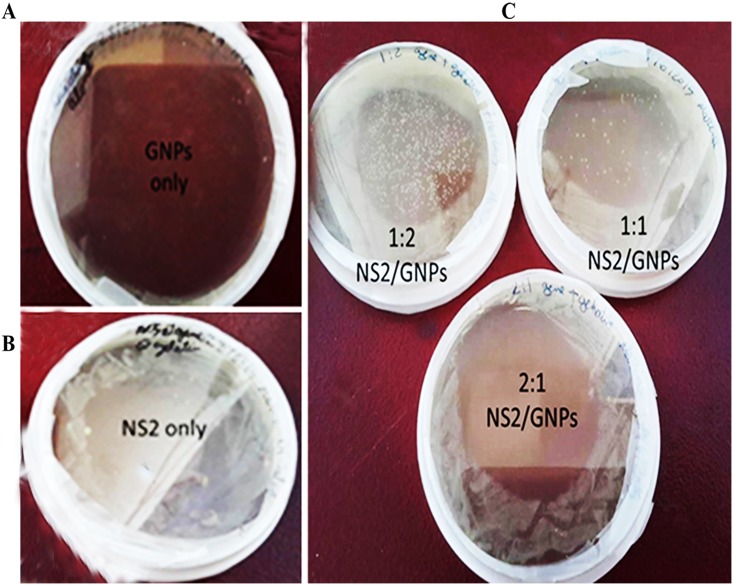
In order to evaluate the efficiency of gelatin nanoparticles as a delivery system for the recombinant NS2 gene, three labeled recombinant NS2/Gel.NPs conjugate samples ratios (w/w); 1:50, 2:50 and 1:100, in addition to the labeled NS2 gene alone and the labeled Gel.NPs samples alone were transformed into DH5-α bacteria which were cultured for 24 h under optimum conditions. Interestingly, the highest transformation efficiency was observed in the plates of the recombinant labeled NS2/Gel.NPs conjugate, (Fig 4C), compared to the plates of labeled NS2 gene alone, (Fig 4B), while no transformation could be detected with the labeled Gel.NPs only (Fig 4A). Moreover; the highest number of bacterial colonies was observed in the plate with bacteria transformed with recombinant NS2/Gel.NPs conjugate with ratio 1:100 compared with the other ratios. The high efficiency of transformation of NS2 conjugated to Gel.NPs was confirmed by imaging bacterial cells using Confocal Laser Scanning Microscope and the number of bacterial replicates was counted using ImageJ software. The results showed that the number of transformed bacterial replicates with NS2/Gel.NPs (1:100) was higher (236± 3.82) (Fig 5C) than other used ratios and than those with NS2 alone (119±3.03) (Fig 5B) and those with Gel.NPs alone (no bacterial growth) (Fig 5A) “S3 and S4 Tables”, “S4 Fig”.
Fig 4. Images of plates containing DH5-α bacteria transformed with A) Gel.NPs only, B) recombinant NS2 gene only, C) labeled conjugate recombinant NS2 Gel.NPs with three ratios (w/w) 1:50, 2:50 and 1:100.
Fig 5. The image of NS2/Gel.NPs transformed DH5-α bacteria by Confocal Laser Scanning Microscope showed high number of colonies in yellow-orange color after laser excitation at 633 nm and 514 nm simultaneously.
The image of Gel.NPs transformed into DH5-α bacteria showed no appearance of any emission colors due to absence of plasmid carrying ampicillin resistant gene, where bacteria died when grown in presence of ampicillin (Fig 5A). NS2 transformed DH5-α bacteria showed lower number of bacterial colonies in green color after laser excitation at 514nm (Fig 5B). The yellow-orange color due to the cross over between the red labeled Gel.NPs and green labeled NS2 gene could be detected in (Fig 5C) which indicates that presence of gelatin nanoparticles enhance transformation efficiency than using naked NS2 in (Fig 5B).
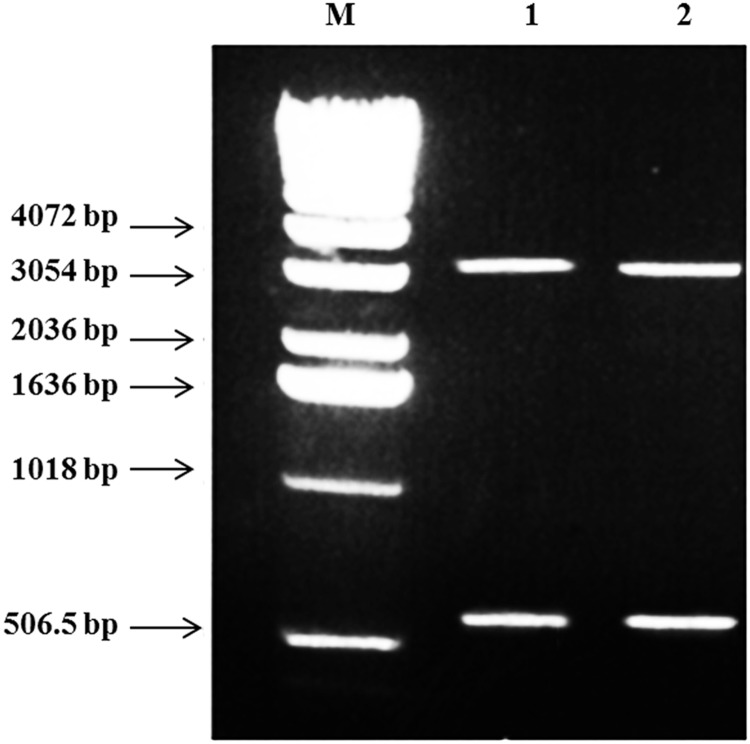
Finally, gel electrophoresis of the miniprep product of the transformed bacteria confirmed that the Gel.NPs didn’t affect the recombinant NS2 gene structure (Fig 6), where the two bands appeared for NS2 insert (650 bp) and the pQE-30 vector (3.4 kb) were exactly at the correct size.
Fig 6. Agarose gel electrophoresis for miniprep samples of the recombinant PQE 30/NS2 plasmid/Gel.NPs double digested (SphI/HindIII) showing two bands; band for NS2 insert (650 bp) and band for pQE-30 vector (3.4 kb) in Lane 1 and 2.
Lane M: 1 kb DNA marker.
Discussion
In vitro and in vivo applications of gelatin in the controlled release devices for bioactive molecules like proteins or plasmid DNA were reviewed [24], and the scientific contributions of gelatin nanoparticles as an ideal carrier system for drug delivery have been increased in the last five years [28–30]. In the present study, we investigated the effectiveness of Gel.NPs as an antigen delivery system against HCV. A previous study has demonstrated that Gel.NPs incorporated in the bronchial epithelial cells and 16HBE14o-cells showed only a few or no cytotoxicity without any inflammatory responses [31]. These findings suggest that Gel.NPs may be a suitable and safe candidate as drug or gene carriers.
Many studies have reported several methods for Gel.NPs synthesis [26,32,33]. Here in, we applied some different processing parameters such as pH, the amount of glutaraldehyde added, and temperature, these variations resulted in synthesis of nanoparticles with particle size and shape smaller than the earlier published work [26,32,33]. Our preparation conditions were optimized using 100μl /gm of glutaraldehyde (GTA) at pH 3, that resulted in synthesis of well dispersed spherical particulate (150±10 nm), which is smaller in size than the previously synthesized gelatin nanoparticles using COESTER method that is used as a reference for all consequent preparations in many studies and usually results in nanoparticles with bigger size (~280 nm) [26]. Previously, cationic Gel.NPs have shown to increase the stability of the nanoparticle system and improve the interaction between the particles and the eye surface of the rabbits and thus increased the potential of ocular drug delivery more than negatively charged Gel.NPs [34]. In addition, cationized Gel.NPs have been proved to act as a highly effective carrier system for immunogenic CpG oligodeoxynucleotides due to its protein based structure that offers many functional groups that can help as anchor of an antigen [35].
In vitro trials have been carried out for using NS2 gene as HCV vaccine but the transformation efficiency still the main challenge. In the present work, we introduced the gelatin nanoparticles as a vehicle for NS2 to facilitate and increase the transformation efficiency as a preliminary step in order to be used as an HCV vaccine delivery system. Stability of our prepared cationic Gel.NPs was improved at +17.6 Mv. compared to -23.1 Mv as mentioned in a previous study [36]. This stability might have played an important role in maintaining the stable electrostatic interaction between Gel.NPs and the negatively charged recombinant NS2 gene to form the intended NS2/Gel.NPs conjugate, in addition to facilitate the penetration of the conjugate into the negatively charged cell membrane and nucleus of the target bacteria which is assumed to enhance the gene delivery process more than the gene alone [34].
Successful transformation was confirmed by imaging the labeled Gel.NPs, the labeled recombinant NS2 gene and the labeled conjugate recombinant NS2/Gel.NPs samples using Confocal Laser Scanning Microscope (CLMS), thus proving that Gel.NPs can be used as a successful gene delivery system into the bacteria as a primary step for testing it later on the human cell line level. To test the transformation efficiency of Gel.NPs, different concentrations of Gel.NPs were used as vehicles to carry the recombinant NS2 gene for intracellular delivery of the DH5-α bacteria, the numbers of transformed bacterial colonies were counted by ImageJ software (National Institutes of Health, Bethesda, MA,USA) after imaging. The present data revealed that recombinant NS2/Gel.NPs conjugated with ratio 1:100induced the highest transformation yield compared to the transformation of NS2 gene alone, thus indicating that Gel.NPs increased the transformation efficiency. Gel.NPs might have protected the recombinant DNA from intracellular degradation or digestion as has been proved in a previous study in which Vu L. Truong-Le and his colleagues demonstrated that DNA conjugated to Gel.NPs is more resistant to nuclease digestion than naked DNA [37]. We could attribute the enhancement in transformation efficiency at 1:100 rather than the other used ratios to the increase in cationic Gel.NPs concentration which in turn increased the possibility of conjugating higher number of negatively charged NS2 molecules [38]. Furthermore, cationic Gel.NPs have more potential to interact with the cell surface and nuclear membrane and thus increased the transfection efficiency [34].
In order to investigate the effect of Gel.NPs on the recombinant DNA, we repeated the Miniprep steps on the transformed bacteria and confirmed that Gel.NPs did not affect the recombinant NS2 gene structure or size and consequently, would not impede NS2 protein expression. Thus, we could implicate Gel.NPs as a safe and an efficient delivery system that can transport recombinant NS2 gene without affecting its structure, and this indicates that NS2/Gel.NPs complex can be expressed successfully within the DH5-α bacterial cell.
Conclusion
Gel.NPs were successfully synthesized with optimum controlled characteristics, conjugated with recombinant NS2 gene and facilitated the delivery of the NS2 gene into the bacterial cell in vitro effectively where it could be amplified and expressed in the bacterial cell without affecting its structure. This study suggests that Gel.NPs could be a useful vaccine and gene delivery system and may be used in conjugation with the NS2 gene for HCV treatment. However, further investigations are required to shed light upon the validity of using such a conjugate as a vaccine against HCV in vivo.
Supporting information
The TEM image of the first preparation procedure (S1 Table; Method 1) showed that these particles have semi spherical shape with average size of 423 nm; illustrated in (S1 Fig A). The TEM image of the second preparation procedure (S1 Table; Method 2) showed that these particles have aggregate complex shape with average size of 350 nm; illustrated in (S1 Fig B). The TEM image of the third preparation procedure (S1 Table; Method 3) showed that these particles have spherical shape with average size of 150.0 ± 2.0 nm; illustrated in (S1 Fig C).
(DOCX)
Particle size distribution measurements was in accordance with TEM imagine, where Particle size image using (Malvern Instruments, UK) of the prepared Gel.NPs (S1 Table; Method 1) showed that these particles have average size of 423 nm with polydispersity index 1.00; illustrated in (S2 Fig A). Particle size image of the prepared Gel.NPs (S1 Table; Method 2) showed that these particles have average size of 350 nm with polydispersity index 0.294; illustrated in (S2 Fig B). Particle size image of the prepared Gel.NPs (S1 Table; Method 3) showed that these particles have average size of 150 nm with polydispersity index 0.109; illustrated in (S2 Fig C).
(DOCX)
It was very important to get information about the capping charges around the Gel.NPs particles where the more positivity the more stability, Zeta potential measurement were done to evaluate the particle surface charge, where Zeta potential using (Malvern Instruments, UK) of the prepared Gel.NPs (S1 Table; Method 1) showed that these particles have +0.3 mV; illustrated in (S3 Fig A). Zeta potential of the prepared Gel.NPs (S1 Table; Method 2) showed that these particles have -21 mV; illustrated in (S3 Fig B). Zeta potential of the prepared Gel.NPs (S1 Table; Method 3) showed that these particles have +17.6 mV; illustrated in (S3 Fig C).
(DOCX)
ImageJ software was used to count bacterial replicates in confocal micrographs. Each bar represents mean ± SE of four independent experiments. As shown the number of bacterial colonies in the plate with bacteria transformed with recombinant NS2+Gel.NPs conjugate was higher than the number of bacterial colonies in the plate with bacteria transformed with recombinant NS2 gene alone. While no bacterial colonies in the plate with bacteria transformed with Gel.NPs alone.
(DOCX)
(DOCX)
(DOCX)
ImageJ software was used to count bacterial replicates in Confocal Laser Scanning Micrograph in Fig 5. Four plates were prepared for each type of transformation and confocal micrograph was captured for each plate.
(DOCX)
ImageJ software was used to count bacterial replicates in confocal micrographs.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Esmat G. Hepatitis C in the Eastern Mediterranean Region. East Mediterr Health J. 2013;19(7):587–8. [PubMed] [Google Scholar]
- 2.Elkady A, Tanaka Y, Kurbanov F, Sugauchi F, Sugiyama M, Khan A, et al. Genetic variability of hepatitis C virus in South Egypt and its possible clinical implication. J Med Virol. 2009;81(6):1015–23. Epub 2009/04/22. doi: 10.1002/jmv.21492 [DOI] [PubMed] [Google Scholar]
- 3.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13(17):2436–41. doi: 10.3748/wjg.v13.i17.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rustgi VK. The epidemiology of hepatitis C infection in the United States. J Gastroenterol. 2007;42(7):513–21. doi: 10.1007/s00535-007-2064-6 [DOI] [PubMed] [Google Scholar]
- 5.D'Ambrosio R, Colombo M. Should surveillance for liver cancer be modified in hepatitis c patients after treatment-related cirrhosis regression? Liver Int. 2016. doi: 10.1111/liv.13106 [DOI] [PubMed] [Google Scholar]
- 6.Kamal SM, El Tawil AA, Nakano T, He Q, Rasenack J, Hakam SA, et al. Peginterferon {alpha}-2b and ribavirin therapy in chronic hepatitis C genotype 4: impact of treatment duration and viral kinetics on sustained virological response. Gut. 2005;54(6):858–66. doi: 10.1136/gut.2004.057182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatti Z, Berenson CS. Adult systemic cat scratch disease associated with therapy for hepatitis C. BMC Infect Dis. 2007;7:8 doi: 10.1186/1471-2334-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris HE, Eldridge KP, Harbour S, Alexander G, Teo CG, Ramsay ME, et al. Does the clinical outcome of hepatitis C infection vary with the infecting hepatitis C virus type? J Viral Hepat. 2007;14(3):213–20. doi: 10.1111/j.1365-2893.2006.00795.x [DOI] [PubMed] [Google Scholar]
- 9.Gad RR, Males S, El Makhzangy H, Shouman S, Hasan A, Attala M, et al. Predictors of a sustained virological response in patients with genotype 4 chronic hepatitis C. Liver Int. 2008;28(8):1112–9. doi: 10.1111/j.1478-3231.2008.01750.x [DOI] [PubMed] [Google Scholar]
- 10.Doss W, Shiha G, Hassany M, Soliman R, Fouad R, Khairy M, et al. Sofosbuvir plus ribavirin for treating Egyptian patients with hepatitis C genotype 4. J Hepatol. 2015;63(3):581–5. doi: 10.1016/j.jhep.2015.04.023 [DOI] [PubMed] [Google Scholar]
- 11.Armstrong GL, Alter MJ, McQuillan GM, Margolis HS. The past incidence of hepatitis C virus infection: implications for the future burden of chronic liver disease in the United States. Hepatology. 2000;31(3):777–82. doi: 10.1002/hep.510310332 [DOI] [PubMed] [Google Scholar]
- 12.Gorzin Z, Gorzin AA, Tabarraei A, Behnampour N, Irani S, Ghaemi A. Immunogenicity evaluation of a DNA vaccine expressing the hepatitis C virus non-structural protein 2 gene in C57BL/6 mice. Iran Biomed J. 2014;18(1):1–7. doi: 10.6091/ibj.1231.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumoulin FL, von dem Bussche A, Li J, Khamzina L, Wands JR, Sauerbruch T, et al. Hepatitis C virus NS2 protein inhibits gene expression from different cellular and viral promoters in hepatic and nonhepatic cell lines. Virology. 2003;305(2):260–6. [DOI] [PubMed] [Google Scholar]
- 14.Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, et al. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 2010;6(12):e1001233 doi: 10.1371/journal.ppat.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popescu C-I, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, et al. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 2011;7(2):e1001278 doi: 10.1371/journal.ppat.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorzin AA, Ramsland PA, Tachedjian G, Gowans EJ. Identification of residues involved in NS2 homodimerization and elucidation of their impact on the HCV life cycle. J Viral Hepat. 2012;19(3):189–98. doi: 10.1111/j.1365-2893.2011.01504.x [DOI] [PubMed] [Google Scholar]
- 17.Lorenz IC. The Hepatitis C Virus Nonstructural Protein 2 (NS2): An Up-and-Coming Antiviral Drug Target. Viruses. 2010;2(8):1635–46. doi: 10.3390/v2081635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vile RG, Hart IR. In vitro and in vivo targeting of gene expression to melanoma cells. Cancer Res. 1993;53(5):962–7. [PubMed] [Google Scholar]
- 19.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949 Pt 1):1465–8. [DOI] [PubMed] [Google Scholar]
- 20.Wolff JA, Budker V. The mechanism of naked DNA uptake and expression. Adv Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X [DOI] [PubMed] [Google Scholar]
- 21.Lehrman S. Virus treatment questioned after gene therapy death. Nature. 1999;401(6753):517–8. doi: 10.1038/43977 [DOI] [PubMed] [Google Scholar]
- 22.Sun JY, Anand-Jawa V, Chatterjee S, Wong KK. Immune responses to adeno-associated virus and its recombinant vectors. Gene Ther. 2003;10(11):964–76. doi: 10.1038/sj.gt.3302039 [DOI] [PubMed] [Google Scholar]
- 23.Choi YS, Lee MY, David AE, Park YS. Nanoparticles for gene delivery: therapeutic and toxic effects. Mol Cell Toxicol. 2014;10(1):1–8. doi: 10.1007/s13273-014-0001-3 [Google Scholar]
- 24.Young S, Wong M, Tabata Y, Mikos AG. Gelatin as a delivery vehicle for the controlled release of bioactive molecules. Journal of Controlled Release. 2005;109(1–3):256–74. doi: 10.1016/j.jconrel.2005.09.023 [DOI] [PubMed] [Google Scholar]
- 25.Ofokansi K, Winter G, Fricker G, Coester C. Matrix-loaded biodegradable gelatin nanoparticles as new approach to improve drug loading and delivery. Eur J Pharm Biopharm. 2010;76(1):1–9. doi: 10.1016/j.ejpb.2010.04.008 [DOI] [PubMed] [Google Scholar]
- 26.Coester CJ, Langer K, van Briesen H, Kreuter J. Gelatin nanoparticles by two step desolvation—a new preparation method, surface modifications and cell uptake. J Microencapsul. 2000;17(2):187–93. doi: 10.1080/026520400288427 [DOI] [PubMed] [Google Scholar]
- 27.Kramer MF, Coen DM. Enzymatic amplification of DNA by PCR: standard procedures and optimization. Curr Protoc Mol Biol. 2001;Chapter 15:Unit-15.1 doi: 10.1002/0471142727.mb1501s56 [DOI] [PubMed] [Google Scholar]
- 28.Foox M, Zilberman M. Drug delivery from gelatin-based systems. Expert Opin Drug Deliv. 2015;12(9):1547–63. doi: 10.1517/17425247.2015.1037272 [DOI] [PubMed] [Google Scholar]
- 29.Su K, Wang C. Recent advances in the use of gelatin in biomedical research. Biotechnol Lett. 2015;37(11):2139–45. doi: 10.1007/s10529-015-1907-0 [DOI] [PubMed] [Google Scholar]
- 30.Shiehzadeh F, Tafaghodi M. Dry powder form of polymeric nanoparticles for pulmonary drug delivery. Curr Pharm Des. 2016. [DOI] [PubMed] [Google Scholar]
- 31.Brzoska M, Langer K, Coester C, Loitsch S, Wagner TO, Mallinckrodt C. Incorporation of biodegradable nanoparticles into human airway epithelium cells-in vitro study of the suitability as a vehicle for drug or gene delivery in pulmonary diseases. Biochemical and biophysical research communications. 2004;318(2):562–70. Epub 2004/05/04. doi: 10.1016/j.bbrc.2004.04.067 [DOI] [PubMed] [Google Scholar]
- 32.Jahanshahi M, Sanati MH, Babaei Z. Optimization of parameters for the fabrication of gelatin nanoparticles by the Taguchi robust design method. Journal of Applied Statistics. 2008;35(12):1345–53. doi: 10.1080/02664760802382426 [Google Scholar]
- 33.Jahanshahi M, Sanati MH, Hajizadeh S, Babaei Z. Gelatin nanoparticle fabrication and optimization of the particle size. physica status solidi (a). 2008;205(12):2898–902. doi: 10.1002/pssa.200824329 [Google Scholar]
- 34.Tseng C-L, Chen K-H, Su W-Y, Lee Y-H, Wu C-C, Lin F-H. Cationic Gelatin Nanoparticles for Drug Delivery to the Ocular Surface:In VitroandIn VivoEvaluation. Journal of Nanomaterials. 2013;2013:1–11. doi: 10.1155/2013/238351 [Google Scholar]
- 35.Zwiorek K, Bourquin C, Battiany J, Winter G, Endres S, Hartmann G, et al. Delivery by cationic gelatin nanoparticles strongly increases the immunostimulatory effects of CpG oligonucleotides. Pharmaceutical research. 2008;25(3):551–62. Epub 2007/10/04. doi: 10.1007/s11095-007-9410-5 [DOI] [PubMed] [Google Scholar]
- 36.Jadhav NR, Tone JS, Irny PV, Nadaf SJ. Development and characterization of gelatin based nanoparticles for targeted delivery of zidovudine. International journal of pharmaceutical investigation. 2013;3(3):126–30. Epub 2013/10/30. doi: 10.4103/2230-973X.119213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truong-Le VL, August JT, Leong KW. Controlled gene delivery by DNA-gelatin nanospheres. Hum Gene Ther. 1998;9(12):1709–17. doi: 10.1089/hum.1998.9.12-1709 [DOI] [PubMed] [Google Scholar]
- 38.Hosseinkhani H, Abedini F, Ou K-L, Domb AJ. Polymers in gene therapy technology. Polymers for Advanced Technologies. 2015;26(2):198–211. doi: 10.1002/pat.3432 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The TEM image of the first preparation procedure (S1 Table; Method 1) showed that these particles have semi spherical shape with average size of 423 nm; illustrated in (S1 Fig A). The TEM image of the second preparation procedure (S1 Table; Method 2) showed that these particles have aggregate complex shape with average size of 350 nm; illustrated in (S1 Fig B). The TEM image of the third preparation procedure (S1 Table; Method 3) showed that these particles have spherical shape with average size of 150.0 ± 2.0 nm; illustrated in (S1 Fig C).
(DOCX)
Particle size distribution measurements was in accordance with TEM imagine, where Particle size image using (Malvern Instruments, UK) of the prepared Gel.NPs (S1 Table; Method 1) showed that these particles have average size of 423 nm with polydispersity index 1.00; illustrated in (S2 Fig A). Particle size image of the prepared Gel.NPs (S1 Table; Method 2) showed that these particles have average size of 350 nm with polydispersity index 0.294; illustrated in (S2 Fig B). Particle size image of the prepared Gel.NPs (S1 Table; Method 3) showed that these particles have average size of 150 nm with polydispersity index 0.109; illustrated in (S2 Fig C).
(DOCX)
It was very important to get information about the capping charges around the Gel.NPs particles where the more positivity the more stability, Zeta potential measurement were done to evaluate the particle surface charge, where Zeta potential using (Malvern Instruments, UK) of the prepared Gel.NPs (S1 Table; Method 1) showed that these particles have +0.3 mV; illustrated in (S3 Fig A). Zeta potential of the prepared Gel.NPs (S1 Table; Method 2) showed that these particles have -21 mV; illustrated in (S3 Fig B). Zeta potential of the prepared Gel.NPs (S1 Table; Method 3) showed that these particles have +17.6 mV; illustrated in (S3 Fig C).
(DOCX)
ImageJ software was used to count bacterial replicates in confocal micrographs. Each bar represents mean ± SE of four independent experiments. As shown the number of bacterial colonies in the plate with bacteria transformed with recombinant NS2+Gel.NPs conjugate was higher than the number of bacterial colonies in the plate with bacteria transformed with recombinant NS2 gene alone. While no bacterial colonies in the plate with bacteria transformed with Gel.NPs alone.
(DOCX)
(DOCX)
(DOCX)
ImageJ software was used to count bacterial replicates in Confocal Laser Scanning Micrograph in Fig 5. Four plates were prepared for each type of transformation and confocal micrograph was captured for each plate.
(DOCX)
ImageJ software was used to count bacterial replicates in confocal micrographs.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.