Abstract
As the tissue macrophages of the CNS, microglia are critically involved in diseases of the CNS. However, it remains unknown what controls their maturation and activation under homeostatic conditions. We observed substantial contributions of the host microbiota to microglia homeostasis, as germ-free (GF) mice displayed global defects in microglia with altered cell proportions and an immature phenotype, leading to impaired innate immune responses. Temporal eradication of host microbiota severely changed microglia properties. Limited microbiota complexity also resulted in defective microglia. In contrast, recolonization with a complex microbiota partially restored microglia features. We determined that short-chain fatty acids (SCFA), microbiota-derived bacterial fermentation products, regulated microglia homeostasis. Accordingly, mice deficient for the SCFA receptor FFAR2 mirrored microglia defects found under GF conditions. These findings suggest that host bacteria vitally regulate microglia maturation and function, whereas microglia impairment can be rectified to some extent by complex microbiota.
Introduction
Microglia are the tissue macrophages of the brain and are crucial for maintaining tissue homeostasis and for scavenging of pathogens, dying cells, and molecules through microbial-associated molecular pattern receptor dependent and independent mechanisms1, 2, 3. In addition to their established role in host defense, microglia are important for proper brain development. In the brain, microglia are also critical for synaptic pruning and remodeling during development and adulthood4.
Given the importance of microglia in physiological brain function, it is not surprising that an increasing number of microglia-related genes have been found to be associated with neuropsychiatric or neurological disorders in humans. These include roles for CD33 in Alzheimer's disease5, 6, TREM2 in frontotemporal dementia7 and CSF1R in hereditary diffuse leukencephalopathy8. In addition, mice lacking CX3CR1, a chemokine receptor expressed on microglia, exhibit defective neuron-microglia signaling, which results in impaired functional brain connectivity and altered social behavior9. There is a critical knowledge gap in the environmental cues that shape microglia function under homeostatic and disease conditions. With the rising incidence of neurodegenerative and neuropsychiatric disorders, ‘microgliopathies’, in which microglia dysfunction is the primary disease-causing condition, are beginning to garner increased attention3.
Despite their localization in the CNS, microglia are ontogenetically and functionally most closely related to the mononuclear phagocytic system in the body, specifically tissue macrophages and circulating myeloid cells10. Similar to other tissue macrophages, microglia develop early during embryogenesis from immature yolk sac progenitors11, 12, 13. In contrast with most of their tissue relatives, microglia persist throughout the entire life of an organism without any substantial input from circulating blood cells as a result of their longevity and self-renewal capacities14, 15. Thus, microglia have several features that are unique from other tissue macrophages that may be attributed to their seclusion behind the blood-brain barrier (BBB), which ensures CNS integrity16.
Despite the brains' separation from the rest of the body, numerous studies have highlighted interactions between the CNS and the gastrointestinal system. Bacterial colonization of the intestine has a major role in the postnatal development and maturation of the endocrine system17, which underpins CNS function and is of particular relevance to neurochemical (for example, serotonergic) system development18. For example, GF mice exhibit exaggerated stress responses19, 20. These results support the idea that brain levels of several neurochemical mediators that are important for neuronal function are under the influence of the host microbiota. However, the interaction of the gut microbiota with the CNS resident immune system, such as microglia, has not been investigated.
The gut microbiota influences the cellular proportions, migration and functions of various immune cell subsets21. Recent studies have provided numerous examples illustrating how gut bacteria modulate innate and adaptive immune responses at mucosal surfaces during infection, inflammation and autoimmunity22, 23. Even outside of the gut, microbiota regulates immune responses that influence organ-specific autoimmunity in animal models of rheumatoid arthritis and type I diabetes24, 25. Autoimmune inflammation of the brain may also be dependent on the presence of a complex microbiota26. In a mouse model of multiple sclerosis, the gut microbiota was essential for triggering peripheral immune processes, leading to a relapsing-remitting autoimmune disease driven by myelin-specific CD4+ T cells26. Gut microbiota metabolites may stimulate CD4+ T cells to achieve their encephalitogenic properties outside of the brain, for example, in the lymphoid tissues of the intestine or in the local draining lymph nodes, and gut bacteria influence innate immune cell population frequencies outside of the CNS. Whether the gut microbiota can shape the maturation and maintenance of the innate immune system in the non-diseased brain is unclear.
We found that a complex gut microbiota promoted the maintenance of microglia under steady-state conditions. The absence of a complex host microbiota led to defects in microglia maturation, differentiation and function. By controlling microglia innate immune function, gut microbiota prepares the brain to rapidly mount immune responses following pathogen or danger encounter, as GF and antibiotic-treated mice were impaired in microglia function. Our results indicate that environmental microbes contribute to actively shaping CNS immunity at its core, the ‘never resting’ microglia.
Results
GF animals display global defects in microglia
Initially, to determine whether host microbiota have any effects on the brain innate immune system under steady-state conditions, we profiled microglia from brains of colonized (specific pathogen free, SPF) and GF animals. In general, GF animals exhibited enlarged caeca with increased weights and dark-colored cecal contents as a consequence of their absent gut microbiome, as described previously27 (Fig. 1a). In flow cytometry–isolated CD11b+ CD45lo microglia from seven biological replicates of SPF and GF mice, we measured the genome-wide mRNA expression profiles by quantitative deep sequencing of the RNA transcripts (RNA-seq; Fig. 1b). We observed marked differences in the mRNA profiles of microglia genes between SPF and GF animals. In total, 198 genes were significantly downregulated and 173 genes were upregulated (P < 0.01 and twofold change) in microglia from GF mice compared with colonized SPF controls. Among the downregulated transcripts in microglia from non-colonized GF animals, we identified many genes that were linked to cell activation such as Mapk8, Fcgr2β, IL-1α, Ly86, Cd86 and Hif1α (Fig. 1c,d and Supplementary Table 1). Genes that are required for signaling of the type I IFN receptors, such as Janus kinase 3 (Jak3) and signal transducer and activator of transcription 1 (Stat1), were also diminished in GF microglia. As a sign of premature immune function of microglia, the MHC class I related β2 microglobulin (B2m) was also found to be reduced. Moreover, TRIM family members such as Trim30α were downregulated. TRIM proteins are involved in pathogen-recognition and regulation of transcriptional pathways in host defense. In contrast, some upregulated genes expressed in microglia from GF mice were inhibitors of transcription, such as Nfkbiα (which encodes IkBα) (Fig. 1b). Notably, the central microglia transcription and survival factors Sfpi1 (encodes PU.1) and Csf1r, which are downregulated in mature adult microglia13, 28, were significantly upregulated in microglia from GF mice (Csf1r, P = 0.0001; Sfpi1, P = 0.0075; Fig. 1e,f, Supplementary Fig. 1a and Supplementary Table 1). Prominently, several genes associated with either activation of cell proliferation (for example, Iqgap1, DNA damage-inducible transcript 4 (Ddit4, also known as Redd1), stimulation of cell cycle (for example, Cdk9 and Ccnd3) or inhibition of apoptosis (Bcl2) were substantially increased under GF conditions (Fig. 1b,e). In fact, the most amplified gene found in microglia from GF mice was Ddit4 (11.2 ± 3.2–fold elevated compared with SPF microglia). DDIT4 fundamentally regulates cell growth, proliferation and survival via inhibition of the activity of the mammalian target of rapamycin complex (mTORC) 1 and activation of mTORC2 (ref. 29). Furthermore, genes related to epigenetic modifications were also found to be dysregulated in microglia from GF mice (Supplementary Fig. 1b).
Figure 1. Altered microglial gene profile and immaturity in GF animals.
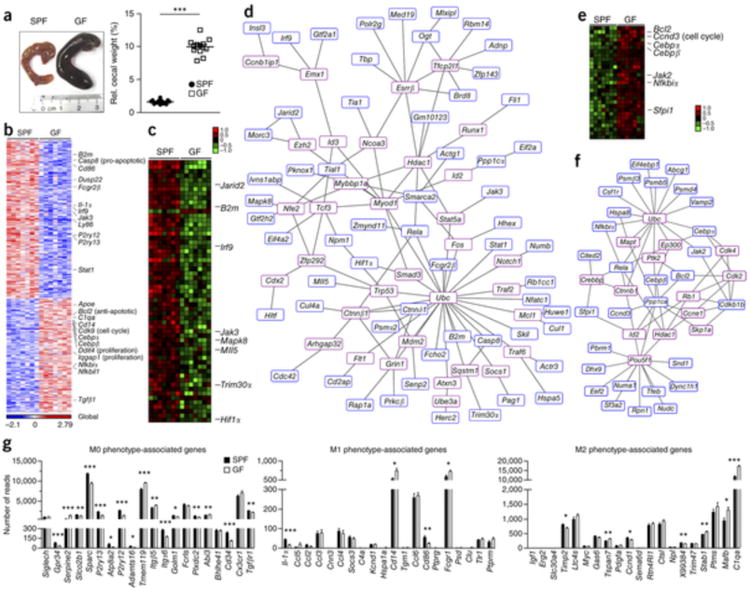
(a) Left, photograph of caeca from SPF (control) and GF mice. Representative pictures are shown, with ruler for scaling. Right, relative cecal weight (to body weight) of SPF and GF mice. Symbols represent individual mice. 12 mice per group were examined. Data are representative of three independent experiments. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test, ***P = 0.0001. (b) RNA-seq analysis. Gene expression data shown was either induced or reduced by a factor of at least 2 (P < 0.01, unpaired t test) in microglia from SPF or GF mice. Representative genes are noted on the right. Each column represents microglia data from one individual mouse, with seven mice per group. Color code presents linear scale. (c) mRNA expression profile of genes that were at least 1.5-fold (P < 0.05, unpaired t test) downregulated in microglia from GF compared to SFP mice. Color code shows linear values. Representative genes are noted on the right. (d) Functional networks of genes that were downregulated (at least 1.5-fold, P < 0.05, unpaired t test) in microglia from GF compared to SPF mice. Blue framed genes were found to be downregulated whereas red framed genes were automatically predicted. (e) Expression profile of genes that were at least 1.5-fold (P < 0.05, unpaired t test) upregulated in microglia from GF compared with SFP mice. Representative genes are noted on the right. Color code presents linear scale. (f) Functional networks of upregulated genes (at least 1.5-fold, P < 0.05, unpaired t test). Blue-framed genes indicate upregulated molecules whereas red-framed genes were automatically predicted. (g) mRNA expression values (number of reads) of genes from microglia in GF (white bars) or SPF (black bars) mice were categorized according to the M0, M1 or M2 phenotypes, as described previously32. Bars represent means ± s.e.m. with seven samples per group. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: Gpr34, 0.0005; Serpine2, <0.0001; Slco2b1, 0.0078; Sparc, <0.0001; P2ry13, 0.0001; Atp8a2, 0.0239; P2ry12, <0.0001; Adamts16, 0.0105; Tmem119, 0.0002; Itgb5, 0.0052; Itga6, 0.0003; Golm1, 0.0125; Plxdc2, 0.006; Abi3, 0.0037; Cd34, 0.0001; Tgfbr1, 0.0035; Il-1α, 0.0002; Cd14, 0.0109; Cd86, 0.004; Fcgr1, 0.0281; Timp2, 0.0144; Tspan7, 0.0074; Ccnd1, 0.0378; X99384, 0.0077; Stab1, 0.0093; Mafb, 0.0257; C1qa, 0.0001.
Microglia can achieve polarized states that are characterized by specific genetic signatures that allow the distinction of M0, M1 and M2 microglia, respectively3, 30, 31, 32. Under GF conditions, we found no bias toward a specific type of microglia polarization (Fig. 1g). M1- and M2-related genes were only marginally changed, whereas most differently regulated genes were found to localize in the M0 cluster, indicating that microglia steady-state condition was severely altered in the absence of microbiota.
We next measured levels of several microglia maturation and activation factors by flow cytometry (Fig. 2). In general, the surface molecules CSF1R, F4/80 and CD31 are strongly developmentally regulated with decreasing levels during maturation13. Notably, the percentage of CSF1R+ (CD115), F4/80+ and CD31+ microglia, as well as the mean fluorescence intensities (MFIs) of CSF1R and F4/80, were increased under GF conditions, whereas the activation molecules CD44, CD62L and MHC class II were not regulated, suggesting that GF microglia had an immature phenotype. In conclusion, comparison between the genome signatures and surface molecules of microglia from colonized (SPF) and non-colonized (GF) animals suggests that commensal microbes are important for shaping microglia maturation and steady-state conditions.
Figure 2. Increased expression of maturation and activation marker in GF microglia.
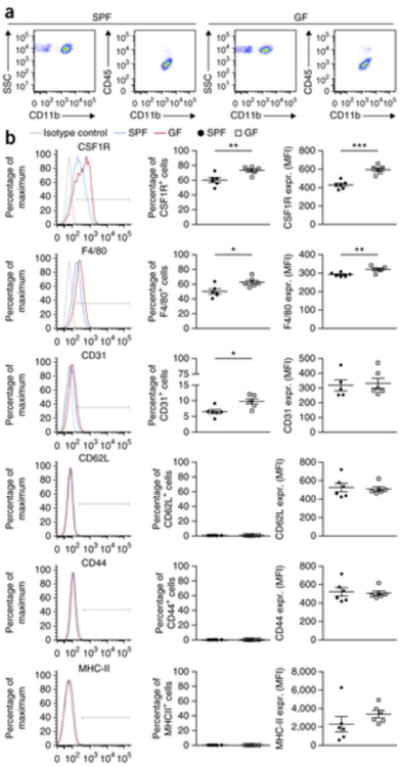
(a) Gating strategy for flow cytometric analysis of CD11 b+ CD45lo microglia from GF and SFP mice. Representative dot plots obtained from three independent experiments are shown. SSC, side scatter. (b) Representative cytometry graphs of the maturation and activation marker CSF1R, F4/80, CD31, CD44, CD62L and MHC class II on microglia from GF mice (red lines), SFP mice (blue lines) and isotype controls (gray lines). In addition, quantifications of the percentages (%) of positively labeled cells and MFIs are depicted. Each symbol represents data from one mouse, with six investigated mice per group. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). Data are representative of three independent experiments. P values: CSF1R (percentage of positive cells), 0.0068; CSF1R (MFI), <0.0001; F4/80 (percentage of positive cells), 0.0134; F4/80 (MFI), 0.0047; CD31 (percentage of positive cells), 0.0103.
To further investigate the mechanism of microglia immaturity when derived from sterile animals, we performed a thorough histopathological analysis of different brain regions from adult GF and SPF mice (Fig. 3a). GF mice exhibited no abnormalities of the CNS gross anatomy on a histological level. Notably, the amount of NeuN+ neurons, GFAP+ astrocytes and Nogo-A+ oligodendrocytes was not altered, indicating a primary effect in the immune cell compartment of the CNS (Supplementary Figs. 2 and 3). Brains of GF mice did not display any obvious lymphocytic infiltrates and were devoid of any perivascular inflammatory cuffs under homeostatic conditions (Supplementary Fig. 4). We did not observe any overt accumulations of microglia. However, there were significantly more Iba-1+ microglia in all of the brain regions investigated in GF mice (cortex, P = 0.0024; corpus callosum, P = 0.0008; hippocampus, P = 0.0073; olfactory bulb, P = 0.0092; cerebellum, P = 0.0246; Fig. 3a,b). Increased microglia cell numbers were not restricted to the gray matter and were also present in subcortical white matter regions (Fig. 3b and Supplementary Fig. 5a). Accordingly, the expression level of the cell cycle and proliferation-promoting gene Ddit4 was increased (Fig. 3c) and the number of proliferating Iba-1+ Ki67+ microglia was elevated under GF conditions (Fig. 3d,e). Semi-automatic quantitative morphometric three-dimensional measurements of microglia revealed significantly longer processes and increased numbers of segments, branching and terminal points. These morphological observations support that marked changes occurred in microglia in GF mice (dendrite length, P = 0.0238; number of segments, P = 0.0238; number of branch points, P = 0.0209; number of terminal points, P = 0.027; volume, P = 0.0413; Fig. 3f,g). GF microglia frequently displayed physical contacts with adjacent microglia and therefore did not respect neighboring cell territories, as observed under SPF conditions (Supplementary Fig. 5b). Because homeostasis and development of microglia are dependent on specific survival factors, such as IL-34, CSF-1, TGFβ1 or the transcription factor PU.1 (refs. 11, 12, 13,32,33), we measured the expression of these molecules in different brain regions of GF mice by quantitative PCR (Supplementary Fig. 5c). In contrast with a recent study34, the absence of a complex microbiota did not result in decreased levels of Csf1 mRNA or other survival factors in the brain. We found that cytokine and growth factor levels were normal or even increased, suggesting that global microglia defects in GF mice are not a result of reduced production of the CSF1R ligands or Tgfβ1 expression by neuroectodermal cells.
Figure 3. Lack of microbes impairs microglia morphology and disturbs cellular network.
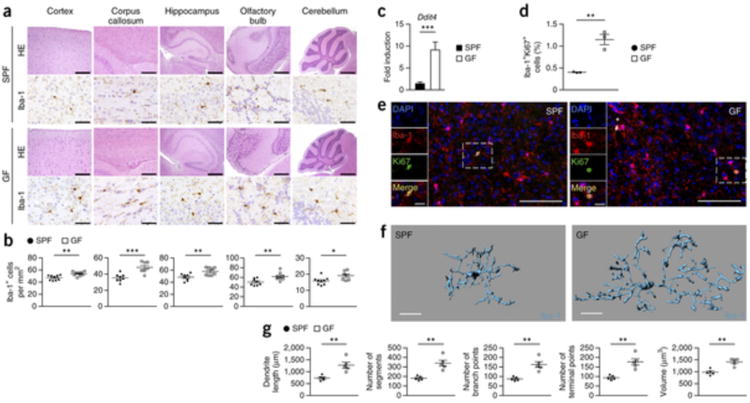
(a) CNS histology of several brain regions that were stained with hematoxylin and eosin (H&E) or subjected to immunohistochemistry for Iba-1 to detect microglia. Scale bars represent 200 μm (H&E, cortex and corpus callosum), 500 μm (H&E, hippocampus and olfactory bulb), 1 mm (H&E, cerebellum) and 50 μm (Iba-1). Representative pictures from nine mice per group are displayed. (b) Number of Iba-1+ ramified parenchymal microglia in different localizations of the CNS. Each symbol represents data from one mouse, with nine mice per group. Three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: cortex, 0.0024; corpus callosum, 0.0008; hippocampus, 0.0073; olfactory bulb, 0.0092; cerebellum, 0.0246. (c) Expression of Ddit4 mRNA measured by qRT-PCR in microglia isolated from SPF (black bar) or GF (white bar) mice. Data are presented as mean ± s.e.m. with five samples in each group. Significant differences were determined by an unpaired t test (***P = 0.0002). Data are representative of two independent experiments. (d) Quantification of proliferating Iba-1+ Ki67+ double-positive parenchymal microglia was performed on cortical brain slices. Each symbol represents one mouse, with three mice per group. Three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (**P = 0.0033). (e) Fluorescence microscopy of Iba-1+ (red) microglia, the proliferation marker Ki67 (green) and DAPI (4′,6-diamidino-2-phenylindole, blue). Overview and magnification are shown. Scale bars represent 100 μm (overview) and 20 μm (inset). (f,g) Three-dimensional reconstruction (scale bars represent 15 μm, f) and Imaris-based automatic quantification of cell morphometry (g) of cortical Iba-1+ microglia. Each symbol represents one mouse with at least three measured cells per mouse. Five mice per group were analyzed. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (**P < 0.01). P values: dendrite length, 0.0035; number of segments, 0.0012; number of branch points, 0.0012; number of terminal points, 0.0012; volume, 0.0011.
Lack of microbes diminishes microglia immune response
Microglia are the first responders to bacterial or viral infections of the brain, mediating early pathogen control and coordinating downstream immune reactions2, 35. To examine the functional consequences of microglial immaturity and malformation in mice lacking microbes, we employed a lipopolysaccharide (LPS) stimulation protocol that leads to a strongly activated CNS innate immune response36. SPF and GF mice were challenged intracerebrally (i.c.) or intraperitoneally (i.p.) with the TLR4 agonist LPS and microglia response was studied 6 h thereafter. First, microglia from i.c. injected mice were isolated and whole-genome gene expression was determined by an Affymetrix Genome 430A 2.0 array. In general, 1,683 genes were similarly modulated under both conditions compared with phosphate-buffered saline (PBS) controls (Fig. 4a). Notably, microglia from SPF animals regulated 481 individual genes, whereas microglia from GF mice exhibited expression changes in 273 unique genes. Statistical analysis of the most differently expressed genes between both housing conditions revealed 21 and 57 significantly (P < 0.01) and at least twofold up- and downregulated genes in microglia from GF and SPF mice, respectively, 6 h after LPS treatment compared with PBS controls (Fig. 4b). Subsequent investigation of the gene ontology enrichment network on differentially expressed genes revealed that mostly cytokine and chemokine pathways were affected in GF microglia, leading to a strongly reduced repertoire of the innate immune response (Supplementary Fig. 6), a finding that was clearly evident in the expression levels of the 78 differently expressed genes (Supplementary Table 2). Gene array data were confirmed by quantitative real-time PCR (qRT-PCR) analysis of the LPS-induced genes (Fig. 4c). The cell cycle gene cyclin D2 (Ccnd2) was only weakly induced in GF microglia compared with SPF controls. In contrast, genes associated with, but not specific for, early myeloid cell activation, such as S100a4, S100a6, S100a8 and S100a10, showed increased expression in GF microglia. Similarly, i.p. injection of LPS resulted in an attenuated induction of several pro-inflammatory genes (Supplementary Fig. 7). Subsequent investigation of microglia morphology in GF mice revealed malformed microglia that failed to display the activated morphology with rounded perikarya and small processes that stimulated SPF microglia exhibit (Fig. 4d,e). However, less-extensive differences were observed as compared with the non-challenged microglia (Fig. 3f,g).
Figure 4. Diminished microglia response to infection under GF conditions.
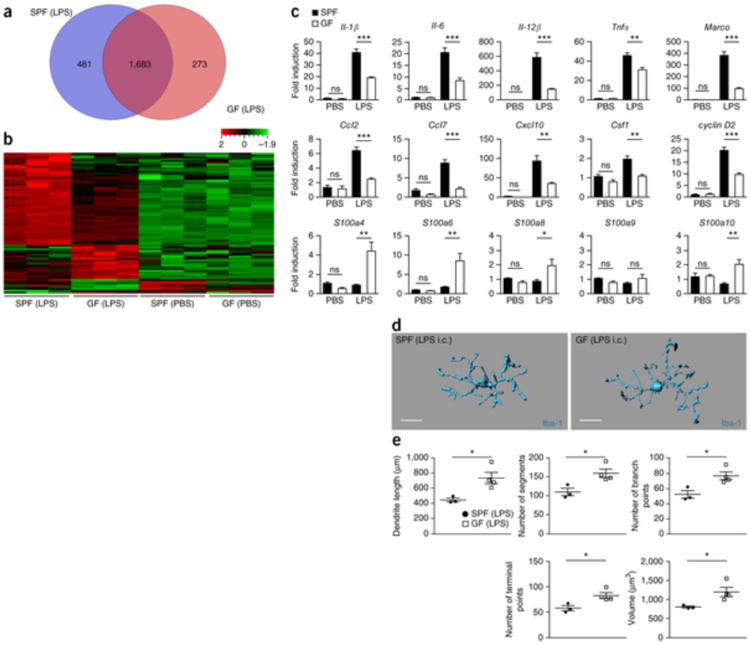
(a) Venn diagram depicting the different regulated and overlapping genes between sorted microglia from GF and SPF animals (P < 0.01) 6 h after LPS treatment compared with PBS-treated controls of the same housing conditions (GF/SPF). (b) Heat map of the mean centered and s.d. scaled expression values for genes that were significantly and at least twofold up- or downregulated in GF compared with SPF microglia 6 h after i.c. treatment with LPS. Only genes that were also significantly up- or downregulated by LPS treatment compared with PBS-treated controls of the same housing conditions (GF and SPF, respectively) were included to account for differences in basal gene regulation. Expression levels exceeding the mean value are colored in red and expression levels below the mean are colored in green (standardized and scaled to linear expression). Values close to the median are colored black. Random variance two-sample t test as implemented in BRB-Tools was performed to test significance at P < 0.01. (c) qRT-PCR in microglia 6 h after i.c. LPS exposure. Data are expressed as the ratio of the mRNA expression compared with endogenous Actb relative to SPF controls and are presented as mean ± s.e.m. At least three mice per group were analyzed. Data are representative of two independent experiments. Significant differences were examined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). ns, not significant. P values: PBS: Il-1β, 0.2324; Il-6, 0.6569; Il-12β, 0.0608; Tnfα, 0.7485; Marco, 0.2415; Ccl2, 0.8035; Ccl7, 0.1757; Cxcl10, 0.2138; Csf1, 0.1224; cyclin D2, 0.8405; S100a4, 0.1279; S100a6, 0.1169; S100a8, 0;1169; S100a9, 0.2677; S100a10, 0.8502. LPS: Il-1β, 0.0001; Il-6, 0.0005; Il-12β, 0.0001; Tnfα, 0.0050; Marco, 0.0001; Ccl2, 0.0001; Ccl7, 0.0003; Cxcl10, 0.0055; Csf1, 0.0028; cyclin D2, 0.0003; S100a4, 0.0033; S100a6, 0.0050; S100a8, 0.0425; S100a9, 0.2136; S100a10, 0.0028. (d,e) Three-dimensional reconstruction (scale bar represents 15 μm, d) and Imaris-based automatic quantification of cell morphometry (e) of cortical Iba-1+ microglia 6 h after i.c. treatment with LPS. Each symbol represents one mouse with at least three measured cells per mouse. Three SPF and four GF animals were investigated. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05). P values: dendrite length, 0.0238; number of segments, 0.0238; number of branch points, 0.0209; number of terminal points, 0.027; volume, 0.0413.
Next, we challenged GF mice with lymphocytic choriomeningitis virus (LCMV) i.c. and examined microglia expansion after 4 d of viral exposure (Fig. 5a). Microglia from SPF mice responded with a robust increase of cell density, whereas the virus-induced microgliosis was less pronounced under GF conditions. Gene expression profile comparisons revealed major differences between GF and SPF microglia (Fig. 5b). Quantitative real-time PCR analysis of LCMV-induced genes confirmed that there was a disturbed innate immune response following viral exposure (Fig. 5c). The expression of regulators of microglia cell proliferation, differentiation, activation and transformation, such as c-Jun, c-Fos, FosB and cyclin B2, and pro-inflammatory molecules, such as Tnf-α and Il-1β, were significantly changed (c-Jun, P = 0.0042; c-Fos, P = 0.0016; FosB, P = 0.0008; cyclin B2, P = 0.0491; Tnfα, P = 0.0072; Il-1β, P = 0.0315). Impairment of microglial immune response under GF condition was accompanied by prominent morphological differences following virus infection (Fig. 5d,e). These data collectively suggest that, in the absence of intestinal microbes, the innate immune response of microglia is strongly diminished and host microbiota promote microglial resistance against bacterial and viral challenge.
Figure 5. Reduced microglia response to viral infection under GF conditions.
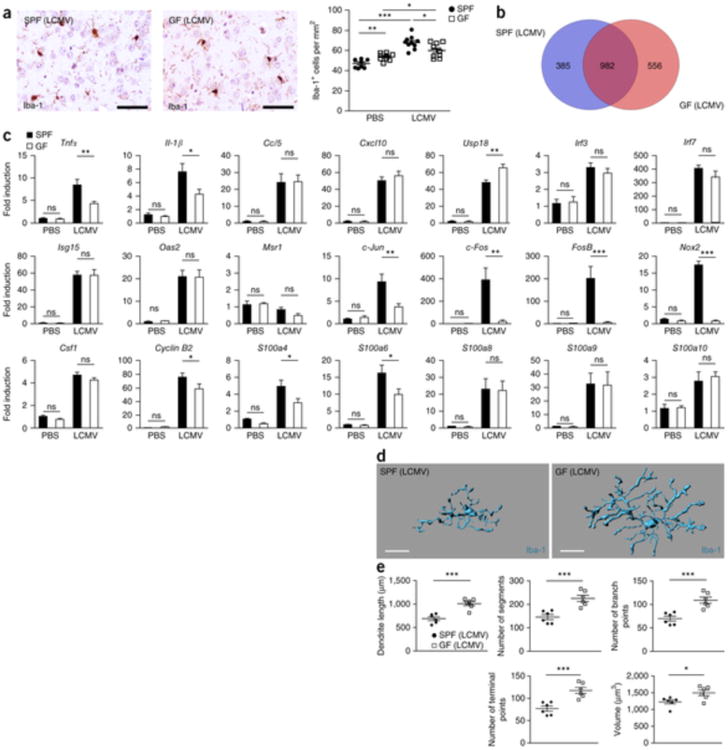
(a) Iba-1 immunhistochemistry depicting cortical microglia response 4 d after i.c. challenge with LCMV (left) and quantification (right). Each symbol represents one mouse, with nine PBS-treated control mice and ten LCMV-treated mice. Three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: SPF (PBS) versus GF (PBS): 0.0024; SPF (PBS) versus SPF (LCMV): 0.0001; GF (PBS) versus GF (LCMV): 0.0323; SPF (LCMV) versus GF (LCMV): 0.0222. Scale bars represent 50 μm. (b) Venn diagram illustrating the differences and overlaps between microglia from GF and SPF animals in significantly (P < 0.01) up- or down-regulated genes 4 d after LCMV treatment compared with PBS-treated controls of the same housing conditions (GF/SPF). (c) qRT-PCR in microglia 4 d after i.c. LCMV exposure. Data are expressed as the ratio of the mRNA expression compared with endogenous Actb relative to SPF controls and show mean ± s.e.m. Ten LCMV-treated and three PBS-control mice were analyzed for both housing conditions, respectively. Data are representative of two independent experiments. Significant differences were examined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). ns, not significant. P values: PBS: Tnfα, 0.5082; Il-1β, 0.5025; Ccl5, 0.7325; Cxcl10, 0.8510; Usp18, 0.8331; Irf3, 0.7942; Irf7, 0.4691; Isg15, 0.5344; Oas2, 0.0738; Msr1, 0.8069; c-Jun, 0.3162; c-Fos, 0.2778; FosB, 0.2491; Nox2, 0.3746; Csf1, 0.1224; cyclin B2, 0.1297; S100a4, 0.1463; S100a6, 0.1296; S100a8, 0.2677; S100a9, 0.4182; S100a10, 0.8502. LCMV: Tnfα, 0.0072; Il-1β, 0.0315; Ccl5, 0.9502; Cxcl10, 0.3839; Usp18, 0.0013; Irf3, 0.3887; Irf7, 0.2704; Isg15, 0.9575; Oas2, 0.9831; Msr1, 0.0579; c-Jun, 0.0042; c-Fos, 0.0016; FosB, 0.0008; Nox2, 0.0001; Csf1, 0.2828; cyclin B2, 0.0491; S100a4, 0.0415; S100a6, 0.0374; S100a8, 0.9149; S100a9, 0.9459; S100a10, 0.6775. (d,e) Three-dimensional reconstruction (scale bars represent 15 μm, d) and Imaris-based automatic quantification of cell morphometry (e) of cortical Iba-1+ microglia 4 d after i.c. LCMV exposure. Each symbol represents one mouse with at three measured cells per mouse. Six animals were analyzed per group. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, ***P < 0.001). P values: dendrite length, 0.0003; number of segments, 0.0009; number of branch points, 0.0008; number of terminal points, 0.0009; volume, 0.0234.
Depletion of intestinal bacteria influences microglia
GF mice have never been exposed to a complex microbiota and therefore microglia prenatal development might also be affected in those mice. To clarify whether microglia in conventionally colonized mice require a continuous input from their resident microbes, we treated SPF mice orally with broad-spectrum antibiotics for 4 weeks. As expected, depletion of the intestinal microbiota induced markedly enlarged caeca (Fig. 6a). Thus, the caeca from mice that were exposed to broad-spectrum antibiotics macroscopically resembled those of GF mice, as noted previously27.
Figure 6. Antibiotic treatment induces immature and malformed microglia.
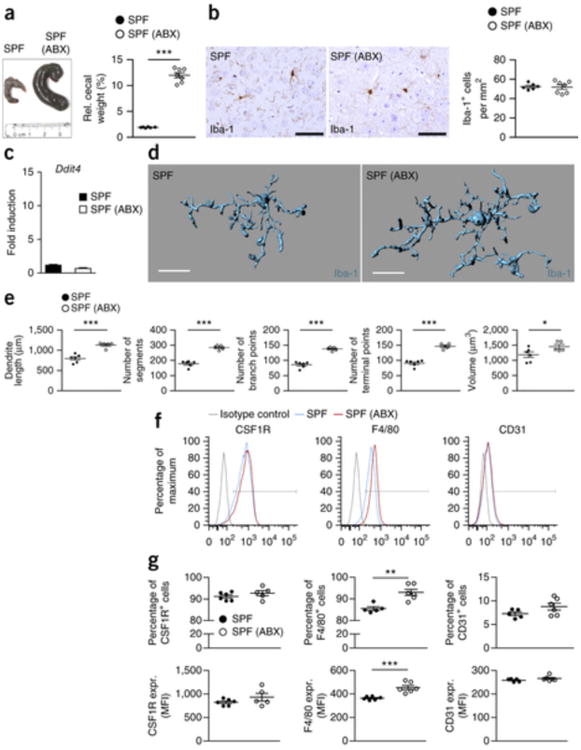
(a) Left, gross morphology of cecum in untreated and antibiotic-treated (ABX) SPF mice. Ruler scale is shown. One representative picture of six investigated control and seven ABX-treated mice is shown. Right, relative cecum weight. Symbols represent individual mice. Data are presented as mean ± s.e.m. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test (***P = 0.0001). (b) Immunhistochemistry for Iba-1+ parenchymal microglia (left) and quantification (right) in the cortex of seven ABX-treated and six untreated (control) SPF mice. Depicted symbols represent individual mice. Three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Data are representative of two independent experiments. Scale bars represent 50 μm. (c) Expression of Ddit4 mRNA measured by qRT-PCR in microglia isolated from untreated SPF (black bar) or antibiotic-treated SPF (white bar) mice. Data are presented as mean ± s.e.m. with at six samples in each group are displayed. No significant differences were detectable by an unpaired t test. Data are representative of two independent experiments. (d,e) Three-dimensional reconstruction (scale bars represent 15 μm, d) and Imaris-based automatic quantification of cell morphometry (e) of cortical Iba-1+ microglia in five ABX-treated and six untreated mice. Each symbol represents one mouse per group with at least three measured cells per mouse. Data are presented as mean ± s.e.m. Significant differences were evaluated by an unpaired t test (*P < 0.05, ***P < 0.001). P values: dendrite length, 0.0001; number of segments, <0.0001; number of branch points, <0.0001; number of terminal points, 0.0001; volume, 0.0486. (f) Representative cytometry graphs of the maturation markers CSF1R, F4/80 and CD31 on microglia from ABX-treated mice (red lines), untreated SFP controls (blue lines) and isotype controls (gray lines). Data are representative of two independent experiments. (g) Percentages and MFI of CSF1R, F4/80 and CD31 on microglia. Each symbol represents one mouse, with six animals per group. Data are presented as mean ± s.e.m. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test. F4/80 (percentage of positive cells), **P = 0.0021; F4/80 (MFI), ***P = 0.0008.
Overall, in antibiotics-treated animals, the immature phenotype of microglia largely mirrored the results obtained in GF animals. However, the number of microglia was not changed by the antibiotic treatment (Fig. 6b). Consistent with this observation, the cell cycle molecule Ddit4 was not altered (Fig. 6c). Direct immunofluorescence visualization of Iba-1–labeled microglia and subsequent semi-automatic morphometric measurements of individual cells showed statistically significant changes of cell morphology, such as dendrite length, number of branching points and segments, and cell volume, reminiscent of the malformed microglia seen in GF animals (dendrite length, P = 0.0001; number of segments, P < 0.0001; number of branch points, P < 0.0001; number of terminal points, P = 0.0001; volume, P = 0.0486; Fig. 6d,e). Furthermore, flow cytometric analysis revealed an increase of the microglia maturation marker F4/80, whereas expression levels for CSF1R and CD31 displayed an increasing trend, but did not reach substantial changes (Fig. 6f,g). Collectively, these data suggest that a continuous contribution of intestinal microbes is critical for the homeostasis of microglia under steady-state conditions.
Reduced complexity of microbiota impairs microglia
We next addressed the question of whether the full repertoire of microbes found in SPF mice is required for microglia homeostasis. Depending on the analysis method, SPF mice usually harbor between 400 and 1,000 bacterial strains in their gut microbiota37. Starting with GF mice, Schaedler and colleagues carried out experiments in the 1960s in which they reintroduced consortia of defined intestinal bacteria38. We examined mice colonized with three strains of the altered Schaedler flora: Bacteroides distasonis (strain ASF 519), Lactobacillus salivarius (strain ASF 361) and Clostridium cluster XIV (ASF 356)37. We took advantage of this tri-colonized mouse line that macroscopically displayed GF-like caeca (Fig. 7a) and examined their microglia. Iba-1+ microglia were readily visible in the cortex of the tri-colonized ASF mice and were, as in GF conditions, present in higher numbers (Fig. 7b). Consistent with this observation Ddit4 mRNA was significantly increased in microglia from the tri-colonized ASF mice (P = 0.0001; Fig. 7c). Similar to our findings in GF mice, microglia from the tri-colonized ASF mice displayed major alterations in their cell morphology, indicative of an immature phenotype (Fig. 7d,e), which suggests that a host microbiota with limited diversity is not sufficient to induce comprehensive microglia maturation.
Figure 7. Only complex microbiota can restore microglia insufficiency.
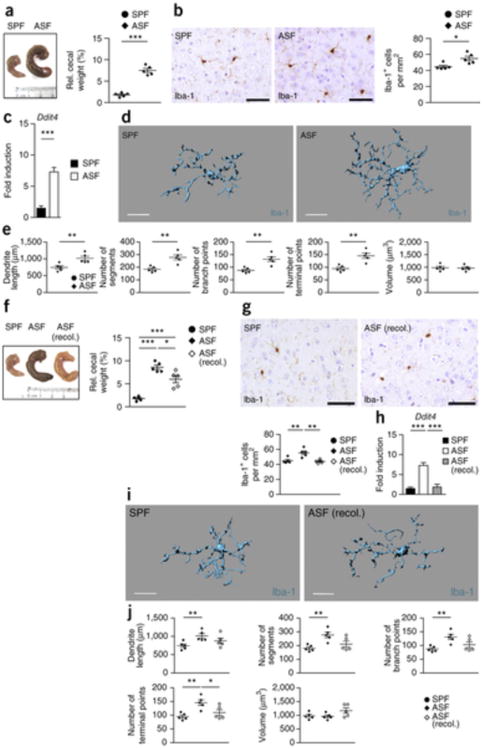
(a) Left, macroscopical view on cecum from SFP animal (control) and an individual with altered Schaedler flora (ASF). Ruler scale is shown. Representative pictures are displayed. Right, relative cecum weight of ASF and control SPF mice. Symbols represent individual mice, with five examined mice per group. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (***P = 0.0001). Data are representative of two independent experiments. (b) Iba-1+ immunohistochemistry (left) in the cortex of ASF and control mice and quantification thereof (right). Every symbol represents one mouse. Five mice per group were investigated and three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Significant differences were evaluated by an unpaired t test and marked with asterisks (*P = 0.0143). Data are representative of two independent experiments. Scale bars represent 50 μm. (c) Ddit4 mRNA measured by qRT-PCR in microglia from SPF (black bar) or ASF (white bar) mice. Data are presented as mean ± s.e.m. with at least five samples in each group. Significant differences were determined by an unpaired t test (***P = 0.0001). Data are representative of two independent experiments. (d,e) Three-dimensional reconstruction (d) of representative Iba-1+ parenchymal microglia from ASF or SPF control mice, respectively. Scale bars represent 15 μm. Automatic Imaris-based quantification of cortical microglia morphology (e) is shown. Each symbol represents one mouse per group with at least three measured cells per mouse. Five animals per group were analyzed. Data are presented as mean ± s.e.m. Significant differences were evaluated by an unpaired t test (**P < 0.01). P values: dendrite length, 0.0099; number of segments, 0.0025; number of branch points, 0.0031; number of terminal points, 0.0022. (f) Left, macroscopical view of cecum from SPF (control), ASF and recolonized ASF mice. Ruler scale is shown. Representative pictures are shown. Right, relative cecum weight of ASF, recolonized ASF and control SPF mice. Symbols represent individual mice, with five mice per group. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, ***P < 0.001). P values: SPF versus ASF, 0.0001; SPF versus ASF (recol.), 0.0008; ASF versus ASF (recol.), 0.0202. Data are representative of two independent experiments. (g) Top, immunohistochemistry for Iba-1+ in the cortex of recolonized ASF and control mice. Bottom, quantification. Three to four sections per mouse were examined. Every symbol represents one mouse, with five mice per group. Data are presented as mean ± s.e.m. (**P < 0.01). P values: SPF versus ASF, 0.0072; ASF versus ASF (recol.), 0.00250. Data are representative of two independent experiments. Scale bars represent 50 μm. (h) Quantitative measurement of Ddit4 mRNA by qRT-PCR in microglia from SPF (black bar), recolonized ASF (gray) and ASF (white bar) mice. Data are presented as mean ± s.e.m. with five samples in each group. Significant differences were determined by an unpaired t test (***P < 0.001). P values: SPF versus ASF, <0.0001; ASF versus ASF (recol.), 0.0001. Data are representative of two independent experiments. (i) Three-dimensional structure of a representative microglia cell of recolonized ASF or SPF control mice. Scale bars represent 15 μm. (j) Imaris-based morphometric measurements of cortical Iba-1+ microglia. Each symbol represents one mouse per group with three measured cells per mouse. Data are presented as mean ± s.e.m. Significant differences were evaluated by an unpaired t test (*P < 0.05, **P < 0.01). P values: dendrite length, SPF versus ASF, 0.0099; number of segments, SPF versus ASF, 0.0025; number of branch points, SPF versus ASF, 0.0031; number of terminal points, SPF versus ASF, 0.0022; ASF versus ASF (recol.), 0.0386.
Defective microglia can be restored by microbiota or SCFA
We investigated whether reconstitution of the intestinal microbiota of ASF mice was sufficient to re-establish microglia features. Tri-colonized ASF mice were exposed to a complex microbiota by co-housing with SPF mice and examined 6 weeks later. Cecal dimensions revealed a SPF-like phenotype macroscopically (Fig. 7f), indicating that this co-housing resulted in colonization with a more complex microbiota. Histological examinations of cortical specimens revealed a normalized microglia cell count (Fig. 7g). Ddit4 mRNA levels returned to levels observed in SPF microglia (Fig. 7h). Similarly, unbiased semi-automatic measurements of microglia cell morphology revealed largely, but not completely, restored defects in microglia proportions, such as almost normal dendrite lengths, numbers of segments and branching points (Fig. 7i,j). To further evaluate the bacterial colonization, we measured the bacterial levels before and after co-housing and compared it with GF conditions or antibiotic treatment (Supplementary Fig. 8). Quantitative measurements of bacterial 16S rRNA gene copies revealed almost no detectable intestinal bacterial species in the caeca of GF and antibiotic-treated SPF mice, whereas both tri-colonized ASF and SPF co-housed tri-colonized ASF mice had a much higher bacterial load (Supplementary Fig. 8a). Targeted quantitative PCR revealed an enrichment of a greater number of bacterial clades in SPF and SPF co-housed tri-colonized ASF mice as compared with tri-colonized ASF mice (Supplementary Fig. 8b), suggesting that bacterial community complexity rather than bacterial biomass is necessary for microglia homeostasis.
What are the signaling pathways by which microbes promote steady-state function and maturation of microglia? Human and mice rely on bacteria to break down indigestible dietary components such as fibers39. Short-chain fatty acids (SCFAs) are bacterial fermentation products that include acetic acid, propionic acid and butyric acid and have recently been shown to be vital for immune cell homeostasis in the colon40. To investigate whether SCFA can restore microglia immaturity and malformation in the absence of host microbiota, we placed a SCFA mix in the drinking water of GF mice for 4 weeks (Fig. 8). Cecal hypertrophic changes were not reversed by SCFA treatment (Fig. 8a). However, histological investigations of cortical specimen revealed a largely normalized microglia density (Fig. 8b) that was accompanied by markedly reduced Ddit4 mRNA levels similar to those observed in SPF levels (Fig. 8c). Measurements of microglia cell morphology showed largely restored defects in microglia proportions, such as approximately normal numbers of segments, branching points, terminal points and cell volumes (Fig. 8d,e). Microglial immaturity, as reflected by high CSFR1 surface expression, was reversed by SCFA supplementation (Fig. 8f), whereas F4/80 and CD31 levels were not decreased. GPR43 (which is encoded by Ffar2) binds SCFA and, through its expression on innate immune cells, mediates resolution of inflammatory responses39. We examined the brains of Ffar2−/− mice and measured microglia cell densities, which were not altered (Fig. 8g). However, Ffar2−/− mice displayed severely malformed microglia with major alterations of dendrite length, number of segments, branching points, terminal points and increased cell volumes (Fig. 8h,i), reminiscent of microglia from GF animals. Given that SCFA can influence microglia features, we asked whether its receptor FFAR2 is expressed on microglia or other CNS cells or whether peripheral myeloid cells harbor this receptor, as reported previously40. We found no Ffar2 mRNA expression on any neuroectodermal CNS cell type, CNS endothelia or microglia, whereas strong signals were observed in the spleen (Fig. 8j). Subsequent immunofluorescence analysis revealed FFAR2 expression on splenic Iba-1+ myeloid cells in the red pulp while parenchymal microglia were devoid of any FFAR2 signal (Fig. 8k). Together, these findings demonstrate that microglial defects are reversible and can be re-established by re-introducing live and complex microbiota or microbial metabolites, for example, SCFA.
Figure 8. SCFA restore microglia malformation and immaturity in GF mice.
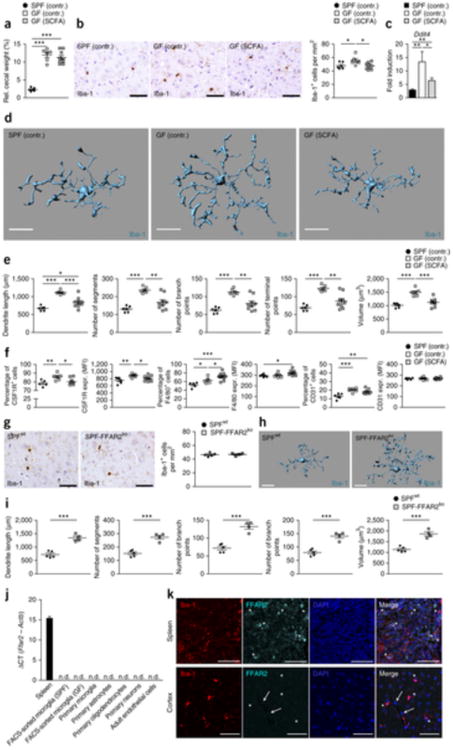
(a) Relative cecal weight of six SPF (SPF contr.), six GF mice fed with sodium-matched water (GF contr.) or nine GF mice treated with SCFAs (GF SCFA) for 4 weeks. Symbols represent data from individual mice. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (***P < 0.001). P values: SPF (contr.) versus GF (contr.), 0.0001; SPF (contr.) versus GF (SCFA.), <0.0001. (b) CNS histology of the cerebral cortex that was subjected to immunohistochemistry for Iba-1 to detect microglia (left) and quantification (right). Scale bars represent 50 μm. Representative pictures of six examined control mice and nine SCFA-treated mice are displayed, respectively. Each symbol represents one mouse and three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05). P values: SPF (contr.) versus GF (contr.), 0.0456; GF (contr.) versus GF (SCFA.), 0.0118. (c) Expression of Ddit4 mRNA measured by qRT-PCR in microglia isolated from SPF (contr.) (black bar), GF (contr.) (white bar) or GF mice treated with SCFA (gray bar). Data are presented as mean ± s.e.m. with six control samples and nine SCFA-treated samples. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01). P values: SPF (contr.) versus GF (contr.), 0.0064; GF (contr.) versus GF (SCFA.), 0.0306; SPF (contr.) versus GF (SCFA.), 0.0056. (d) Morphology of representative cortical microglia (scale bars represent 15 μm, d) and Imaris-based quantification of cellular parameters (e). Each symbol displays one mouse with three measured cells per animal. Six SPF and GF control animals and nine SCFA-treated GF animals were investigated. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: dendrite length SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0008; SPF (contr.) versus GF (SCFA), 0.0247; number of segments SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0021; number of branch points SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0027; number of terminal points SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0018; volume SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0004. (f) Quantifications of the percentages of positively labeled cells and MFIs for CSF1R, F4/80 and CD31 on microglia from six SPF (contr.), six GF (contr.) and nine GF (SCFA) mice. Each symbol represents one mouse. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: CSF1R (percentage of positive cells) SPF (contr.) versus GF (contr.), 0.0094; GF (contr.) versus GF (SCFA), 0.0156; CSF1R (MFI) SPF (contr.) versus GF (contr.), 0.0076; GF (contr.) versus GF (SCFA), 0.0427; F4/80 (percentage of positive cells) SPF (contr.) versus GF (contr.), 0.0204; GF (contr.) versus GF (SCFA), 0.0367; SPF (contr.) versus GF (SCFA), 0.0002; F4/80 (MFI) SPF (contr.) versus GF (SCFA), 0.0194; CD31 (percentage of positive cells) SPF (contr.) versus GF (contr.), 0.0009; SPF (contr.) versus GF (SCFA), 0.0023. (g) Iba-1 immunhistochemistry (left) and quantification (right) of cortical sections from four FFAR2-deficient (SPF-FFAR2ko) or five competent (SPFwt) mice. Scale bars represent 50 μm. Representative pictures are displayed. Each symbol represents one mouse. Three to four sections per mouse were examined. Data are presented as mean ± s.e.m. (h,i) Imaris-based three-dimensional reconstruction of representative microglia from SPF-FFAR2ko and (SPFwt) mice (scale bars represent 15 μm) and quantification of the cellular parameters (i). Each symbol shows one mouse with at least three measured cells per animal. Four FFAR2-deficient (SPF-FFAR2ko) and five competent (SPFwt) mice were analyzed. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (***P < 0.001). P values: dendrite length, 0.0007; number of segments, 0.0007; number of branch points, 0.0006; number of terminal points, 0.0008; volume, 0.0008. (j) Ffar2 mRNAs levels measured by qRT-PCR for the indicated cells and tissues, respectively. Data are expressed as ΔCT ratio of Ffar2 mRNA expression versus endogenous Actb and exhibited as mean ± s.e.m. Bar represents means ± s.e.m. with at least three samples in each group. n.d., not detectable. (k) Immunofluorescence images of FFAR2 (aqua green) on Iba-1+ (red) microglia or macrophages of wild-type mice. Asterisks indicate double-positive cells in the spleen (upper row) and FFAR2-negative microglia in the cortex (lower row) of wild-type mice. Arrows highlight FFAR2-negative endothelial cells. Nuclei were stained with DAPI (blue). Scale bars represent 50 μm.
Microbial-associated molecular patterns (MAMPs) have been shown to modulate various aspects of the host immune response41, 42. Furthermore, MyD88 (an adaptor for recognition of many MAMPs) was recently shown to promote myelomonocyte expansion and differentiation43. Accordingly, we sought to address whether microbiota-derived MAMPs are involved in the maintenance of microglia under homeostatic conditions. We examined mouse mutants lacking Toll-like receptor (TLR)2, TLR3, TLR4, TLR7 and TLR9, which all primarily use MyD88- or TRIF-dependent pathways following engagement44. Given that these microglia are unresponsive to most bacterial MAMPs (such as bacterial lipoproteins (TLR2), double-stranded RNA (TLR3), LPS (TLR4), RNA (TLR7) and CpG-DNA (TLR9)), we would anticipate an absence of activation by the microbiota. Nevertheless, Tlr2−/−; Tlr3−/−; Tlr4−/−; Tlr7−/−; Tlr9−/− mice displayed normal cecal dimensions (Supplementary Fig. 9a) and normal appearance, density, morphology and an unchanged maturation status of parenchymal microglia (Supplementary Fig. 9b–f), suggesting that these MAMPs are not necessary for the maintenance of microglia under steady-state conditions. Collectively, our data indicate that microglia maturation and function are tightly controlled by the host microbiota and that complex molecular signals promote microglia differentiation, ensuring their function as sentinels for the early detection and control of microbial danger signals. Furthermore, TLR2, TLR3, TLR4, TLR7 and TLR9 were not required for microglia maintenance under non-diseased conditions, and impaired microglia maturation observed under GF conditions could successfully be rescued by re-establishment of a complex microbiota or by supplementation with SCFA.
Discussion
We examined interactions between the gut microbiota and the brain's endogenous macrophages, microglia. Colonization with a complex microbiota modulated microglia activation and maturation status during homeostatic conditions. Constitutive (as found in GF mice) or induced (antibiotics) depletion of a host's resident microbes markedly compromised microglia maturation and cell shape, leading to blunted early responses following microbial-associated molecular pattern or pathogen encounter such as exposure to LPS or LCMV.
The host microbiota is necessary for maintaining systemic populations of neutrophils in the circulation by modulating hematopoietic stem cell and myeloid precursors in the bone marrow45. Consistent with these observations, bone marrow–derived splenic macrophage and monocyte populations were found to be reduced under GF conditions34. In both studies, paucity of myelomonocytic cells was ascribed to impaired expansion and development of myeloid precursors in the bone marrow, possibly as a result of reduced levels of the myeloid survival factor CSF1 (ref. 34). In contrast with these reports, we found increased levels of parenchymal macrophages in the CNS. In addition, the expression levels of the CSF1R ligands Il-34 and Csf1 were not diminished in several brain regions, suggesting that there are other mechanisms of microbiota-driven maintenance of tissue macrophages in the CNS. Several molecules positively regulating cell cycle, such as Dit4, were elevated in the absence of microbiota. Dit4 is induced for example, upon cellular stress and nutrient deficiency29. Consistent with the increased microglia cell number in GF and tri-colonized ASF mice, we observed enhanced levels of Ddit4, which is an actor in the mTor signaling cascade.
How commensal microbes (presumably in the gut) are able to control microglia maturation and innate immune reaction in distant sites such as the brain remains incompletely understood. Mice deficient in MyD88 signaling display reductions in systemic myeloid cell populations and myeloid precursors43. Given that microbial ligands have been detected in systemic sites, including the bone marrow42, microbiota-derived MAMPs that originate in the gut may mediate steady-state myelopoiesis in primary immune sites. For microglia, however, we did not observe any changes in microglia maturation and morphology when the MAMPs recognized by TLR2, TLR3, TLR3, TLR7 and TLR9 were absent, suggesting that there are other mechanisms for microbiota-driven microglia maturation. Whether MyD88-deficient animals have an altered quantity and maturation of microglia has not been determined.
We found that bacterial products or metabolites from members of the gut microbiota, such as SCFAs, are key molecules that modulate microglia maturation, morphology and function. SCFAs may translocate from the intestinal mucosa to the systemic circulation, where they could affect immune regulation and CNS function46. Our expression study revealed no expression of the SCFA receptor FFAR2 in the CNS, instead finding it only on peripheral myeloid cells, which is in contrast with a recent study that suggested direct SCFA effects on brain endothelial cells47. However, receptors are not necessary for SCFA cell entry into the CNS46.
In addition, the gut communicates to the brain via hormonal signaling pathways that involve the release of gut peptides with modulatory functions from enteroendocrine cells46. Studies in GF mice suggest that the gut microbiota mediates and regulates the release of gut peptides48. Microbiota can elicit signals via the vagal nerve to the brain and vice versa. The behavioral effects mediated by some probiotic strains in rodents were dependent on intact vagal nerve activation49. In addition, indirect effects of the gut microbiota on the innate immune system can result in alterations in the circulating levels of pro-inflammatory and anti-inflammatory cytokines that directly affect microglia function.
Our study indicates that microglia are particularly affected when a complex microbiota is absent. However, we cannot completely rule out the possibility that other CNS cells, such as neurons, astrocytes or endothelia, are affected as well and that microglia receive signals via those cells. However, our detailed histological analyses, including unbiased automated measurement of cell structures, revealed substantial changes only in microglia in GF mice, whereas other cell populations were virtually unaltered.
When does the host microbiota shape microglia functions? Microglia are derived from early c-kit+ erythromyeloid precursors in the yolk sac around 8.0 days post conception (dpc) that subsequently mature into macrophages and populate the developing neuroectoderm starting from 9.5 dpc3, 13. It is generally accepted that all microglia in the adult brain are derived from this early wave of development and represent an extremely long-lived cell population without any input from the circulation after birth15. In turn, intrauterine environment and fetus are sterile until delivery and a first encounter with germs occurs in the first days, when the BBB is already established46. On the other hand, prenatal immune activation by exposure of mothers to the viral mimetic poly(I:C) has recently been suggested to increase the vulnerability of the pubescent offspring to stress, resulting in behavior abnormalities and aggravated neuroimmune responses in mice that are characterized by increased numbers of activated microglia in the hippocampus50. Our results, however, support the idea that, even during adulthood, a continuous input from complex microbiota is necessary for microglia maintenance. Notably, even preservation of the maturation status of microglia is obviously an active process controlled by microbiota-derived signals. If these input signals are lost, for example, by antibiotics-induced eradication of bacteria, microglia regain an immature status reminiscent of developing juvenile microglia. Notably, this juvenile phenotype of microglia can be overcome by recolonization of the gut with complex microbiota, indicating a great plasticity of the gut-microglia connection.
In summary, we found that host microbiota are crucial for microglia maturation and activation during health and disease, and we provide evidence for a constitutive activity of microbes on the brains innate immune system. These results may assist our understanding and treatment of microglia-mediated inflammatory, neuropsychiatric and oncologic diseases of the CNS.
Methods
Mice
Conventional SPF mice on C57BL/6 background were purchased from Janvier Labs. GF mice were a kind gift from A. Diefenbach (University of Mainz) or purchased from the University Hospital Bern. All mice (mixed gender) were used at 6–10 weeks of age. ASF and recolonized ASF mice37 (females) were provided by B. Stecher (Max-von-Pettenkofer Institute, LMU Munich, German Center for Infection Research). ASF mice were housed under GF conditions in a flexible film isolator with HEPA-filtered air and autoclaved chow and bedding. For recolonization, ASF mice were co-housed with conventional SPF donor mice in the same cage for 6 weeks. For substantial depletion of the microbiota, SPF mice were provided with drinking water containing 1 mg ml−1 cefoxitin (Santa Cruz Biotechnology), 1 mg ml−1 gentamicin (Sigma-Aldrich), 1 mg ml−1 metronidazole (Sigma-Aldrich), and 1 mg ml−1 vancomycin (Hexal) for 4 weeks ad libitum. Antibiotics were renewed every other day. 0.05 g of feces (2–3 pellets) was solubilized in 1 ml PBS and was plated on BHI and MHPB plates to track the success of the treatment. For treatment with SCFAs, 25 mM sodium propionate, 40 mM sodium butyrate and 67.5 mM sodium acetate (Sigma-Aldrich) were added to drinking water for 4 weeks as described previously40. Sodium matched water was applied to control mice. Water solutions were prepared and changed weekly. FFAR2-deficient mice and wild-type controls (females) were provided by W.S. Garrett (Harvard School of Public Health). TLR2,3,4,7,9-deficient mice51 (mixed gender) were provided by T. Buch (TU Munich). All animal experiments were approved by the Ministry for Nature, Environment and Consumers' Protection of the state of Baden-Württemberg and were performed in accordance to the respective national, federal and institutional regulations.
Histology
Histology was performed as described recently52, 53. Brains were removed and fixed in 4% buffered formalin. Then brain tissue was dissected and parasagittal sections were embedded in paraffin before staining with H&E, Iba-1 (cat. no. 019-19741, Wako)15 for microglia, GFAP (cat. no. Z0334, Dako)54 for astrocytes, NeuN (cat. no. MAB377, Merck Millipore)54 for neurons, Nogo-A (kindly provided by M.E. Schwab, Laboratory of Neural Regeneration and Repair, University and ETH Zurich)54 for oligodendrocytes, CD3 (cat. no. MCA 1477, Serotec)15 for T cells and B220 (cat. no. 557390, BD Bioscience)15 for B cells. At least 3–4 parasagittal brain sections per mouse were evaluated using analySIS software (Olympus). For Ki67 immunohistochemistry 14-μm frontal and parasagittal cryo sections from adult brain tissue were prepared and stained first with the anti-Ki67 antibody (cat. no. ab15580, Abcam)13 for 24 h (dilution 1:500 at 4 °C), followed by Alexa-Fluor-647–conjugated secondary antibody (cat. no. A31573, Life technologies) staining (2 h, dilution 1:500, 20–22 °C). Second the specimens were stained with the anti-Iba-1 antibody (cat. no. 019-19741, Wako, Osaka, Japan)15 for 24 h (dilution 1:500 at 4 °C), subsequently Alexa-Fluor-568–conjugated secondary antibody (cat. no. A11011, Life technologies) was added at a dilution of 1:500 for 2 h at 20–22 °C. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, cat. no. 236276, Boehringer). The pictures were taken with BZ-9000 Biorevo microscope (Keyence) and the number of positive cells was determined using BZ-II Analyzer (Keyence). At least three brain sections per mouse were counted. For FFAR2 immunohistochemistry 4-μm parasagittal FFPE sections from adult brain and splenic tissue were prepared. To improve accessibility of antibodies heat mediated antigen retrieval (pH 6.0) was carried out. First, slices were stained with the anti-FFAR2 antibody (cat. no. ab118449, Abcam)40 for 24 h (dilution 1:200 at 4 °C), followed by Alexa-Fluor-647-conjugated secondary antibody (cat. no. A31573, Life technologies) staining for 2 h at 20–22 °C (dilution 1:500). Second sections were incubated with the anti-Iba-1 antibody (cat. no. 019-19741, Wako)15 for 24 h (dilution 1:500 at 4 °C), followed by the Alexa Fluor-568–conjugated secondary antibody (cat. no. A11011, Life technologies) staining, which was added at a dilution of 1:500 for 2 h at 20–22 °C. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, cat. no. 236276, Boehringer). Representative pictures were taken with BZ-9000 Biorevo microscope (Keyence).
Three-dimensional reconstruction of microglia
30-μm parasagittal cryo sections from adult brain tissue were stained with anti-Iba-1 (cat. no. 019-19741, Wako)15 for 48 h (dilution 1:500 at 4 °C), followed by Alexa Fluor 568–conjugated secondary antibody (cat. no. A11011, Life technologies) staining, which was added at a dilution of 1:500 overnight at 4 °C. Nuclei were counterstained with DAPI. Imaging was performed on an Olympus Fluoview 1000 confocal laser scanning microscope (Olympus) using a 20× 0.95 NA objective. Z stacks were done with 1.14-μm steps in z direction, 1,024 × 1,024 pixel resolution were recorded and analyzed using IMARIS software (Bitplane). Three cortical cells were reconstructed per analyzed mouse.
Microglia isolation and flow cytometry
Adult microglia were harvested using density gradient separation and were prepared as described before53. In short, Cells were stained with primary antibodies directed against CD11b (cat. no. 17-0112-83)13, CD45 (cat. no. 48-0451-82)13, CD115 (cat. no. 12-1152-82)13, F4/80 (cat. no. 12-4801-82)13, CD31 (cat. no. 12-0311-81, eBioscience)13, CD44 (cat. no. 553133)13, CD62L (cat. no. 553150, BD Bioscience)13, MHC class II (cat. no. 107630, Biolegend)13 at 4 °C for 15 min. Cells were washed and analyzed using a BD LSRFortessa (Becton Dickinson) or were sorted with a MoFlo Astrios (Beckman Coulter) or BD FACSAria III (Becton Dickinson) and further processed. Viable cells were gated by forward and side scatter pattern. Data were acquired with FACSdiva software (Becton Dickinson). Post-acquisition analysis was performed using FlowJo software (Tree Star).
RNA-seq
For RNA-seq 1000 microglia (stained with antibodies against CD11b cat. no. 17-0112-83 and CD45 cat. no. 12-0451-82)13 cells per brain were FACS-sorted directly into an 96-well plate containing 4-μl lysis buffer (RNase-free H2O, 0.2% Triton-X (Roth) and 0.4 U μl−1 RNasin (Promega)) as described above. Thereafter, the plate was centrifuged, snap frozen on dry ice and stored at −80 °C. RNA-seq library production, following sample preparation and analysis were carried out as described previously55.
Gene expression analysis
FACS-sorted microglial cell populations were collected directly in cell lysis buffer and subsequently RNA was isolated with the Arcturus Pico Pure RNA Isolation Kit (Life Technologies) according to the manufacturer's protocol. Reverse transcription and real-time PCR analysis were performed using high capacity RNA-to-cDNA-Kit and Gene Expression Master Mix reagents (Applied Biosystems) according to the manufacturer's recommendations. RT-PCRs were analyzed with a LightCycler 480 (Roche). For gene expression analysis, we used the following TaqMan Gene Expression Assays: Actb (Mm01205647_g1), Ccl2 (Mm00441242_m1), Ccl5 (Mm01302427_m1), Ccl7 (Mm00443113_m1), Cxcl10 (Mm99999072_m1), Il-1β (Mm00434228_m1), Il-6 (Mm00446191_m1), Il-12β (Mm00434174_m1), Tnfα (Mm00443258_m1), Marco (Mm00440265_m1), Csf1 (Mm00432686_m1), cyclin D2 (Mm00438070_m1), cyclin B2 (Mm01171453_m1), Usp18 (Mm00449453_m1), Irf3 (Mm00516784_m1), Irf7 (Mm00516793_g1), Isg15 (Mm01705338_s1), Oas2 (Mm00460961_m1), Msr1 (Mm00446214_m1), c-Jun (Mm00495062_s1), c-Fos (Mm00487425_m1), FosB (Mm00500401_m1), Nox2 (Mm01287743_m1), S100a4 (Mm00803372_g1), S100a6 (Mm00771682_g1), S100a8 (Mm00496696_g1), S100a9 (Mm00656925_g1), S100a10 (Mm00501458_g1) Ddit4 (Mm00512504_g1) and Ffar2 (Mm01176527_m1).
Brain tissue was removed and stored for 2–3 d in RNAlater (Qiagen) at 4 °C. Different brain regions were dissected under a Bino microscope. Afterwards tissue was homogenized and RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Reverse transcription and real-time PCR analysis was performed as described above. Following TaqMan Gene Expression Assays were used: Actb (Mm01205647_g1), Csf1 (Mm00432686_m1), Csf2 (Mm01290062_m1), Tgfβ1 (Mm01178820_m1), Il-34 (Mm01243248_m1) and Sfpi1 (Mm00488142_m1).
Splenic tissue was removed and stored at −80 °C. Primary cell cultures of microglia, astrocytes, oligodendrocytes and neurons were prepared as described previously54. For endothelial cell cultures the cerebrum was isolated and dissected without meninges. The brain was homogenized and was centrifuged at 4,000g for 5 min. The pellet was resuspended in 18% dextran solution (molecular weight: 64,000–76,000, Sigma-Aldrich) in DMEM (Gibco, Life Technologies) and centrifuged at 6,000g for 10 min. After removing the supernatant and myelin debris, the pellet was resuspended in DMEM containing 1 mg ml−1 collagenase/dispase (Roche), 40 μg ml−1 DNase 1 (Roche), and 0.147 μg ml−1 tosyllysine chloromethyl ketone (Sigma-Aldrich) and incubated at 37 °C for 75 min with occasional agitation to free endothelial cells from pericytes, perivascular macrophages and remains of the basement membrane. The cell suspension was centrifuged at 4,000g for 5 min, the supernatant was discarded, and cells were washed and seeded in 6-well plates coated with mouse collagen IV (Becton Dickinson). Cells were subsequently grown in DMEM-F12 (Gibco, Life Technologies) supplemented with 10% fetal bovin serum (PAA Laboratories), 2 mM L-Glutamine, 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin, 15 U ml−1 heparin (Biochrom) and 30 μg ml−1 of endothelial cell growth supplement (Sigma-Aldrich). 4 μg ml−1 puromycin (Sigma-Aldrich) were added for 48 h to deplete non-endothelial cells. RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Reverse transcription and real-time PCR analysis was performed as described above. Following TaqMan gene expression assays were used: Actb (Mm01205647_g1) and Ffar2 (Mm01176527_m1).
qPCR for intestinal bacteria
Cecum tissue samples were first homogenized and then bacterial DNA was extracted with the powersoil DNA isolation kit (MoBio). SYBR green-based quantitative real-time PCR (qPCR) was performed with universal eubacterial56 or group-specific primers for 16S rRNA targeting strains of the Altered Schaedler Flora and common bacterial taxa of the mouse intestinal tract, as described previously56. Primers used: universal eubacteria (ACTCCTACGGGAGGCAGCAGT, ATTACCGCGGCTGCTGGC), Parabacteroides (TTGCCGTTGAAACTGGTTGA, GGAGTTCTGCGTGATATCTATGCA), Lactobacillus (AGCAGTAGGGAATCTTCCA, CACCGCTACACATGGAG) and Clostridium (GATGCCTCCTAAGAACCGTATGC, GCGGACGGGTGAGTAACGT), mouse intestinal Bacteroides (CCAGCAGCCGCGGTAATA, CGCATTCCGCATACTTCTC) and Enterobacteriaceae (GTGCCAGCMGCCGCGGTAA, GCCTCAAGGGCACAACCTCCAAG). For total bacterial load eubacterial 16S rRNA gene copies were normalized to DNA content. For detection of bacterial groups in recolonized mice a Ct value >35 cycles was used as a cut-off.
Microarray analysis
Transcriptional profiles of FACS-sorted microglia were assessed using Affymetrix GeneChip Arrays (Mouse Gene 2.1 ST Arrays). Affymetrix GeneChip array data was pre-processed using Affymetrix Expression Console and normalized through the Robust Multi-array Average (RMA) implementation in the Expression Console. Sample processing and Affymetrix microarray hybridization were carried out at a genomics core facility: Center of Excellence for Fluorescent Bioanalytics (University of Regensburg, Germany). Further analyses were performed using BRB-ArrayTools developed by R. Simon and BRB-ArrayTools Development Team. Gene Ontology analysis and identification of differentially expressed genes belonging to specific pathways was performed through the DAVID online service and Gorilla (Gene Ontology enRIchment anaLysis and visuaLizAtion tool) online service57, 58, 59. Venn diagrams were created by intersection of gene lists exported from BRB Tools into a Webtool provided by the Bioinformatics & Evolutionary Genomics group of Y. van de Peer (University of Ghent).
LPS challenge
For acute inflammatory challenges, either 50 μg LPS (Sigma-Aldrich) were diluted in 150 μl sterile PBS and applied i.p. or 5 μg LPS were diluted in 10 μl PBS and applied intracranially under isofluoran anesthesia. Animals returned to their cages once they had recovered their righting reflex. 6 h later, animals were analyzed. Control animals were injected with 150 μl sterile PBS i.p. or 10 μl sterile PBS i.c.
LCMV challenge
LCMV strain WE was propagated and titrated as plaque forming units (PFU) on L929 cells as described before60. PFU were multiplied by the factor 10 to be converted into infectious units (IU). Mice were infected by intracerebral inoculation of 103 IU into the right hemisphere.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software, Version 6.0, La Jolla, USA). In general, chosen sample sizes are similar to those reported in previous puplications15. All data were tested for normality applying the Kolmogorov-Smirnov test. If normality was given, an unpaired t test was applied. Differences were considered significant when P < 0.05. To obtain unbiased data, experimental mice were all processed together by technicians and cell quantifications were performed blinded by two scientists independently and separately.
A Supplementary Methods Checklist is available.
Supplementary Material
Supplementary Figure 1: Gene profile from microglia in GF mice. (64 KB), mRNA expression values (number of reads) of genes from microglia in GF animals (white bars) or SPF mice (black bars). (a) Prototypical microglia genes are presented and compared to the non-microglia genes such as Gfap, Mog, Tubb3 and Rbfox3. (b) Epigenetic signature of microglia was examined by investigating genes (number of reads) related to histone acetylation, deacetylation, methylation and demethylation, respectively. Bars represent means ± s.e.m. with seven samples in each group. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). P values: Csf1r: 0.0001, Sfpi1: 0.0075, Hexb: 0.0060. Myst3: 0.0121, Hdac1: 0.0191, Sirt2: 0.0113, Sirt7: 0.0441, Mll3: 0.0489, Mll5: 0.0105, Setd1a: 0.0360, Setd7: 0.0134, Kdm4a: 0.0476, Kdm6b: 0.0005, Jarid2: <0.0001.
Supplementary Figure 2: Absence of microbes does not alter the amount of neurons, astrocytes and oligodendrocytes. (151 KB) CNS histology of the neuroectodermal compartment of 6-8 weeks old GF or control SPF mice. NeuN labels mature neurons, Nogo-A oligodendrocytes and glial fibrillary acidic protein (GFAP) astrocytes. Representative pictures are shown. Scale bars: NeuN: 100 μm, Nogo-A and GFAP: 50 μm.
Supplementary Figure 3: Absence of microbes does not alter the amount of neurons, astrocytes and oligodendrocytes. (84 KB) (a,b) Quantitative assessment of the neuroectodermal compartment of 6-8 weeks old GF or control SPF mice. Hippocampal NeuN+ cells were counted in the gyrus dentatus. Cerebellar NeuN+ cells were counted in the granular cell layer. Each symbol represents one mouse, with five samples per group. Means ± s.e.m. are indicated. No significant differences could be determined by using an unpaired t test. Representative pictures are shown. n.d. = not detectable. Data are representative of two independent experiments. Scale bars: H&E: 200 um.
Supplementary Figure 4: Lymphocytic cells are indistinguishable in the brains of SPF and GF mice. (82 KB), Immunohistochemistry for T cells (CD3) and B cells (B220) in different brain regions of SPF and GF mice, respectively. Insets depict spleen sections as positive controls. Scale bars: overviews: cortex, corpus callosum and cerebellum (CD3): 50 μm; olfactory bulb (CD3): 20 μm; hippocampus (CD3), cortex, corpus callosum, hippocampus, olfactory bulb and cerebellum (B220): 100 μm. Inset: 50 μm (CD3) and 100 μm (B220).
Supplementary Figure 5: Increased microglia number in cerebellar grey and white matter of GF mice. (77 KB), (a) Quantification of Iba-1+ parenchymal microglia from either SPF (black symbols) or GF (white symbols) mice was performed on cerebellar brain slices. Each symbol represents one mouse. Three to four sections per mouse were examined. Means ± s.e.m. are shown. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, ***P < 0.001). P values: granular cell layer: 0.0008, white matter: 0.0287. Representative pictures of nine examined mice per group are displayed. (b) Imaris-based 3-D reconstruction of the cortical microglia networks under SPF (left) or GF (right) conditions. GF microglia have no distances to neighbouring microglia and show partial physical contacts to adjacent cells. Inset exhibits such an interaction. Scale bars: 20 μm (overview), 3 μm (inset). (c) Quantitative RT-PCR for the indicated mRNAs. Data are expressed as ratio of mRNA expression versus endogenous Actb relative to SPF conditions and exhibited as mean ± s.e.m. Bars (black: SPF, white: GF) represent means ± s.e.m. with at least four samples in each group. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). n.d. = not detectable. P values: Il-34: hippocampus: 0.0031. Csf1: cortex: 0.0071, cerebellum: 0.0009. Tgfß1: cortex: 0.0253, cerebellum: 0.031. Sfpi1: cortex: 0.0302, corpus callosum: 0.0290, cerebellum: 0.0345.
Supplementary Figure 6: Decreased inflammatory repertoire of microglia from GF mice upon intracerebral LPS challenge. (85 KB) Gene ontology enrichment network on differentially expressed genes in microglia from SPF (a) or GF (b) mice on the basis of an Affymetrix DNA microarray analysis. Diagram depicts functional results of GO clustering through GORilla. Only very highly significantly overrepresented GO terms are included with P values ranging from P < 10-9 (yellow) to P < 10-24 (red). White framed terms depict not significantly overrepresented parent terms of the colored terms.
Supplementary Figure 7: Diminished inflammatory response of microglia from GF mice upon peripheral LPS challenge. (69 KB) Quantitative RT-PCR of LPS-induced factors in microglia 6 hrs after intraperitoneal LPS challenge. Data are expressed as ratio of the mRNA expression compared to endogenous Actb relative to control and show mean ± s.e.m. At least three mice per group were analyzed. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). ns: not significant. P values: PBS: Il1- b:0.3720, Il-6: 0.5625, Il12 b: 0.2442, Tnfα p=0.5955, Marco: 0.0642, Ccl2: 0.6608, Ccl7: 0.3316, Cxcl10: 0.0867, Csf1: 0.1194, cyclin D2: 0.1135, S100a4: 0.5357, S100a6: 0.4281, S100a8: 0.0642, S100a9: 0.4583, S100a10: 0.1771. LPS: Il-1 b: 0.0023, Il-6: 0.1329, Il-12 b: <0.0001, Tnfα 0.0008, Marco: 0.0001, Ccl2: 0.0673, Ccl7: 0.2708, Cxcl10:.1522, Csf1: 0.1828, cyclin D2: 0.0344, S100a4: 0.0064, S100a6: 0.0341, S100a8: 0.0009, S100a9: 0.5782, S100a10: 0.0352.
Supplementary Figure 8: Determination of intestinal bacterial loads and complexity. (37 KB), (a) qPCR analysis for eubacterial 16S rRNA could not detect significant amounts of bacteria in the caeca of GF and antibiotic-treated SPF animals. Representative data from fifteen SPF, five GF, six SPF (ABX), five ASF and five ASF (recol.) mice are depicted. Gene copy numbers were normalized to DNA content. Each symbol represents one mouse. Means ± s.e.m. are indicated. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01). P values: SPF vs. GF: 0.0151, SPF vs. SPF (ABX): 0.0046, SPF vs. ASF (recol.): 0.0280. (b) Recolonized ASF mice (ASF recol.) harbour a greater microbial diversity as detected by qPCR using group specific primers. Besides Parabacteroides (ASF357), Lactobacillus (ASF 361) and Clostridium (ASF356), mouse intenstinal Bacteroides (mib) and Enterobacteriaceae groups are detectable in the caecum of SPF and re-colonised ASF animals using group-specific primers, respectively.
Supplementary Figure 9: Microglia maturation is independent from the TLRs2,3,4,7,9. (73 KB), (a) Left: View of caecum from a SPF-TLRwt (control, left) and SPF-TLR2,3,4,7,9-deficient (SPF-TLRko) mouse (right). Ruler scale is shown. Representative pictures are shown. Right: Relative caecum weight of control SPF-TLRwt and SPF-TLRko mice. Symbols represent individual mice, with six animals per group. Means ± s.e.m. are shown. No statistically significant differences were found by an unpaired t test. (b) Iba-1 immunohistochemistry in the cortex of SPF-TLRwt and SPF-TLRko animals (left) and quantification thereof (right). Every symbol represents one mouse. Six animals per group were analyzed and three to four sections per mouse were examined. Data are representative of two independent experiments. Means ± s.e.m. are indicated. No significant differences could be determined by an unpaired t test. (c) Ddit4 mRNA measured by qRT-PCR in microglia from SPF-TLRwt (black bar) or SPF-TLRko (white bar) mice. Means ± s.e.m. with six samples in each group are displayed. No significant differences could be determined by an unpaired t test. Data are representative of two independent experiments. (d) Three-dimensional reconstruction (Scale bars: 15μm) and Imaris-based automatic quantification of cell morphometry (e) of Iba-1+ microglia. Individual symbols represent one mouse per group with at least three measured cells per mouse. Six mice per group were analyzed. Means ± s.e.m. are indicated. No significant differences were detected by an unpaired t test. (f) Flow cytometry of isolated microglia from control six SPF-TLRwt and six SPF-TLRko individuals. Percentages and mean fluorescence intensities (MFI) of the indicated surface molecules on microglia are shown. Each symbol represents one mouse. Means ± s.e.m. are indicated. Significant differences were evaluated by an unpaired t test and labelled with asterisks (*P < 0.05). P value: CD31 (MFI): 0.0419. Data are representative of two independent experiments.
Supplementary Table 1: Altered microglial gene profile and immaturity in germ-free animals. (33 KB) Significantly down- (upper table) and up- (lower table) regulated genes subjected to functional GO cluster analysis. The significance of genes is indicated by the given P values.
Supplementary Table 2: Reduced gene induction in microglia from non-colonized GF animals. (42 KB) Affymetrix Genome 430A 2.0 array-based gene induction of microglia from SPF mice compared to GF individuals. 78 genes significantly differentially expressed in GF vs. SPF microglia 6 hr after i.c. LPS treatment are shown. Only genes also significantly up- or downregulated by LPS treatment compared to PBS treated controls of the same housing conditions (GF and SPF respectively) are included to account for differences in basal gene regulation. (P < 0.01).
Acknowledgments
This work is dedicated to our former friend Uwe-Karsten Hanisch, past Professor for Neuroscience at the Paul-Flechsig-Institute for Brain Research, Leipzig, who devoted his whole life to the exploration of microglia function. We thank M. Oberle, M. Ditter and T. el Gaz for excellent technical assistance. We are grateful to J. Bodinek-Wersing and G. Rappl for cell sorting and to A. Spies for providing cDNA from cell cultures. M.P. was supported by the BMBF-funded competence network of multiple sclerosis (KKNMS), the Sobek Foundation, the DFG (SFB 992, FOR1336, PR 577/8-1), the Fritz-Thyssen Foundation and the Gemeinnützige Hertie Foundation (GHST). B.S. was supported by the SPP1656 “Intestinal Microbiota” (STE 1971/4-1).
Footnotes
Contributions: D.E., A.L.H.d.A., P.W., D.J., K.J., T.M., B.S., O.U., I.A., H.K.-S., E.D. and K.D.M. conducted the experiments. I.A. and O.S. analyzed the RNA sequencing and microarray data, respectively. T.B., B.S., A.D., P.S., V.S., E.C. and W.S.G. provided mice and designed experiments. M.P. supervised the project and wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
Contributor Information
Daniel Erny, Institute of Neuropathology, University of Freiburg, Freiburg, Germany.
Anna Lena Hrabě de Angelis, Institute of Neuropathology, University of Freiburg, Freiburg, Germany.
Diego Jaitin, Lab for ImmunoGenomics, Weizmann Institute of Science, Rehovot, Israel.
Peter Wieghofer, Faculty of Biology, University of Freiburg, Freiburg, Germany.
Ori Staszewski, Institute of Neuropathology, University of Freiburg, Freiburg, Germany.
Eyal David, Lab for ImmunoGenomics, Weizmann Institute of Science, Rehovot, Israel.
Hadas Keren-Shaul, Lab for ImmunoGenomics, Weizmann Institute of Science, Rehovot, Israel.
Tanel Mahlakoiv, Department of Virology, University of Freiburg, Freiburg, Germany.
Kristin Jakobshagen, Institute for Medical Microbiology, Immunology and Hygiene & Center for Molecular Medicine Cologne (CMMC), University of Cologne, Cologne, Germany.
Thorsten Buch, Institute for Medical Microbiology, Immunology, and Hygiene, Technische Universität München, Munich, Germany.
Vera Schwierzeck, Institute of Medical Microbiology and Hygiene, University of Mainz Medical Centre, Mainz, Germany.
Olaf Utermöhlen, Institute for Medical Microbiology, Immunology and Hygiene & Center for Molecular Medicine Cologne (CMMC), University of Cologne, Cologne, Germany.
Eunyoung Chun, Department of Immunology and Infectious Diseases, Harvard School of Public Health, Boston, Massachusetts, USA.
Wendy S Garrett, Department of Immunology and Infectious Diseases, Harvard School of Public Health, Boston, Massachusetts, USA.
Kathy D McCoy, Mucosal Immunology Lab, Department of Clinical Research, University of Bern, Bern, Switzerland.
Andreas Diefenbach, Institute of Medical Microbiology and Hygiene, University of Mainz Medical Centre, Mainz, Germany.
Peter Staeheli, Department of Virology, University of Freiburg, Freiburg, Germany.
Bärbel Stecher, Max-von-Pettenkofer Institute, LMU Munich, German Center for Infection Research (DZIF), partner site LMU Munich, Munich, Germany.
Ido Amit, Lab for ImmunoGenomics, Weizmann Institute of Science, Rehovot, Israel.
Marco Prinz, Institute of Neuropathology, University of Freiburg, Freiburg, Germany; BIOSS Centre for Biological Signaling Studies, University of Freiburg, Freiburg, Germany.
References
- 1.Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- 2.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 3.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 4.Schafer DP, Stevens B. Phagocytic glial cells: sculpting synaptic circuits in the developing nervous system. Curr Opin Neurobiol. 2013;23:1034–1040. doi: 10.1016/j.conb.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naj AC, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingworth P, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerreiro RJ, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 2013;70:78–84. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rademakers R, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet. 2012;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan Y, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014;17:400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- 10.Prinz M, Mildner A. Microglia in the CNS: immigrants from another world. Glia. 2011;59:177–187. doi: 10.1002/glia.21104. [DOI] [PubMed] [Google Scholar]
- 11.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 13.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 14.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmann T, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 16.Prinz M, Tay TL, Wolf Y, Jung S. Microglia: unique and common features with other tissue macrophages. Acta Neuropathol. 2014;128:319–331. doi: 10.1007/s00401-014-1267-1. [DOI] [PubMed] [Google Scholar]
- 17.Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Front Physiol. 2011;2:94. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspar P, Cases O, Maroteaux L. The developmental role of serotonin: news from mouse molecular genetics. Nat Rev Neurosci. 2003;4:1002–1012. doi: 10.1038/nrn1256. [DOI] [PubMed] [Google Scholar]
- 19.Diaz Heijtz R, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bercik P, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141609:599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 21.Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 23.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markle JG, et al. gammadelta T cells are essential effectors of type 1 diabetes in the nonobese diabetic mouse model. J Immunol. 2013;190:5392–5401. doi: 10.4049/jimmunol.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 27.Reikvam DH, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS ONE. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kierdorf K, Prinz M. Factors regulating microglia activation. Front Cell Neurosci. 2013;7:44. doi: 10.3389/fncel.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu IM, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butovsky O, et al. Identification of a unique TGF-beta–dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greter M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khosravi A, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prinz M, et al. Microglial activation by components of gram-positive and -negative bacteria: distinct and common routes to the induction of ion channels and cytokines. J Neuropathol Exp Neurol. 1999;58:1078–1089. doi: 10.1097/00005072-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 36.Cunningham C, Wilcockson DC, Campion S, Lunnon K, Perry VH. Central and systemic endotoxin challenges exacerbate the local inflammatory response and increase neuronal death during chronic neurodegeneration. J Neurosci. 2005;25:9275–9284. doi: 10.1523/JNEUROSCI.2614-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stecher B, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaedler RW, Dubos RJ. The fecal flora of various strains of mice. Its bearing on their susceptibility to endotoxin. J Exp Med. 1962;115:1149–1160. doi: 10.1084/jem.115.6.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bindels LB, Dewulf EM, Delzenne NM. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol Sci. 2013;34:226–232. doi: 10.1016/j.tips.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke G, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 43.Fiedler K, Kokai E, Bresch S, Brunner C. MyD88 is involved in myeloid as well as lymphoid hematopoiesis independent of the presence of a pathogen. Am J Blood Res. 2013;3:124–140. [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deshmukh HS, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borre YE, et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med. 2014;20:509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Braniste V, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schéle E, et al. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology. 2013;154:3643–3651. doi: 10.1210/en.2012-2151. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Burgos A, et al. Psychoactive bacteria Lactobacillus rhamnosus (JB-1) elicits rapid frequency facilitation in vagal afferents. Am J Physiol Gastrointest Liver Physiol. 2013;304:G211–G220. doi: 10.1152/ajpgi.00128.2012. [DOI] [PubMed] [Google Scholar]
- 50.Giovanoli S, et al. Stress in puberty unmasks latent neuropathological consequences of prenatal immune activation in mice. Science. 2013;339:1095–1099. doi: 10.1126/science.1228261. [DOI] [PubMed] [Google Scholar]
- 51.Conrad ML, et al. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med. 2009;206:2869–2877. doi: 10.1084/jem.20090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dann A, et al. Cytosolic RIG-I-like helicases act as negative regulators of sterile inflammation in the CNS. Nat Neurosci. 2012;15:98–106. doi: 10.1038/nn.2964. [DOI] [PubMed] [Google Scholar]
- 53.Mildner A, et al. Microglia in the adult brain arise from Ly-6C(hi)CCR2(+) monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 54.Raasch J, et al. I{kappa}B kinase 2 determines oligodendrocyte loss by non-cell-autonomous activation of NF-{kappa}B in the central nervous system. Brain. 2011;134:1184–1198. doi: 10.1093/brain/awq359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herz J, et al. Acid sphingomyelinase is a key regulator of cytotoxic granule secretion by primary T lymphocytes. Nat Immunol. 2009;10:761–768. doi: 10.1038/ni.1757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Gene profile from microglia in GF mice. (64 KB), mRNA expression values (number of reads) of genes from microglia in GF animals (white bars) or SPF mice (black bars). (a) Prototypical microglia genes are presented and compared to the non-microglia genes such as Gfap, Mog, Tubb3 and Rbfox3. (b) Epigenetic signature of microglia was examined by investigating genes (number of reads) related to histone acetylation, deacetylation, methylation and demethylation, respectively. Bars represent means ± s.e.m. with seven samples in each group. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). P values: Csf1r: 0.0001, Sfpi1: 0.0075, Hexb: 0.0060. Myst3: 0.0121, Hdac1: 0.0191, Sirt2: 0.0113, Sirt7: 0.0441, Mll3: 0.0489, Mll5: 0.0105, Setd1a: 0.0360, Setd7: 0.0134, Kdm4a: 0.0476, Kdm6b: 0.0005, Jarid2: <0.0001.
Supplementary Figure 2: Absence of microbes does not alter the amount of neurons, astrocytes and oligodendrocytes. (151 KB) CNS histology of the neuroectodermal compartment of 6-8 weeks old GF or control SPF mice. NeuN labels mature neurons, Nogo-A oligodendrocytes and glial fibrillary acidic protein (GFAP) astrocytes. Representative pictures are shown. Scale bars: NeuN: 100 μm, Nogo-A and GFAP: 50 μm.
Supplementary Figure 3: Absence of microbes does not alter the amount of neurons, astrocytes and oligodendrocytes. (84 KB) (a,b) Quantitative assessment of the neuroectodermal compartment of 6-8 weeks old GF or control SPF mice. Hippocampal NeuN+ cells were counted in the gyrus dentatus. Cerebellar NeuN+ cells were counted in the granular cell layer. Each symbol represents one mouse, with five samples per group. Means ± s.e.m. are indicated. No significant differences could be determined by using an unpaired t test. Representative pictures are shown. n.d. = not detectable. Data are representative of two independent experiments. Scale bars: H&E: 200 um.
Supplementary Figure 4: Lymphocytic cells are indistinguishable in the brains of SPF and GF mice. (82 KB), Immunohistochemistry for T cells (CD3) and B cells (B220) in different brain regions of SPF and GF mice, respectively. Insets depict spleen sections as positive controls. Scale bars: overviews: cortex, corpus callosum and cerebellum (CD3): 50 μm; olfactory bulb (CD3): 20 μm; hippocampus (CD3), cortex, corpus callosum, hippocampus, olfactory bulb and cerebellum (B220): 100 μm. Inset: 50 μm (CD3) and 100 μm (B220).
Supplementary Figure 5: Increased microglia number in cerebellar grey and white matter of GF mice. (77 KB), (a) Quantification of Iba-1+ parenchymal microglia from either SPF (black symbols) or GF (white symbols) mice was performed on cerebellar brain slices. Each symbol represents one mouse. Three to four sections per mouse were examined. Means ± s.e.m. are shown. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, ***P < 0.001). P values: granular cell layer: 0.0008, white matter: 0.0287. Representative pictures of nine examined mice per group are displayed. (b) Imaris-based 3-D reconstruction of the cortical microglia networks under SPF (left) or GF (right) conditions. GF microglia have no distances to neighbouring microglia and show partial physical contacts to adjacent cells. Inset exhibits such an interaction. Scale bars: 20 μm (overview), 3 μm (inset). (c) Quantitative RT-PCR for the indicated mRNAs. Data are expressed as ratio of mRNA expression versus endogenous Actb relative to SPF conditions and exhibited as mean ± s.e.m. Bars (black: SPF, white: GF) represent means ± s.e.m. with at least four samples in each group. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). n.d. = not detectable. P values: Il-34: hippocampus: 0.0031. Csf1: cortex: 0.0071, cerebellum: 0.0009. Tgfß1: cortex: 0.0253, cerebellum: 0.031. Sfpi1: cortex: 0.0302, corpus callosum: 0.0290, cerebellum: 0.0345.
Supplementary Figure 6: Decreased inflammatory repertoire of microglia from GF mice upon intracerebral LPS challenge. (85 KB) Gene ontology enrichment network on differentially expressed genes in microglia from SPF (a) or GF (b) mice on the basis of an Affymetrix DNA microarray analysis. Diagram depicts functional results of GO clustering through GORilla. Only very highly significantly overrepresented GO terms are included with P values ranging from P < 10-9 (yellow) to P < 10-24 (red). White framed terms depict not significantly overrepresented parent terms of the colored terms.
Supplementary Figure 7: Diminished inflammatory response of microglia from GF mice upon peripheral LPS challenge. (69 KB) Quantitative RT-PCR of LPS-induced factors in microglia 6 hrs after intraperitoneal LPS challenge. Data are expressed as ratio of the mRNA expression compared to endogenous Actb relative to control and show mean ± s.e.m. At least three mice per group were analyzed. Data are representative of two independent experiments. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). ns: not significant. P values: PBS: Il1- b:0.3720, Il-6: 0.5625, Il12 b: 0.2442, Tnfα p=0.5955, Marco: 0.0642, Ccl2: 0.6608, Ccl7: 0.3316, Cxcl10: 0.0867, Csf1: 0.1194, cyclin D2: 0.1135, S100a4: 0.5357, S100a6: 0.4281, S100a8: 0.0642, S100a9: 0.4583, S100a10: 0.1771. LPS: Il-1 b: 0.0023, Il-6: 0.1329, Il-12 b: <0.0001, Tnfα 0.0008, Marco: 0.0001, Ccl2: 0.0673, Ccl7: 0.2708, Cxcl10:.1522, Csf1: 0.1828, cyclin D2: 0.0344, S100a4: 0.0064, S100a6: 0.0341, S100a8: 0.0009, S100a9: 0.5782, S100a10: 0.0352.
Supplementary Figure 8: Determination of intestinal bacterial loads and complexity. (37 KB), (a) qPCR analysis for eubacterial 16S rRNA could not detect significant amounts of bacteria in the caeca of GF and antibiotic-treated SPF animals. Representative data from fifteen SPF, five GF, six SPF (ABX), five ASF and five ASF (recol.) mice are depicted. Gene copy numbers were normalized to DNA content. Each symbol represents one mouse. Means ± s.e.m. are indicated. Significant differences were determined by an unpaired t test and marked with asterisks (*P < 0.05, **P < 0.01). P values: SPF vs. GF: 0.0151, SPF vs. SPF (ABX): 0.0046, SPF vs. ASF (recol.): 0.0280. (b) Recolonized ASF mice (ASF recol.) harbour a greater microbial diversity as detected by qPCR using group specific primers. Besides Parabacteroides (ASF357), Lactobacillus (ASF 361) and Clostridium (ASF356), mouse intenstinal Bacteroides (mib) and Enterobacteriaceae groups are detectable in the caecum of SPF and re-colonised ASF animals using group-specific primers, respectively.
Supplementary Figure 9: Microglia maturation is independent from the TLRs2,3,4,7,9. (73 KB), (a) Left: View of caecum from a SPF-TLRwt (control, left) and SPF-TLR2,3,4,7,9-deficient (SPF-TLRko) mouse (right). Ruler scale is shown. Representative pictures are shown. Right: Relative caecum weight of control SPF-TLRwt and SPF-TLRko mice. Symbols represent individual mice, with six animals per group. Means ± s.e.m. are shown. No statistically significant differences were found by an unpaired t test. (b) Iba-1 immunohistochemistry in the cortex of SPF-TLRwt and SPF-TLRko animals (left) and quantification thereof (right). Every symbol represents one mouse. Six animals per group were analyzed and three to four sections per mouse were examined. Data are representative of two independent experiments. Means ± s.e.m. are indicated. No significant differences could be determined by an unpaired t test. (c) Ddit4 mRNA measured by qRT-PCR in microglia from SPF-TLRwt (black bar) or SPF-TLRko (white bar) mice. Means ± s.e.m. with six samples in each group are displayed. No significant differences could be determined by an unpaired t test. Data are representative of two independent experiments. (d) Three-dimensional reconstruction (Scale bars: 15μm) and Imaris-based automatic quantification of cell morphometry (e) of Iba-1+ microglia. Individual symbols represent one mouse per group with at least three measured cells per mouse. Six mice per group were analyzed. Means ± s.e.m. are indicated. No significant differences were detected by an unpaired t test. (f) Flow cytometry of isolated microglia from control six SPF-TLRwt and six SPF-TLRko individuals. Percentages and mean fluorescence intensities (MFI) of the indicated surface molecules on microglia are shown. Each symbol represents one mouse. Means ± s.e.m. are indicated. Significant differences were evaluated by an unpaired t test and labelled with asterisks (*P < 0.05). P value: CD31 (MFI): 0.0419. Data are representative of two independent experiments.
Supplementary Table 1: Altered microglial gene profile and immaturity in germ-free animals. (33 KB) Significantly down- (upper table) and up- (lower table) regulated genes subjected to functional GO cluster analysis. The significance of genes is indicated by the given P values.
Supplementary Table 2: Reduced gene induction in microglia from non-colonized GF animals. (42 KB) Affymetrix Genome 430A 2.0 array-based gene induction of microglia from SPF mice compared to GF individuals. 78 genes significantly differentially expressed in GF vs. SPF microglia 6 hr after i.c. LPS treatment are shown. Only genes also significantly up- or downregulated by LPS treatment compared to PBS treated controls of the same housing conditions (GF and SPF respectively) are included to account for differences in basal gene regulation. (P < 0.01).


