Abstract
The role of high-potential iron sulfur protein (HiPIP) in donating electrons to the photosynthetic reaction center in the halophilic γ-proteobacterium Halorhodospira halophila was studied by EPR and time-resolved optical spectroscopy. A tight complex between HiPIP and the reaction center was observed. The EPR spectrum of HiPIP in this complex was drastically different from that of the purified protein and provides an analytical tool for the detection and characterization of the complexed form in samples ranging from whole cells to partially purified protein. The bound HiPIP was identified as iso-HiPIP II. Its Em value at pH 7 in the form bound to the reaction center was ≈100 mV higher (+140 ± 20 mV) than that of the purified protein. EPR on oriented samples showed HiPIP II to be bound in a well defined geometry, indicating the presence of specific protein–protein interactions at the docking site. At moderately reducing conditions, the bound HiPIP II donates electrons to the cytochrome subunit bound to the reaction center with a half-time of ≤11 μs. This donation reaction was analyzed by using Marcus's outer-sphere electron-transfer theory and compared with those observed in other HiPIP-containing purple bacteria. The results indicate substantial differences between the HiPIP- and the cytochrome c2-mediated re-reduction of the reaction center.
Keywords: electron transfer, photosynthesis, electron paramagnetic resonance, redox potential, reaction center
High-potential iron sulfur proteins (HiPIPs) are soluble electron carriers containing a single cubane [Fe4S4] cluster [for reviews, see refs. 1 and 2] found in both photosynthetic (3–6) and nonphotosynthetic (7, 8) proteobacteria, as well as in members of the so-called FBC group (flexibacter, bacteroides, cytophaga group) (9). Despite three decades of detailed biophysical and biochemical characterization, the role of HiPIPs as physiological electron donors to the reaction center (RC) in photosynthesis and to oxidase in respiration was not demonstrated until 1995 (10–12).
HiPIPs are commonly regarded as exotic substitutes for the “normal” soluble carrier cytochrome c. This notion is based on the presence of soluble cytochromes in mitochondrial respiration, in cyanobacterial photosynthesis, and in a variety of other prokaryotic energy conserving systems. Moreover, the usual “workhorses” in the study of purple bacterial photosynthesis, Rhodobacter sphaeroides, Rhodobacter capsulatus, and Blastochloris viridis, do not contain HiPIP. Instead, cytochrome c2, together with its various iso-forms, functions in their bioenergetic chains. Several decades of studies on these species have resulted in an understanding of the structural and functional details of electron donation from cytochrome c2 to the photosynthetic RC (see, for example, refs. 13–19). In contrast, the interaction between HiPIP and RC is still poorly understood. Except for the case of Rubrivivax gelatinosus (20–23), only scarce data are available (24, 25). Surveys of photosynthetic electron transfer among proteobacterial species, however, show that the participation of HiPIP instead of soluble cytochrome c is the rule rather than the exception (26). For an exhaustive description of electron donation to the RC in proteobacteria, therefore, detailed understanding of the interaction of HiPIP with the RC is needed.
Among the species containing HiPIPs, one stands out for its apparent “weirdness,” i.e., Halorhodospira (H.) halophila. This halophilic γ-proteobacterium has long been known to contain two iso-HiPIP proteins. Both differ from the bulk of HiPIPs by their low redox potentials (+50 mV and +110 mV) (27) compared with the range of +260 mV to +450 mV typically found. These low redox potentials apparently argue against an implication of these two HiPIPs in photosynthetic electron transport. In addition, in vivo flash-induced absorbance changes have been interpreted to suggest the implication of a cytochrome rather than a HiPIP in photosynthesis in this organism (26). In this work, we have characterized the interaction of HiPIP and the RC in H. halophila in detail. It turns out that this reportedly strange species allows profound insights into the functional characteristics of HiPIP and its redox partner.
Experimental Procedures
Purification of HiPIPs. Cultures of H. halophila were grown photosynthetically and anaerobically at 30°C in ATCC 1448 medium in a 20-liter fermenter.
Cells were suspended in 50 mM 3-[morpholino]propanesulfonic acid (Mops) at pH 7 (buffer A) and broken by passing through a French press. Unbroken cells were eliminated by centrifugation at low speed, and a subsequent ultracentrifugation separated the “total soluble fraction” in the supernatant from the “membrane-fragments fraction” in the pellet. This total soluble fraction was dialyzed to remove the remaining salt of the growth medium and used to purify the so-called “soluble HiPIP I and II.” The membrane-fragment fraction recovered after the ultracentrifugation step was resuspended in buffer A. This fraction contains almost exclusively closed vesicles that can retain the so-called “membrane-bound HiPIP.” The membrane fragments were twice sonicated and ultracentrifuged. The supernatants from both centrifugations were pooled and used to purify the membrane-bound HiPIP. Both soluble and membrane-attached HiPIP were purified following a protocol derived from Meyer (28). The sample was adsorbed onto a DEAE-52 column equilibrated with buffer A. The column was eluted with a step-gradient from 0–0.6 M NaCl. The photoactive yellow protein was eluted at 0.12 M NaCl, the major part of the c551 at 0.14, a new cytochrome c at 0.17–0.2 M (the characterization of which will be presented elsewhere), HiPIP I at 0.24 M, and HiPIP II together which some cytochrome c′ at 0.28 M. Fractions containing HiPIP I or II were dialyzed, each charged onto a second DEAE-52 column and eluted with a step-gradient from 0.2–0.24 M NaCl for HiPIP I and 0.24–0.28 M NaCl for HiPIP II. Fractions containing HiPIP I or II were dialyzed, charged onto Sephadex G-100 size-exclusion columns, and eluted with buffer A, 100 mM NaCl.
Preparation of Oriented Samples. Oriented membrane multilayers were obtained by partial dehydration of the membrane fragments on Mylar as described by Rutherford and Sétif (29). Ascorbate-reduced and ferricyanide-oxidized oriented samples were prepared as described by Lieutaud et al. (22).
Spectroscopic Methods. EPR spectra were recorded on a Bruker (Karlsruhe, Germany) ESP 300 X-band spectrometer fitted with an Oxford Instruments (Eynsham, England) liquid helium cryostat and temperature control system. Illumination was performed on whole cells resuspended in growth medium, on membrane fragments diluted in buffer A, 130 g/liter NaCl (pH 7) or on membrane multilayers. During illumination, the EPR tube was kept in a water/ice bath to minimize heating of the sample.
Optical spectra were recorded on a Cary (Victoria, Australia) 5E spectrophotometer.
Redox Titrations. Redox titrations were performed on membrane samples (after washing in 5 mM ferricyanide/5 mM EDTA) or purified proteins at 15°C as described by Dutton (30) in buffer A with or without 130 g/liter NaCl, in the presence of the following redox mediators at 100 μM for EPR or 15 μM for optical titrations: 1,4 p-benzoquinone; 2,5-dimethyl-p-benzoquinone; 2-hydroxy 1,2-naphthoquinone; 1,4-naphthoquinone; dimethyl-1,4-naphthoquinone; 2,5-dihydroxy-p-benzoquinone; dihydroxy-1,4-naphthoquinone; and anthraquinone-2-sulfonate. Ferricyanide was present at 10 μM. Reductive titrations were carried out by using sodium dithionite, and oxidative titrations were carried out by using potassium hexachloroiridate (IV).
Light-Induced Absorption Changes. Membrane fragments were diluted in 50 mM Mops (pH 7), 130 g/liter NaCl. Laser flash-induced absorption changes were measured by a laboratory-built spectrophotometer as described (23).
Results
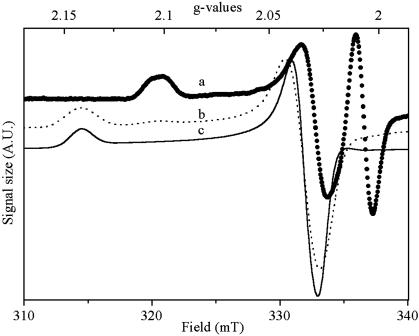
The EPR Spectrum of Photooxidized HiPIP in Whole Cells Differs from That of the Purified Proteins. The EPR spectrum of HiPIP observed on membranes or illuminated whole cells of H. halophila has previously been reported by Leguijt et al. (31) and Menin et al. (26). Astonishingly, these spectra differ substantially from those published for purified HiPIP iso-I and iso-II (see, for example, refs. 32 and 33) with a g1 peak significantly shifted toward higher fields, i.e., peaking at 2.10 rather than at 2.14, a characteristic that has not previously been noticed. We therefore repeated EPR measurements on all types of HiPIP samples. Fig. 1, traces b and c, shows spectra recorded on purified soluble HiPIP I and II, which we identified by N-terminal sequencing (not shown). HiPIP I yielded a “complex” EPR spectrum with a dominant g1 signal at 2.14 and a minor g1 signal at 2.10 (Fig. 1, trace b) (see also ref. 32), whereas a “simple” EPR spectrum with a unique g1 at 2.14 (Fig. 1, trace c) was recorded on isoform II (see also ref. 33). The spectrum obtained after illumination of whole cells (Fig. 1, trace a) is similar to that published by Menin et al. (26), except for a resolved splitting of the g1 signal at 2.095 and 2.105, probably due to use of a smaller modulation amplitude. This result confirms the spectral differences between purified HiPIPs and the paramagnetic species photooxidized in vivo.
Fig. 1.
EPR spectra recorded on illuminated whole cells (trace a) and on chemically oxidized purified HiPIP I (trace b) and HiPIP II (trace c) from H. halophila. Instrument settings: microwave frequency, 9.44 GHz; temperature, 15 K; microwave power, 6.3 mW; modulation 1 mT.
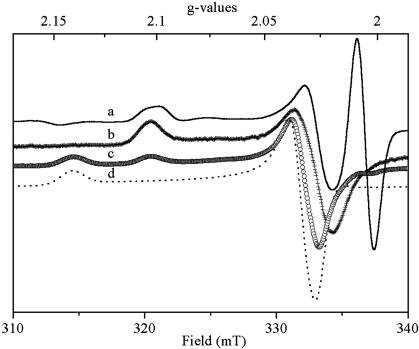
H. halophila Contains a HiPIP Firmly Bound to the Membrane with a Spectrum Similar to That Detected in Vivo. EPR spectra of chemically oxidized membrane fragments from H. halophila (Fig. 2, trace a) feature signals similar to those of illuminated whole cells. Because HiPIPs are periplasmic proteins, their copurification with the membrane fraction is a priori unexpected, but comparable results have been obtained with Rubrivivax gelatinosus (22). Trapping of soluble HiPIPs in chromatophores might explain this finding. In the case of Rubrivivax gelatinosus, however, the absence of invaginations in the cytoplasmic membrane renders trapping highly unlikely and indicates genuine physical association between HiPIP and its membrane-bound redox partners. Salt treatment was sufficient to dissociate the HiPIP from the membranes (22). Unfortunately, corresponding experiments to assay whether HiPIP is specifically bound to membranes or fortuitously trapped cannot be performed in H. halophila. Firstly, H. halophila membranes form almost entirely closed vesicles upon French Press treatment, and, secondly, the extremely halophilic character of this strain renders salt-induced disruption of specific protein–protein interactions unlikely. Nevertheless, other lines of experimental evidence (see below) clearly demonstrated that the membrane-bound HiPIP in H. halophila is bound to the membrane in a specific electron transfer competent conformation.
Fig. 2.
EPR spectra recorded on membrane fragments and on increasingly enriched fractions of the membrane-bound HiPIP. Membrane fragments (spectrum a) were oxidized by addition of 2 mM ferricyanide in 50 mM Mops (pH 7), sedimented by ultracentrifugation, and resuspended in 50 mM Mops (pH 7). Spectrum b was recorded on a supernatant fraction obtained after sonication of these membranes. Spectrum c was recorded on the HiPIP fraction collected after DEAE-column chromatography of the supernatant fraction described above. Spectrum d was recorded on HiPIP collected from the size exclusion column at the end of the purification. Instrument settings were as in Fig. 1.
Identification of the Membrane-Bound HiPIP. To clarify whether the membrane-bound HiPIP represents a third, as yet undetected, HiPIP, or whether it corresponds to a modified form of HiPIP I or II, we purified this protein. A “solubilized” fraction of this HiPIP was obtained by repeatedly sonicating the membrane fragments in buffer A and removing membrane fragments by ultracentrifugation. The EPR spectrum recorded on this crude fraction containing “solubilized” HiPIP was similar to that obtained on membrane fragments (Fig. 2, trace b) except for the g1 signal splitting. This crude fraction was subsequently purified (see Experimental Procedures). EPR spectra taken during the purification (Fig. 2) showed that the g1 = 2.10 signal was transformed into a g1 = 2.14 peak with increasing purity of HiPIP. In the crude extract, a cytochrome component was observed that was increasingly depleted during subsequent purification steps. Quantification of HiPIP during the purification demonstrated that the g = 2.14 species observed at the end of the purification (Fig. 2, trace d) roughly corresponded to the entire population showing the g = 2.10 signal before purification. N-terminal sequencing of the purified protein (GLPDDVEDLPK...) unambiguously identified this protein as HiPIP II. HiPIP II as prepared from the soluble fraction and the protein isolated from the membranes have identical N-terminal sequences. Note that, in our sequences, the fifth residue is D rather than G as published by Tedro et al. (34) and Van Driessche et al. (6).
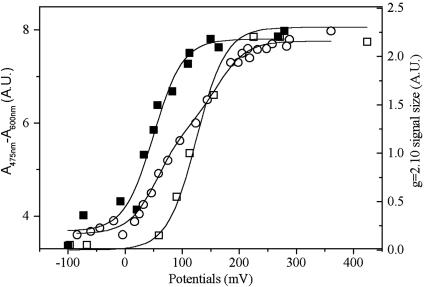
The Redox Potential of HiPIP II Bound to the Membrane Differs Significantly from That of the Isolated Form. Repetitive EPR redox titrations at pH 7 of the membrane-bound HiPIP yield an average Em of +140 ± 20 mV (Fig. 3, open squares, for an example) irrespective of the presence or absence of salt. This value is significantly higher than that published for purified HiPIP II at low salt concentration [Em7 = +50 mV (21)] and which we confirmed by cyclic voltammetry (not shown), UV/Vis titration (Fig. 3, closed squares), and EPR (not shown).
Fig. 3.
Redox titrations of HiPIP II in membrane fragments and in the soluble form. EPR titration on membrane fragments (open squares) was performed in the dark in 50 mM Mops, 130 g/liter NaCl (pH 7). The amplitude of the g = 2.10 peak measured on EPR spectra recorded as in Fig. 1 is plotted versus the ambient redox potential. For this particular experiment, the data points are fitted by an n = 1 Nernst curve with Em =+125 mV. Optical redox titrations of purified HiPIP II were performed in 50 mM Mops (pH 7) in the absence (closed squares) or the presence (open circles) of 2.5 M NaCl. The A475 nm - A600 nm difference is plotted versus the ambient redox potential. The best fit in the absence of NaCl is obtained with Em =+50 mV (n = 1). In the presence of NaCl, two equivalent components with n = 1 and Em = +50 mV and +160 mV were required.
Because H. halophila grows at 130 g/liter salt and because both iso-HiPIPs from this species have so far only been studied under low salt conditions, we have performed optical redox titrations on the soluble, purified HiPIP II in the presence and absence of 130 g/liter NaCl. Fig. 3 (open circles) shows that these physiological salt conditions induced a strong modification of the titration curve. Two equivalent n = 1 components with Em = +50 mV and +160 mV were required. This result can be interpreted as a shift in redox potential in half of the HiPIP II population toward +160 ± 10 mV. Because EPR titrations have an intrinsically higher error than optical titrations, yet HiPIPs cannot be titrated optically in membrane samples, the apparent difference between the Em values at high salt of the membrane associated (+140 ± 20 mV) and the purified (+160 ± 10 mV) form of HiPIP II may not be significant. However, the EPR spectra of HiPIP II did not change in the range of 0.3–2.5 M NaCl and were devoid of a g1 = 2.10 signal. The membrane-bound form of HiPIP II is thus genuinely different from the soluble form at all salt concentrations, arguing for interaction with a membranous protein as the likely source of the observed modifications of spectral redox properties. We therefore studied the association of HiPIP II to the membrane in more detail.
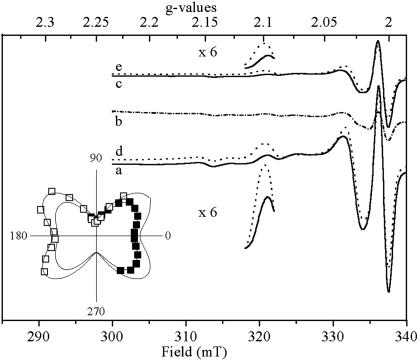
HiPIP II Is Firmly and Specifically Bound to, and Photooxidized by, the RC. Fig. 4 shows EPR spectra of ordered membranes. The spectrum of a ferricyanide-washed sample (Fig. 4, trace a) closely resembles the spectra presented in Figs. 1 and 2 and again shows the splitting (although less pronounced) of the g1 signal. Reduction of the sample by ascorbate strongly decreased but never completely abolished the spectrum arising from the membrane-bound HiPIP (Fig. 4, trace b), in line with its moderately low redox potential (Em7 = +140 ± 20 mV). It is noteworthy that ascorbate only marginally reduces the purified HiPIP II (Em7 = +50 mV). When the sample was thawed, illuminated at 4°C and refrozen under illumination, roughly 30% of HiPIP underwent photo-induced oxidation (Fig. 4, trace c).
Fig. 4.
EPR spectra recorded on partially ordered membrane fragments from H. halophila. Spectra obtained on the chemically oxidized membranes show HiPIP signals with significant anisotropy as illustrated by selected spectra (trace a at 90° and trace d at 30°). Spectrum b obtained on ascorbate-reduced samples shows largely diminished HiPIP signals. The anisotropic signal reappears after illumination at 4°C of these ascorbate reduced membranes (spectra c and e). The additional lines at 315 mT and at 325 mT are due to manganese. (Inset) Polar plot evaluation of the dependence of the g1 signal amplitude at 2.10 versus angle of the magnetic field with respect to the membrane plane. Filled squares represent data points of the chemically oxidized sample, and open squares represent data points of the photooxidized sample. Instrument settings are as in Fig. 1.
EPR spectra were recorded on oriented ferricyanide-oxidized or ascorbate-treated/illuminated membranes from H. halophila with magnetic fields pointing from -40° to +110° with respect to the plane of the membrane. Signal amplitudes of spectra of both the chemically oxidized and the 4°C-illuminated membranes were strongly anisotropic (Fig. 4, compare spectrum a with d or c with e). As can be seen by the shift of the g1 signal between spectrum e and c or between spectrum d and a, the g = 2.105 and the g = 2.095 peaks are not oriented in strictly the same way. The angle difference, however, was too small to be detectable in a polar plot evaluation (Fig. 4 Inset). The g1 signal at 2.1 in spectra both from the ascorbate-reduced/4°C-illuminated and from chemically oxidized membranes was maximal at 30° with respect to the membrane. HiPIP II from H. halophila differs in this respect from the Rubrivivax gelatinosus HiPIP, a considerable fraction of which was observed in orientations differing from those of the photooxidized fraction (22). It is noteworthy that a splitting (already visible in Rubrivivax gelatinosus; figure 4 in ref. 22) is visible in both chemically and photooxidized samples ranging from whole cells to dehydrated membranes. The splitting disappears upon detachment of HiPIP from membranes. At present, we have no explanation for this doublet splitting.
The first article dealing with the RC of H. halophila reported the presence of a tetraheme subunit associated to the RC core (35). This finding was more recently challenged (36). Our observation of low temperature photooxidation of heme in H. halophila (37), however, provides clear evidence for the presence of such a subunit. Electron donation from HiPIP to the photooxidized pigment (P+) must therefore transit through the cytochrome subunit, and so we measured the kinetics of absorption changes due to redox reactions of primary donor pigment (P) and the cytochrome subunit in membrane vesicles. Spectral contributions arising from HiPIP's redox transitions are too weak to be detected directly.
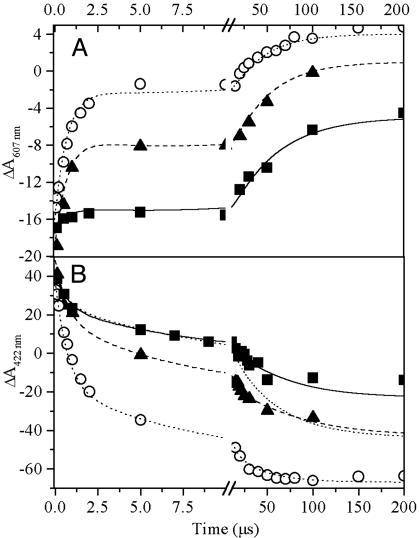
Absorption changes induced by single-turnover laser flashes were recorded at 607 nm (reflecting mainly changes in the redox state of P) and at 422 nm (predominantly due to cytochrome c redox reactions) at defined redox potentials. The 607-nm kinetics obtained at -40 mV, i.e., where the cytochrome subunit is close to fully reduced (37), were triphasic, with a largely dominating fast monoexponential decay phase of t1/2 = 210 ns (Fig. 5A, open circles). This decay phase of P+ was exactly paralleled by a phase of cytochrome oxidation (confirmed by kinetically resolved spectra, not shown) observed at 422 nm (Fig. 5B, open circles). The presence of a submicroseconds, monoexponential P+ re-reduction phase is characteristic for electron donation from a cytochrome subunit. The amplitude ratio of the fast phase measured at 422 nm and 607 nm was 4, in line with the typical ratio of the extinction coefficients of the Soret band of a c-type heme redox difference and of Qx-band bleaching of BChla special pairs (Table 1, which is published as supporting information on the PNAS web site). Increasing the ambient potential to values where the low potential hemes are predominantly oxidized (+100 mV, filled squares) results in an almost complete loss of the fast phase of P+ re-reduction, which is replaced by significantly slower kinetics with a t1/2 of 18 μs. This slow phase is also observable at -40 mV as a minor component. Kinetic traces recorded at intermediate potentials provided evidence for a transition from a predominantly fast phase to almost entirely slow kinetics of P+ reduction with increasing oxidation of the low-potential pair of hemes. The t1/2 of the μs phase increased from 11 to 18 μs between -40 and +100 mV. In contrast to the fast phase, the amplitude of the μs-cytochrome oxidation measured at 422 nm shows a conspicuous absorption deficit. This “missing cytochrome oxidation,” evident at all ambient potentials studied, is visualized by the dotted trace in Fig. 5B, representing the kinetics expected at 422 nm if the amplitude ratio were that of the fast phase at +100 mV. Because we know from the EPR experiments that HiPIP II donates to photo-induced P+, we included a cytochrome re-reduction phase to account for electron donation from HiPIP into our kinetic fits. To reproduce the observed traces, this kinetic phase must have a t1/2 ≤ 11 μs and concern ≈50% of the centers.
Fig. 5.
Kinetics of laser flash-induced absorption changes recorded on membrane fragments in 50 mM Mops, 130 g/liter NaCl (pH 7) at 607 nm (A) and at 422 nm (B). The ambient redox potential was poised to +100 mV (filled squares), +10 mV (filled triangles), or -40 mV (open circles). Kinetics were fitted by using four components with t1/2 = 210 ns, 2.1 μs, 11–18 μs, and 11 μs. The relative amplitudes of the four phases vary as a function of ambient redox potential. The dotted simulation of the μs-phase at high redox potentials was obtained assuming an amplitude ratio between 607 nm and 422 nm kinetics equal to that of the fast (ns-) phase.
These observations taken together demonstrate that 30–50% of the firmly associated HiPIP II donate to the RC at moderately reducing conditions, through a low-potential heme, as discussed below, with t1/2 ≤11 μs. For the fully reduced cytochrome subunit, the low-potential heme remains photooxidized due to the unfavorable equilibrium constant for electron transfer from HiPIP to this heme. This behavior is similar to that observed in systems where a cytochrome c2 re-reduces the RC. As shown above, the rate of electron donation from the cytochrome subunit to P+ strongly decelerates upon oxidation of the low-potential pair of hemes. Similar effects have been observed for other purple bacterial reaction centers (38). A third, intermediate kinetic phase is present in all traces shown. In contrast to the cytochrome-donation phases, this phase has negative amplitudes both at 422 and 607 nm. Kinetically resolved spectra confirmed that it does not arise from P+ reduction by heme.
Discussion
HiPIP II Is the Physiological Electron Donor to the Photosynthetic RC in H. halophila. As detailed above, a significant fraction of HiPIP present in the periplasm of H. halophila cosediments with the membrane during preparation of chromatophores. This HiPIP has been identified as HiPIP II. The facts that this HiPIP (i) can be photooxidized in immobilized samples and (ii) is bound to the membranes in a well defined conformation in photo-oxidized or chemically oxidized samples, demonstrate that part if not all of HiPIP II forms a tight and electron transfer competent complex with the RC. The kinetics of light-induced absorption changes corroborate the existence of this complex and show that the HiPIP to P+ electron transfer is mediated by a cytochrome subunit. A docking site for HiPIP II on this subunit is in line with the observation that crude extracts of HiPIP II containing a cytochrome component retain the g1 = 2.10 signal. The kinetic data yield a t1/2 of ≤11 μs for electron donation from HiPIP to the cytochrome subunit.
The results therefore demonstrate that HiPIP II is the electron carrier re-reducing the RC in H. halophila by means of a cytochrome subunit (at present there is no evidence for involvement of another electron carrier). Both the optical and the EPR experiments show a substoichiometric oxidation of the RC-bound HiPIP. A major part of this “missing” HiPIP photooxidation can be attributed to the equilibrium constant between P+ and HiPIP. The observed strong deceleration of the donation reaction from the cytochrome subunit to P+ and the decrease of the extent of P+ oxidation increasing the ambient redox potential provides evidence for a significantly lower ΔEm between P+, heme, and HiPIP in the coupled system (at ambient potentials where the high potential pair of hemes is reduced) than expected from equilibrium titrations. Effects of electrostatic interactions on kinetic reactions in RCs have been detailed in ref. 38.
HiPIP II from H. halophila Shows Strongly Variable EPR and Redox Properties. The RC-bound HiPIP II has EPR spectra significantly different from those published and confirmed in this work for the soluble HiPIP II. Ironically, the shift of g1 from 2.10 in the RC-bound state to 2.14 in the purified protein is documented in two previous articles (26, 31) but has previously been overlooked. Among all HiPIPs studied in the past, the soluble HiPIP II from H. halophila stands out by its spectral simplicity. Although HiPIPs typically show “complicated” EPR spectra probably arising from the superposition of two or more paramagnetic centers, the spectrum of soluble H. halophila HiPIP II can be simulated by a single paramagnetic species with g1 at 2.14, g2 at 2.034, and g3 at 2.024 (33). NMR data suggest that the “simplicity” of the HiPIP II EPR species arises from a single positioning of the mixed valence pair in the oxidized cluster (39), in contrast to two or more permutations of this pair among the four iron atoms of the cubane in other HiPIPs. As shown above, this single paramagnetic species in the purified protein differs from the single paramagnetic species in the RC-bound HiPIP II. Interaction of HiPIP II with the cytochrome subunit thus most probably turns a different mixed valence pair distribution into the lowest energy state, resulting in distinguishable paramagnetic species in soluble and RC-bound HiPIP. An attribution of g-tensor to molecular axes has been determined for HiPIP II based on electron-nuclear double resonance (ENDOR) results (33). The likely change of mixed valence localization, however, unfortunately precludes a structural interpretation of the g-tensor orientations determined in our work.
In summary, although the molecular bases for the observed Em and paramagnetic variabilities are not clear at present, they nevertheless (i) provide an excellent tool for analyzing the interactions of HiPIP II with its redox partners and (ii) permit us to understand the functional energetics of the H. halophila electron transfer chain (see next section).
The Solution to the “Redox Enigma.” Previously, the low Em values of the two iso-HiPIPs from H. halophila (+50 mV and +110 mV; determined at low salt and in the isolated state) (40) were taken as evidence against their implication in photosynthetic electron transfer. The Em values of P/P+ in the RC and the Rieske center in the bc1 complex were tacitly assumed to be in the typical range for purple bacteria (i.e., +400 to +500 mV for P/P+ and +250 to +340 mV for Rieske). In this work, the Em of HiPIP II at physiological salt concentrations and in the complexed state was determined to be +140 ± 20 mV. In our studies of H. halophila, we measured the redox potentials of P/P+ and the hemes present in the cytochrome subunit of the photosynthetic RC, as well as the redox potential of the Rieske cluster of the bc1 complex (37). The Em values of P/P+ and of the Rieske center in H. halophila were found to be +270 mV and +120 ± 20 mV, respectively, at physiological salt conditions. Electron transfer from the Qo-site of the bc1 complex to the photosynthetic RC thus occurs in the range between +100 mV and +300 mV, i.e., >100 mV below the values encountered in other purple bacterial systems. Moreover, the potential of HiPIP II in physiological salt conditions is at least 70 mV more positive than that determined previously under standard conditions. These two corrections to previously assumed potentials render HiPIP II well suited for shuttling electrons between the bc1 complex and the RC. So far, none of our results indicate an implication of HiPIP I in photosynthetic electron transfer. The functional role of HiPIP I thus remains to be elucidated.
Kinetics and Energetics of the Donation Reaction from HiPIP to the RC. Our kinetic data indicate that electron transfer between HiPIP and RC proceeds with a t1/2 below 11 μs, in the same order of magnitude as the fast phase of electron transfer to the RC reported for Rhodoferax fermentans (2.4 μs) (41) but substantially faster than the t1/2 = 300 μsor480 μs of electron transfer determined in whole cells for Rubrivivax gelatinosus or Rhodocyclus tenuis, respectively (10, 42). In Rubrivivax gelatinosus (22), just as in H. halophila, HiPIP forms a complex with the RC, and the slower time constant can therefore not be explained by a diffusion controlled process. The large differences between these two kinetic processes however, can be rationalized by taking account of the differences in free energies of the two reactions. Marcus's theory of electron transfer, together with the empirical determination of the relevant parameters, allows calculation of the electron transfer rate (ket) between two cofactors as a function of the edge-to-edge distance (R) between the two cofactors, the free energy (ΔG), and the reorganization energy (λ) of the reaction (43). All structural models proposed so far suggest that HiPIP interacts with the outermost heme of the subunit (heme 1 or H1), through its hydrophobic surface (20–25). Solving the Marcus equation by using an Em value of +140 mV mentioned above for the docked HiPIP II and +60 mV determined for H1 and by adopting the docking geometry assumed by Alric et al. (23) for the case of Rubrivivax gelatinosus, we arrive at a t1/2 for electron transfer of ≈11 μs. To be able to reproduce the experimentally determined t1/2 of 300 μs in the Rubrivivax gelatinosus case, Alric et al. (23) interchanged the previous attribution of Em-values (44) to heme H1 and H4 (the second low potential heme). Stimulated by our results on H. halophila, we determined the Em value of the Rubrivivax gelatinosus HiPIP in its membrane-bound state. This Em value was also found to increase (37). Taking this shift in redox potential into account, the initially proposed H1 potential yields a correct prediction of the observed electron transfer rates (see Table 2, which is published as supporting information on the PNAS web site). In the case of Rhodoferax fermentans, the redox potentials of the soluble HiPIP and H1 are +351 mV and +79 or 0 mV (the arrangements of the two low potential hemes has not yet been determined), respectively (45, 46). The published value of t1/2 = 2.4 μs value for electron transfer from HiPIP to the RC in this species can be rationalized only if either the redox potential of the complexed HIPIP is significantly lower than that of the soluble form or if the HiPIP binds significantly closer to H1 of the tetraheme subunit than in the other species mentioned above.
The HiPIP Electron Shuttle in Proteobacteria: Merely a Substitute for Soluble Cytochromes? Because the first reports demonstrated that HiPIPs can play a functional role equivalent to that of soluble cytochrome c2 (10, 11), a number of studies have (i) shown the widespread occurrence of HiPIPs in proteobacterial electron transfer chains (26) and (ii) stimulated the characterization of the kinetic parameters of the HiPIP/RC electron donation reaction (10, 41, 42). The bulk of the data obtained to date has been interpreted with the HiPIPs functioning like soluble cytochromes c2 (with slightly slower electron transfer rates) and are therefore alternatives to the soluble cytochromes just like the small copper proteins in some proteobacteria or in oxygenic photosynthesis. However, evidence is accumulating indicating that the electron transfer mediated by HiPIPs may be substantially different from that of cytochome c2.
Site-directed mutagenesis experiments on the Rubrivivax gelatinosus tetraheme subunit (18, 20, 21, 23) have already indicated that the docking of HiPIP to the cytochrome subunit involved hydrophobic rather than electrostatic interactions as is the case for cytochrome c2.
The interaction between HiPIP and its membranous partner is strong. HiPIP II is routinely retained during preparation of membranes, and specific efforts must be made to fully deplete the membrane fraction of HiPIP molecules. This tight binding is not restricted to the case of HiPIP II in H. halophila. We have in fact observed copurification of HiPIPs with membranes in all of the proteobacterial species that we have studied so far (22, 37). A significant affinity toward the membrane-integral reaction partners thus seems to be a common feature of proteobacterial HiPIPs. This behavior contrasts with the situation encountered for cytochrome c2, which, although frequently retained in sealed vesicles during the preparation of chromatophores, does not show a particularly strong tendency to copurify with the membrane fraction. Our finding of strong association of HiPIP with the RC in all proteobacteria studied in fact poses a kinetic problem, because a tight complex is expected to hinder rapid exchange of a photooxidized HiPIP for another reduced HiPIP. Further experiments will be required to solve this electron transfer riddle.
A third particularity of HiPIPs, observed also with HiPIP II from H. halophila, consists in their strong tendency to multimerize in solution (37). It seems unavoidable to us that the high local concentration of HiPIP in the periplasm must increase its tendency to form multimers. Either such multimers have physiological relevance during electron transfer or another unspecified protein keeps the HiPIP from multimerizing in whole cells.
In summary, the results reported in this work together with previously published data indicate that the detailed functional mechanism of HiPIP electron shuttling may substantially deviate from that used by cytochrome c2.
Supplementary Material
Acknowledgments
We thank A. Cornish-Bowden and M.-L. Cardenas for critically reading the manuscript and correcting the English.
Author contributions: C.L., J.A., W.N., and B.S.-C. performed research; W.N. and B.S.-C. designed research; W.N. and B.S.-C. analyzed data; M.B. performed bacteria cultures; and W.N. and B.S.-C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HiPIP, high-potential iron sulfur protein; RC, reaction center.
References
- 1.Beinert, H. (2000) J. Biol. Inorg. Chem. 5, 2-15. [DOI] [PubMed] [Google Scholar]
- 2.Carter, C. W., Jr. (2001) in Handbook of Metalloproteins, eds. Messerschmidt, A., Huber, R., Poulos, T. & Wieghardt, K. (Wiley, Chichester, U.K.), pp. 602-609.
- 3.Bartsch, R. G. (1991) Biochim. Biophys. Acta 1058, 28-30. [DOI] [PubMed] [Google Scholar]
- 4.Meyer, T. E. (1995) in Anoxygenic Photosynthetic Bacteria, eds. Blankenship, R. E., Madigan, M. T. & Bauer, C. E. (Kluwer, Dordrecht, The Netherlands), Chap. 36, pp. 775-805.
- 5.Schoepp, B., Brugna, M., Lebrun, E. & Nitschke, W. (1999) Adv. Inorg. Chem. 47, 335-360. [Google Scholar]
- 6.Van Driessche, G., Vandenberghe, I., Devreese, B., Samyn, B., Meyer, T. E., Leigh, R., Cusanovich, M. A., Bartsch, R. G., Fischer, U. & Van Beeumen, J. J. (2003) J. Mol. Evol. 57, 181-199. [DOI] [PubMed] [Google Scholar]
- 7.Tedro, S. M., Meyer, T. E. & Kamen, M. D. (1977) J. Biol. Chem. 252, 7826-7833. [PubMed] [Google Scholar]
- 8.Cavazza, C., Guigliarelli, B., Bertrand, P. & Bruschi, M. (1995) FEMS Microbiol. Lett. 130, 193-200. [Google Scholar]
- 9.Peireira, M. M., Antunes, A. M., Nunes, O. C., da Costa, M. S. & Teixeira, N.M. (1994) FEBS Lett. 352, 327-330. [DOI] [PubMed] [Google Scholar]
- 10.Schoepp, B., Parot, P., Menin, L., Gaillard, J., Richaud, P. & Verméglio, A. (1995) Biochemistry 34, 11736-11742. [DOI] [PubMed] [Google Scholar]
- 11.Hochkoeppler, A., Kofod, P. & Zannoni, D. (1995) FEBS Lett. 355, 70-74. [DOI] [PubMed] [Google Scholar]
- 12.Hochkoeppler, A., Kofod, P. & Zannoni, D. (1995) FEBS Lett. 375, 197-200. [DOI] [PubMed] [Google Scholar]
- 13.Overfield, R. E., Wraight, C. A. & Devalt, D. (1979) FEBS lett. 105, 137-142. [DOI] [PubMed] [Google Scholar]
- 14.Adir, N., Axelrod, H. L., Beroza, P., Isaacsn, R. A., Rongey, S. H., Okamura, M. Y. & Feher, G. (1996) Biochemistry 35, 2535-2547. [DOI] [PubMed] [Google Scholar]
- 15.Drepper, F. & Mathis, P. (1997) Biochemistry 36, 1428-1440. [DOI] [PubMed] [Google Scholar]
- 16.Garcia D., Richaud, P. & Verméglio, A. (1993) Biochim. Biophys. Acta 1144, 295-301. [Google Scholar]
- 17.Ortega, J. M., Drepper, F. & Mathis, P. (1999) Photosynth. Res. 59, 147-157. [Google Scholar]
- 18.Osyczka, A., Nagashima, K. V. P., Sogabe, S., Miki, K., Shimada, K. & Katsumi, M. (2001) J. Biol. Chem. 276, 24108-24112. [DOI] [PubMed] [Google Scholar]
- 19.Miyashita, O., Onuchic, J. N. & Okamura, M. Y. (2003) Biochemistry 42, 1151-11660. [DOI] [PubMed] [Google Scholar]
- 20.Osyczka, A., Nagashima, K. V. P., Shimada, K. & Matsuura, K. (1999) Biochemistry 38, 2861-2865. [DOI] [PubMed] [Google Scholar]
- 21.Osyczka, A., Nagashima, K. V. P., Sogabe, S., Miki, K., Shimada, K. & Matsuura, K. (1999) Biochemistry 38, 15779-15790. [DOI] [PubMed] [Google Scholar]
- 22.Lieutaud, C., Nitschke, W., Vermglio, A., Parot, P. & Schoepp-Cothenet, B. (2003) Biochim. Biophys. Acta 1557, 83-90. [DOI] [PubMed] [Google Scholar]
- 23.Alric, J., Yoshida, M., Nagashima, K. V., Hienerwadel, R., Parot, P., Verméglio, A., Chen, S. W. & Pellequer, J. L. (2004) J. Biol. Chem. 279, 32545-32553. [DOI] [PubMed] [Google Scholar]
- 24.Venturoli, G., Mamedov, M. D., Mansy, S. S., Musiani, F., Strocchi, M., Francia, F., Semenov, A. Y., Cowan, J. A. & Ciurli, S. (2004) Biochemistry 43, 437-445. [DOI] [PubMed] [Google Scholar]
- 25.Nogi, T., Fathir, I., Kobayashi, M., Nozawa, T. & Miki, K. (2000) Proc. Natl. Acad. Sci. USA 97, 1361-13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menin, L., Gaillard, J., Parot, P., Schoepp, B., Nitschke, W. & Verméglio, A. (1998) Photosynth. Res. 55, 343-348. [Google Scholar]
- 27.Przysiecki, C., Meyer, T. E. & Cusanovich. (1985) Biochemistry 24, 2542-2549. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, T. E. (1985) Biochim. Biophys. Acta 806, 175-183. [DOI] [PubMed] [Google Scholar]
- 29.Rutherford, A. W. & Sétif, P. (1990) Biochim. Biophys. Acta 1019, 128-132. [Google Scholar]
- 30.Dutton, P. L. (1971) Biochim. Biophys. Acta 226, 63-80. [DOI] [PubMed] [Google Scholar]
- 31.Leguijt, T., Engels, P. W., Crielaard, W., Albracht, S. P. J. & Hellingwerf, K. J. (1993) J. Bacteriol. 175, 1629-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dilg, A. W. E., Mincione, G., Achterhold, K., Iakovleva, O., Mentler, M., Luchinat, C., Bertini, I. & Parak, F. G. (1999) J. Biol. Inorg. Chem. 4, 727-741. [DOI] [PubMed] [Google Scholar]
- 33.Kappl, R., Ciurli, S., Luchinat, C. & Hüttermann, J. (1999) J. Am. Chem. Soc. 121, 1925-1935. [Google Scholar]
- 34.Tedro, S. M., Meyer, T. E. & Kamen, M. D. (1985) Arch. Biochem. Biophys. 241, 656-664. [DOI] [PubMed] [Google Scholar]
- 35.Lefebvre, S., Picorel, R., Cloutier, Y. & Gingras, G. (1984) Biochemistry 23, 5279-5288. [Google Scholar]
- 36.Leguijt, T. & Hellingwerf, K. J. (1991) Biochim. Biophys. Acta 1057, 353-360. [Google Scholar]
- 37.Lieutaud, C. (2004) Ph.D. thesis, Université de la Méditerranée, Marseille, France.
- 38.Alric, J., Cuni, A., Maki, H., Nagashima, K. V., Verméglio, A. & Rappaport, F. (2004) J. Biol. Chem. 279, 47849-47855. [DOI] [PubMed] [Google Scholar]
- 39.Banci, L., Bertini, I., Ciurli, S., Ferretti, S., Luchinat, C. & Piccioli, M. (1993) Biochemistry 32, 9387-9397. [DOI] [PubMed] [Google Scholar]
- 40.Heering, H. A., Bulsink, Y. B. M., Hagen, W. R. & Meyer, T. E. (1995) Biochemistry 34, 14675-14686. [DOI] [PubMed] [Google Scholar]
- 41.Hochkoeppler, A., Zannoni, D., Ciurli, S., Meyer, T. E., Cusanovich, M. A. & Tollin, G. (1996) Proc. Natl. Acad. Sci. USA 93, 6998-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menin, L., Schoepp, B., Parot, P. & Verméglio, A. (1997) Biochemistry 36, 12183-12188. [DOI] [PubMed] [Google Scholar]
- 43.Moser, C. C., Keslke, J. M., Warncke, K., Farid, R. S. & Dutton, P. L. (1992) Nature 355, 796-802. [DOI] [PubMed] [Google Scholar]
- 44.Nitschke, W., Agalidis, I. & Rutherford, A. W. (1992) Biochim. Biophys. Acta 1100, 49-57. [Google Scholar]
- 45.Hochkoeppler, A., Kofod, P., Ferro, G. & Ciurli, S. (1995) Arch. Biochem. Biophys. 322, 313-318. [DOI] [PubMed] [Google Scholar]
- 46.Hochkoeppler, A., Zannoni, D. & Venturoli, G. (1995) Biochim. Biophys. Acta 1229, 81-88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.