Significance
Sequence variants that create or eliminate splice sites are often clinically classified as variants of unknown significance (VUS) due to imperfect understanding of RNA splice signals and cumbersome functional assays. In autosomal dominant disorders caused by haploinsufficiency, variants that alter normal splicing of one allele are pathogenic. We developed enhanced computational tools to prioritize potential splice-altering VUS and used a minigene assay to functionally confirm splice-altering sequence variants. In studying all reported variants across LMNA and MYBPC3, two known heart disease genes, we demonstrate that ∼5% of VUS from affected patients alter splicing and are undetected disease-causing variants. This strategy improves clinical detection of pathogenic variants and should be broadly relevant to other human disorders that are caused by haploinsufficiency.
Keywords: VUS, splicing, cardiomyopathy, LMNA, MYBPC3
Abstract
Genetic variants that cause haploinsufficiency account for many autosomal dominant (AD) disorders. Gene-based diagnosis classifies variants that alter canonical splice signals as pathogenic, but due to imperfect understanding of RNA splice signals other variants that may create or eliminate splice sites are often clinically classified as variants of unknown significance (VUS). To improve recognition of pathogenic splice-altering variants in AD disorders, we used computational tools to prioritize VUS and developed a cell-based minigene splicing assay to confirm aberrant splicing. Using this two-step procedure we evaluated all rare variants in two AD cardiomyopathy genes, lamin A/C (LMNA) and myosin binding protein C (MYBPC3). We demonstrate that 13 LMNA and 35 MYBPC3 variants identified in cardiomyopathy patients alter RNA splicing, representing a 50% increase in the numbers of established damaging splice variants in these genes. Over half of these variants are annotated as VUS by clinical diagnostic laboratories. Familial analyses of one variant, a synonymous LMNA VUS, demonstrated segregation with cardiomyopathy affection status and altered cardiac LMNA splicing. Application of this strategy should improve diagnostic accuracy and variant classification in other haploinsufficient AD disorders.
DNA sequence analysis of patient DNA has been used to clinically diagnose associated inherited diseases, leading to the identification of thousands of rare sequence variants in affected and unaffected individuals. Interpreting the medical significance of these variants has proven to be a significant challenge. The annotation of nonsense, frameshift, insertion, and deletion variants that cause a highly predictable outcome [i.e., loss of function (LoF) of proteins encoded by disease genes] allows these to be clinically classified as pathogenic variants in more than 4,000 genes that cause disease by haploinsufficiency (1). Classification of rare synonymous and missense variants in these genes, however, has been more difficult. Although bioinformatic algorithms predict that some variants may damage the encoded protein, these annotations are imperfect, and many rare variants remain unclassified and are clinically designated as variants of unknown significance (VUS) (2).
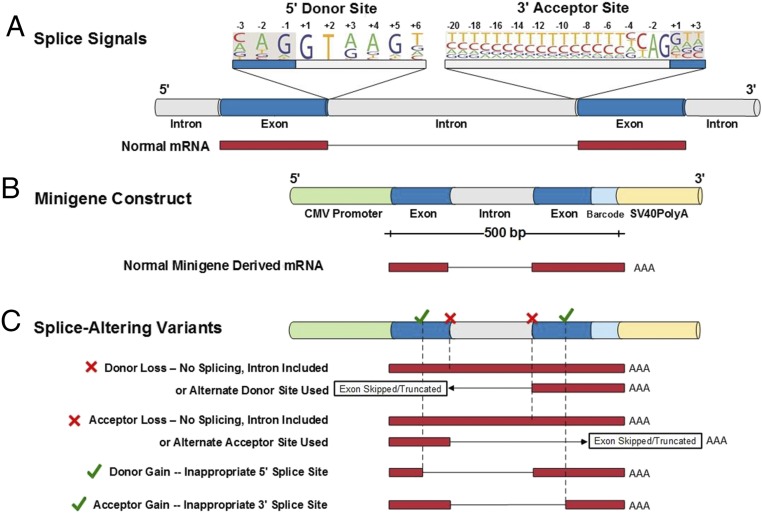
Splicing signals reside at all exon–intron junctions and include a 9-bp 5′ splice donor sequence with an invariant GT dinucleotide and a 23-bp 3′ splice acceptor sequence with an invariant AG dinucleotide (3) (Fig. 1A). Variants that alter the canonical GT and AG dinucleotides in haploinsufficient autosomal dominant (AD) genes are diagnostically classified as pathogenic, whereas substitutions of the remaining seven residues within the donor sequence, or the 21 residues within the acceptor sequence, are often classified as VUS (4). VUS that alter these sequences, however, have the potential to disrupt normal RNA splicing (5) (resulting in donor or acceptor loss; Fig. 1C). Similarly, variants within a nearby intron or exon may sufficiently mimic the consensus donor or acceptor sites to generate new, inappropriate splice signals (donor or acceptor gain; Fig. 1C). Although existing in vitro splicing assays can identify variants that alter splicing and thereby identify pathogenic variants, cost and time constraints limit their application to clinical gene-based diagnosis (6, 7).
Fig. 1.
Schematics of gene splice signals and a functional assay of potential splice-altering variants. (A) A gene segment containing two exons (blue) and flanking introns (gray) with consensus 9-bp splice donor and 23-bp splice acceptor sequences is shown. The nucleotide letter sizes (drawn using Pictogram; genes.mit.edu/pictogram.html) denote use of that base in splicing across the genome. Dinucleotides GT (donor site) and AG (acceptor site) are invariant. Normally spliced transcript (red) that excludes intron sequence is depicted. (B) Functional assay of splicing candidates used a 1,200-bp minigene construct comprised of CMV promoter, exon–intron–exon sequence, a 2-bp barcode to identify the construct, and SV40polyA sequence. Exon–intron–exon test sequences were encoded on a synthetic 500-bp oligonucleotide that limited the inclusion of intron sequences to 200 bp (further details provided in Materials and Methods and Datasets S3 and S4). A normally spliced minigene-derived transcript (red) that excludes intron sequence is depicted. (C) Splice-altering variants detected by functional assays. Variants that destroy donor or acceptor signals prevent normal splicing (“x”) and yield transcripts with included introns, partial deletions, or entirely skipped exons. [Note that analyses reported excluded all variants that altered the invariant GT (donor site) and AG (acceptor site) sequences.] Variants that create a novel splice signal (donor or acceptor gain, “✓”) inappropriately delete sequences from the 5′ or 3′ exon. Novel splice sites that occur within introns and insert sequences are not depicted.
Results
LMNA and MYBPC3 Variants That Affect RNA Splicing.
LoF variants in LMNA and MYBPC3 are prevalent causes of dilated cardiomyopathy (DCM) with associated conduction defects (8) and hypertrophic cardiomyopathy (HCM) (9, 10), respectively. Databases report 948 unique LMNA and 2,029 unique MYBPC3 variants observed across cardiomyopathy patients and normal subjects (Materials and Methods). Among these, we sought to identify splice-altering variants. After excluding all LoF variants (nonsense, frameshifts, and variants that alter canonical donor GT and acceptor AG splice sequences) and common variants (allele frequency >0.003; LMNA n = 133; MYBPC3 n = 454, Table S1), we identified 815 LMNA and 1575 MYBPC3 rare missense, synonymous, and intronic variants for study.
First, we prioritized variants using a computational algorithm to calculate MaxEnt scores (11), which are computed as the “distance” from splice signals found in human genes (Materials and Methods and Fig. S1). Based on the calculated MaxEnt scores for 815 LMNA (Dataset S1) and 1,575 MYBPC3 variants (Dataset S2), we selected 57 LMNA and 139 MYBPC3 splicing candidates for further assessment.
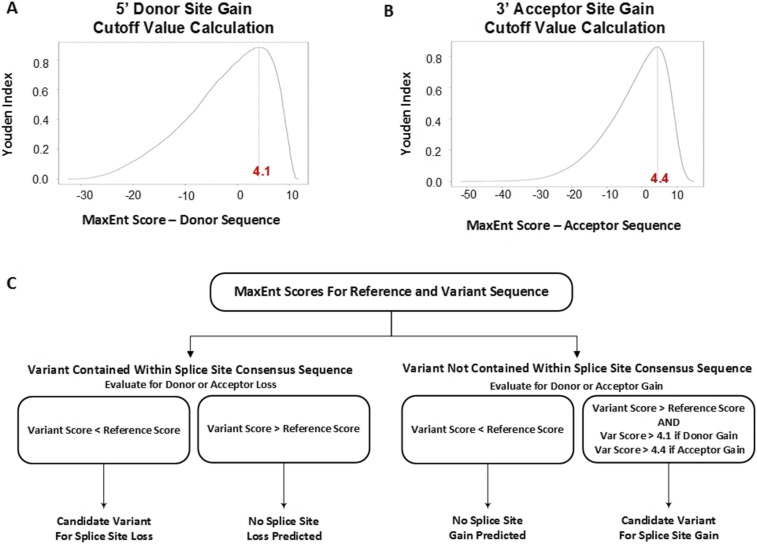
Fig. S1.
Thresholds for selection of candidate splice altering variants (Materials and Methods). Youden’s index (index = sensitivity + specificity − 1) was maximized to set a MaxEnt score value below which variants were not predicted to lead to 5′ donor or 3′ acceptor splice site gain. (A) At the 5′ donor splice site, Youden’s index was maximized when MaxEnt score = 4.1. (B) At the 3′ acceptor splice site, Youden’s index was maximized when MaxEnt score = 4.4. (C) A decision tree depicts the algorithm used for selection of candidate splice-altering variants. Variants predicted to cause loss of a donor or acceptor splice site (contained within splice site, ΔMaxEnt < 0) were selected for cell splicing assays. Variants predicted to create a gain of donor splice site (outside of splice site, ΔMaxEnt > 0) and had MaxEntvar score > 4.1 were selected for the minigene splicing assay. Similarly, variants predicted to create a gain of acceptor splice site (outside of splice site, ΔMaxEnt > 0) and had MaxEntvar score > 4.4 were selected for the minigene assay.
We developed a cell-based minigene assay to test all splicing candidates (Fig. S2). The assay compared RNA transcripts extracted from HEK293 cells transfected with pairs of minigene constructs (Fig. 1B) that included 500 bp of a functional exon–intron–exon containing either the reference or variant sequence (Datasets S3 and S4). Using this assay we confirmed that a previously described MYBPC3 pathogenic HCM variant (12, 13) significantly altered splicing in comparison with the reference sequence (P = 2.3e-06, two-sided Fisher’s exact test, Dataset S6). Similarly, we demonstrated significantly altered splicing (P < 0.001, two-sided Fisher’s exact test) in 14 of 57 LMNA splicing candidates (Dataset S5) and in 39 of 139 MYBPC3 splicing candidates (Fig. 2A and Dataset S6).
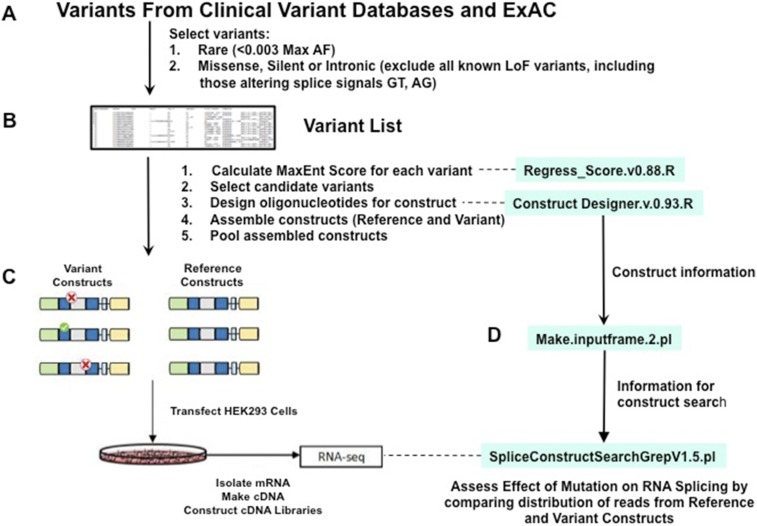
Fig. S2.
The workflow of the entire minigene splicing assay. All scripts are available online at https://github.com/SplicingVariant/SplicingVariants_Beta. (A) Variants are extracted from the clinical diagnostic and ExAC databases. Variants were initially screened to exclude all variants with allele frequencies >0.003, as well as variants that led to nonsense, frameshift, or altered the canonical GT or AG bases in consensus splice sites. (B) Variant positions in a given transcript were retrieved from Biomart database and MaxEnt scores for both reference allele and variant allele were calculated. The decision tree to select candidate splice altering variants based on MaxEnt scoring is depicted in Fig. S1C. Candidate variants were used to design mutant and reference constructs as described in Materials and Methods. (C) Reference and variant minigene constructs are assembled using a combination of synthetic oligonucleotides and PCR. Minigene constructs are pooled and transfected into HEK293 cells. After 24 h, RNA is extracted, converted into a cDNA library, and sequenced. (D) The sequence data are demultiplexed and analyzed for quantitative assessment of no splice, normal splice, and aberrant splicing. The status of splicing is determined comparing statistics between reference- and variant-containing minigenes.
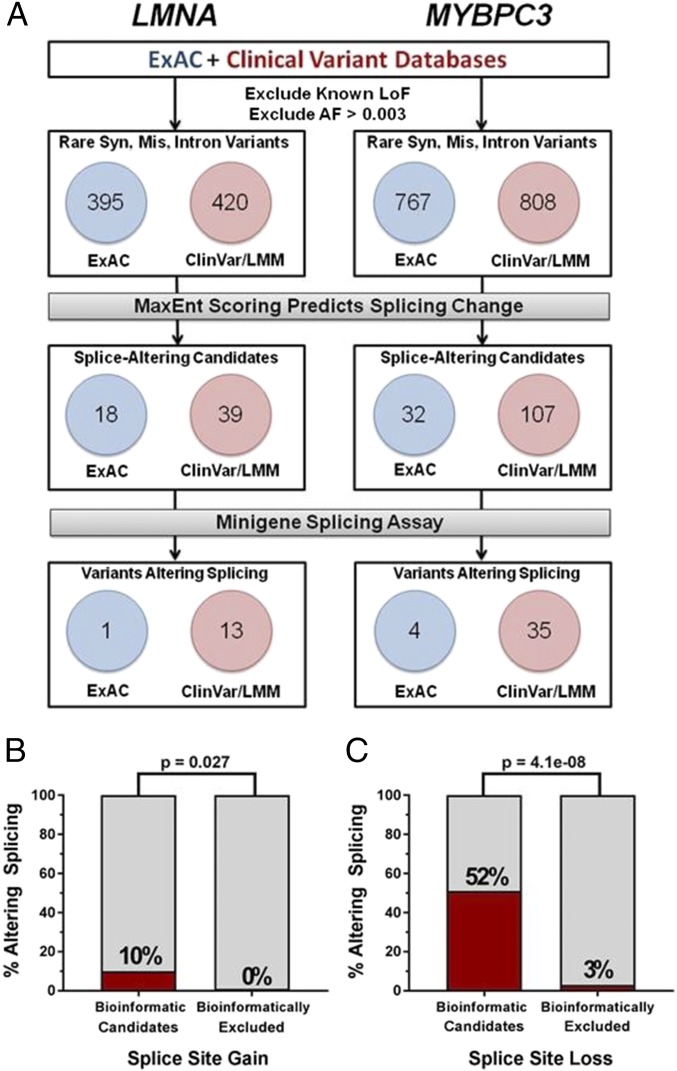
Fig. 2.
Prioritization and functional assessment of synonymous, missense, or intron variants in LMNA and MYBPC3. (A) Rare (allele frequency <0.003) variants in LMNA (n = 815) and MYBPC3 (n = 1,575) variants reported in the ExAC and clinical diagnostic databases were assessed by computational algorithms to prioritize LMNA (n = 57) and MYBPC3 (n = 139) variants for functional assays. Fourteen LMNA and 39 MYBPC3 variants altered splicing in the cell-based assay. (B) Percent of variants successfully conferring splice site gain among bioinformatically prioritized candidates and bioinformatically excluded variants. A total of 12 out of 117 variants predicted to alter splicing did so in the minigene assay. Of 39 variants tested with MaxEnt scores below the threshold set for creation a new splice site, zero altered splicing (P = 0.03). (C) Percent of variants leading to splice site loss among bioinformatically prioritized candidates and bioinformatically excluded variants. A total of 41 out of 79 variants predicted to lead to splice site loss did so in the minigene assay. Of 35 variants predicted to have no effect on the splice site, one was found to abrogate normal splicing (P = 4e-08).
Analyses of Variants Determined to Possess Low Probability to Alter Splicing.
To confirm that MaxEnt-prioritized selection of splicing candidates was robust we performed cell-based assays for LMNA and MYBPC3 variants with MaxEnt scores that did not meet our selection thresholds, because these were computationally predicted to have no effect on splicing (Dataset S7). Among 39 potential splice-site gain variants with low MaxEnt scores none altered minigene splicing, whereas 12 of 117 splice-site gain variants with MaxEnt scores that predicted new splice sites altered normal splicing (P = 0.03, one-sided Fisher’s exact test, Fig. 2B). Parallel analyses of 35 computationally excluded splice site loss variants identified one that abrogated the existing splice site, compared with 41 of 79 variants that passed our computational filter (P = 4e-08, one-sided Fisher’s exact test, Fig. 2C).
Clinical Phenotypes Associated with Splice-Altering Variants.
The proportions of minigene-validated splice-altering variants differed for those variants reported in cardiomyopathy patients and those observed in “normal” subjects. Approximately 0.4% (LMNA: 1/395, MYBPC3: 4/767) of rare synonymous, missense, or intronic variants reported only in normal subjects altered splicing, whereas ∼4% (LMNA: 13/420; MYBPC3: 35/808) of such variants reported in cardiomyopathy patients altered splicing (P = 1e-09, two-sided Fisher’s exact test, Fig. 2A). The 10-fold enrichment of functional splicing variants identified in the clinical databases compared with the normal subject database strongly supports the conclusion that rare synonymous, missense, and intronic LMNA and MYBPC3 variants that alter splicing are pathogenic.
Cardiomyopathy Caused by a Synonymous LMNA Variant.
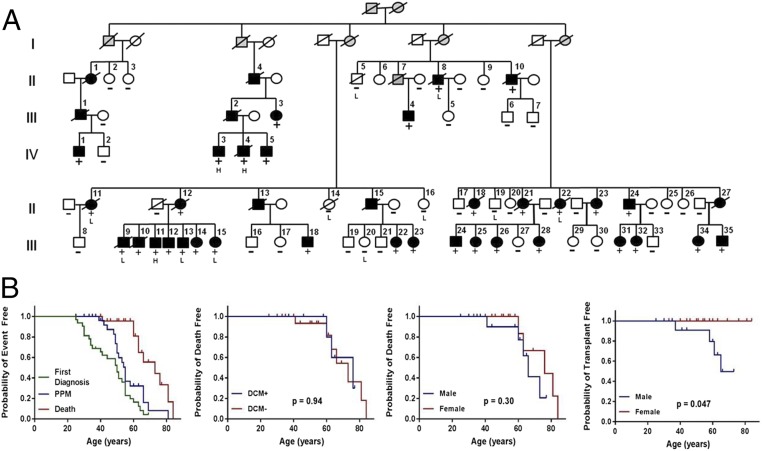
One splice-altering LMNA VUS was identified in a previously described DCM family (14). Clinical reevaluations demonstrated an AD trait (Fig. 3A and Table S2) in 31 individuals with progressive atrioventricular block, 16 individuals with pacemaker devices (implanted at ages 39–66 y), 10 individuals with DCM, and 4 with cardiac transplantations (at age 37–65 y). Four study participants and five other relatives (ages 21–76 y) died of sudden cardiac death. Affected men and women had comparable survival (median life expectancy with pacemaker, 74.5 y; without pacemaker, 60 y, Fig. 3B).
Fig. 3.
A rare synonymous LMNA variant segregates with DCM and conduction system disease. (A) The four-generation pedigree demonstrated autosomal dominant inheritance of the LMNA c.768 G > A variant (LOD = 20.02, θ = 0) and cardiomyopathy. (Solid symbols, affected; open symbols, unaffected; +, LMNA variant present; and −, LMNA variant absent). Analyses of lymphocyte (“L”) and cardiac (“H”) RNA was from the individuals indicated. (B) Kaplan–Meier curves of LMNA c.768G > A variant carriers, including (i) event-free survival from first diagnosis of the symptom, pacemaker implantation, and death; (ii) death-free survival comparing those with DCM to those without DCM; (iii) death-free survival; and (iv) transplant-free survival comparing male to female. P value was calculated by log-rank test.
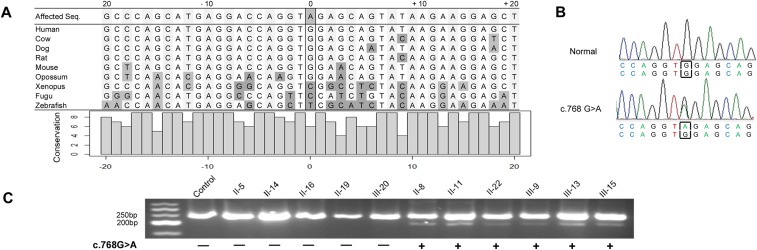
The disease locus mapped to chr1q21.3–q23.3 [Materials and Methods, maximum logarithm of the odds (LOD) = 4.7, θ = 0; data available upon request], where the LMNA gene is encoded. Although pathogenic LMNA LoF variants cause the phenotypes exhibited by these affected family members, sequence analyses showed no nonsynonymous variants, and only one synonymous variant (LMNA c.768 G > A) that segregated with affection status (Fig. 3A; LOD score = 20.02, θ = 0). The LMNA c.768 G > A variant is absent from the Exome Aggregation Consortium (ExAC) database of 60,000 individuals and alters a highly conserved nucleotide across all mammalian species (Fig. S3A).
Fig. S3.
LMNA sequence conservation and aberrant splicing caused by LMNA c.768G > A mutation. Related to Fig. 4. (A) A snapshot of LMNA sequence conservation. The position denotes the distance from the c.768 G > A mutation. Dark gray-filled nucleotides signify sequences differing from human reference. The bar graph represents the conservation of each individual nucleotide. Note that “9” indicates identity across all listed species). (B) Sanger sequencing confirmed heterozygous LMNA c.768 G > A mutation in the DNA of affected subject. (C) Gel-fractionation of RT-PCR products of lymphocyte-derived RNA from several individuals in Family MAE/MAN, showing those with the c.768G > A synonymous variant display an additional smaller species consistent with ∼45-bp deletion of part of the LMNA transcript.
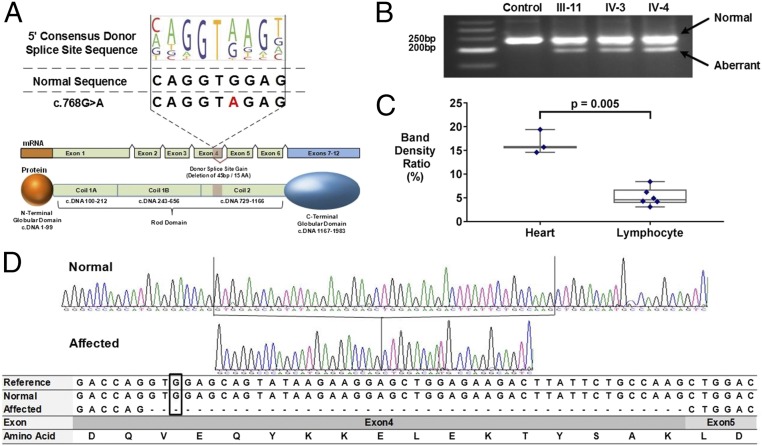
Computational assessment of the LMNA c.768 G > A variant (MaxEnt = 7.1) predicted this would be a more effective splice donor site than the reference sequence (reference MaxEnt = 2.6, ΔMaxEnt = +4.5), which we confirmed in the minigene splice assay (P = 0.0007, two-sided Fisher’s exact test, Fig. S4 and Dataset S5). RNA analyses of cardiac tissues from affected family members (Fig. 4 A, B, and D and Fig. S3B) validated this conclusion: Premature splicing of exon 4 deleted the terminal 45 bps and resulted in the loss of 15 amino acids in the rod domain of LMNA. Misspliced LMNA was observed in mutation-carrying individuals’ cardiac and lymphocyte RNA but not in lymphocyte or cardiac RNA from subjects without the mutation (Fig. 4C and Fig. S3C).
Fig. S4.
LMNA c.768 G > A confirmed to lead to donor site gain in the minigene splice assay. In addition to quantifying the number of reads with no splicing, normal splicing, and aberrant splicing for each reference- and variant-containing minigene, all reads were also aligned to the original minigene sequence using STAR, and splice patterns for each construct were directly visualized using the IGV to confirm computations. (A) A visual schematic of a standard minigene construct used in the splicing assay containing the CMV promoter, exon–intron–exon sequence, and the SV40PA signal. (B) Reads derived from the reference construct are depicted. Independent RNA sequencing reads uniformly demonstrate normal splicing out of the intron. (C) Reads from the variant construct demonstrate both the normal splice and aberrant splice due to a novel donor site. Aberrant splicing deleted 45 bp from the end of the first exon.
Fig. 4.
A rare synonymous LMNA variant creates a novel splice donor sequence in Exon 4. (A) Comparison of the 5′ donor consensus splice signal sequence with the LMNA reference (c.768 G, MaxEnt score = 2.6) and variant (c.768 A; MaxEnt score = 7.1). A ΔMaxEnt score (+4.5) predicted the gain of a novel splice donor in LMNA exon 4. A schematic showing premature splicing of the terminal end of exon 4 corresponds to a 15-aa deletion within coil 2 of the LMNA rod domain. (B) Gel fractionation of RT-PCR products of cardiac RNA detects a normal and smaller transcript in synonymous variant carriers. (C) The ratio of normal and aberrant splicing quantified by densitometric analyses of the gel-fractionated RT-PCR products in cardiac tissue (n = 3) and lymphocyte (n = 6) RNA demonstrates threefold more misspliced RNA in cardiac tissue (P = 0.005, two-sided t test) than in lymphocytes. (D) The sequence traces of RT-PCR products from a normal and an affected individual as well as aligned sequences of RT-PCR products to the reference mRNA of LMNA. The normal band from mRNA of LMNA c.768 G > A subject includes all nucleotides present in the reference, and the aberrant band deletes 45 bp in the end of exon 4.
Discussion
We conclude that computational prioritization of rare variants combined with functional assessment in a minigene splicing assay provides evidence that 13 LMNA variants and 35 MYBPC3 variants in cardiomyopathy patients alter splicing. Although over half of these are currently classified as VUS (Table 1), our data predict these to be pathogenic. Current clinical databases report 22 LMNA variants and 72 MYBPC3 variants that alter the canonical GT/AG splice signals. The inclusion of variants that are functionally validated to alter splicing yielded a 50% increase in pathogenic splicing variants seen in cardiomyopathy patients (Table S1).
Table 1.
Rare LMNA and MYBPC3 missense, synonymous, and intronic variants that alter RNA splicing
| All reported variants | LMM and ClinVar | ExAC only | |||||||
| Gene | Predictedeffect | Total variants | Splice altering candidates | Variants affecting splicing | Pathogenic | Likely pathogenic | VUS | Benign | Variants affecting splicing |
| LMNA | Acceptor loss | 45 | 5 | 5 | 1 | 1 | 3 | 0 | 0 |
| Donor loss | 25 | 8 | 3 | 1 | 0 | 2 | 0 | 0 | |
| Acceptor gain | 745 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Donor gain | 19 | 5 | 3 | 1 | 1 | 0 | 1 | ||
| Total | 815 | 57 | 13 | 5 | 2 | 6 | 0 | 1 | |
| MYBPC3 | Acceptor loss | 126 | 30 | 11 | 1 | 3 | 7 | 0 | 2 |
| Donor loss | 69 | 36 | 19 | 6 | 3 | 10 | 0 | 1 | |
| Acceptor gain | 1,380 | 32 | 3 | 0 | 1 | 2 | 0 | 0 | |
| Donor gain | 41 | 2 | 0 | 0 | 2 | 0 | 1 | ||
| Total | 1,575 | 139* | 35 | 7 | 7 | 21 | 0 | 4 | |
Of the 815 rare LMNA variants, 57 were computationally predicted to alter normal splicing patterns. Among 14 of 57 LMNA splice candidates that altered splicing in the cell assay, 13 are reported in clinical databases as classified as indicated. One functional variant reported only in the ExAC database is not clinically classified. Of the 1,575 rare MYBPC3 variants, 139 were computationally predicted to alter normal splicing. Among 39 MYBPC3 splice candidates that altered splicing in the cell assays, 35 are reported in clinical databases as classified as indicated. Four functional variants reported only in the ExAC database are not clinically classified.
MYBPC3 c.2905+5G > T was predicted to cause multiple splicing changes. Independent constructs and cell assays were performed to evaluate whether the variant caused a donor loss or acceptor gain (Dataset S4). Cell assays only showed the variant functioned as a donor site loss that significantly altered splicing.
We recognize three potential limitations to this approach for identification of similar pathogenic splice-altering variants. First, the minigene splice assay may not identify the precise splice alteration that occurs in vivo. However, of the 53 splice-altering variants identified here, 16 variants were independently characterized by RT-PCR in patient tissues (15–25), alter splicing, and are pathogenic (Table S3). Moreover, the fact that 48 of 53 VUS were identified in patients with an AD cardiomyopathy provides a very strong likelihood that the aberrant splicing observed in the minigene assay occurs in vivo. Second, despite multiplex sequencing analyses, the cost of the assay (∼$200 per variant tested) would be prohibitive for analyses of all rare variants identified in clinical samples, especially without the initial bioinformatic prioritization. Third, the minigene splice assay used kidney cells (HEK293), and some variants might have different impact in distinct cell lines, even though HEK293 have historically been used to evaluate sequence features that cause abnormal splicing (26). Further offsetting this concern, we observed high concordance between splicing consequences of >50 variants studied in both HEK293 cells and an iPS cell line (data available upon request).
We expect that the application of these methodologies to AD disorders caused by haploinsufficiency will produce a similar 5% increase in the detection of pathogenic splice variants. In addition, we speculate that the severity of clinical phenotypes may correspond to degrees of normal and aberrant splicing, as well as cellular requirements for a given protein and/or tolerance for a mutant peptide. Determining whether the proportions of misspliced transcripts caused by different splice variants (Datasets S5 and S6) affects the age of onset, severity, and progression of disease (27, 28) would enhance the clinical utility of sequence variant annotation. These data would inform emerging therapies that aim to suppress disease phenotypes by modulating the expression of normal or mutated genes.
Materials and Methods
Identification of Rare Variants in LMNA and MYBPC3.
LMNA and MYBPC3 variants identified in cardiomyopathy patients reported before 2015 by the Laboratory for Molecular Medicine (LMM) of Partners Healthcare and/or reported in ClinVar (29), an NIH-sponsored database of human variants associated with clinical phenotypes, were included in this study. In parallel we studied rare LMNA and MYBPC3 variants identified in the ExAC, a reference sequence database reflecting sequence analyses of over 60,000 unrelated individuals. The aggregation of all unique variants across the three databases included 948 LMNA variants and 2,029 MYBPC3 variants (Table S1). From these we included only single-nucleotide variants, exclusive of variants with allele frequencies >0.003, or those predicted to be pathogenic LoF (including variants disrupting the invariant GT and AG sequences in canonical splice sites). The remaining 815 rare LMNA (Dataset S1) and 1,575 rare MYBPC3 variants (Dataset S2) from the clinical (LMM, ClinVar) or the normal population (ExAC) databases were evaluated for consequences on RNA splicing.
Computational Prioritization of Variants for Splicing Assays.
We used MaxEntScan scoresplice (11) to analyze 9-bp sequences as potential splice donor sites (scoresplice5) and 23-bp sequences as splice acceptor sites (scoresplice3). Using an in-house script we calculated and ranked the MaxEnt scores for each variant and reference sequence (see Regress_Score.v0.88.R, posted online at https://github.com/SplicingVariant/SplicingVariants_Beta). These were filtered as follows. Variants within a known functional 9-bp or 23-bp splice-site sequence (retrieved from Biomart database) were deemed potential 5′ donor or 3′ acceptor splice-site loss when the MaxEntvariant was less than MaxEntreference (ΔMaxEnt = MaxEntvar − MaxEntref < 0). Variants with scores greater than the reference score (ΔMaxEnt > 0) were deemed unlikely to affect the reference splice site.
Variants located outside of canonical splice sequences were deemed potential 5′ donor or 3′ acceptor gains when the MaxEntvar was greater than MaxEntref (ΔMaxEnt > 0). Due to the large number of these variants, and the large proportion that had only marginal increases in MaxEnt score, we defined a threshold above which a sequence variant was reliably predicted to lead to a new functional splice site. We defined the threshold for splice-creating sequences based on the Youden index.
Calculating the Youden Index to Define Splice-Site Gain Threshold.
Using a training set of 25,321 splice donor or acceptor sequences in >1,200 human genes and 540,606 sequences that resemble, but do not function as, splice donor or acceptor sequences (genes.mit.edu/burgelab/maxent/ssdata/MEMset/), we calculated MaxEnt scores for all sequences in these two datasets. We then defined the MaxEnt values that maximized Youden’s index, using the following calculation: Y = sensitivity + specificity − 1, with sensitivity and specificity defined as
For potential 9-bp donor site sequences the Youden index was maximum when the MaxEnt score = 4.1 (Y = 0.88, Fig. S1A). For potential 23-bp acceptor site sequences the Youden index was maximum when the MaxEnt score = 4.4 (Y = 0.86, Fig. S1B). Because these thresholds were maximally sensitive and specific for identifying functional splice sites, sequences with values above this threshold were strong candidates for novel splice site gain. Alternatively, sequences below this threshold value were denoted as low probability for altering splicing.
From all rare LMNA and MYBPC3 variants we used a decision tree (Fig. S1C) to select variants for functional assays. For variants that potentially caused a donor splice-site gain we prioritized those with MaxEntvar greater than MaxEntref (ΔMaxEnt > 0) and the MaxEntvar > 4.1. For variants predicted to create an acceptor site, we prioritized those with MaxEntvar greater than MaxEntref (ΔMaxEnt > 0) and a MaxEntvar > 4.4. We also selected any variants within an existing 9-bp donor or 23-bp acceptor splice site in which ΔMaxEnt < 0, because this predicted splice site loss. This selection algorithm identified 57 candidate variants in LMNA and 139 variants in MYBPC3 that were studied in functional assays (Table 1).
Minigene Splicing Assay.
Design of gBlocks.
Minigene splicing assays tested a ∼1,200 bp DNA construct containing a CMV promoter, the relevant intron flanked by exon fragments, and a 2 bp barcode (Fig. 1B). 500 bp oligonucleotides were designed to contain the relevant exon–intron–exon features, and were ordered as gBlock Gene Fragments (Integrated DNA Technologies). Each 500 bp gBlock was designed using in-house scripts (see Construct Designer.v.0.93.R, posted online at https://github.com/SplicingVariant/SplicingVariants_Beta) from variant information. A reference construct and variant construct differing solely by the single nucleotide variant and a 2 bp barcode were designed for each candidate variant. Full gBlock sequences for each reference and alternate sequence are reported in Datasets S3 and S4.
In brief, four types of gBlocks were designed:
-
i)For exonic variants predicted to create a 5′ splice donor change, the script assigned
-
•85 bp to the first exon before the variant
-
•85 bp to the first exon after the variant
-
•85 bp to the first exon before the first exon–intron boundary
-
•85 bp to the intron following the first exon–intron boundary
-
•55 bp to the intron before the second exon–intron boundary
-
•40 bp to the second exon following the second exon–intron boundary
-
•
-
ii)For intronic variants predicted to create a 5′ splice change, the script assigned
-
•85 bp to the first exon
-
•85 bp to the intron following the first exon–intron boundary
-
•85 bp to the intron before the variant
-
•85 bp to the intron after the variant
-
•55 bp to the intron before the second exon–intron boundary
-
•40 bp to the second exon following the second exon–intron boundary
-
•
-
iii)For exonic variants predicted to create a 3′ splice acceptor change, the script assigned
-
•40 bp to the first exon before the first exon–intron boundary
-
•55 bp to the intron after the first exon–intron boundary
-
•85 bp to the intron before the second exon–intron boundary
-
•85 bp to the exon following the second exon–intron boundary
-
•85 bp to the second exon before the variant
-
•85 bp to the second exon after the variant
-
•
-
iv)For intronic variants predicted to create a 3′ splice acceptor change, the script assigned
-
•40 bp to the first exon before the first exon–intron boundary
-
•55 bp to the intron after the first exon–intron boundary
-
•85 bp to the intron before the variant
-
•85 bp to the intron after the variant
-
•85 bp to the intron before the second exon–intron boundary
-
•85 bp to the second exon following the second exon–intron boundary
-
•
When the length of any segment was shorter than an assigned length, the remaining length was added to the following segment. Flanking the exon–intron–exon sequence, we included shared adapter sequences in each gBlock. The 5′ adapter sequence included a 24-bp-long complementary sequence of the CMV promoter. The 3′ adapter sequence comprised a 2-bp-long barcode sequence and 39-bp-long complementary sequence of SV40-polyA (altogether, 500 bp). Because gBlock synthesis fails on high-CG-content sequences, high-GC intron sequences were deleted. Finally, we note that when constructing smaller 1,000-bp minigene constructs encoding only 250 bp of the exon–intron–exon sequence, 50% of constructs failed to splice appropriately in this assay, thus requiring the larger size used.
DNA template assembly.
CMV-promoter and SV40-polyA sequences were attached to 500-bp-long gBlocks using overlapping adapter sequences by two rounds of PCR. In the first PCR, the gBlock and SV40-polyA sequence were attached using primer sequences, 5′-ACGCCAAGTTATTTAGGTGACA-3′ and 5′-TAAGATACATTGATGAGTTTGGACAAACC-3′. In the second PCR, the CMV promoter was attached to the gBlock/SV40-polyA using primer sequences 5′-TAGTAATCAATTACGGGGTCATTAGTTCATA-3′ and 5′-TAAGATACATTGATGAGTTTGGACAAACC-3′ to form the final construct (Fig. 1B). Products were purified using Agencourt AMPure XP (Beckman Coulter) and the concentration was measured on TapeStation D5000 Screen Tape (Agilent Technologies).
HEK293 cell transfection and RNA isolation.
HEK293 cells (ATCC) were cultured in DMEM (Gibco) with 10% FBS, l-glutamine, and gentamycin on cell-culture plates (FALCON). For transfection, 0.3 million cells were seeded into a six-well plate. Using lipofectamine 2,000 (Invitrogen), up to 24 reference and variant constructs (100 ng of each construct) were transfected into one well. Twenty-four hours after transfection, cells were harvested, total RNA was extracted (TRIzol reagent; Ambion), and RNA integrity number values were assessed to ensure quality and quantity using TapeStation RNA screen tape (Agilent Technologies).
Reverse transcription and construction of Illumina DNA sequencing libraries.
mRNA was isolated from total RNA by Dynabeads Oligo dT (Ambion), and cDNA was synthesized by Super Script III (Invitrogen). Quantitative PCR (qPCR) was performed on cDNA to determine the number of PCR cycles permitted to avoid saturation. cDNA was then amplified using primers specific for the head and tail regions of the gBlocks. PCR products were end-repaired using End-it DNA End-Repair kit (Epicentre) and A-tailing was performed using Klenow fragment (New England Biolabs), followed by adaptor ligation. Final amplification was conducted using multiplex primer 1.0 (Illumina) and in-house indexing primers. Each intermediate library product was purified by Agencourt AMPure XP (Beckman Coulter).
Illumina MiSeq sequencing and statistical analyses.
Using scripts (see Make.inputframe.2.pl and SpliceConstructSearchGrepV1.5.pl, posted at https://github.com/SplicingVariant/SplicingVariants_Beta) we quantified the number of reads without splicing, normal splicing, and aberrant splicing for each reference and variant-containing construct. A minimum of 100 analyzable reads from both the reference and the variant construct (identified by barcode) was required to assess consequences of splicing. The number of analyzable reads was normalized to 100 and P value was calculated by two-sided Fisher’s exact test. All reference construct and variant construct reads were also aligned to the original exon–intron–exon nucleotide sequence using STAR (30), and splice patterns for each construct were directly visualized using the Integrative Genomics Viewer (IGV) (31) to confirm computations (Fig. S4). As positive controls we demonstrated that 10 different variants known to alter splicing also abrogated normal splicing in this assay.
Clinical Evaluation of LMNA Family.
A family reported by Lynch et al. (14) described as “Family S” was reevaluated as Family MAE/MAN. Written informed consent was obtained in accordance with the requirements of the Partners Human Research Committee for all study participants. Clinical evaluations, performed without knowledge of genotype status, included personal health histories, physical examinations, 12-lead electrocardiography, and transthoracic echocardiography as described (32). Subjects were classified as affected when electrophysiology demonstrated atrioventricular conduction system abnormalities and/or echocardiography showed unexplained increased left ventricular dimensions and abnormal ejection fraction using standard diagnostic criteria (33). Disease status of deceased family members was determined from review of medical records. Deaths were classified as disease-related or as due to noncardiac causes as described previously (34). Family members with the lamin A/C variant underwent further evaluations, including a rereview of medical record.
DNA Sequencing and Genotyping for MAE/MAN Family.
Genome-wide linkage analyses.
Samples were genotyped on the Illumina HumanOmniExpress-12v1. Genome-wide SNP multipoint LOD scores were calculated with Vitesse (35) using five-marker sliding windows. The parameters SNP allele frequencies, penetrance, disease frequency, and phenocopy rate were set at 0.5 for both major and minor allele, 95%, 0.001, and 0.00, respectively. Subsequently, two-point linkage analyses were performed assessing the likelihood of association between the rare synonymous LMNA variant and disease (FASTLINK v4.1P) (36). SNP allele frequency (major allele frequency/minor allele frequency), penetrance, disease frequency, and phenocopy rate were set at 0.999/0.001, 95%, 0.001, and 0.00, respectively.
Sanger sequencing of the LMNA variant.
Exon 4 of LMNA was amplified from genomic DNA from all study subjects using PCR primers 5′-AGCTGCGTGAGACCAAGC-3′ and 5′-AAGTCTTCTCCAGCTCCTTCTT-3′. PCR products were gel-purified using Qiagen QIAquick PCR purification kit and submitted to GENEWIZ, Inc. for dideoxy sequencing.
Analyses of LMNA splicing in cardiac and lymphocyte tissues.
Total RNA was extracted from cardiac tissue or lymphocytes using the Qiagen RNeasy mini kit. Ten nanograms of total RNA was used for RT-PCR using a Qiagen one-step RT-PCR kit. The number of cycles required for RT-PCR was first determined by qPCR to avoid signal saturation. PCR primer sequences, 5′-GCAGACCATGAAGGAGGAAC-3′ and 5′-GACTGCCTGGCATTGTCC-3′, were designed to span from exon 3 to exon 5. The PCR fragments were gel-purified and Sanger-sequenced by Genewiz, Inc.
Data Availability.
Variants used for study are publicly available online at ClinVar (www.ncbi.nlm.nih.gov/clinvar/) and ExAC (exac.broadinstitute.org/). Variant lists from the Laboratory for Molecular Medicine are available on request from the corresponding author (J.G.S.). The data generated from this study (list of assessed variants with genomic positions, calculated MaxEnt scores, sequences of synthetic oligonucleotide constructs, read counts derived from each reference, and variant construct used for statistical analysis) are all available in Supporting Information.
Code Availability.
All computer code used in methods and analysis are available for public use at https://github.com/SplicingVariant/SplicingVariants_Beta.
Supplementary Material
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute, NIH Grants 1R01HL080494 and 1R01HL084553 (to J.G.S. and C.E.S.), the Fondation Leducq (J.G.S. and C.E.S.), the Howard Hughes Medical Institute (S.R.D., K.I., and C.E.S.), the Banyu Fellowship Program and the Uehara Research Fellowship Program (K.I.), a fellowship from the Sarnoff Cardiovascular Research Foundation (P.N.P.), and the National Health and Medical Research Council of Australia (D.F.).
Footnotes
Conflict of interest statement: C.E.S. and J.G.S. are founders of and own shares in Myokardia Inc., a startup company that is developing therapeutics that target the sarcomere.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707741114/-/DCSupplemental.
References
- 1.Medicine M-NIoG 2017. Online Mendelian Inheritance in Man (Johns Hopkins Univ, Baltimore)
- 2.Richards S, et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baralle D, Baralle M. Splicing in action: Assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krawczak M, et al. Single base-pair substitutions in exon-intron junctions of human genes: Nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 6.Gaildrat P, et al. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- 7.Vreeswijk MP, et al. Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat. 2009;30:107–114. doi: 10.1002/humu.20811. [DOI] [PubMed] [Google Scholar]
- 8.Wolf CM, et al. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. J Mol Cell Cardiol. 2008;44:293–303. doi: 10.1016/j.yjmcc.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marston S, et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 10.van Dijk SJ, et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: Haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 11.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 12.Waldmüller S, et al. Novel deletions in MYH7 and MYBPC3 identified in Indian families with familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2003;35:623–636. doi: 10.1016/s0022-2828(03)00050-6. [DOI] [PubMed] [Google Scholar]
- 13.Dhandapany PS, et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch HT, et al. Hereditary progressive atrioventricular conduction defect. A new syndrome? JAMA. 1973;225:1465–1470. [PubMed] [Google Scholar]
- 15.Chrestian N, et al. A novel mutation in a large French-Canadian family with LGMD1B. Can J Neurol Sci. 2008;35:331–334. doi: 10.1017/s031716710000891x. [DOI] [PubMed] [Google Scholar]
- 16.Benedetti S, et al. Phenotypic clustering of lamin A/C mutations in neuromuscular patients. Neurology. 2007;69:1285–1292. doi: 10.1212/01.wnl.0000261254.87181.80. [DOI] [PubMed] [Google Scholar]
- 17.Otomo J, et al. Electrophysiological and histopathological characteristics of progressive atrioventricular block accompanied by familial dilated cardiomyopathy caused by a novel mutation of lamin A/C gene. J Cardiovasc Electrophysiol. 2005;16:137–145. doi: 10.1046/j.1540-8167.2004.40096.x. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, et al. Two novel mutations of the MYBPC3 gene identified in Chinese families with hypertrophic cardiomyopathy. Can J Cardiol. 2010;26:518–522. doi: 10.1016/s0828-282x(10)70464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarikas A, et al. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc Res. 2005;66:33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Helms AS, et al. Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2014;7:434–443. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrier L, et al. Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res. 1997;80:427–434. [PubMed] [Google Scholar]
- 23.Jääskeläinen P, et al. Mutations in the cardiac myosin-binding protein C gene are the predominant cause of familial hypertrophic cardiomyopathy in eastern Finland. J Mol Med (Berl) 2002;80:412–422. doi: 10.1007/s00109-002-0323-9. [DOI] [PubMed] [Google Scholar]
- 24.Marston S, Copeland O, Gehmlich K, Schlossarek S, Carrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J Muscle Res Cell Motil. 2012;33:75–80, and erratum (2012) 33:81. doi: 10.1007/s10974-011-9268-3. [DOI] [PubMed] [Google Scholar]
- 25.Watkins H, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 26.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Møller LB, et al. Similar splice-site mutations of the ATP7A gene lead to different phenotypes: Classical Menkes disease or occipital horn syndrome. Am J Hum Genet. 2000;66:1211–1220. doi: 10.1086/302857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nissim-Rafinia M, Kerem B. Splicing regulation as a potential genetic modifier. Trends Genet. 2002;18:123–127. doi: 10.1016/s0168-9525(01)02619-1. [DOI] [PubMed] [Google Scholar]
- 29.Landrum MJ, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita H, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mestroni L, et al. Heart Muscle Disease Study Group Familial dilated cardiomyopathy: Evidence for genetic and phenotypic heterogeneity. J Am Coll Cardiol. 1999;34:181–190. doi: 10.1016/s0735-1097(99)00172-2. [DOI] [PubMed] [Google Scholar]
- 34.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- 36.Becker A, Geiger D, Schäffer AA. Automatic selection of loop breakers for genetic linkage analysis. Hum Hered. 1998;48:49–60. doi: 10.1159/000022781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Variants used for study are publicly available online at ClinVar (www.ncbi.nlm.nih.gov/clinvar/) and ExAC (exac.broadinstitute.org/). Variant lists from the Laboratory for Molecular Medicine are available on request from the corresponding author (J.G.S.). The data generated from this study (list of assessed variants with genomic positions, calculated MaxEnt scores, sequences of synthetic oligonucleotide constructs, read counts derived from each reference, and variant construct used for statistical analysis) are all available in Supporting Information.