Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the most frequent pancreatic cancer type and is characterized by a dismal prognosis due to late diagnosis, local tumor invasion, frequent distant metastases and poor sensitivity to current therapy. In this context, circulating tumor cells and circulating tumor DNA constitute easily accessible blood‐borne tumor biomarkers that may prove their clinical interest for screening, early diagnosis and metastatic risk assessment of PDAC. Moreover these markers represent a tool to assess PDAC mutational landscape. In this review, together with key biological findings, we summarize the clinical results obtained using “liquid biopsies” at the different stages of the disease, for early and metastatic diagnosis as well as monitoring during therapy.
Keywords: Circulating tumor cells, Circulating tumor DNA, Liquid biopsy, Pancreatic cancer
Highlights
Epithelial CTC detection rate is usually low in PDAC, due epithelial–mesenchymal transition.
KRAS mutations are very frequent in PDAC and are mostly used for ctDNA detection and quantification.
Neither CTC nor ctDNA can be used for PDAC screening or diagnosis, because of limited sensitivity.
CTC, the “seeds” of metastases, are of prognostic significance in non‐metastatic PDAC.
ctDNA might be more accurate than CA19.9 for metastatic relapse prediction and therapy management.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a rare tumor that displays a very aggressive behavior. Its annual worldwide incidence is around 1 to 10 cases per 100,000. Pancreatic cancer accounts for only 3% of all cancers, but is responsible of 7% of cancer deaths (Jemal et al., 2011). The American Cancer Society estimates that in 2015 about 49,000 new cases of pancreatic cancer will be diagnosed in the US, causing 40,000 deaths. PDAC is by far the most common type, representing around 80% of all pancreatic cancers (Siegel et al., 2015). PDAC originates from the epithelial cells of the pancreatic duct as well as from the gland‐like structures and occurs in the head of the pancreas in approximately 60–70% of cases (Modolell et al., 1999).
Currently, surgery plays a crucial role in the treatment of localized PDAC. Unfortunately, due to the paucity of symptoms, the diagnosis is often delayed and only 10–20% of tumors are amenable to resection at initial diagnosis (Poruk et al., 2013). Another critical point is represented by the complex anatomical relationships that pancreas displays with other organs and major vessels, which may contribute to early dissemination of tumor cells in distant organs. However, patients who underwent a surgical resection have a 20–25% 5‐year survival rate (Vincent et al., 2011).
In this context, early diagnosis of PDAC can have a dramatic impact on survival. However, screening methods currently used have not shown to be effective. Among proteomic serum markers, CEA and CA19.9 are used to monitor early recurrences in patients affected by PDAC, but their low sensitivity and specificity prevent any use as a screening tool in healthy people (Goonetilleke and Siriwardena, 2007). In addition, imaging techniques fail to detect early lesions or to distinguish between benign and malignant lesions (Capurso et al., 2015). Besides these screening issues, cytological analysis of pancreatic punctures has a high false negative rate and requires repeated sampling. At metastatic stage, discordant data on predictive value of CA19.9 have been reported, serum tumor marker changes during therapy being marginally associated with survival (Hess et al., 2008; Ishii et al., 1997).
In this context, new effective and reliable biomarkers are necessary not only to improve the early detection and diagnosis of PDAC but also to monitor treatment response and guide therapeutic choices. Circulating tumor cell (CTC) and circulating tumor DNA (ctDNA) could fulfill this need. This review summarizes how non‐invasive “liquid biopsy” approaches could improve PDAC diagnosis, monitoring and treatment decisions.
2. Mutational landscape of PDAC
The most common genetic alterations in pancreatic adenocarcinoma are activating mutations of KRAS and inactivating mutations of CDKN2A, TP53, SMAD4 and BRCA2 (Biankin et al., 2012). A recent study identified other frequent somatic alterations in genes implicated in chromatin regulation or modification (MLL, ARID1A), which may be associated to a better prognosis (Sausen et al., 2015).
KRAS mutations are detected in around 90% of PDAC. This detection rate is much higher than in any other tumor type (Almoguera et al., 1988). KRAS activating point mutations impair proliferation, differentiation and cell metabolism. Several studies have detected KRAS mutations in premalignant pancreatic lesions. More than 90% of pancreatic intraepithelial neoplasms (PanINs) harbor KRAS mutations and the mutation rate is directly correlated to the PanIN grade (Kanda et al., 2012). Other mutations, such as CDKN2A, TP53 and SMAD4, occur with increasing frequency in high grade intraepithelial neoplasms (Hustinx et al., 2005). These observations suggest that KRAS mutation is an early oncogenic event, while subsequent mutations that contribute to tumor progression might display more intra‐ and inter‐patient heterogeneity.
KRAS mutations are located in recurrent hotspots (e.g., codon 12 and 13) and involve single nucleotide variations. These ubiquitous and recurrent mutations are therefore ideal targets to detect and quantify the presence of tumor DNA in a sample (Croce, 2008; Sinn et al., 2014). KRAS mutation detection has thus been investigated in pancreatic juice and stool. Mutation detection was successful in 60–80% of pancreatic juice samples from PDAC patients, but collecting such sample requires uneasy endoscopic procedures (Van Laethem et al., 1998; Watanabe et al., 1999; Wilentz et al., 1998). In the stool, KRAS mutations were detected in only 20–55% of PDAC patients; this low sensitivity prevents the use as a screening or diagnostic test in clinics (Caldas et al., 1994; Wenger et al., 1999). Blood‐based “liquid biopsies” can outperform these deceiving results, and may prove to be clinically relevant.
3. ctDNA release and detection
3.1. ctDNA biology
In 1948, two French biochemists reported that circulating nucleic acids are physiologically present in the serum (Mandel and Metais, 1948). cfDNA primarily originates from apoptotic and necrotic cells (Jahr et al., 2001), but the exact biological mechanisms underlying the release of these 70–200 base pair‐long DNA fragments remain to be fully elucidated. A recent study showed that in healthy people most of cfDNA derive from bone‐marrow and other organs such as liver (Sun et al., 2015). cfDNA has a short half‐life ranging from 15 min to few hours and is cleared away by liver and kidney (Fleischhacker and Schmidt, 2007).
Tumor cells also release fragments of DNA as a result of their high turnover (circulating tumor DNA, ctDNA) and, in cancer patients, ctDNA represents a variable fraction of cfDNA. ctDNA is distinguished from normal cfDNA by the presence of cancer‐related mutations, as ctDNA fragments released by the tumor harbor the same genetic alterations. Indeed, several reports showed high concordance between ctDNA mutations, when detectable, and matched tumor mutations (Douillard et al., 2014; Kinugasa et al., 2015; Lebofsky et al., 2015). It is noteworthy that the ctDNA fraction is extremely variable, and can represent from 0.01% to more than 50% of the cfDNA (Diehl et al., 2008). Several studies have positively correlated ctDNA levels to the tumor burden (Allen et al., 2004; Fleischhacker and Schmidt, 2007; Madic et al., 2012; Schwarzenbach et al., 2008). A recent study analyzed the level of ctDNA in a large cohort of patients affected by tumors of different histologies and stages. The percentage of detectable ctDNA was related to the stage, being lower in localized than in metastatic disease (49–78% vs. 86–100%) (Bettegowda et al., 2014). ctDNA is also believed to be released from all the tumor deposits in a given patient and is less impacted by intratumor heterogeneity than a single specimen of tumor tissue (Diaz and Bardelli, 2014). Therefore, “liquid biopsies”, sampling of which can be repeated and is minimally invasive, hold the promise to detect and monitor the whole cancer mutational landscape in a given PDAC patient.
3.2. ctDNA detection
As mentioned above, the quantity of ctDNA detectable in the blood is mainly influenced by the tumor burden. However, it also depends on the tumor type and other potential mechanisms such as the activity of plasma nucleases which degrade DNA fragments (Barra et al., 2015; El Messaoudi et al., 2013). Although rarely observed in PDAC, response to systemic therapy has been associated with low or undetectable ctDNA levels in several cancer types, suggesting that ctDNA is more likely detected at the time of tumor progression. Besides this biological variability, which remains to be fully explained, understanding the technical aspects of ctDNA detection is necessary to fully understand the clinical value of each ctDNA‐related report.
First, pre‐analytical conditions appear critical. The volume of blood directly impacts the total amount of cfDNA (and therefore of ctDNA) and limited blood volume (<5–10 ml of blood) lower the sensitivity of the whole procedure, whatever the technique used. It is also critical to avoid contamination by normal blood cells DNA in the sampling tube as it dilutes the ctDNA fraction and ultimately impacts the sensitivity of ctDNA detection. Plasma (supernatant obtained in the absence of blood clotting) is therefore most suited for ctDNA detection than serum, in which some DNA is released by clotted normal blood cells. The time elapsed between the blood draw and plasma isolation by centrifugation is also critical when standard tubes are used. Ideally tubes must be processed within less than 1 h. To be able to store tubes for several days before processing and shipped them to a central laboratory, several companies have developed tubes that contain fixatives.
Following DNA extraction, the detection of ctDNA consists of another pre‐analytical step that is not fully standardized yet. Recent techniques and approaches allow for the detection and quantification of a minority of mutated alleles in a background of normal DNA. ctDNA detection can be addressed both by targeting known mutations (previously characterized on tumor tissue or highly recurrent mutation such as KRAS) or by looking for de novo genetic alterations investigating multiple genes of interest. For the first aim, recent PCR‐based approaches have reached high levels of sensibility, ranging from 0.1 to 0.01% (i.e., making it possible to detect up to 1 mutated allele out of 100,000 normal alleles, Figure 1). Among PCR‐based techniques, digital PCR (dPCR, which include droplet dPCR and its technical variant “BEAMing”) appears as the most promising approach for detection of highly recurrent hotspot mutations, with high sensitivity and specificity. Using this approach, Bettegowda and colleagues reported a sensitivity of 87.2% and a specificity of 99.2% to detect KRAS mutation in colorectal cancer (Bettegowda et al., 2014). In our hands, >95% sensitivity and 99% specificity was reached for KRAS mutation detection by droplet‐dPCR (ddPCR) in more than 120 metastatic colorectal cancer patients entering a first line trial and who were not pre‐exposed to anti‐EGFR drugs (unpublished data). Because of the very high incidence of KRAS mutations, most ctDNA studies conducted in PDAC patients used these PCR‐based methods.
Figure 1.
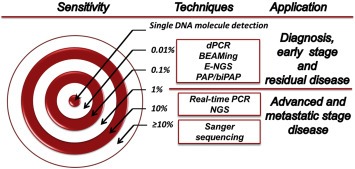
Different ctDNA detection techniques and their sensitivity. Techniques are ranked according to their usual sensitivity towards minor allele detection. The optimal technique would allow the detection of a single mutated DNA molecule whatever the normal DNA background. Digital PCR (dPCR), and its variants, droplet‐dPCR and BEAMing (“Beads, Emulsion, Amplification, Magnetic”), can be considered as the current standard for hotspot mutation detection (typically, KRAS mutations) in ctDNA, with 0.01%–0.1% sensitivity. Pyrophosphorolysis‐activated polymerization (PAP)‐PCR and bidirectional‐PAP (biPAP)‐PCR techniques have similar theoretical 0.01%–0.1% sensitivity (although not tested on KRAS). Modified next generation sequencing procedures (CAPP‐Seq, Safe‐Seq, etc, called “E‐NGS” here) exhibited at least 0.1% sensitivity.
In contrast to these approaches targeting hotspot mutations, next generation sequencing (NGS) enables to study several genes and therefore to identify ctDNA tumor‐derived genetic lesions without knowing a priori the genotype of the tumor. It generates extensive information about the mutation landscape and can reveal new emergent mutations, which could be involved in drug resistance and/or the disease progression. Zill and colleagues analyzed a cohort of 26 patients with PDAC or biliary cancer. They showed that NGS of a gene panel on cfDNA can identify 90% of tumor tissue mutations across the five more frequent identified mutated genes (KRAS, TP53, APC, FBXW7 and SMAD4) with an excellent specificity (Zill et al., 2015). NGS‐based approaches are however restricted by an overall lower sensitivity than dPCR (0.1% at best, 1% usually), while modified ultrasensitive NGS approaches (e.g., SafeSeq (Kinde et al., 2011)) are currently too complex for clinical laboratories.
4. CTC release and detection
4.1. CTC biology
The existence of circulating tumor cells (CTCs) has been shown for the first time in 1869 (Ashworth, 1869). These tumor cells released by the tumor masses (primary tumor or metastases) circulate in the bloodstream. CTCs can be passively shed from the tumor, or can be the result of an active intravasation process during which tumor cells invade the tissue stroma and blood vessels, and enter the bloodstream. CTCs have been broadly studied as prognostic factor in breast, prostate and colorectal cancer but their role in pancreatic cancer has not extensively been explored yet (Bidard et al., 2014; Cohen et al., 2008; de Bono et al., 2008). Early CTCs detection could identify patients with increased risk of metastasis, giving the chance to customize adjuvant therapy for patients with operable or locally advance PDAC. Moreover the characterization of CTCs could help to decipher the metastatic process and identify new treatment targets.
Importantly, among carcinomas, PDAC cells are most prone to epithelial–mesenchymal transition (EMT), a mechanism by which carcinoma cells undergo reprogramming, lose or reduce their epithelial phenotype while expressing mesenchymal markers (see below). The mechanism by which cancer cells undergo EMT is not fully understood. High proliferation and hypoxic stimuli might lead to a loss of cancer cell polarity, which is one of the first steps toward EMT. The acquisition of a mesenchymal phenotype is associated with increased cellular invasion and mobility, and eventually increases cancer cell stemness (Labelle et al., 2011; Thiery, 2002; Yu et al., 2013). As EMT strongly modifies the expression pattern of membrane proteins, it strongly influences CTC detection in PDAC.
4.2. CTC detection
CTCs are extremely rare and lost in a large number of normal blood cells, with a ratio of about 1 CTC per 106–108 leukocytes, making their detection challenging. CTC detection techniques are usually made of two steps: CTC enrichment followed by CTC detection.
CTC enrichment can be performed with different techniques:
-
‐
Antibody‐based enrichment uses antibodies directed against cell surface markers. Positive immunoselection relies on antibodies directed against cancer cell surfaces markers. Most CTC detection techniques rely on Epithelial Cell Adhesion Molecule (EpCAM) positive immunoselection. Negative selections deplete leukocytes from the blood sample and usually use CD45‐binding antibodies.
-
‐
Physical/biological assays isolate CTCs on the basis of cell size or bioelectric features. Most CTCs exhibit a larger size and different density, electromagnetic charge and motility than normal blood cells. This allows their separation by dedicated devices (filters, dielectrophoresis …), sometimes integrated in microfluidic chips (Autebert et al., 2015; Bobek et al., 2014).
The second step, CTC detection, can be achieved by proteomic, transcriptomic or genomic approaches. Immunocytostaining can detect proteins associated with a pancreatic or epithelial origin. Combined with a morphological examination of the stained cells, cytological approaches are the gold standard for CTC detection. Among them, the CellSearch® technique combines an immunomagnetic EpCAM‐based enrichment followed by immunocytofluorescence detection. This method identifies and enumerates CTCs according to the expression of epithelial cytokeratins, negative CD45 staining and the presence of a nucleus. The United State Food and Drug Administration cleared this technique for the management of metastatic breast, colorectal and prostate cancers. It was also used to detect CTCs in other tumor types, such as PDAC (Table 1), although in PDAC the level of EpCAM expression might be lowered by EMT. Interestingly, another report using this technology found a very high CTC detection rate in patients with gastro‐intestinal neuro‐endocrine tumors, including some pancreatic neuro‐endocrine tumors (not discussed in this review) (Khan et al., 2011).
Table 1.
CTC detection by the CellSearch® system in PDAC patients.
| Reference | Patients | Stage | CTC detection rate (and threshold) | Findings |
|---|---|---|---|---|
| (Allard et al., 2004) | N = 16 | Metastatic disease | 37%(≥2CTC/7.5 ml) | – |
| (Bidard et al., 2013) | N = 79 | Locally advanced disease | 11%(≥1CTC/7.5 ml) | CTC‐positivity correlated with poor tumor differentiation and shorter OS. |
| (Khoja et al., 2012) | N = 54 | Locally advanced and metastatic disease | 40%(≥1CTC/7.5 ml) | ISET (filter‐based assay) vs. CellSearch: higher number of CTC and mesenchymal CTCs detected by ISET. |
| (Bissolati et al., 2015) | N = 20 | Localized disease | 45%(≥1CTC/7.5 ml) | CTC positivity not correlated with OS. Portal vein CTCs associated to higher rate of liver metastases. |
| (Kurihara et al., 2008) | N = 26 | Localized, locally advanced and metastatic disease | 42%(≥1CTC/7.5 ml) | CTC‐positivity correlated with shorter OS. |
| (Earl et al., 2015) | N = 45 | Localized, locally advanced and metastatic disease | 20%(≥1CTC/7.5 ml) | CTC‐positivity correlated with shorter OS. |
OS: Overall Survival.
Transcriptomic approaches are based on the detection of mRNA associated with a pancreatic or epithelial origin (e.g., EpCAM, CK19, CEA …). In the absence of morphological control, some concerns were raised about the specificity of these approaches, especially when investigating EMT‐related mRNA (which can be found in normal blood cells). Despite interesting preliminary results in PDAC (see below) (Hoffmann et al., 2007; Sergeant et al., 2011; Soeth et al., 2005), their development has been mostly discontinued. The third detection approach consists in retrieving cancer‐specific mutations, typically KRAS mutations, in the DNA of enriched cells. This demonstrates that isolated CTCs are clonally related to the primary tumor with a theoretical 100% specificity. However, these mutation‐based approaches require time‐consuming single‐cell analysis, which currently prevents any use in clinics.
Overall, these studies showed that CTCs can be isolated in localized and metastatic PDAC with various techniques. Detection rates ranged from 10% to 80%, depending on the technique and disease stage (Allard et al., 2004; Bidard et al., 2013; Kurihara et al., 2008; Mataki et al., 2004).
5. Clinical applications
5.1. Early detection and differential diagnosis
Beyond imaging, there is no standardized PDAC screening tool, even for patients with high risk (e.g., Peutz‐Jeghers syndrome). PDAC must be distinguished from other preneoplastic conditions that can eventually lead to PDAC (e.g., intraductal papillary mucinous neoplasms) and from other benign pancreatic masses. On imaging, benign lesions could be however easily mistaken for tumor and vice versa, while pancreatic punctures are invasive and hard to perform. In that context, any non‐invasive screening or diagnostic tool might become extremely useful.
Very few data are available on the incidence of CTC in early PDAC. In a recent study, CTC was evaluated as prediagnostic biomarker. Using a microfluidic technology, they tested 11 patients with PDAC, at all stages, 21 patients with benign pancreatic cystic lesions and 19 negative controls. They identified 73%, 33% and 0 patients positive for CTCs (threshold ≥ 3 CTCs) in the three groups respectively (Rhim et al., 2014). The pancreatic origin of isolated CTC was demonstrated by Pdx‐1 staining, a pancreas specific transcription factor. However, no correlation was found between CTC count and cyst size, tumor stage, CEA or CA19.9. As a conclusion, CTC might be present in the bloodstream of patients with non‐invasive pancreatic lesions and limits the use of CTC count as an aid for differential diagnosis. Using the CellSearch® system, Mudan and colleges detected 4 CTCs in a patient with a pancreatic mass which turned out to be a pancreatic cancer (Mudan et al., 2010). However, using the same technique, only 5% of 79 cytologically‐proven locally‐advanced non‐metastatic PDAC patients had one or more CTC detected at the time of treatment initiation (Bidard et al., 2013), highlighting a major lack of sensitivity of this technique. In that regard, a preliminary study on nine operable patients showed a better sensitivity of filter‐based assay, with one or more CTC detected in six patients (66%), although the specificity was limited (Bobek et al., 2014). Such difference between CellSearch® and filter‐based techniques was also seen in non‐operable PDAC patients (93% detection rate vs. 40% for CellSearch®), supporting the importance of EpCAM downregulation by the EMT process (Khoja et al., 2012). In light of these results, the use of CTC detection as a screening tool appears of limited interest with EpCAM‐based detection techniques, while filter‐based techniques deserve more investigation.
In contrast, the very high frequency of KRAS mutations in PDAC could theoretically facilitate setting up a ctDNA‐based screening. Bettegowda and colleagues used dPCR to detect ctDNA in 640 plasma samples of patients with different type and stage of cancers, including 155 PDAC. ctDNA detection rate in patients without distant metastases was 48% and the fraction of patients with detectable ctDNA increased according to the clinical stage (Bettegowda et al., 2014). This data were confirmed by another study with the same technical approach in 51 patients with resectable PDAC (detection rate 43% (22/51) with a specificity > 99.9%) (Sausen et al., 2015). Uemura et al., detected KRAS mutations in plasma samples of 35% of patients before surgery, using the limited in sensitivity PCR‐RFLP approach (polymerase chain reaction combined to restriction fragment length polymorphism) (Uemura et al., 2004). In our hands, an attempt of studying ctDNA kinetics before and after pancreatectomy did not retrieve any KRAS mutation in the plasma of 7 Ukrainian patients with operable PDAC, despite the use of KRAS‐targeted ddPCR (Dronov, Khomenko, & Bidard, unpublished data). Taken together, these reports suggest that KRAS‐oriented ctDNA detection is feasible in some but not all patients with localized PDAC at the time of diagnosis.
The question of whether KRAS mutations are present in preneoplastic conditions and other differential diagnoses and whether these mutations can be detected in the peripheral blood is currently unclear. In 1998, using a technique of limited sensitivity, a first study in four patients with chronic pancreatitis and detectable KRAS mutations reported that all these patients were later diagnosed with PDAC (Mulcahy et al., 1998). These findings were not confirmed in a larger study by Maire et al., which reported detectable KRAS mutations in cell‐free DNA of 4 out of 31 patients with chronic pancreatitis (Maire et al., 2002). None of the four patients was diagnosed with PDAC after a mean follow‐up of 36 months. Some studies focusing on KRAS mutation detection have used patients with benign pathology as negative controls. In three cohorts of limited size, KRAS mutation was not detected in the cfDNA of patients with benign pancreatic disease (Däbritz et al., 2009; Dianxu et al., 2002; Yamada et al., 1998). In two other cohorts, KRAS mutations were found at a lower frequency than in PDAC patients: Kinugasa et al., found KRAS mutation in the serum of 1/20 healthy patients and in 4/20 patients with chronic pancreatitis (Kinugasa et al., 2015); Castells et al., detected in 2/37 patients with chronic pancreatitis (Castells et al., 1999). Further comparison of these cohorts is difficult due to inconsistent designs of the studies and ctDNA detection methods. In summary, the specificity of KRAS mutation detection in plasma appears limited, while current ctDNA detection techniques did not demonstrate a better sensitivity than CTC in the context of early PDAC diagnosis or screening.
5.2. Operable PDAC and post‐surgery follow‐up
Surgical resection is hindered by the close anatomical relationships that pancreas displays with major vessels and other organs. As a consequence, less than 20% of patients are amenable to surgery at presentation. Whatever the procedure, pancreatectomy is uneasy and associated with a perioperative mortality of 2–5% that is directly correlated to the surgeon's experience (Mamidanna et al., 2015). Even if macroscopically complete, PDAC resection is often incomplete, with microscopically positive margins (R1) (Konstantinidis et al., 2013). While neoadjuvant and adjuvant studies are currently testing new chemotherapy regimens such as FOLFIRINOX or gemcitabine plus nab‐paclitaxel, the current management of operable PDAC consists in curative surgical resection followed by adjuvant gemcitabine for 6 months (Maeda et al., 2008; Neoptolemos et al., 2010; Oettle et al., 2013). Adjuvant chemoradiotherapy is not a standard (Neoptolemos et al., 2004), but is discussed in some patients with positive margins. In that clinical setting, the identification of patients with increased risk of recurrence might prove useful to customize pre‐ and post‐surgical treatments and follow‐ups.
Uemura et al., using mutation‐specific mismatch ligation assay, examined KRAS status in plasma of 28 patients before and after surgery. Nine patients had positive pre‐surgery liquid biopsy. After curative excision, 4 of 9 subjects were still positive for KRAS mutations. No patient had a metastatic relapse in the 8 months of follow‐up (Uemura et al., 2004). In contrast, using an allele‐specific PCR method, Yamada et al., found that patients with persistently detectable KRAS ctDNA mutations after resection were likely to have a poorer prognosis. They also showed, in this cohort of 21 patients, a correlation between ctDNA detection and tumor size (Yamada et al., 1998). In a recent work, Sausen et al., used ddPCR to analyze 51 plasma samples and showed that patients with PDAC and detectable ctDNA after surgery were more likely to relapse. Disease relapses were detected about 6 months earlier by ctDNA than by standard imaging (Sausen et al., 2015). This early detection of metastatic disease could theoretically lead to an earlier start of first line therapy. However, the real clinical benefit of such strategy is unclear because of the limited efficacy of current chemotherapy regimens.
Concerning CTCs, results are controversial. Mataki et al., using RT‐PCR based assay to analyze CEA mRNA levels, showed that increasing levels of CEA mRNA are early indexes of recurrence. In fact, they detected increased levels of CEA mRNA in 30% (6/20) of patients undergoing curative surgery. Five of these 6 patients developed metastasis compared to 2/12 with negative CEA mRNA (Mataki et al., 2004).
On the other hand, Hoffmann et al., and Sergeant et al., who used RT‐qPCR assay to detect CK19 and EpCAM mRNA before and after surgery, did not find any prognostic impact in two cohorts (N = 37 and 48 patients) (Hoffmann et al., 2007; Sergeant et al., 2011). Using the CellSearch® system, Bissolati et al., did not find any prognostic impact of CTC detection in the peripheral blood of 20 operable patients. CTC detection in the portal vein, during surgery, was marginally associated with the occurrence of later liver metastases (Bissolati et al., 2015).
5.3. Locally advanced PDAC
Locally advanced PDAC (LAPDAC) are unresectable PDAC that have spread locally and are not amenable to surgical resection although no overt distant metastasis can be detected by imaging (CT‐scan, mostly). LAPDAC represent about 20–30% of all PDAC cases at diagnosis, and are treated with chemotherapy, using the same regimens than in metastatic disease. In the LAP07 phase III trial, chemoradiotherapy did not demonstrate a significant benefit in patients without tumor progression after four months of induction chemotherapy (Hammel P. et al., 2013). Thirty percent of LAPDAC‐related deaths are attributable to a locally destructive tumor progression, while the growth of distant metastases account for the rest (70%) (Iacobuzio‐Donahue et al., 2009). Therefore, two key questions are of utmost interest in LAPDAC:
Is it possible to distinguish LAPDAC that have already released distant micrometastases from those in which tumor cells are still confined to the pancreatic mass? In the former scenario, systemic therapy should remain the cornerstone of the treatment while radiation therapy might play a role in the treatment of the latter LAPDAC cases. For this clinical issue, ctDNA has probably no role to play, as there is no way to track the origin of circulating mutated DNA fragments back to the organ that released them. In contrast, the detection of CTC is very likely to be associated with an ongoing dissemination process and to the onset of distant metastasis. We reported the only large CTC study in that precise setting using the CellSearch® technique to detect CTC at inclusion and after 2 months of gemcitabine in LAPDAC patients enrolled in the LAP07 trial. The overall detection rate was low, 11%, using ≥1/7.5 ml of blood threshold in 79 patients. CTC positivity was correlated with poor tumor differentiation and demonstrated an independent prognostic impact on overall survival. No relationship was observed between CTC changes during therapy and the LAPDAC response to therapy (Bidard et al., 2013). This first result can be considered as a proof of principle but should be confirmed by further studies. Moreover, in the next trials, it might be interesting to use other CTC detection techniques, in order to increase the CTC detection rate. Indeed, five out of seven LAPDAC patients were found to be CTC‐positive with a filter‐based technique (Kulemann et al., 2015).
While several chemotherapy regimens are used to reduce the primary tumor size and eventually allow surgery of the primary tumor, is it possible to predict the sensitivity of the tumor to any of the current chemotherapies? In that regard, liquid biopsy approaches may allow an early assessment of the therapy efficacy. In a small study, Olsen et al., showed that the persistence of KRAS ctDNA after treatment was associated with a worse prognosis. Conversely, patients with undetectable KRAS mutations after gefitinib and chemoradiotherapy had a better overall survival (Olsen et al., 2009). A recent study presented at ASCO 2015 used a quantitative PCR‐NGS mutation enrichment assay to detect KRAS mutations in a mixed cohort of 182 patients with LAPDAC or metastatic disease. The results showed that patients with high KRAS ctDNA levels at diagnosis had a worse prognosis. ctDNA levels were monitored during therapy: patients with persistent low levels of KRAS ctDNA had a longer survival (Johansen et al., 2015).
5.4. Metastatic disease
Patients with metastatic PDAC have a very limited life expectancy, despite some incremental progresses obtained with FOLFIRINOX and the gemcitabine nab–paclitaxel combination (Conroy et al., 2011; Von Hoff et al., 2013). In that setting and in the absence of any predictive marker of chemotherapy efficacy, studies focused on the detection rate and the prognostic value of CTC and ctDNA at baseline and during therapy.
Z'graggen et al., detected tumor cells by immunocytostaining both in blood (CTCs) and bone marrow (disseminated tumor cells) of 105 PDAC patients. CTC‐positivity was more frequent among patients with metastatic PDAC (12/31, 39%) than in patients with LAPDAC (12/42, 28%) or with resectable PDAC (3/32, 9%). However, CTC detection was only marginally associated with disease progression in this first report (Z'graggen et al., 2001). De Albuquerque et al., evaluated the expression of KRT19, MUC1, EPCAM, CEACAM5 and BIRC5 in CTCs by immunomagnetic separation followed by RT‐qPCR assay. Before starting the treatment, 47% of pancreatic patients showed at least one detectable tumor‐associated transcript, these patients experiencing significantly shorter progression‐free survival (de Albuquerque et al., 2012). Kurihara et al., used the CellSearch® to detect CTCs in 26 patients with PDAC, 24 of which had a metastatic disease. Eleven patients (42%) had a CTC count ≥1/7.5 ml of blood. The median overall survival of CTC‐positive patients was significantly shorter (110 vs. 376 days. p = 0.001) (Kurihara et al., 2008).
Changes of CTC during therapy to distinguish good vs. poor responders to therapy (and eventually guide therapeutic decisions) have been less investigated. Preclinical data reported by Torphy et al., in patient‐derived xenografts highlighted the correlation between CTC count (detected by microfluidic chip) and tumor burden variations (Torphy et al., 2014). Ren et al., evaluated changes in CTC count (immunostaining) during chemotherapy in 41 patients with LAPDAC or metastatic PDAC. Blood samples were collected before and after seven days on 5FU chemotherapy regimen. They reported a reduction in the number of CTC‐positive patients during therapy (12 vs. 33) together with an increase of apoptotic CTCs (detected by a TUNEL assay). However, the clinical consequences of these changes were not investigated (Ren et al., 2011).
As previously mentioned, Bettegowda et al., detected ctDNA by dPCR in >75% of 34 patients with metastatic PDAC (Bettegowda et al., 2014). Similar detection rates were previously reported in a smaller study (81% of 21 patients) (Mulcahy et al., 1998). Chen et al., studied the prognostic value of KRAS mutations detected by direct sequencing in plasma samples. With this technique of very limited sensitivity, only 33% (30/91) of metastatic patients harbored KRAS ctDNA, but these patients had a significantly worse survival (Chen et al., 2010). In the recent study of Kinugasa et al., median overall survival was also significantly shorter in patients harboring KRAS mutations in plasma (176 vs. 489 days. p = 0.003), while the mutational status in the tumor did not demonstrate any prognostic impact (Kinugasa et al., 2015). A similar prognostic impact of high ctDNA yield was reported in 14 metastatic PDAC patients, of which 10 had a KRAS mutation detected in plasma (Tjensvoll et al., 2015). Interestingly, Zill et al., showed that NGS‐based analysis of cfDNA allows for a thorough analysis of the mutational landscape of metastatic PDAC (Zill et al., 2015). An interesting two‐step approach for NGS‐based mutational screening has been also reported by Takai et al.,: a first screen based on ddPCR allowed selecting the 48 PDAC patients who had ≥1% mutant allele frequencies of KRAS in plasma cfDNA, out of 259 patients (18%). KRAS mutations were found in 40 other patients (15%) at <1% allele frequency, which was deemed insufficient for NGS for further analysis of ctDNA by NGS (i.e. below NGS standard sensitivity). The 48 selected plasma samples were then subjected to standard targeted NGS focusing on potentially actionable mutations, which were found in 14 patients (Takai et al., 2015). This large study is a very good example of the potential and pitfalls of ctDNA‐based targetable mutation discovery in metastatic PDAC: (i) ctDNA is present, detected in a third of metastatic patients, but at a lower than expected allelic frequency when compared with other tumor types (e.g., breast or lung cancers); (ii) targeted NGS is technically feasible and can lead to valuable result as long the ctDNA allelic frequency is over 1%; (iii) at the bottom line, this heroic, labor‐intensive work retrieved a mutation of potential interest in only 5% of metastatic PDAC patients.
Finally, in a recent face‐to‐face study, Earl et al., examined both KRAS mutation by dPCR and CTCs by CellSearch® in a cohort of 45 patients with PDAC (14 with localized disease, 13 with LAPDAC and 18 with metastatic disease). They detected KRAS mutation in 8 patients (4 with metastatic disease) and ≥1CTC/7.5 ml of blood in seven patients (all but one with metastatic disease). Overall survival was statistically and significantly poorer in CTC‐positive or KRAS mutated patients, with no overt superiority of one technique over the other (Earl et al., 2015).
6. Biological findings
When isolated with sensitive techniques, CTC represent tumor material that can be obtained from PDAC patients more easily than by tumor biopsy or puncture. Performing genomic, transcriptomic and proteomic profiling, on either single CTC or a population of CTCs, hold the promise for deciphering biological mechanisms involved in PDAC growth and metastasis.
The existence of EMT features in PDAC has been repeatedly reported (Hanahan and Weinberg, 2011; Yu et al., 2013) and is thought to contribute to the aggressiveness of this cancer type. EMT is thought to be inducible by the tumor microenvironment (e.g., following the release of growth factors) and hypoxia, leading to an increased spread of individual cancer cells. In that regards, it is interesting to note that, using an antigen‐independent filter‐based detection technique, CTC aggregates, also called clusters, were found at a lower frequency in PDAC patients than in other epithelial tumors (Cho et al., 2012). Epithelial cells that have undergone EMT may express mesenchymal markers while reducing the level of expression of epithelial markers such as EpCAM (Thiery, 2002). The exact way epithelial cells turn into migratory mesenchymal cells is not yet known, but this process generates CTCs that can give rise to metastasis. The reverse process (mesenchymal to epithelial transition MET), which would happen once the cells find a suitable site to generate metastasis, is even less clear (Figure 2). When investigating EMT on CTC, it is critical to separate CTC from leukocytes as normal blood cells are of mesenchymal origin, and to use an antigen‐free enrichment method. Another approach is to combine several capture antigens, e.g., EpCAM and MUC1 (Thege et al., 2014). Using an epitope‐independent microfluidic technique followed by RNA sequencing of the isolated CTCs, Haber and colleagues found that CTC from mice models and PDAC patients overexpress Wnt2 and the Wnt signaling pathways (Yu et al., 2012). This original observation was confirmed by functional studies, which demonstrated the role of Wnt2 in resistance to anchorage‐independent survival and in the enhancement of metastatic propensity in vivo. With a similar approach, the same group reported more recently that pancreatic CTC expression profiles clustered separately from those of matched primary tumors. CTCs display less proliferation markers, increased ALDH1 gene expression, considered as a stemness marker, and the co‐expression of epithelial and mesenchymal genes (Ting et al., 2014).
Figure 2.
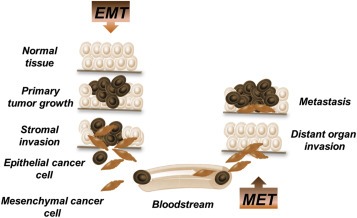
EMT and MET process in the development of PDAC metastasis. Cancer cells undergoing epithelial–mesenchymal transition (EMT) acquire extra motility, invasive features and are more resistant to apoptosis. These cells can intravasate, circulate in the blood (as CTC) and eventually extravasate in distant organs, leading to the later development of distant metastases. The reverse process (mesenchymal–epithelial transition, MET) is thought to happen during the invasion of the distant site, eventually induced by the new microenvironment of disseminated tumor cells.
7. Conclusion
In the context of PDAC, a life‐threatening cancer with limited treatment options, that is hardly accessible to tissue biopsy, liquid biopsy is of critical interest to improve early diagnosis, disease monitoring and treatment options. As reviewed here, CTC and ctDNA hold the promise of providing different and complementary data, although current levels of evidence supporting their use in clinics remain currently low, because of the cohort limited sizes and lack of technical standardization.
The major role played by EMT in PDAC growth and metastatic process certainly limited the accuracy of many EpCAM‐based CTC detection techniques, which have been mostly used over the past decade. Further improvements are eagerly awaited from CTC detection techniques that can detect both standard epithelial CTC and post‐EMT CTC; these cells may deliver critical biological and therapeutic information on the biological processes underlying (or predicting) PDAC chemoresistance, spread and metastasis.
While the high prevalence of KRAS mutations in PDAC is a theoretically favorable setting to implement ctDNA detection and quantification, reports showed lower than expected ctDNA detection rates (50–75% of metastatic patients), when compared to other cancer types, such as metastatic breast or lung cancer (>70–80%, usually). Biological processes behind ctDNA release and degradation in blood are currently poorly understood and further implementation in PDAC would benefit from biological studies on ctDNA biology. Notwithstanding these potential biological issues, ctDNA hold the promise of providing a non‐invasive, dynamic overview of the mutational landscape of PDAC, including potentially targetable mutations.
Authors' disclosures of potential conflicts of interest
The authors declare that they have no conflict of interest. The Circulating Tumor Biomarkers laboratory at Institut Curie has received support from the Innovative Medicines Initiative joint undertaking under grant agreement no. 115749 (project Cancer‐ID).
Riva Francesca, Dronov Oleksii I., Khomenko Dmytro I., Huguet Florence, Louvet Christophe, Mariani Pascale, Stern Marc-Henri, Lantz Olivier, Proudhon Charlotte, Pierga Jean-Yves, Bidard Francois-Clement, (2016), Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.01.006.
This is a contribution to the special issue edited by Klaus Pantel and Catherine Alix‐Panabieres, Liquid Biopsies.
References
- Allard, W.J. , Matera, J. , Miller, M.C. , Repollet, M. , Connelly, M.C. , Rao, C. , Tibbe, A.G.J. , Uhr, J.W. , Terstappen, L.W.M.M. , 2004. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 10, 6897–6904. 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- Allen, D. , Butt, A. , Cahill, D. , Wheeler, M. , Popert, R. , Swaminathan, R. , 2004. Role of cell-free plasma DNA as a diagnostic marker for prostate cancer. Ann. N. Y. Acad. Sci. 1022, 76–80. 10.1196/annals.1318.013 [DOI] [PubMed] [Google Scholar]
- Almoguera, C. , Shibata, D. , Forrester, K. , Martin, J. , Arnheim, N. , Perucho, M. , 1988. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 53, 549–554. [DOI] [PubMed] [Google Scholar]
- Ashworth, T.R. , 1869. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med. J. Aust. 14, 146–149. [Google Scholar]
- Autebert, J. , Coudert, B. , Champ, J. , Saias, L. , Guneri, E.T. , Lebofsky, R. , Bidard, F.-C. , Pierga, J.-Y. , Farace, F. , Descroix, S. , Malaquin, L. , Viovy, J.-L. , 2015. High purity microfluidic sorting and analysis of circulating tumor cells: towards routine mutation detection. Lab. Chip. 15, 2090–2101. 10.1039/c5lc00104h [DOI] [PubMed] [Google Scholar]
- Barra, G.B. , Santa Rita, T.H. , Vasques, J. de A. , Chianca, C.F. , Nery, L.F.A. , Costa, S.S.S. , 2015. EDTA-mediated inhibition of DNAses protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin. Biochem. 10.1016/j.clinbiochem.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Bettegowda, C. , Sausen, M. , Leary, R.J. , Kinde, I. , Wang, Y. , Agrawal, N. , Bartlett, B.R. , Wang, H. , Luber, B. , Alani, R.M. , Antonarakis, E.S. , Azad, N.S. , Bardelli, A. , Brem, H. , Cameron, J.L. , Lee, C.C. , Fecher, L.A. , Gallia, G.L. , Gibbs, P. , Le, D. , Giuntoli, R.L. , Goggins, M. , Hogarty, M.D. , Holdhoff, M. , Hong, S.-M. , Jiao, Y. , Juhl, H.H. , Kim, J.J. , Siravegna, G. , Laheru, D.A. , Lauricella, C. , Lim, M. , Lipson, E.J. , Marie, S.K.N. , Netto, G.J. , Oliner, K.S. , Olivi, A. , Olsson, L. , Riggins, G.J. , Sartore-Bianchi, A. , Schmidt, K. , Shih, le-M. , Oba-Shinjo, S.M. , Siena, S. , Theodorescu, D. , Tie, J. , Harkins, T.T. , Veronese, S. , Wang, T.-L. , Weingart, J.D. , Wolfgang, C.L. , Wood, L.D. , Xing, D. , Hruban, R.H. , Wu, J. , Allen, P.J. , Schmidt, C.M. , Choti, M.A. , Velculescu, V.E. , Kinzler, K.W. , Vogelstein, B. , Papadopoulos, N. , Diaz, L.A. , 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 10.1126/scitranslmed.3007094 224ra24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin, A.V. , Waddell, N. , Kassahn, K.S. , Gingras, M.-C. , Muthuswamy, L.B. , Johns, A.L. , Miller, D.K. , Wilson, P.J. , Patch, A.-M. , Wu, J. , Chang, D.K. , Cowley, M.J. , Gardiner, B.B. , Song, S. , Harliwong, I. , Idrisoglu, S. , Nourse, C. , Nourbakhsh, E. , Manning, S. , Wani, S. , Gongora, M. , Pajic, M. , Scarlett, C.J. , Gill, A.J. , Pinho, A.V. , Rooman, I. , Anderson, M. , Holmes, O. , Leonard, C. , Taylor, D. , Wood, S. , Xu, Q. , Nones, K. , Fink, J.L. , Christ, A. , Bruxner, T. , Cloonan, N. , Kolle, G. , Newell, F. , Pinese, M. , Mead, R.S. , Humphris, J.L. , Kaplan, W. , Jones, M.D. , Colvin, E.K. , Nagrial, A.M. , Humphrey, E.S. , Chou, A. , Chin, V.T. , Chantrill, L.A. , Mawson, A. , Samra, J.S. , Kench, J.G. , Lovell, J.A. , Daly, R.J. , Merrett, N.D. , Toon, C. , Epari, K. , Nguyen, N.Q. , Barbour, A. , Zeps, N. , Australian Pancreatic Cancer Genome Initiative, Kakkar, N. , Zhao, F. , Wu, Y.Q. , Wang, M. , Muzny, D.M. , Fisher, W.E. , Brunicardi, F.C. , Hodges, S.E. , Reid, J.G. , Drummond, J. , Chang, K. , Han, Y. , Lewis, L.R. , Dinh, H. , Buhay, C.J. , Beck, T. , Timms, L. , Sam, M. , Begley, K. , Brown, A. , Pai, D. , Panchal, A. , Buchner, N. , De Borja, R. , Denroche, R.E. , Yung, C.K. , Serra, S. , Onetto, N. , Mukhopadhyay, D. , Tsao, M.-S. , Shaw, P.A. , Petersen, G.M. , Gallinger, S. , Hruban, R.H. , Maitra, A. , Iacobuzio-Donahue, C.A. , Schulick, R.D. , Wolfgang, C.L. , Morgan, R.A. , Lawlor, R.T. , Capelli, P. , Corbo, V. , Scardoni, M. , Tortora, G. , Tempero, M.A. , Mann, K.M. , Jenkins, N.A. , Perez-Mancera, P.A. , Adams, D.J. , Largaespada, D.A. , Wessels, L.F.A. , Rust, A.G. , Stein, L.D. , Tuveson, D.A. , Copeland, N.G. , Musgrove, E.A. , Scarpa, A. , Eshleman, J.R. , Hudson, T.J. , Sutherland, R.L. , Wheeler, D.A. , Pearson, J.V. , McPherson, J.D. , Gibbs, R.A. , Grimmond, S.M. , 2012. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 491, 399–405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard, F.C. , Huguet, F. , Louvet, C. , Mineur, L. , Bouché, O. , Chibaudel, B. , Artru, P. , Desseigne, F. , Bachet, J.B. , Mathiot, C. , Pierga, J.Y. , Hammel, P. , 2013. Circulating tumor cells in locally advanced pancreatic adenocarcinoma: the ancillary CirCe 07 study to the LAP 07 trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO. 24, 2057–2061. 10.1093/annonc/mdt176 [DOI] [PubMed] [Google Scholar]
- Bidard, F.-C. , Peeters, D.J. , Fehm, T. , Nolé, F. , Gisbert-Criado, R. , Mavroudis, D. , Grisanti, S. , Generali, D. , Garcia-Saenz, J.A. , Stebbing, J. , Caldas, C. , Gazzaniga, P. , Manso, L. , Zamarchi, R. , de Lascoiti, A.F. , De Mattos-Arruda, L. , Ignatiadis, M. , Lebofsky, R. , van Laere, S.J. , Meier-Stiegen, F. , Sandri, M.-T. , Vidal-Martinez, J. , Politaki, E. , Consoli, F. , Bottini, A. , Diaz-Rubio, E. , Krell, J. , Dawson, S.-J. , Raimondi, C. , Rutten, A. , Janni, W. , Munzone, E. , Carañana, V. , Agelaki, S. , Almici, C. , Dirix, L. , Solomayer, E.-F. , Zorzino, L. , Johannes, H. , Reis-Filho, J.S. , Pantel, K. , Pierga, J.-Y. , Michiels, S. , 2014. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 15, 406–414. 10.1016/S1470-2045(14)70069-5 [DOI] [PubMed] [Google Scholar]
- Bissolati, M. , Sandri, M.T. , Burtulo, G. , Zorzino, L. , Balzano, G. , Braga, M. , 2015. Portal vein-circulating tumor cells predict liver metastases in patients with resectable pancreatic cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 36, 991–996. 10.1007/s13277-014-2716-0 [DOI] [PubMed] [Google Scholar]
- Bobek, V. , Gurlich, R. , Eliasova, P. , Kolostova, K. , 2014. Circulating tumor cells in pancreatic cancer patients: enrichment and cultivation. World J. Gastroenterol. WJG. 20, 17163–17170. 10.3748/wjg.v20.i45.17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas, C. , Hahn, S.A. , Hruban, R.H. , Redston, M.S. , Yeo, C.J. , Kern, S.E. , 1994. Detection of K-ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res. 54, 3568–3573. [PubMed] [Google Scholar]
- Capurso, G. , Signoretti, M. , Valente, R. , Arnelo, U. , Lohr, M. , Poley, J.-W. , Delle Fave, G. , Del Chiaro, M. , 2015. Methods and outcomes of screening for pancreatic adenocarcinoma in high-risk individuals. World J. Gastrointest. Endosc. 7, 833–842. 10.4253/wjge.v7.i9.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells, A. , Puig, P. , Móra, J. , Boadas, J. , Boix, L. , Urgell, E. , Solé, M. , Capellà, G. , Lluís, F. , Fernández-Cruz, L. , Navarro, S. , Farré, A. , 1999. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 17, 578–584. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Tu, H. , Meng, Z.Q. , Chen, Z. , Wang, P. , Liu, L.M. , 2010. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 36, 657–662. 10.1016/j.ejso.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Cho, E.H. , Wendel, M. , Luttgen, M. , Yoshioka, C. , Marrinucci, D. , Lazar, D. , Schram, E. , Nieva, J. , Bazhenova, L. , Morgan, A. , Ko, A.H. , Korn, W.M. , Kolatkar, A. , Bethel, K. , Kuhn, P. , 2012. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys. Biol. 9, 016001 10.1088/1478-3975/9/1/016001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S.J. , Punt, C.J.A. , Iannotti, N. , Saidman, B.H. , Sabbath, K.D. , Gabrail, N.Y. , Picus, J. , Morse, M. , Mitchell, E. , Miller, M.C. , Doyle, G.V. , Tissing, H. , Terstappen, L.W.M.M. , Meropol, N.J. , 2008. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 26, 3213–3221. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- Conroy, T. , Desseigne, F. , Ychou, M. , Bouché, O. , Guimbaud, R. , Bécouarn, Y. , Adenis, A. , Raoul, J.-L. , Gourgou-Bourgade, S. , de la Fouchardière, C. , Bennouna, J. , Bachet, J.-B. , Khemissa-Akouz, F. , Péré-Vergé, D. , Delbaldo, C. , Assenat, E. , Chauffert, B. , Michel, P. , Montoto-Grillot, C. , Ducreux, M. , Groupe Tumeurs Digestives of Unicancer, PRODIGE Intergroup2011. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825. 10.1056/NEJMoa1011923 [DOI] [PubMed] [Google Scholar]
- Croce, C.M. , 2008. Oncogenes and cancer. N. Engl. J. Med. 358, 502–511. 10.1056/NEJMra072367 [DOI] [PubMed] [Google Scholar]
- Däbritz, J. , Preston, R. , Hänfler, J. , Oettle, H. , 2009. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas. 38, 534–541. 10.1097/MPA.0b013e31819f6376 [DOI] [PubMed] [Google Scholar]
- de Albuquerque, A. , Kubisch, I. , Breier, G. , Stamminger, G. , Fersis, N. , Eichler, A. , Kaul, S. , Stölzel, U. , 2012. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. Oncology. 82, 3–10. 10.1159/000335479 [DOI] [PubMed] [Google Scholar]
- de Bono, J.S. , Scher, H.I. , Montgomery, R.B. , Parker, C. , Miller, M.C. , Tissing, H. , Doyle, G.V. , Terstappen, L.W.W.M. , Pienta, K.J. , Raghavan, D. , 2008. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 14, 6302–6309. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- Dianxu, F. , Shengdao, Z. , Tianquan, H. , Yu, J. , Ruoqing, L. , Zurong, Y. , Xuezhi, W. , 2002. A prospective study of detection of pancreatic carcinoma by combined plasma K-ras mutations and serum CA19-9 analysis. Pancreas. 25, 336–341. [DOI] [PubMed] [Google Scholar]
- Diaz, L.A. , Bardelli, A. , 2014. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 32, 579–586. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, F. , Schmidt, K. , Choti, M.A. , Romans, K. , Goodman, S. , Li, M. , Thornton, K. , Agrawal, N. , Sokoll, L. , Szabo, S.A. , Kinzler, K.W. , Vogelstein, B. , Diaz, L.A. , 2008. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard, J.-Y. , Ostoros, G. , Cobo, M. , Ciuleanu, T. , Cole, R. , McWalter, G. , Walker, J. , Dearden, S. , Webster, A. , Milenkova, T. , McCormack, R. , 2014. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer. 9, 1345–1353. 10.1097/JTO.0000000000000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl, J. , Garcia-Nieto, S. , Martinez-Avila, J.C. , Montans, J. , Sanjuanbenito, A. , Rodríguez-Garrote, M. , Lisa, E. , Mendía, E. , Lobo, E. , Malats, N. , Carrato, A. , Guillen-Ponce, C. , 2015. Circulating tumor cells (ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. 15, 10.1186/s12885-015-1779-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi, S. , Rolet, F. , Mouliere, F. , Thierry, A.R. , 2013. Circulating cell free DNA: preanalytical considerations. Clin. Chim. Acta Int. J. Clin. Chem. 424, 222–230. 10.1016/j.cca.2013.05.022 [DOI] [PubMed] [Google Scholar]
- Fleischhacker, M. , Schmidt, B. , 2007. Circulating nucleic acids (CNAs) and cancer–a survey. Biochim. Biophys. Acta. 1775, 181–232. 10.1016/j.bbcan.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Goonetilleke, K.S. , Siriwardena, A.K. , 2007. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 33, 266–270. 10.1016/j.ejso.2006.10.004 [DOI] [PubMed] [Google Scholar]
- Hammel, P. , Huguet, F. , Van Laethem, J.L. , Goldstein, D. , Glimelius, B. , Artru, P. , Borbath, I. , Bouche, O. , Shannon, J. , André, T. , Mineur, L. , Chibaudel, B. , Bonnetain, F. , Louvet, C. , 2013. Comparison of chemoradiotherapy (CRT) and chemotherapy (CT) in patients with a locally advanced pancreatic cancer (LAPC) controlled after 4 months of gemcitabine with or without erlotinib: Final results of the international phase III LAP 07 study. J. Clin. Oncol. 31, (suppl; abstr LBA4003) [Google Scholar]
- Hanahan, D. , Weinberg, R.A. , 2011. Hallmarks of cancer: the next generation. Cell. 144, 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hess, V. , Glimelius, B. , Grawe, P. , Dietrich, D. , Bodoky, G. , Ruhstaller, T. , Bajetta, E. , Saletti, P. , Figer, A. , Scheithauer, W. , Herrmann, R. , 2008. CA 19-9 tumour-marker response to chemotherapy in patients with advanced pancreatic cancer enrolled in a randomised controlled trial. Lancet Oncol. 9, 132–138. 10.1016/S1470-2045(08)70001-9 [DOI] [PubMed] [Google Scholar]
- Hoffmann, K. , Kerner, C. , Wilfert, W. , Mueller, M. , Thiery, J. , Hauss, J. , Witzigmann, H. , 2007. Detection of disseminated pancreatic cells by amplification of cytokeratin-19 with quantitative RT-PCR in blood, bone marrow and peritoneal lavage of pancreatic carcinoma patients. World J. Gastroenterol. WJG. 13, 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustinx, S.R. , Leoni, L.M. , Yeo, C.J. , Brown, P.N. , Goggins, M. , Kern, S.E. , Hruban, R.H. , Maitra, A. , 2005. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 18, 959–963. 10.1038/modpathol.3800377 [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue, C.A. , Fu, B. , Yachida, S. , Luo, M. , Abe, H. , Henderson, C.M. , Vilardell, F. , Wang, Z. , Keller, J.W. , Banerjee, P. , Herman, J.M. , Cameron, J.L. , Yeo, C.J. , Halushka, M.K. , Eshleman, J.R. , Raben, M. , Klein, A.P. , Hruban, R.H. , Hidalgo, M. , Laheru, D. , 2009. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 27, 1806–1813. 10.1200/JCO.2008.17.7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, H. , Okada, S. , Sato, T. , Wakasugi, H. , Saisho, H. , Furuse, J. , Ishikawa, O. , Matsuno, S. , Yokoyama, S. , 1997. CA 19-9 in evaluating the response to chemotherapy in advanced pancreatic cancer. Hepatogastroenterology. 44, 279–283. [PubMed] [Google Scholar]
- Jahr, S. , Hentze, H. , Englisch, S. , Hardt, D. , Fackelmayer, F.O. , Hesch, R.D. , Knippers, R. , 2001. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665. [PubMed] [Google Scholar]
- Jemal, A. , Bray, F. , Center, M.M. , Ferlay, J. , Ward, E. , Forman, D. , 2011. Global cancer statistics. CA. Cancer J. Clin. 61, 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- Johansen, J.S. , Vibat, C.R.T. , Calatayud, D. , Jensen, B.V. , Hasselby, J.P. , Collisson, E.A. , Lu, T. , Poole, J.C. , Erlander, M. , 2015. Comparative circulating tumor DNA levels for KRAS mutations in patients with nonresectable pancreatic cancer. J. Clin. Oncol. 33, (suppl 3; abstr 288) [Google Scholar]
- Kanda, M. , Matthaei, H. , Wu, J. , Hong, S.-M. , Yu, J. , Borges, M. , Hruban, R.H. , Maitra, A. , Kinzler, K. , Vogelstein, B. , Goggins, M. , 2012. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 142, 10.1053/j.gastro.2011.12.042 730–733.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.S. , Tsigani, T. , Rashid, M. , Rabouhans, J.S. , Yu, D. , Luong, T.V. , Caplin, M. , Meyer, T. , 2011. Circulating tumor cells and EpCAM expression in neuroendocrine tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 17, 337–345. 10.1158/1078-0432.CCR-10-1776 [DOI] [PubMed] [Google Scholar]
- Khoja, L. , Backen, A. , Sloane, R. , Menasce, L. , Ryder, D. , Krebs, M. , Board, R. , Clack, G. , Hughes, A. , Blackhall, F. , Valle, J.W. , Dive, C. , 2012. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br. J. Cancer. 106, 508–516. 10.1038/bjc.2011.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde, I. , Wu, J. , Papadopoulos, N. , Kinzler, K.W. , Vogelstein, B. , 2011. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 108, 9530–9535. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa, H. , Nouso, K. , Miyahara, K. , Morimoto, Y. , Dohi, C. , Tsutsumi, K. , Kato, H. , Matsubara, T. , Okada, H. , Yamamoto, K. , 2015. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 10.1002/cncr.29364 [DOI] [PubMed] [Google Scholar]
- Konstantinidis, I.T. , Warshaw, A.L. , Allen, J.N. , Blaszkowsky, L.S. , Castillo, C.F.-D. , Deshpande, V. , Hong, T.S. , Kwak, E.L. , Lauwers, G.Y. , Ryan, D.P. , Wargo, J.A. , Lillemoe, K.D. , Ferrone, C.R. , 2013. Pancreatic ductal adenocarcinoma: is there a survival difference for R1 resections versus locally advanced unresectable tumors? what is a “true” R0 resection?. Ann. Surg. 257, 731–736. 10.1097/SLA.0b013e318263da2f [DOI] [PubMed] [Google Scholar]
- Kulemann, B. , Pitman, M.B. , Liss, A.S. , Valsangkar, N. , Fernández-Del Castillo, C. , Lillemoe, K.D. , Hoeppner, J. , Mino-Kenudson, M. , Warshaw, A.L. , Thayer, S.P. , 2015. Circulating tumor cells found in patients with localized and advanced pancreatic cancer. Pancreas. 44, 547–550. 10.1097/MPA.0000000000000324 [DOI] [PubMed] [Google Scholar]
- Kurihara, T. , Itoi, T. , Sofuni, A. , Itokawa, F. , Tsuchiya, T. , Tsuji, S. , Ishii, K. , Ikeuchi, N. , Tsuchida, A. , Kasuya, K. , Kawai, T. , Sakai, Y. , Moriyasu, F. , 2008. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J. Hepatobiliary. Pancreat. Surg. 15, 189–195. 10.1007/s00534-007-1250-5 [DOI] [PubMed] [Google Scholar]
- Labelle, M. , Begum, S. , Hynes, R.O. , 2011. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 20, 576–590. 10.1016/j.ccr.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky, R. , Decraene, C. , Bernard, V. , Kamal, M. , Blin, A. , Leroy, Q. , Rio Frio, T. , Pierron, G. , Callens, C. , Bieche, I. , Saliou, A. , Madic, J. , Rouleau, E. , Bidard, F.-C. , Lantz, O. , Stern, M.-H. , Le Tourneau, C. , Pierga, J.-Y. , 2015. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol. Oncol. 9, 783–790. 10.1016/j.molonc.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madic, J. , Piperno-Neumann, S. , Servois, V. , Rampanou, A. , Milder, M. , Trouiller, B. , Gentien, D. , Saada, S. , Assayag, F. , Thuleau, A. , Nemati, F. , Decaudin, D. , Bidard, F.-C. , Desjardins, L. , Mariani, P. , Lantz, O. , Stern, M.-H. , 2012. Pyrophosphorolysis-activated polymerization detects circulating tumor DNA in metastatic uveal melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 18, 3934–3941. 10.1158/1078-0432.CCR-12-0309 [DOI] [PubMed] [Google Scholar]
- Maeda, A. , Boku, N. , Fukutomi, A. , Kondo, S. , Kinoshita, T. , Nagino, M. , Uesaka, K. , 2008. Randomized phase III trial of adjuvant chemotherapy with gemcitabine versus S-1 in patients with resected pancreatic cancer: Japan Adjuvant Study Group of Pancreatic Cancer (JASPAC-01). Jpn. J. Clin. Oncol. 38, 227–229. 10.1093/jjco/hym178 [DOI] [PubMed] [Google Scholar]
- Maire, F. , Micard, S. , Hammel, P. , Voitot, H. , Lévy, P. , Cugnenc, P.-H. , Ruszniewski, P. , Puig, P.L. , 2002. Differential diagnosis between chronic pancreatitis and pancreatic cancer: value of the detection of KRAS2 mutations in circulating DNA. Br. J. Cancer. 87, 551–554. 10.1038/sj.bjc.6600475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidanna, R. , Ni, Z. , Anderson, O. , Spiegelhalter, S.D. , Bottle, A. , Aylin, P. , Faiz, O. , Hanna, G.B. , 2015. Surgeon volume and Cancer Esophagectomy, Gastrectomy, and pancreatectomy: a population-based study in England. Ann. Surg. 10.1097/SLA.0000000000001490 [DOI] [PubMed] [Google Scholar]
- Mandel, P. , Metais, P. , 1948. Comptes Rendus Séances Société Biol. Ses Fil. 142, 241–243. [PubMed] [Google Scholar]
- Mataki, Y. , Takao, S. , Maemura, K. , Mori, S. , Shinchi, H. , Natsugoe, S. , Aikou, T. , 2004. Carcinoembryonic antigen messenger RNA expression using nested reverse transcription-PCR in the peripheral blood during follow-up period of patients who underwent curative surgery for biliary-pancreatic cancer: longitudinal analyses. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 10, 3807–3814. 10.1158/1078-0432.CCR-03-0130 [DOI] [PubMed] [Google Scholar]
- Modolell, I. , Guarner, L. , Malagelada, J.R. , 1999. Vagaries of clinical presentation of pancreatic and biliary tract cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO. 10, (Suppl. 4) 82–84. [PubMed] [Google Scholar]
- Mudan, S. , Giakoustidis, A. , Thillainayagam, A.V. , Jacob, J. , Stebbing, J. , 2010. Clinical utility of circulating tumor cell measurement in the diagnosis of indeterminate lesions of the pancreas. Future Oncol. 6, 177–179. 10.2217/fon.09.156 [DOI] [PubMed] [Google Scholar]
- Mulcahy, H.E. , Lyautey, J. , Lederrey, C. , qi Chen, X. , Anker, P. , Alstead, E.M. , Ballinger, A. , Farthing, M.J. , Stroun, M. , 1998. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 4, 271–275. [PubMed] [Google Scholar]
- Neoptolemos, J.P. , Stocken, D.D. , Bassi, C. , Ghaneh, P. , Cunningham, D. , Goldstein, D. , Padbury, R. , Moore, M.J. , Gallinger, S. , Mariette, C. , Wente, M.N. , Izbicki, J.R. , Friess, H. , Lerch, M.M. , Dervenis, C. , Oláh, A. , Butturini, G. , Doi, R. , Lind, P.A. , Smith, D. , Valle, J.W. , Palmer, D.H. , Buckels, J.A. , Thompson, J. , McKay, C.J. , Rawcliffe, C.L. , Büchler, M.W. , European Study Group for Pancreatic Cancer2010. Adjuvant chemotherapy with fluorouracil plus folinic acid vs. gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 304, 1073–1081. 10.1001/jama.2010.1275 [DOI] [PubMed] [Google Scholar]
- Neoptolemos, J.P. , Stocken, D.D. , Friess, H. , Bassi, C. , Dunn, J.A. , Hickey, H. , Beger, H. , Fernandez-Cruz, L. , Dervenis, C. , Lacaine, F. , Falconi, M. , Pederzoli, P. , Pap, A. , Spooner, D. , Kerr, D.J. , Büchler, M.W. , European Study Group for Pancreatic Cancer2004. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 350, 1200–1210. 10.1056/NEJMoa032295 [DOI] [PubMed] [Google Scholar]
- Oettle, H. , Neuhaus, P. , Hochhaus, A. , Hartmann, J.T. , Gellert, K. , Ridwelski, K. , Niedergethmann, M. , Zülke, C. , Fahlke, J. , Arning, M.B. , Sinn, M. , Hinke, A. , Riess, H. , 2013. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 310, 1473–1481. 10.1001/jama.2013.279201 [DOI] [PubMed] [Google Scholar]
- Olsen, C.C. , Schefter, T.E. , Chen, H. , Kane, M. , Leong, S. , McCarter, M.D. , Chen, Y. , Mack, P. , Eckhardt, S.G. , Stiegmann, G. , Raben, D. , 2009. Results of a phase I trial of 12 patients with locally advanced pancreatic carcinoma combining gefitinib, paclitaxel, and 3-dimensional conformal radiation: report of toxicity and evaluation of circulating K-ras as a potential biomarker of response to therapy. Am. J. Clin. Oncol. 32, 115–121. 10.1097/COC.0b013e318180baa3 [DOI] [PubMed] [Google Scholar]
- Poruk, K.E. , Firpo, M.A. , Adler, D.G. , Mulvihill, S.J. , 2013. Screening for pancreatic cancer: why, how, and who?. Ann. Surg. 257, 17–26. 10.1097/SLA.0b013e31825ffbfb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, C. , Han, C. , Zhang, J. , He, P. , Wang, D. , Wang, B. , Zhao, P. , Zhao, X. , 2011. Detection of apoptotic circulating tumor cells in advanced pancreatic cancer following 5-fluorouracil chemotherapy. Cancer Biol. Ther. 12, 700–706. 10.4161/cbt.12.8.15960 [DOI] [PubMed] [Google Scholar]
- Rhim, A.D. , Thege, F.I. , Santana, S.M. , Lannin, T.B. , Saha, T.N. , Tsai, S. , Maggs, L.R. , Kochman, M.L. , Ginsberg, G.G. , Lieb, J.G. , Chandrasekhara, V. , Drebin, J.A. , Ahmad, N. , Yang, Y.-X. , Kirby, B.J. , Stanger, B.Z. , 2014. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 146, 647–651. 10.1053/j.gastro.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen, M. , Phallen, J. , Adleff, V. , Jones, S. , Leary, R.J. , Barrett, M.T. , Anagnostou, V. , Parpart-Li, S. , Murphy, D. , Kay Li, Q. , Hruban, C.A. , Scharpf, R. , White, J.R. , O'Dwyer, P.J. , Allen, P.J. , Eshleman, J.R. , Thompson, C.B. , Klimstra, D.S. , Linehan, D.C. , Maitra, A. , Hruban, R.H. , Diaz, L.A. , Von Hoff, D.D. , Johansen, J.S. , Drebin, J.A. , Velculescu, V.E. , 2015. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 6, 7686 10.1038/ncomms8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach, H. , Stoehlmacher, J. , Pantel, K. , Goekkurt, E. , 2008. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann. N. Y. Acad. Sci. 1137, 190–196. 10.1196/annals.1448.025 [DOI] [PubMed] [Google Scholar]
- Sergeant, G. , Roskams, T. , van Pelt, J. , Houtmeyers, F. , Aerts, R. , Topal, B. , 2011. Perioperative cancer cell dissemination detected with a real-time RT-PCR assay for EpCAM is not associated with worse prognosis in pancreatic ductal adenocarcinoma. BMC Cancer. 11, 47 10.1186/1471-2407-11-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.L. , Miller, K.D. , Jemal, A. , 2015. Cancer statistics, 2015. CA. Cancer J. Clin. 65, 5–29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- Sinn, B.V. , Striefler, J.K. , Rudl, M.A. , Lehmann, A. , Bahra, M. , Denkert, C. , Sinn, M. , Stieler, J. , Klauschen, F. , Budczies, J. , Weichert, W. , Stenzinger, A. , Kamphues, C. , Dietel, M. , Riess, H. , 2014. KRAS mutations in codon 12 or 13 are associated with worse prognosis in pancreatic ductal adenocarcinoma. Pancreas. 43, 578–583. 10.1097/MPA.0000000000000077 [DOI] [PubMed] [Google Scholar]
- Soeth, E. , Grigoleit, U. , Moellmann, B. , Röder, C. , Schniewind, B. , Kremer, B. , Kalthoff, H. , Vogel, I. , 2005. Detection of tumor cell dissemination in pancreatic ductal carcinoma patients by CK 20 RT-PCR indicates poor survival. J. Cancer Res. Clin. Oncol. 131, 669–676. 10.1007/s00432-005-0008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, K. , Jiang, P. , Chan, K.C.A. , Wong, J. , Cheng, Y.K.Y. , Liang, R.H.S. , Chan, W.-K. , Ma, E.S.K. , Chan, S.L. , Cheng, S.H. , Chan, R.W.Y. , Tong, Y.K. , Ng, S.S.M. , Wong, R.S.M. , Hui, D.S.C. , Leung, T.N. , Leung, T.Y. , Lai, P.B.S. , Chiu, R.W.K. , Lo, Y.M.D. , 2015. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512. 10.1073/pnas.1508736112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, E. , Totoki, Y. , Nakamura, H. , Morizane, C. , Nara, S. , Hama, N. , Suzuki, M. , Furukawa, E. , Kato, M. , Hayashi, H. , Kohno, T. , Ueno, H. , Shimada, K. , Okusaka, T. , Nakagama, H. , Shibata, T. , Yachida, S. , 2015. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 5, 18425 10.1038/srep18425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thege, F.I. , Lannin, T.B. , Saha, T.N. , Tsai, S. , Kochman, M.L. , Hollingsworth, M.A. , Rhim, A.D. , Kirby, B.J. , 2014. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab. Chip. 14, 1775–1784. 10.1039/c4lc00041b [DOI] [PubMed] [Google Scholar]
- Thiery, J.P. , 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2, 442–454. 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- Ting, D.T. , Wittner, B.S. , Ligorio, M. , Vincent Jordan, N. , Shah, A.M. , Miyamoto, D.T. , Aceto, N. , Bersani, F. , Brannigan, B.W. , Xega, K. , Ciciliano, J.C. , Zhu, H. , MacKenzie, O.C. , Trautwein, J. , Arora, K.S. , Shahid, M. , Ellis, H.L. , Qu, N. , Bardeesy, N. , Rivera, M.N. , Deshpande, V. , Ferrone, C.R. , Kapur, R. , Ramaswamy, S. , Shioda, T. , Toner, M. , Maheswaran, S. , Haber, D.A. , 2014. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8, 1905–1918. 10.1016/j.celrep.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjensvoll, K. , Lapin, M. , Buhl, T. , Oltedal, S. , Steen-Ottosen Berry, K. , Gilje, B. , Søreide, J.A. , Javle, M. , Nordgård, O. , Smaaland, R. , 2015. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 10.1016/j.molonc.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torphy, R.J. , Tignanelli, C.J. , Kamande, J.W. , Moffitt, R.A. , Herrera Loeza, S.G. , Soper, S.A. , Yeh, J.J. , 2014. Circulating tumor cells as a biomarker of response to treatment in patient-derived xenograft mouse models of pancreatic adenocarcinoma. PloS One. 9, e89474 10.1371/journal.pone.0089474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T. , Hibi, K. , Kaneko, T. , Takeda, S. , Inoue, S. , Okochi, O. , Nagasaka, T. , Nakao, A. , 2004. Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J. Gastroenterol. 39, 56–60. 10.1007/s00535-003-1245-1 [DOI] [PubMed] [Google Scholar]
- Van Laethem, J.L. , Bourgeois, V. , Parma, J. , Delhaye, M. , Cochaux, P. , Velu, T. , Devière, J. , Cremer, M. , 1998. Relative contribution of KI-RAS gene analysis and brush cytology during ERCP for the diagnosis of biliary and pancreatic diseases. Gastrointest. Endosc. 47, 479–485. [DOI] [PubMed] [Google Scholar]
- Vincent, A. , Herman, J. , Schulick, R. , Hruban, R.H. , Goggins, M. , 2011. Pancreatic cancer. Lancet. 378, 607–620. 10.1016/S0140-6736(10)62307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff, D.D. , Ervin, T. , Arena, F.P. , Chiorean, E.G. , Infante, J. , Moore, M. , Seay, T. , Tjulandin, S.A. , Ma, W.W. , Saleh, M.N. , Harris, M. , Reni, M. , Dowden, S. , Laheru, D. , Bahary, N. , Ramanathan, R.K. , Tabernero, J. , Hidalgo, M. , Goldstein, D. , Van Cutsem, E. , Wei, X. , Iglesias, J. , Renschler, M.F. , 2013. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 369, 1691–1703. 10.1056/NEJMoa1304369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, H. , Ha, A. , Hu, Y.X. , Ohtsubo, K. , Yamaguchi, Y. , Motoo, Y. , Okai, T. , Toya, D. , Tanaka, N. , Sawabu, N. , 1999. K-ras mutations in duodenal aspirate without secretin stimulation for screening of pancreatic and biliary tract carcinoma. Cancer. 86, 1441–1448. [PubMed] [Google Scholar]
- Wenger, F.A. , Zieren, J. , Peter, F.J. , Jacobi, C.A. , Müller, J.M. , 1999. K-ras mutations in tissue and stool samples from patients with pancreatic cancer and chronic pancreatitis. Langenbecks Arch. Surg. Dtsch. Ges. Für Chir. 384, 181–186. [DOI] [PubMed] [Google Scholar]
- Wilentz, R.E. , Chung, C.H. , Sturm, P.D. , Musler, A. , Sohn, T.A. , Offerhaus, G.J. , Yeo, C.J. , Hruban, R.H. , Slebos, R.J. , 1998. K-ras mutations in the duodenal fluid of patients with pancreatic carcinoma. Cancer. 82, 96–103. [DOI] [PubMed] [Google Scholar]
- Yamada, T. , Nakamori, S. , Ohzato, H. , Oshima, S. , Aoki, T. , Higaki, N. , Sugimoto, K. , Akagi, K. , Fujiwara, Y. , Nishisho, I. , Sakon, M. , Gotoh, M. , Monden, M. , 1998. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 4, 1527–1532. [PubMed] [Google Scholar]
- Yu, M. , Bardia, A. , Wittner, B.S. , Stott, S.L. , Smas, M.E. , Ting, D.T. , Isakoff, S.J. , Ciciliano, J.C. , Wells, M.N. , Shah, A.M. , Concannon, K.F. , Donaldson, M.C. , Sequist, L.V. , Brachtel, E. , Sgroi, D. , Baselga, J. , Ramaswamy, S. , Toner, M. , Haber, D.A. , Maheswaran, S. , 2013. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 339, 580–584. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. , Ting, D.T. , Stott, S.L. , Wittner, B.S. , Ozsolak, F. , Paul, S. , Ciciliano, J.C. , Smas, M.E. , Winokur, D. , Gilman, A.J. , Ulman, M.J. , Xega, K. , Contino, G. , Alagesan, B. , Brannigan, B.W. , Milos, P.M. , Ryan, D.P. , Sequist, L.V. , Bardeesy, N. , Ramaswamy, S. , Toner, M. , Maheswaran, S. , Haber, D.A. , 2012. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 487, 510–513. 10.1038/nature11217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Z'graggen, K. , Centeno, B.A. , Fernandez-del Castillo, C. , Jimenez, R.E. , Werner, J. , Warshaw, A.L. , 2001. Biological implications of tumor cells in blood and bone marrow of pancreatic cancer patients. Surgery. 129, 537–546. 10.1067/msy.2001.113819 [DOI] [PubMed] [Google Scholar]
- Zill, O.A. , Greene, C. , Sebisanovic, D. , Siew, L.M. , Leng, J. , Vu, M. , Hendifar, A.E. , Wang, Z. , Atreya, C.E. , Kelley, R.K. , Van Loon, K. , Ko, A.H. , Tempero, M.A. , Bivona, T.G. , Munster, P.N. , Talasaz, A. , Collisson, E.A. , 2015. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 5, 1040–1048. 10.1158/2159-8290.CD-15-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]


