Abstract
Periampullary adenocarcinomas can be of two histological subtypes, intestinal or pancreatobiliary. The latter is more frequent and aggressive, and characterized by a prominent desmoplastic stroma, which is tightly related to the biology of the cancer, including its poor response to chemotherapy. Whereas miRNAs are known to regulate various cellular processes and interactions between cells, their exact role in periampullary carcinoma remains to be characterized, especially with respect to the prominent stromal component of pancreatobiliary type cancers. The present study aimed at elucidating this role by miRNA expression profiling of the carcinomatous and stromal component in twenty periampullary adenocarcinomas of pancreatobiliary type. miRNA expression profiles were compared between carcinoma cells, stromal cells and normal tissue samples. A total of 43 miRNAs were found to be differentially expressed between carcinoma and stroma of which 11 belong to three miRNA families (miR‐17, miR‐15 and miR‐515). The levels of expression of miRNAs miR‐17, miR‐20a, miR‐20b, miR‐223, miR‐10b, miR‐2964a and miR‐342 were observed to be higher and miR‐519e to be lower in the stromal component compared to the carcinomatous and normal components. They follow a trend where expression in stroma is highest followed by carcinoma and then normal tissue. Pathway analysis revealed that pathways regulating tumor–stroma interactions such as ECM interaction remodeling, epithelial–mesenchymal transition, focal adhesion pathway, TGF‐beta, MAPK signaling, axon guidance and endocytosis were differently regulated. The miRNA‐mRNA mediated interactions between carcinoma and stromal cells add new knowledge regarding tumor‐stroma interactions.
Keywords: Periampullary adenocarcinoma, Tumor‐stroma interaction, miRNA/mRNA expression profiling, Stromal reaction, Pathway analysis, Tumor microenvironment, Bioinformatics and statistical analyses
1. Introduction
Adenocarcinomas originating from the pancreatic duct, the distal common bile ducts and the ampulla are collectively referred to as periampullary adenocarcinomas (PAs), all located in the pancreatic head. They are treated by the same surgical treatment and their diagnostic distinction may be difficult clinically, radiologically and morphologically. Recently, two histological subtypes of PAs were described: intestinal and pancreatobiliary (Westgaard et al., 2008). As implicated by the names, the latter has the typical histomorphology of pancreatic or distal bile duct cancer, and shares with these also the poor outcome. According to the Norwegian Cancer Registry (2014), the 5‐year survival rate for pancreatic ductal adenocarcinomas (PDAC), which is the most common of PAs, lies at approximately 5% and has not improved during the last 10 years. Despite a large number of studies that aimed at improving survival for PDAC, treatment strategies have remained largely unsuccessful (Oettle, 2014). The high levels of heterogeneity between individual tumors as well as the extensive stromal component of the cancer (up to 80% of tumor mass) are considered main obstacles to effective treatment (Oettle, 2014). The stroma also plays an active role in disease progression and resistance to chemotherapy (Feig et al., 2012) and represents a relevant target for cancer treatment. It constitutes the microenvironment of the cancer cells and consists of fibroblasts, stellate cells, extracellular matrix, vasculature, peripheral nerves and immune cells (De Wever and Mareel, 2003). Studies have shown that miRNAs play an important role in regulating the tumor microenvironment (Zhang et al., 2013, 2014). It is regulated by different mechanisms like epithelial mesenchyme transition (EMT), extracellular matrix (ECM) remodeling, recruitment of metastasis‐promoting stromal cells, immune escape, hypoxia, angiogenesis and modulation of key effectors such as cancer‐associated fibroblasts (CAF), cell adhesion molecules (CAM) and matrix metalloproteinases (Zhang et al., 2013, 2014).
Studies have compared miRNA expression in pancreatic cancer with normal tissue and pancreatitis (Lee et al., 2007, 2007). Recently, we performed an integrated expression analysis of mRNAs and miRNAs, comparing PAs with normal tissues (Sandhu et al., 2015). Lately, research focus has been shifted toward the interaction between cancer cells and its stromal components; more specifically to understand early stromal activity and the effect of stellate cells in carcinogenesis (Erkan et al., 2012). Studies on the role of miRNA and tumor microenvironment in different cancers have revealed that miRNAs are not only regulators of various cellular processes but also affect the interaction between carcinoma and stromal cells (Zhang et al., 2014). To the best of our knowledge, none of the published studies have used expression profiling of miRNAs of both the carcinomatous and stromal components from the same PAs. However, mRNA expression profiling of cancer and stromal cells has previously been compared in pancreatic cancer cell lines and stromal fibroblast induced by co‐culture (Sato et al., 2004). Stromal cells have been associated with the promotion of cancer cell growth, invasion, metastatic progression, de‐differentiation, and resistance to therapy (Erkan et al., 2012). Furthermore, recent data from mouse model studies uncovered a potentially host‐protective role of the stroma in PDAC. Indeed, stromal depletion resulted in more aggressive, undifferentiated PDAC with enhanced EMT, profiles of infiltrating immune cells were altered, increased proliferation of cancer cells, attenuated angiogenesis and increased response to immune checkpoint blockade (Rhim et al., 2014, 2014; Gore and Korc, 2014).
In the present study, we identified differentially expressed miRNAs in stromal versus carcinoma cells in formalin fixed paraffin embedded (FFPE) tissues from patients with PA (n = 20). In addition, carcinoma and stromal tissue profiles were compared with those from normal pancreatic tissues (n = 8) that were contained in the same specimens. Pathway analysis was carried out using mRNAs that were anti‐correlated and were predicted targets of miRNAs. The aim was to identify the role of these miRNAs and the pathways that may be crucial in tumor–stroma interactions.
2. Materials and methods
2.1. Patients and specimens
A total of 20 patients with PA (10 PDACs, 9 bile duct adenocarcinomas and 1 ampullary adenocarcinoma, all of pancreatobiliary type) treated at Oslo University Hospital, were included in the present study. The median age of the patients was 69 years (range 38–79 years), seven were males and 13 females. The macroscopic and microscopic pathology examination followed a standardized protocol. The clinicopathologic characteristics of the patients are presented in Table 1. All PAs were stage T3, except for one PDAC that was limited to the pancreas (T1) and one of the bile duct adenocarcinomas that invaded adjacent structures (T4). One of the PDACs had metastasized to liver (M1 metastasis status). The present study was approved by the Regional Ethics Committee, and written informed consent was obtained from each patient.
Table Table 1.
Clinicopathologic characteristics of study series.
| Number (n = 20) | Percentage | Median | ||
|---|---|---|---|---|
| Gender | Female | 13 | 65 | – |
| Male | 7 | 35 | – | |
| Differentiation | Well | 0 | 0 | – |
| Moderate | 11 | 55 | – | |
| Poor | 8 | 40 | – | |
| Undifferentiated | 1 | 5 | – | |
| pT | T1 | 1 | 5 | – |
| T2 | 0 | 0 | – | |
| T3 | 18 | 90 | – | |
| T4 | 1 | 5 | – | |
| N | N0 | 6 | 30 | – |
| N1 | 14 | 70 | – | |
| M | M0 | 19 | 95 | – |
| M1 | 1 | 5 | – | |
| R | R0 | 8 | 40 | – |
| R1 | 12 | 60 | – | |
| Vessel infiltration | No | 9 | 45 | – |
| Yes | 11 | 55 | – | |
| Perineural infiltration | No | 1 | 5 | – |
| Yes | 19 | 95 | – | |
| KRAS codons 12/13 | Mutated | 15 | 75 | – |
| Wild type | 5 | 25 | – | |
| Age (in years) | – | – | 69.00 | |
| Disease free survival (DFS) (in months) | – | – | 17.34 | |
| Overall survival (OS) (in months) | – | – | 21.25 | |
2.2. Sampling of carcinoma and stromal tissues
Hematoxylin and eosin (HE) stained sections from FFPE tissue blocks were examined by a pathologist, who marked areas with carcinoma, stroma and normal cells. Core tissue biopsies (diameter 1.0 mm; height 1–2 mm) were sampled from the selected areas of carcinoma and stroma, using Beecher Instruments' Manual Tissue Arrayer (MTA‐1) from Estigen. The stroma located in close vicinity of the tumor was labeled. Using microdissected FFPE tissues enabled us to get very clean samples of stromal cells. However, the pancreatic cancer samples are heterogeneous and grow in a highly dispersed fashion. The tumor samples were sampled from regions with the higher tumor burden, preferentially >50–75% carcinoma cells. They may contain some lower percentages of stromal cells and inflammatory cells but most of the samples were predominated by carcinoma cells (Supplementary Table S1). The selected regions were examined and marked by trained pathologists. In each case new HE sections were made from these regions and the pathologist ensured that the proper tissues had been sampled from the marked areas. Two carcinoma and two stromal samples were discarded from the analysis since the contents of carcinoma or stromal cells were <10%. Paired set of carcinoma/stroma samples could be taken from 16 patients (n = 32 samples), while either only stroma samples (n = 2) or carcinoma samples (n = 2) were taken from two additional patients. Four of the 36 carcinoma samples were borderline with the content of carcinoma cells ranging from 30 to 50%. The normal samples (n = 8) were mainly composed of pancreatic acinar parenchyma cells with normal epithelium and some normal stroma. Areas of fibrosis were avoided when collecting the samples.
2.3. RNA extraction
RNA was extracted from the FFPE tissue samples by using the miRNeasy FFPE kit (Qiagen) as described by the manufacturer.
2.4. miRNA array profiling of carcinoma and stromal tissues
The tissue core biopsies of carcinoma (n = 18), stroma (n = 18) and normal (n = 8) tissues were subjected to miRNA array profiling by Exiqon Services, Denmark. The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. The quality control (QC) report of the samples is available in Supplementary Figure S1. A total of 700 ng RNA from both the sample and the universal reference was labeled with Hy3™ and Hy5™ fluorescent labels, respectively, using the miRCURY LNA™ microRNA Hi‐Power Labeling Kit, Hy3™/Hy5™ (Exiqon, Denmark) as described by the manufacturer. The Hy3TM‐labeled samples and a Hy5TM‐labeled reference RNA sample were mixed pair‐wise and hybridized to the miRCURY LNA™ microRNA Array 7th (Exiqon, Denmark), which captures probes targeting all miRNAs for human tissues (n = 2042) that are registered in the miRBASE 18.0 (Griffiths‐Jones et al., 2006). Hybridization was performed according to the miRCURY LNA™ microRNA Array Instruction manual using a Tecan HS4800™ hybridization station (Tecan, Austria). Following hybridization, the microarray slides were scanned and stored in an ozone‐free environment (ozone level below 2.0 ppb) in order to prevent potential bleaching of the fluorescent dyes. The miRCURY LNA™ microRNA Array slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., USA), and the image analysis was carried out using ImaGene® 9 (miRCURY LNA™ microRNA Array Analysis Software, Exiqon, Denmark). The miRNA expression data is accessible through GEO accession number GSE71533.
2.5. miRNA expression analysis, pre‐processing and normalization
The quantified signals from FFPE carcinoma, stromal and normal tissues were background corrected using the Normexp background correction method with offset value 10 (Ritchie et al., 2007). The samples were normalized using the quantile normalization method, which is found to produce the best between‐slide normalization to minimize the intensity‐dependent differences between samples. The threshold of detection was calculated for each individual microarray slide as 1.2 times the 25th percentile of the overall signal intensity of the slide. The miRNAs with intensities above threshold observed in less than 20% of the samples were removed from the final data set that was used for the expression analysis. For the present data set, a total of 1405 probes were discarded by this filtering procedure. A total of 677 miRNAs were used for further expression analysis.
2.6. mRNA expression data
The mRNA expression data for fresh frozen PAs (n = 14) and normal samples (n = 12), available in GEO with accession number GSE60979 were used in this study (Sandhu et al., 2015). The mRNA data was background corrected and quantile normalized. The moderated t‐test (Smyth, 2004; Smyth et al., 2005) was used for identifying differentially expressed mRNAs between tumor and normal samples at adjusted P value < 0.05 (Benjamini and Hochberg, 1995).
2.7. Bioinformatics and statistical analyses
2.7.1. Unsupervised analysis
The unsupervised analysis was done using hierarchical clustering and sparse principal component analysis.
2.7.1.1. Hierarchical clustering
Unsupervised hierarchical clustering of 18 carcinoma samples, 18 stromal tissues and eight normal samples was carried out using the most variant miRNAs (n = 183) having standard deviation (s.d) > 0.8. The samples were clustered using the complete linkage method. Pearson's correlation coefficient and Spearman's rank correlation method were used as a distance measure for miRNAs and samples, respectively. The heatmap is plotted using the gplot library in R.
2.7.1.2. Sparse principal component analysis
Sparse Principal Component Analysis (SPCA) is an extension to the traditional principal component analysis (Jolliffe, 1986) that is more concise and gives a more interpretable set of principal components for large genomic data by applying Penalized Matrix Decomposition (PMD) (Witten and Tibshirani, 2009). It thus reduces the dimensions of large data sets and therefore represents a useful way of exploring the naturally arising sample classes based on expression profile. SPCA of all samples was carried out using the 183 most variable miRNAs. Firstly, cross validation was performed on first Sparse Principal Component (SPC) using the SPC.cv function in R. This helps in selecting the tuning parameter for SPCA, which involves applying PMD to a data matrix with lasso penalty on the columns and no penalty on the rows. The tuning parameter controls the sum of absolute values of the elements of SPCA. After tuning the parameters the SPCA was performed using the PMA library in R with parameters (sumabsv = 4 and K = 1) tuned using the SPC cross validation function (Witten et al., 2009, 2009). The SPCA plot was plotted using ggplot2 library in R.
2.7.2. Identification of differentially expressed miRNAs in carcinoma and stroma
The Moderated t‐tests (Smyth, 2004; Smyth et al., 2005) were carried out to identify miRNAs that are differentially expressed, between carcinoma and stromal tissues; carcinoma and normal tissues; stromal and normal tissues at Benjamini & Hochberg (BH) adjusted P < 0.05 (Benjamini and Hochberg, 1995) using the Limma module of the Bioconductor package in R. The results were confirmed using the non‐parametric Mann–Whitney test (Mann et al., 1947) at BH‐adjusted P < 0.05. Analysis of Variance (One‐way ANOVA) was carried out to identify relative miRNA expression patterns in carcinoma, stromal and normal tissues with BH‐adjusted P < 0.05 using ANOVA package in R. The results were confirmed using non‐parametric Kruskal–Wallis ANOVA (Kruskal and Wallis, 1952) at BH‐adjusted P < 0.05.
2.7.3. Correlations between miRNA and mRNA levels and target site prediction
Firstly, all miRNAs differentially expressed in carcinomas and stromal samples (n = 43) were compared to all mRNAs differentially expressed in tumor and normal samples in order to identify negative correlations. A Pearson correlation test was applied for this purpose, with significance threshold P < 0.05 and 95% CI for both stromal and carcinoma miRNA expression versus tumor sample (mRNA). Secondly, the miRNA‐mRNA pairs found in the first step were further predicted by miRanda (John et al., 2003, 2004) in microRNA.org “http://www.microrna.org/microrna/” August 2010 release (Betel et al., 2008) and Targetscan release 6.2 (Lewis et al., 2005, 2003).
All the miRNA‐mRNA pairs in this two‐step curation were reported and further checked against experimentally validated interactions in the miRTarbase4 database (release 4.5, Nov, 2013) (Hsu et al., 2011, 2014).
2.7.4. Gene set enrichment analysis
The Gene set enrichment analysis was carried out using the WebGestalt tool (Zhang et al., 2013, 2005). WebGestalt uses an hypergeometric test for enrichment evaluation analysis at P < 0.05 after BH correction. The minimum number of genes required for a pathway to be considered significant is set to 10. The WebGestalt analysis gives pathway enriched genes, number of genes enriched, raw P value (rawP) from the Hypergeometric test and Benjamin and Hochberg corrected P value (adjP). The genes significantly associated with differentially expressed miRNAs in carcinoma and stroma after two‐step curation in the previous step was used for the enrichment analysis.
2.7.5. Plotting the miRNA‐mRNA‐pathways interactions
The candidate miRNA target interactions in the enriched pathways were plotted using the tool Gephi version beta 0.8.2 (Bastian et al., 2009).
3. Results
An overview of the analysis pipeline carried out using the miRNA carcinoma/stroma and the mRNA expression data is summarized in Figure 1.
Figure 1.
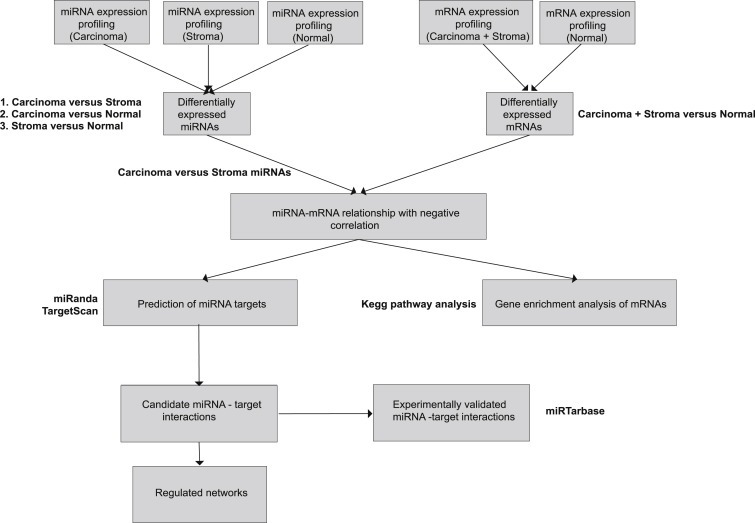
Analysis pipeline of the PA samples using miRNA and mRNA expression profiling.
3.1. Unsupervised clustering analysis of expression data
Unsupervised hierarchical clustering of miRNAs (n = 183) showing intrinsically variable expression revealed separation of carcinoma, stromal and normal tissues (Figure 2a). The carcinoma and stromal samples clustered separately at P = 9.071e‐09 (Fisher's exact test). SPCA was carried out to identify clusters based on the explained variance. The unsupervised analyses of the 183 most variable miRNAs revealed separation of the samples into carcinoma, stroma and normal (Figure 2b). The normal samples clustered separately from the carcinoma and the stromal samples. An overlap between the carcinoma and stromal samples was observed in the heatmap for four of the samples, whereas, SPCA clustered carcinoma, stroma and normal samples separately.
Figure 2.
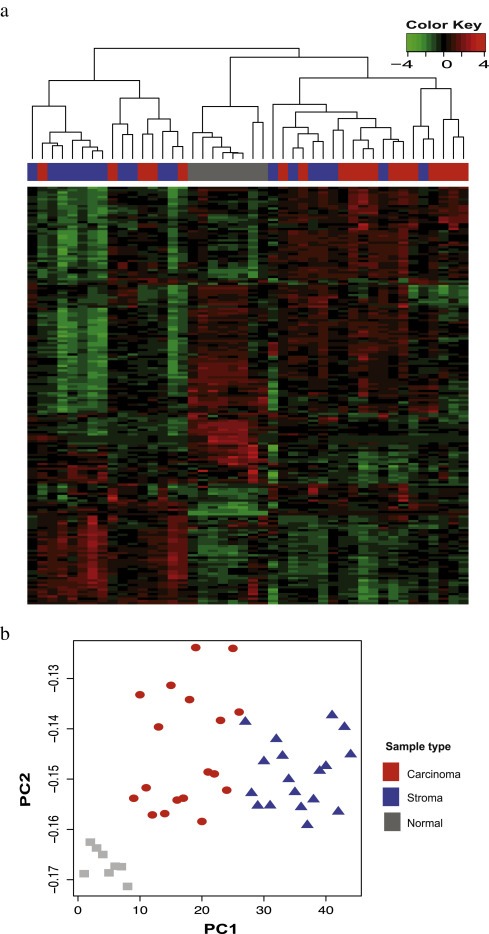
2a) The figure shows the heatmap of all the samples with the column representing the samples and the row representing the miRNAs. The samples were clustered using the complete linkage method. Pearson's correlation coefficient and Spearman's rank correlation were used as a distance measure for miRNAs and samples, respectively. The heatmap shows the separation of carcinoma, stromal and normal samples based on expression levels of the most intrinsically variable miRNAs (n = 183). The carcinoma and stromal tissues clustered separately at P = 9.071e‐09 for Fisher's exact test. 2b) The figure shows the scatterplot of the first two principal components identified by the SPCA performed on all the samples and on 183 miRNAs with s.d > 0.8. The color and shape of the points refer to type of the tissues, and the x‐axis represents the first principal component and the y‐axis represents the second principal component of the samples.
3.2. miRNA expression analysis
The analysis using the Moderated t‐test (modT) found that the numbers of differentially expressed miRNAs between carcinoma versus stromal tissues, stromal versus normal tissues and carcinoma versus normal tissues were 43, 287 and 331, respectively at P < 0.05 and adjusted for FDR correction < 5% using BH correction. The Venn diagram (Figure 3) shows the number of differentially expressed miRNAs that are common and unique in the different comparisons. There is a high number (n = 228) of differentially expressed miRNAs common to both carcinoma and stromal tissues as compared to the normal tissue (P = 2.2e‐44).
Figure 3.
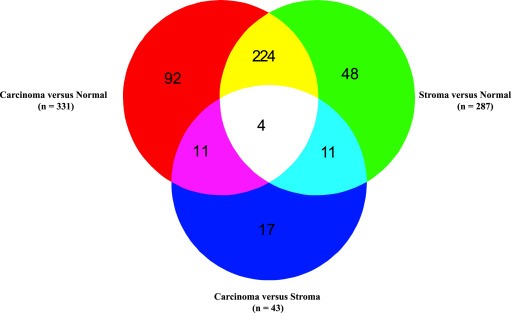
The Venn diagram shows the numbers of differentially expressed miRNAs with respect to comparisons between carcinoma, stroma and normal tissues.
The miRNAs differentially expressed in stromal tissue versus carcinoma at BH‐adjusted P < 0.05 are enlisted in Table 2. A total of 26 and 17 miRNAs were upregulated and downregulated in stromal tissue, respectively. Some of the differentially expressed miRNAs relate to known miRNA families; miR‐17, miR‐15 and miR‐515. The miR‐17 (miR‐17, miR‐20a, miR‐20b, miR‐106a) and miR‐15 (miR‐15a, miR‐15b, miR‐16, miR‐195) families were upregulated in stroma compared to carcinoma. miR‐515 (miR‐518a, miR‐519d, miR‐519e) family members were downregulated in stroma compared to carcinoma. These miRNA families were curated from mirbase database (release 21, June 2014) (www.mirbase.org), (Griffiths‐Jones et al., 2006).
Table Table 2.
(a–b): List of differentially expressed miRNAs in stroma versus carcinoma.
| 2a. miRNAs upregulated in stroma compared to carcinoma samples at FDR adjusted P < 0.05. | ||||||
|---|---|---|---|---|---|---|
| No. | miRNA | Log FC | Average expression | Calculated value of t statistic | P value | FDR |
| 1 | miR‐144‐3p | −2.019 | 7.119 | −6.135 | 0.000 | 0.000 |
| 2 | miR‐451a | −2.613 | 10.244 | −5.730 | 0.000 | 0.000 |
| 3 | miR‐126‐3p | −1.298 | 9.379 | −5.037 | 0.000 | 0.002 |
| 4 | miR‐140‐3p | −1.089 | 7.194 | −4.683 | 0.000 | 0.004 |
| 5 | miR‐126‐5p | −0.873 | 7.619 | −4.320 | 0.000 | 0.010 |
| 6 | miR‐223‐3p | −1.268 | 8.682 | −4.304 | 0.000 | 0.010 |
| 7 | miR‐16‐5p | −1.230 | 9.828 | −4.218 | 0.000 | 0.011 |
| 8 | miR‐10b‐5p | −0.661 | 7.655 | −4.188 | 0.000 | 0.011 |
| 9 | miR‐142‐5p | −1.232 | 7.286 | −4.126 | 0.000 | 0.012 |
| 10 | miR‐342‐3p | −0.922 | 7.075 | −3.853 | 0.000 | 0.021 |
| 11 | miR‐20b‐5p | −0.821 | 7.091 | −3.743 | 0.001 | 0.025 |
| 12 | miR‐191‐5p | −0.853 | 7.782 | −3.665 | 0.001 | 0.027 |
| 13 | miR‐195‐5p | −1.102 | 8.682 | −3.635 | 0.001 | 0.027 |
| 14 | miR‐130a‐3p | −0.941 | 8.265 | −3.613 | 0.001 | 0.027 |
| 15 | miR‐17‐5p | −0.966 | 7.419 | −3.593 | 0.001 | 0.027 |
| 16 | miR‐15b‐5p | −0.675 | 7.501 | −3.586 | 0.001 | 0.027 |
| 17 | miR‐15a‐5p | −0.736 | 8.357 | −3.566 | 0.001 | 0.027 |
| 18 | miR‐139‐5p | −0.420 | 6.284 | −3.557 | 0.001 | 0.027 |
| 19 | miR‐106a‐5p | −0.752 | 6.685 | −3.454 | 0.001 | 0.033 |
| 20 | miR‐20a‐5p | −0.829 | 8.089 | −3.434 | 0.001 | 0.034 |
| 21 | miR‐4301 | −1.137 | 10.638 | −3.327 | 0.002 | 0.041 |
| 22 | miR‐140‐5p | −0.694 | 6.967 | −3.320 | 0.002 | 0.041 |
| 23 | miR‐142‐3p | −1.130 | 9.648 | −3.316 | 0.002 | 0.041 |
| 24 | miR‐19b‐3p | −0.763 | 7.800 | −3.201 | 0.003 | 0.049 |
| 25 | miR‐2964a‐5p | −0.492 | 10.990 | −3.178 | 0.003 | 0.049 |
| 26 | miR‐378a‐3p | −0.364 | 7.698 | −3.133 | 0.003 | 0.049 |
| 2b. miRNAs downregulated in stroma compared to carcinoma samples at FDR adjusted P < 0.05 | ||||||
| No. | miRNA | Log FC | Average expression | Calculated value of t statistic | P value | FDR |
| 1 | miR‐519e‐3p | 0.515 | 7.052 | 3.987 | 0.000 | 0.017 |
| 2 | miR‐519d | 1.107 | 8.433 | 3.919 | 0.000 | 0.019 |
| 3 | miR‐615‐3p | 0.513 | 7.887 | 3.819 | 0.000 | 0.021 |
| 4 | miR‐4633‐5p | 0.742 | 10.231 | 3.697 | 0.001 | 0.027 |
| 5 | miR‐551b‐5p | 0.539 | 6.155 | 3.635 | 0.001 | 0.027 |
| 6 | miR‐4506 | 0.630 | 6.826 | 3.488 | 0.001 | 0.031 |
| 7 | miR‐4436b‐5p | 0.602 | 9.532 | 3.355 | 0.002 | 0.041 |
| 8 | miR‐518a‐5p/527 | 0.843 | 6.789 | 3.303 | 0.002 | 0.041 |
| 9 | miR‐636 | 0.415 | 6.376 | 3.260 | 0.002 | 0.045 |
| 10 | miR‐5010‐3p | 0.341 | 6.983 | 3.207 | 0.002 | 0.049 |
| 11 | miR‐596 | 0.542 | 6.779 | 3.183 | 0.003 | 0.049 |
| 12 | miR‐3676‐5p | 0.492 | 10.425 | 3.170 | 0.003 | 0.049 |
| 13 | miR‐4288 | 0.563 | 9.374 | 3.164 | 0.003 | 0.049 |
| 14 | miR‐5571‐5p | 0.934 | 7.865 | 3.146 | 0.003 | 0.049 |
| 15 | miR‐4742‐3p | 0.385 | 8.269 | 3.136 | 0.003 | 0.049 |
| 16 | miR‐302c‐5p | 0.508 | 7.561 | 3.127 | 0.003 | 0.049 |
| 17 | miR‐1298 | 0.553 | 7.741 | 3.123 | 0.003 | 0.049 |
The miRNAs that were identified by the modT test and the Mann–Whitney test in stromal versus carcinoma tissues at BH‐adjusted P < 0.05 are enlisted in Table 2. The miRNAs that were identified to be differentially expressed between carcinoma versus normal tissues and stromal versus normal tissues using the Moderated t‐test are enlisted in the Supplementary Tables (S2a and S2b, respectively).
ANOVA test was performed on all the miRNAs to identify relative miRNA expression patterns in the three groups i.e. carcinoma, stromal and normal tissues. The test identified eight miRNAs that were upregulated in stromal as compared to carcinoma and normal tissue samples at BH‐adjusted P < 0.05. The relative miRNA expression of six out of these eight miRNAs follows a decreasing trend, from stromal to carcinoma to normal tissues (Figure 4), while only one miRNA (miR‐519e‐3p) shows an increasing trend from stromal to carcinoma to normal tissues. miRNA‐2964a‐5p exhibits a unique pattern, as it is highly overexpressed in both stroma and carcinoma as compared to normal tissue. It is noted that the boxplots show moderate deviation from symmetry for expression of miRNAs in stroma and carcinoma as compared to normal tissue. Compared to carcinoma, the stroma shows a higher degree of dispersion towards the upper limit. The results are consistent with the Kruskal–Wallis rank sum test at BH‐adjusted P < 0.05 (Supplementary Table S2c). The boxplots of 35/43 differentially expressed miRNAs between carcinoma versus stromal tissues are available in Supplementary Figure S2.
Figure 4.
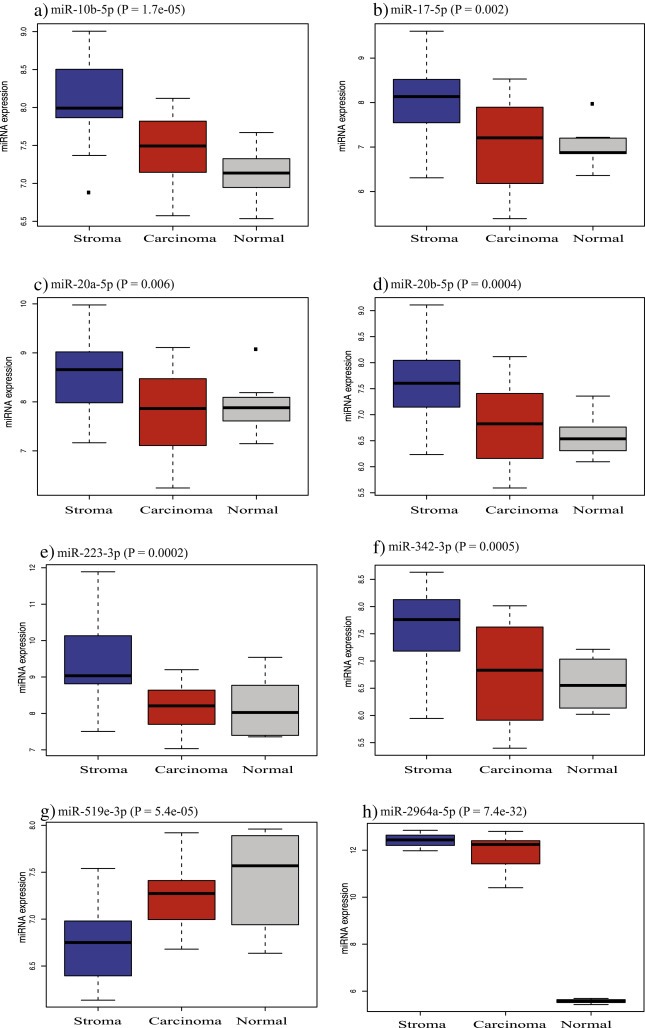
The boxplots show the relative expression of the eight most differentially expressed miRNAs in stroma (blue), carcinoma (red) and normal tissues (grey) at BH‐adjusted P < 0.05.
3.3. Anti‐correlation analysis and miRNA target prediction
The total of 8670 mRNAs were found differentially expressed between tumor and normal samples (Supplementary Table S3). These differentially expressed mRNAs were used for anti‐correlation analysis against 43 miRNAs differentially expressed between carcinoma and stroma. The two‐step analyses for candidate miRNA‐mRNA target networks (see Materials and method section 2.7.3) found 2219 carcinoma miRNA‐mRNA and 1363 stroma miRNA‐mRNA interactions. The total of 104 and 163 interactions were found to be experimentally validated between the carcinoma miRNA‐mRNA and stroma miRNA‐mRNA, respectively (Supplementary Tables S4a and S4b).
3.4. Pathway analysis
The list of mRNAs significantly associated with differentially expressed miRNAs between carcinoma and stroma samples were used for pathway analysis using the WebGestalt tool. Ten pathways were found to be enriched in the analysis (Table 3).
Table Table 3.
Pathway analysis using mRNAs anti‐correlated to miRNAs differentially expressed between carcinoma versus stromal expression and predicted as miRNA target sites by miRanda and Targetscan tools.
| No. | Pathways | P value | Adjusted P value | Predicted miRNA‐mRNA interactions (in carcinoma) | Predicted miRNA‐mRNA interactions (in stroma) |
|---|---|---|---|---|---|
| 1 | Metabolic pathways | 1.42e‐15 | 2.58e‐13 | 83 | 31 |
| 2 | Endocytosis | 1.21e‐12 | 1.10e‐10 | 14 | 40 |
| 3 | Pathways in cancer | 2.52e‐11 | 1.53e‐09 | 48 | 79 |
| 4 | Axon guidance | 3.02e‐10 | 1.38e‐08 | 18 | 34 |
| 5 | MAPK signaling | 7.87e‐09 | 2.86e‐07 | 43 | 42 |
| 6 | Cell cycle | 2.98e‐08 | 9.04e‐07 | 5 | 52 |
| 7 | Focal adhesion | 6.42e‐08 | 1.67e‐06 | 31 | 40 |
| 8 | ECM receptor remodelling | 1.24e‐07 | 2.82e‐06 | 17 | 13 |
| 9 | TGF beta signaling | 1.12e‐07 | 2.30e‐06 | 11 | 15 |
| 10 | p53 signaling pathway | 2.24e‐06 | 4.08e‐05 | 5 | 31 |
3.5. Proliferation associated miRNAs and mRNAs
We identified five of 11 proliferation genes included in the PAM50 gene expression signature (Parker et al., 2009) to be differentially expressed between tumors and normal tissues. The proliferation genes were anti‐correlated to the carcinoma miRNA expressions and predicted by the miRNA target prediction softwares (Figure 5). Interactions between miR‐20a and CCNB1, miR‐140 and UBE2C, miR‐16 and UBE2C, miR‐17 and UBE2C, miR‐20a and UBE2C, miR‐16 and MKI67 and miR‐16 and CEP55 were found to be experimentally validated interactions.
Figure 5.

The figure shows the heatmap of the miRNAs and mRNAs associated with proliferation of the cells. 5a) The heatmap shows expression of the miRNA anti‐correlated to the proliferation genes in the carcinoma and stromal samples 5b) The heatmap shows expression of proliferation genes in the periampullary adenocarcinoma samples. 5c) The miRNA‐mRNA pairs anti‐correlated at adjusted P value < 0.05.
3.6. The miRNA‐mRNA pathway network
The miRNA‐mRNA interactions for the deregulated pathways were plotted for the stromal (Figure 6a) and carcinoma (Figure 6b) samples. The network showed closed interactions between the miR‐17 and the miR‐15 family members. They share many common targets. The miRNA‐mRNA pairs with significant anti‐correlation at P < 0.05 for all deregulated pathways (Table 3) are reported in Supplementary Tables S5a–j for the carcinoma and the stromal samples.
Figure 6.
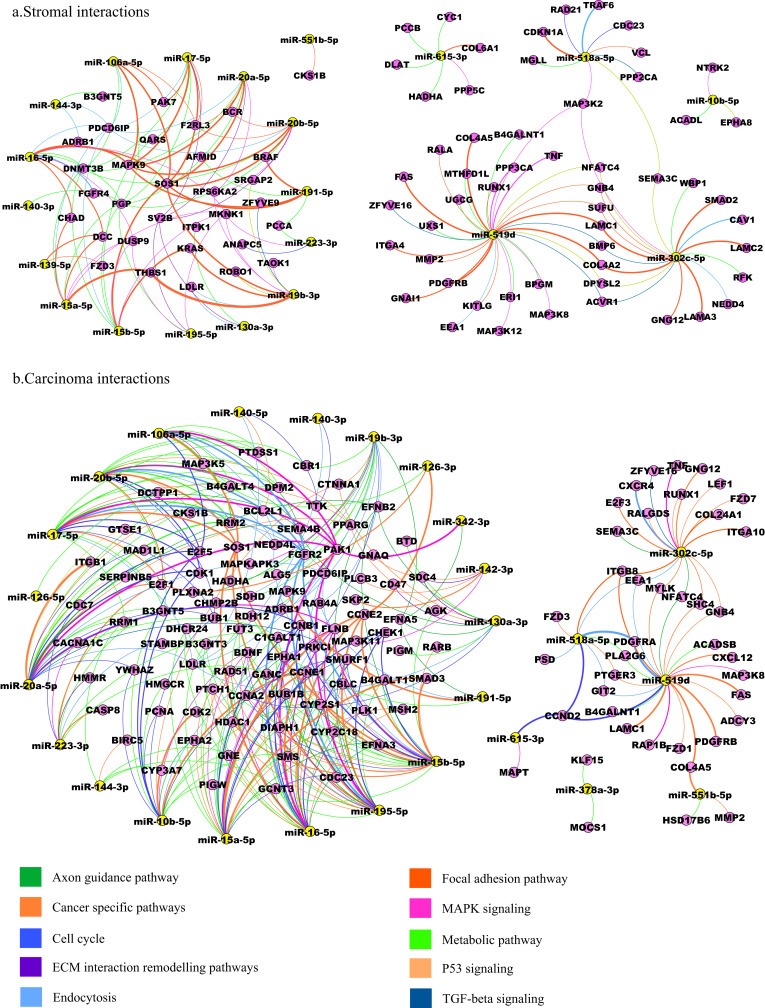
The figure shows the interaction between the miRNAs and mRNAs in the deregulated pathways in the stroma and carcinoma samples. 6a) The miRNA‐mRNA‐pathway network shows the miRNA and the targeted mRNAs in the deregulated pathways in the stromal samples. 6b) The miRNA‐mRNA‐pathway network shows the miRNA and the targeted mRNAs in the deregulated pathways in the carcinoma samples.
All the scripts and codes used in this project are available in Supplementary file S1.
4. Discussion
The present study is to our knowledge the first to analyze miRNA expression profiling of carcinoma, stromal and normal tissues from patients with PAs. The aim of the study was to identify pathways and miRNAs that may be crucial in interactions between carcinoma and associated stroma through the analysis of differentially expressed miRNAs and mRNAs.
The most differentially expressed miRNAs, miR‐144 and miR‐451a (Table 2) are located on chromosome 17 at less than 10 kb in distance from each other and are positional clusters. The positional clusters of miRNAs are known to have sequence similarity in the seed region and functional similarity with respect to the target genes and corresponding pathways (Becker et al., 2012). The miR‐17 and miR‐15 families were upregulated in stroma compared to carcinoma and the miR‐515 family was downregulated in stroma compared to carcinoma samples. The members of the miR‐17 family (miR‐20a, miR‐20b, miR‐19b and miR‐106a) and the miR‐15 family (miR‐15, miR‐16a and miR‐195) share many common target genes and are closely interacting (Figure 6a and 6b). The members of these families have been associated with key regulatory roles of carcinogenesis. Members of both the miR‐17 and the miR‐15 family targets proliferation genes (Figure 5). The miR‐15 family members, miR‐15a and miR‐16, are also known to be associated with regulation of cancer‐associated fibroblasts as well as with tumor growth in prostate cancer (Bonci et al., 2008) and angiogenesis in multiple myeloma (Sun et al., 2013).
The miRNAs that were found differentially expressed between carcinoma and stroma play significant roles in various cancers. Briefly, miR‐191 and miR‐130a are associated with cancer progression in breast and colorectal cancer, respectively (Nagpal et al., 2013; Liu et al., 2013). miR‐223 is involved in migration and invasion by targeting the Mef2c gene in breast cancer (Yang et al., 2011). It is also linked to increased migration, invasion and decreased cell adhesion in gastric cancer (Li et al., 2011). Both miR‐126 and miR‐378 have roles in angiogenesis and in repressing the recruitment of mesenchymal stem cells in various cancers (Chou et al., 2013; Lee et al., 2007a). miR‐17, miR‐19b, miR‐451a, and miR‐139 all have roles in the regulation of cancer‐associated fibroblasts by targeting various downstream cytoskeleton regulatory proteins, cell–cell adhesion molecules and cell‐matrix molecules (Valastyan and Weinberg, 2011). We identified miR‐17 and miR‐19 in focal adhesion and in the ECM pathways. miR‐195 has a function in proliferation, migration and cell adhesion to the extracellular matrix in endocrine cancers (Jain et al., 2014). In pancreatic cancer, miR‐106a suppresses the translation of CDKN1A, PTEN, TGFBR2 and KLF11, activates proliferation‐signaling pathways, and induces EMT (Jung et al., 2012). miR‐144 induces EMT by downregulating the Notch‐1 gene through an miR‐144 regulated mechanism (Sureban et al., 2011). In breast cancer, miR‐10b induces EMT by targeting TGF‐β1 (Han et al., 2014). Upregulation of the miR‐515 family members, miR‐515‐3p, miR‐519d and miR‐518e, is associated with metastatic colonization of melanoma cells (Mueller et al., 2009). The miR‐10b, miR‐155 and miR‐106a were recently identified in bile and plasma of pancreatic cancer patients and have been reported as prognostic markers (Cote et al., 2014). We found miR‐10b and miR‐106b (miR‐106a and miR‐106b are both members of the miR‐17 family) to be upregulated in the stroma, which may be the source of these miRNAs in plasma.
The pathways that are dysregulated in PAs include various pathways (Table 3). Pathways like MAPK, TGF‐beta, p53 and cell cycle signaling have previously been reported in an integrated whole genome analysis of mRNAs and miRNAs in PA samples (Sandhu et al., 2015). Other pathways dysregulated in the present study include the ECM interaction, focal adhesion molecule signaling, endocytosis and axon guidance. The integrated analysis of differentially expressed miRNA (candidate miRNAs) between carcinoma and stromal cells, and differentially expressed mRNA between carcinoma and normal samples identified pathways that may be important in regulating tumor–stroma interactions and that facilitate cell proliferation. The predicted miRNA‐mRNA interactions in carcinoma and stromal samples identified a number of genes from different pathways. Known oncogenes and tumor suppressor genes like KRAS, FAS, DCC, SMAD2, MAPK9 and CDKN1A were anti‐correlated to miRNA expressions in the stroma and were predicted as potential targets. While, in the carcinoma, genes like CCNE1, FAS, E2F1, FGFR were anti‐correlated to miRNA expressions in the carcinoma cells and were predicted as potential targets.
EMT is an important mechanism of regulating tumor stroma interaction (Zhang et al., 2014; Chou et al., 2013). Various signaling pathways like MAPK, TGF‐beta, PI3K‐AKT, JNK can cooperate to induce full EMT progression (Lamouille et al., 2014). We found TGF‐beta signaling and MAPK signaling pathway enriched in our samples. We identified the genes SMURF1, SMAD2/3 and BMP6 as targets of candidate miRNAs in the TGF‐beta pathways. SMURFs have a key role in EMT, cell migration, fibrosis and cancer (Izzi and Attisano, 2006) and SMAD2/3 form complex with SMAD4 and induces EMT (Xu et al., 2009). Growth factor receptors like FGFR2, PDGFRA were found to be associated candidate miRNA targets in the MAPK signaling pathway. These genes regulate EMT by MAPK signaling (Xu et al., 2009). In addition we found the PAK1, MAPK9 (JNK2) and TRAF6 genes in the MAPK signaling pathway, which are important genes in activation of JNK mediated EMT signaling (Lamouille et al., 2014; Alcorn et al., 2008).
Besides EMT regulation by MAPK, TGF‐beta and JNK, we found ECM interaction and the focal adhesion pathway to be dysregulated, which also regulate tumor‐stroma interaction (Zhang et al., 2013, 2014). ECM interactions not only regulate cell differentiation, its protein domains bind cell adhesion molecule, transduce signals and also regulate microenvironment of cells (Hynes, 2009). The deregulated ECM dynamics can lead to deregulated cell proliferation, migration, loss of cell differentiation and survival (Hynes, 2009). Various collagens (COL4A2, COL4A5 and COL24A1), laminins (LAMC1 and LAMC2) and integrins (ITGB8, ITGB1 and ITGA10) were differentially expressed and found to be candidate miRNA targets in focal adhesion and ECM interaction pathways. The desmoplasia was induced in pancreatic cancer by TGF‐beta signaling, collagens of ECM and the PDGF gene in pancreatic cancer (Lohr et al., 2001). The interaction analysis indicates that the TGF‐beta signaling pathway, ECM and PDGF gene in the focal adhesion pathway might be regulated by candidate miRNAs.
The endocytosis pathway helps in material uptake, regulation of signal transduction and morphogenetic processes like cell adhesion and migration. The deregulated endocytic pathway in human tumors lead to disunion of cell–cell junctions like adherence and tight junctions and transform to malignant cells by delaying endocytosis mediated inactivation of growth factor receptors (Mosesson et al., 2008). We found alterations of the RAB family gene RAB4A and the cytoskeleton genes LDLR, FGFR2, CAV1 associated with the endocytosis pathway as target of candidate miRNAs. The gene CXCR4 has previously been associated with tumor‐stroma interaction in pancreatic cancer (Singh et al., 2012), and CXCR4 was identified as a candidate target gene in the endocytosis network pathway in our study as well.
The axon guidance pathway is known to be dysregulated in pancreatic cancer (Biankin et al., 2012) and it was also enriched in our samples. Recently, studies have shown the role of nerve cells in promoting tumor progression. The mechanism involves release of neurotransmitters directly into the vicinity of tumor and stroma cells to activate corresponding membrane receptors in pancreatic cancer (Kim‐Fuchs et al., 2015, 2014). We found that genes associated with the axon guidance pathway (CXCR4, DCC, PAK7, ROBO1 and KRAS) were enriched in the interaction analysis.
A challenge in the current study was to obtain carcinoma and stroma samples with satisfactory purity. This is difficult due to the fact that pancreatobiliary PAs grow in a highly dispersed fashion. A degree of heterogeneity can therefore not be excluded and is probably reflected in the heatmap depicting hierarchical clustering (Figure 2), where four of the stromal samples clustered with their respective carcinomas.
miRNAs found to be differentially expressed between carcinoma and stromal cells may contribute to an oncogenic phenotype, and the molecular mechanisms through which they operate should be further investigated. The role of the tumor stroma in PAs is complex, while some studies indicated that it contributes to the aggressiveness of the cancer (Maehara et al., 2001), others recently suggested a protective role (Rhim et al., 2014; Ozdemir et al., 2014). In this study, miRNAs that are differentially expressed in carcinoma and stroma were identified, along with their associated pathways. The findings indicate that these miRNAs are involved in tumor–stroma interactions, which may make them attractive as potential therauptic targets.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
Supported by grants from the South‐Eastern Regional Health Authority, Hole's Foundation, The Radium Hospital Foundation, Oslo University Hospital and Telemark University College. We thank all the patients who participated in the study.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.10.011.
Sandhu V., Bowitz Lothe I.M., Labori K.J., Skrede M.L., Hamfjord J., Dalsgaard A.M., Buanes T., Dube G., Kale M.M., Sawant S., Kulkarni-Kale U., Børresen-Dale A.-L., Lingjærde O.C., Kure E.H., (2016), Differential expression of miRNAs in pancreatobiliary type of periampullary adenocarcinoma and its associated stroma, Molecular Oncology, 10, doi: 10.1016/j.molonc.2015.10.011.
References
- Alcorn, J.F. , Guala, A.S. , van der Velden, J. , McElhinney, B. , Irvin, C.G. , Davis, R.J. , 2008. Jun N-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-beta1. J. Cell Sci. 121, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian, M. , Heymann, S. , Jacomy, M. , 2009. Gephi: Anopen source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media [Google Scholar]
- Becker, L.E. , Lu, Z. , Chen, W. , Xiong, W. , Kong, M. , Li, Y. , 2012. A systematic screen reveals microRNA clusters that significantly regulate four major signaling pathways. PloS One. 7, e48474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , Hochberg, Y. , 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 57, 289–300. [Google Scholar]
- Betel, D. , Wilson, M. , Gabow, A. , Marks, D.S. , Sander, C. , 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36, D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin, A.V. , Waddell, N. , Kassahn, K.S. , Gingras, M.C. , Muthuswamy, L.B. , Johns, A.L. , 2012. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 491, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci, D. , Coppola, V. , Musumeci, M. , Addario, A. , Giuffrida, R. , Memeo, L. , 2008. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 14, 1271–1277. [DOI] [PubMed] [Google Scholar]
- Chou, J. , Shahi, P. , Werb, Z. , 2013. microRNA-mediated regulation of the tumor microenvironment. Cell Cycle. 12, 3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote, G.A. , Gore, A.J. , McElyea, S.D. , Heathers, L.E. , Xu, H. , Sherman, S. , 2014. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am. J. Gastroenterol. 109, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wever, O. , Mareel, M. , 2003. Role of tissue stroma in cancer cell invasion. J. Pathol. 200, 429–447. [DOI] [PubMed] [Google Scholar]
- Enright, A.J. , John, B. , Gaul, U. , Tuschl, T. , Sander, C. , Marks, D.S. , 2003. MicroRNA targets in Drosophila. Genome Biol. 5, R1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkan, M. , Hausmann, S. , Michalski, C.W. , Fingerle, A.A. , Dobritz, M. , Kleeff, J. , 2012. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat. Rev. Gastroenterol. Hepatol. 9, 454–467. [DOI] [PubMed] [Google Scholar]
- Feig, C. , Gopinathan, A. , Neesse, A. , Chan, D.S. , Cook, N. , Tuveson, D.A. , 2012. The pancreas cancer microenvironment. Clin. Cancer Res. Off J. Am. Assoc. Cancer Res. 18, 4266–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore, J. , Korc, M. , 2014. Pancreatic cancer stroma: friend or foe?. Cancer Cell. 25, 711–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S. , Grocock, R.J. , van Dongen, S. , Bateman, A. , Enright, A.J. , 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X. , Yan, S. , Weijie, Z. , Feng, W. , Liuxing, W. , Mengquan, L. , 2014. Critical role of miR-10b in transforming growth factor-beta1-induced epithelial-mesenchymal transition in breast cancer. Cancer Gene Ther. 21, 60–67. [DOI] [PubMed] [Google Scholar]
- Hsu, S.D. , Lin, F.M. , Wu, W.Y. , Liang, C. , Huang, W.C. , Chan, W.L. , 2011. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39, D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, S.D. , Tseng, Y.T. , Shrestha, S. , Lin, Y.L. , Khaleel, A. , Chou, C.H. , 2014. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 42, D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R.O. , 2009. The extracellular matrix: not just pretty fibrils. Science. 326, 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi, L. , Attisano, L. , 2006. Ubiquitin-dependent regulation of TGFbeta signaling in cancer. Neoplasia. 8, 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, M. , Zhang, L. , Boufraqech, M. , Liu-Chittenden, Y. , Bussey, K. , Demeure, M.J. , 2014. ZNF367 inhibits cancer progression and is targeted by miR-195. PloS One. 9, e101423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling, P. , Pundavela, J. , Oliveira, S.M. , Roselli, S. , Walker, M.M. , Hondermarck, H. , 2015. Nerve-cancer cell cross-talk: a novel promoter of tumor progression. Cancer Res. 75, 1777–1781. [DOI] [PubMed] [Google Scholar]
- John, B. , Enright, A.J. , Aravin, A. , Tuschl, T. , Sander, C. , Marks, D.S. , 2004. Human microRNA targets. PLoS Biol. 2, e363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe, I.T. , 1986. Principal component analysis Springer-Verlag; 978-0-387-95442-4 487 10.1007/b98835 [Google Scholar]
- Jung, C.J. , Iyengar, S. , Blahnik, K.R. , Jiang, J.X. , Tahimic, C. , Torok, N.J. , 2012. Human ESC self-renewal promoting microRNAs induce epithelial–mesenchymal transition in hepatocytes by controlling the PTEN and TGFbeta tumor suppressor signaling pathways. Mol. Cancer Res. MCR. 10, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Fuchs, C. , Le, C.P. , Pimentel, M.A. , Shackleford, D. , Ferrari, D. , Angst, E. , 2014. Chronic stress accelerates pancreatic cancer growth and invasion: a critical role for beta-adrenergic signaling in the pancreatic microenvironment. Brain Behav. Immun. 40, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal, W. , Wallis, W.A. , 1952. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, (260) 583–621. 10.1080/01621459.1952.10 483441 [Google Scholar]
- Lamouille, S. , Xu, J. , Derynck, R. , 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.Y. , Deng, Z. , Wang, C.H. , Yang, B.B. , 2007. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. U.S.A. 104, 20350–20355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E.J. , Gusev, Y. , Jiang, J. , Nuovo, G.J. , Lerner, M.R. , Frankel, W.L. , 2007. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. J. Int. du Cancer. 120, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P. , Burge, C.B. , Bartel, D.P. , 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P. , Shih, I.H. , Jones-Rhoades, M.W. , Bartel, D.P. , Burge, C.B. , 2003. Prediction of mammalian microRNA targets. Cell. 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhang, Y. , Zhang, H. , Liu, X. , Gong, T. , Li, M. , 2011. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. MCR. 9, 824–833. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Nie, J. , Chen, L. , Dong, G. , Du, X. , Wu, X. , Tang, Y. , Han, W. , 2013. The oncogenic role of microRNA-130a/301a/454 in human colorectal cancer via targeting Smad4 expression. PLoS One. 8, (2) e55532 Epub 2013 Feb 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr, M. , Schmidt, C. , Ringel, J. , Kluth, M. , Muller, P. , Nizze, H. , 2001. Transforming growth factor-beta1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 61, 550–555. [PubMed] [Google Scholar]
- Maehara, N. , Matsumoto, K. , Kuba, K. , Mizumoto, K. , Tanaka, M. , Nakamura, T. , 2001. NK4, a four-kringle antagonist of HGF, inhibits spreading and invasion of human pancreatic cancer cells. Br. J. Cancer. 84, 864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, Henry, B. , Whitney, Donald, R. , 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, (1) 50–60. 10.1214/aoms/1177730491 MR 22058. Zbl 0041.26103 [Google Scholar]
- Mosesson, Y. , Mills, G.B. , Yarden, Y. , 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 8, 835–850. [DOI] [PubMed] [Google Scholar]
- Mueller, D.W. , Rehli, M. , Bosserhoff, A.K. , 2009. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Invest. Dermatol. 129, 1740–1751. [DOI] [PubMed] [Google Scholar]
- Nagpal, N. , Ahmad, H.M. , Molparia, B. , Kulshreshtha, R. , 2013. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 34, 1889–1899. [DOI] [PubMed] [Google Scholar]
- Oettle, H. , 2014. Progress in the knowledge and treatment of advanced pancreatic cancer: from benchside to bedside. Cancer Treat. Rev. 40, 1039–1047. [DOI] [PubMed] [Google Scholar]
- Ozdemir, B.C. , Pentcheva-Hoang, T. , Carstens, J.L. , Zheng, X. , Wu, C.C. , Simpson, T.R. , 2014. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 25, 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.S. , Mullins, M. , Cheang, M.C. , Leung, S. , Voduc, D. , Vickery, T. , 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim, A.D. , Oberstein, P.E. , Thomas, D.H. , Mirek, E.T. , Palermo, C.F. , Sastra, S.A. , 2014. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 25, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E. , Silver, J. , Oshlack, A. , Holmes, M. , Diyagama, D. , Holloway, A. , 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 23, 2700–2707. [DOI] [PubMed] [Google Scholar]
- Sandhu, V. , Bowitz Lothe, I.M. , Labori, K.J. , Lingjaerde, O.C. , Buanes, T. , Dalsgaard, A.M. , 2015. Molecular signatures of mRNAs and miRNAs as prognostic biomarkers in pancreatobiliary and intestinal types of periampullary adenocarcinomas. Mol. Oncol. 9, 758–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N. , Maehara, N. , Goggins, M. , 2004. Gene expression profiling of tumor-stromal interactions between pancreatic cancer cells and stromal fibroblasts. Cancer Res. 64, 6950–6956. [DOI] [PubMed] [Google Scholar]
- Singh, A.P. , Arora, S. , Bhardwaj, A. , Srivastava, S.K. , Kadakia, M.P. , Wang, B. , 2012. CXCL12/CXCR4 protein signaling axis induces sonic hedgehog expression in pancreatic cancer cells via extracellular regulated kinase- and Akt kinase-mediated activation of nuclear factor kappaB: implications for bidirectional tumor-stromal interactions. J. Biol. Chem. 287, 39115–39124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. , 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. , Michaud, J. , Scott, H.S. , 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 21, 2067–2075. [DOI] [PubMed] [Google Scholar]
- Sun, C.Y. , She, X.M. , Qin, Y. , Chu, Z.B. , Chen, L. , Ai, L.S. , 2013. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 34, 426–435. [DOI] [PubMed] [Google Scholar]
- Sureban, S.M. , May, R. , Lightfoot, S.A. , Hoskins, A.B. , Lerner, M. , Brackett, D.J. , 2011. DCAMKL-1 regulates epithelial–mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 71, 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska, A.E. , Davison, T.S. , John, J. , Cannon, T. , Sipos, B. , Maghnouj, A. , 2007. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 26, 4442–4452. [DOI] [PubMed] [Google Scholar]
- Valastyan, S. , Weinberg, R.A. , 2011. Roles for microRNAs in the regulation of cell adhesion molecules. J. Cell Sci. 124, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Duncan, D. , Shi, Z. , Zhang, B. , 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. Web Server issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgaard, A. , Tafjord, S. , Farstad, I.N. , Cvancarova, M. , Eide, T.J. , Mathisen, O. , 2008. Pancreatobiliary versus intestinal histologic type of differentiation is an independent prognostic factor in resected periampullary adenocarcinoma. BMC cancer. 8, 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten, D.M. , Tibshirani, R. , Hastie, T. , 2009. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 10, 515–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten, D.M. , Tibshirani, R.J. , 2009. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat. Appl. Genet. Mol. Biol. 8, Article28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Lamouille, S. , Derynck, R. , 2009. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Chen, J. , Su, F. , Yu, B. , Su, F. , Lin, L. , 2011. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer. 10, 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Kirov, S.A. , Snoddy, J.R. , 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748. Web Server issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, P. , Wang, X.F. , 2014. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol. 24, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Web links
- Cancer registry of Norway: http://www.kreftregisteret.no/en/.
- miRbase: http://www.mirbase.org/ (release 21, June, 2014).
- miRNA.org: http://www.microrna.org/microrna/home.do (release August, 2010).
- miRTarBase: http://mirtarbase.mbc.nctu.edu.tw/ (Release 4.5, November 1, 2013).
- TargetScan: http://www.targetscan.org/ (Release 6.2, June 2012).
- WebGestalt: http://bioinfo.vanderbilt.edu/webgestalt/ (Update 24 May, 2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


