Abstract
Genome and antigenome synthesis of negative-strand RNA viruses is initiated at promoters located in inverted terminal repeats (ITR). The ITR of Borna disease virus (BDV), a persisting neurotropic virus with a nuclear replication phase, are exceptional in that they appear to be noncomplete. Our analysis showed that the vast majority of genomic and antigenomic RNA molecules of BDV lack four 5′-terminal nucleotides required for perfect complementarity with the 3′ ITR. By using a previously undescribed reverse genetics system, we investigated whether the structure of the ITR would affect virus propagation. BDV rescued from cDNA encoding complete ITR (rBDVc) showed wild-type virulence, whereas virus rescued from cDNA encoding a viral genome with noncomplete ITR (rBDVnc) was strongly attenuated. Both recombinant viruses expressed similar RNA and protein levels in persistently infected cells. However, rBDVnc particles were less infectious, indicating that complete ITR are required for high viral replicase but not transcriptase activity. Interestingly, genomic RNA from purified rBDVc particles lacked 5′-terminal nucleotides like authentic BDV, strongly suggesting programmed genome truncation. By specifically trimming its genome at the 5′ terminus, BDV seems to limit viral genome amplification, which may favor noncytolytic viral persistence.
Keywords: genome truncation, viral RNA termini, viral persistence, neurotropic
Borna disease virus (BDV) can persist in cultured cells as well as in neurons and some other brain cells of infected animals without causing a cytopathic effect (1–3). On the basis of several unique genetic and biological properties, BDV has been classified into a separate virus family (Bornaviridae) in the order Mononegavirales. It is an enveloped neurotropic virus with a nonsegmented, negative-strand RNA genome (4). The genome of BDV is replicated and transcribed in the nucleus of infected cells (5, 6). To achieve a balanced expression of the six viral proteins from only three transcription units, BDV employs a variety of strategies that include the use of overlapping ORFs, read-through of transcriptional signals, and alternative splicing of polycistronic transcripts (7, 8).
A broad range of warm-blooded animals can be infected with BDV (1), and serological evidence suggests that BDV or a BDV-like virus also infects humans (9). Infection of newborn Lewis rats results in neurodevelopmental and behavioral abnormalities in the absence of inflammation, reminiscent of mood disorders, schizophrenia, and autism in humans (10). In contrast, infection of adult Lewis rats frequently induces an immune-mediated neurological disorder (11), characterized by partial ataxia of the hind legs, uncoordinated movement, and massive weight loss. BDV thus provides an important model for studying mechanisms of viral persistence and immune-mediated CNS pathology as well as for the development of virus-induced neuropsychiatric disease.
Because of the multiplication strategy of negative-strand RNA viruses, their naked genomic RNA by itself is not infectious. Replication and transcription of the viral genome requires proteins which together with viral genomic RNA form the active ribonucleoprotein complex. First, basic information on the composition and regulation of the BDV polymerase complex recently resulted from the use of viral minireplicon systems (12, 13), which revealed that functional BDV ribonucleoprotein complexes are composed of the viral polymerase (L), the nucleoprotein (N), and the phosphoprotein (P).
The genomes of negative-strand RNA viruses contain inverted terminal repeats (ITR), which have the potential to form a panhandle structure with perfectly matching 5′ and 3′ termini. The ITR include the promoters for viral transcription and replication. For members of Orthomyxoviridae (14, 15) and Bunyaviridae (16, 17), the formation of panhandle structures is required for efficient initiation of viral genome replication. In the case of the Mononegavirales, it is unclear whether the panhandle structure is indeed formed during viral replication (18) or whether the terminal complementarity of the genome simply reflects conserved structures of the genomic and antigenomic promoters (19).
Previous work indicated that the ITR of BDV are exceptional. Although data from different laboratories did not match satisfactorily, they were in agreement on the view that the vast majority of negative-sense RNA molecules (vRNA) of BDV lacks complete ITR (4, 20, 21). The relevance of this unexpected finding could originally not be tested, because no appropriate assays were available. When minireplicons for BDV became available (12, 13), it became clear that artificial genomes with noncomplete ITR work very well in these artificial systems. However, a more rigorous molecular analysis of the ITR function was hampered by the lack of a system for the recovery of recombinant BDV from cDNA.
We report here that the structure of the BDV genome is even more peculiar then previously appreciated. We found that the majority of both vRNA and positive-sense genomic RNA (cRNA) molecules possesses trimmed 5′ termini. This unusual genome structure also was observed in recombinant BDV derived from cDNA encoding viral RNA with perfectly complementary termini, indicating that the viral genome is actively truncated during replication. BDV thus appears to employ a unique genome trimming strategy for the control of replication, which may facilitate noncytolytic virus persistence.
Materials and Methods
Plasmid Constructions. The full-length BDV antigenome was assembled in vector pBRT7-MV(+), a derivative of vector pCDV3 (22). The BDV antigenome was amplified by RT-PCR from RNA isolated from cells persistently infected with BDV strain He/80. Three G residues were inserted at the transcriptional start site of the T7 polymerase to enhance promoter activity. T7 transcripts further carried the hammerhead sequence 5′-GTTGTTAACGCAACACTGATGAGGCCGAAAGGCCGAAACTCCGTAAGGAGTCTGTTGCGTTAACAAC-3′ (BDV antigenome underlined). The overall structure of the rescue plasmid is shown in Fig. 2 A. PCR primer sequence information and details on the cloning strategy are available on request. Complete sequence information for the rescue plasmids is available from the GenBank database (accession nos. AY705791 and AY705792).
Fig. 2.
Rescue of BDV from cDNA. (A) Schematic drawing of the T7 RNA polymerase-driven expression cassettes used for generating viral cRNA. To enhance transcription, three G nucleotides were inserted immediately downstream of the T7 promoter. A T7 terminator sequence (ϕ) was included in the construct. To produce correct 5′ and 3′ termini of the viral cRNA, a self-cleavage hammerhead ribozyme (H) and the hepatitis delta virus ribozyme (δ) sequence were inserted. The position of a diagnostic XhoI restriction site (created by introducing a silent point mutation) is indicated. Positions of primers used for diagnostic RT-PCR amplification are indicated by arrows. (B) Schematic drawing of potential base pairing between the 5′ and 3′ ITR of viral cRNA. The four bases present in construct rBDVc, but not rBDVnc, are shown in red. (C) RT-PCR analysis of authentic and recombinant BDV. Diagnostic PCR fragments were incubated with XhoI as indicated. DNA size markers are shown in lane 1. (D and E) Nucleotide sequences at the extreme termini of vRNA and cRNA from rBDVc (D) and rBDVnc (E). Terminal sequences were determined as described in the Fig. 1 legend. Antiparallel alignment of vRNA and cRNA strands are indicated schematically.
Cells and Transfections. Cells were maintained in DMEM supplemented with 10% FCS. Transfection of BSR-T7 cells in 35-mm (six-well) dishes was performed as described in ref. 13.
Virus Rescue. Semiconfluent BSR-T7 cells in 35-mm dishes were transfected with 1.5 μg of either plasmid pBRT7-HrBDVc or pBRT7-HrBDVnc in combination with 0.5 μg of pCA-N, 0.05 μg of pCA-P, and 0.1 μg of pCA-L (13). Three days after transfection, the cells were trypsinized and seeded into 94-mm dishes together with 106 G418-resistant Vero cells. The cocultures were kept under continuous G418 selection. Every 3rd day, the cells were split in a ratio of 1:3, and samples were seeded onto coverslips for analysis.
Virus Stocks. Authentic and recombinant BDV used for subsequent infection experiments was isolated as described in ref. 5. Resulting virus stocks were dialyzed for 2 days against PBS and titrated on Vero cells.
Fluorescence Microscopy. Cells were seeded onto coverslips, fixed for 10 min in 3% paraformaldehyde, and permeabilized by incubation for 5 min in PBS containing 0.5% Triton X-100. Virus antigen was detected as described in ref. 13.
RNA Preparation and Northern Blot Analysis. Total RNA for RT-PCR analysis and Northern blotting was prepared from 6-well dishes or from purified virus stocks with the peqGOLD TriFast reagent (PeqLab Biotechnologie, Erlangen, Germany) as recommended by the manufacturer. Northern blot analysis using 10-μg samples of total RNA was performed as described in ref. 13. DNA probes for the detection of RNAs derived from the N and X/P genes or the M, G, and L genes were amplified by PCR using primer pairs 976(+)/1749(-) and 4014(+)/4729(-), respectively. The numbering refers to the position and orientation of the primers on the BDV antigenome. PCR products were radioactively labeled by using a Prime-It II random primer labeling kit (Stratagene).
RT-PCR Analysis. RNA was reverse-transcribed by using hexamer primers and the H Minus First Strand cDNA synthesis kit (Fermentas, St. Leon-Roth, Germany), according to the manufacturer's protocol. To distinguish between wild-type (wt) and recombinant viruses, we used the PCR primer pair 34(+)/658(-). For analysis of viral genome content in purified particles, the PCR primer pair was 8027(+)/8908(-).
Western Blot Analysis. Cells in 6-well dishes were lysed in 200 μl of RIPA buffer (50 mM Tris, pH 8.0/62.5 mM EDTA/1% Nonidet P-40/0.4% deoxycholate) supplemented with 8 μl of a 25× stock of complete protease inhibitor mixture (Roche) and 0.4 μl of benzonase (Merck). Proteins (10 μg) were size-fractionated by SDS/PAGE and blotted onto a poly(vinylidene difluoride) membrane (Millipore). The membrane was treated with blocking solution (Genosys, The Woodlands, TX) and incubated overnight at 4°C with rabbit antisera against the indicated BDV proteins (13, 23, 24). The blots was developed by using peroxidase-coupled donkey anti-rabbit serum and the ECL+ chemiluminescence (Amersham Pharmacia).
Determination of 5′-Terminal Sequences of BDV vRNA and cRNA. A synthetic RNA oligonucleotide (5′-CGACTGGAGCACGAGGACACTGACATGGACTGAAGGAGTAGAAA-3′) was ligated to 1 μg of RNA recovered from purified viral particles by using the GeneRacer kit (Invitrogen). Modified RNA was reverse-transcribed by using hexamer random primers, and the 5′ end of vRNA was amplified by PCR (30 cycles) by using primers 8468(+) and GeneRacer 5′ primer (Invitrogen). Nested PCR (30 cycles) was performed by using 2 μl of PCR samples and primers 8680(+) and GeneRacer 5′ nested primer. The 5′ end of cRNA was amplified accordingly, by using primers 440(-) and GeneRacer 5′ primer for the first round of PCR (30 cycles) and primer 280(-) and GeneRacer 5′ nested primer for nested PCR (30 cycles).
Determination of 3′-Terminal Sequences of BDV vRNA and cRNA. RNA recovered from purified viral particles (1 μg) was tailed with C residues by using Escherichia coli poly(A) polymerase as described in ref. 20. The 3′ end of the C-tailed vRNA was amplified by PCR (30 cycles) using primers 440(-) and the abridged anchored primer (3′-RACE kit, GIBCO). Nested PCR (30 cycles) was performed by using 2 μl of PCR sample and primers 280(-) and AUAP (3′-RACE kit, GIBCO). The 3′ end of the C-tailed cRNA was amplified accordingly by using primers 8468(+) and the abridged anchored primer for the first round of PCR (30 cycles) and primers 8680(+) and AUAP for nested PCR (30 cycles).
Results
BDV-Derived vRNA and cRNA Have Recessed 5′ Termini. Available results on the structure of the BDV genome at the termini did not reveal a consistent picture (20). We found that the 5′-terminal sequence of the majority of BDV-derived vRNA molecules was 5′-GCGC..., whereas the 3′-terminal sequence of the majority of vRNA molecules was... ACGCAACA-3′ (Fig. 1). In negative-strand RNA viruses, cRNA should be the exact mirror image of vRNA. Our analysis showed, however, that the majority of BDV-derived cRNA molecules started with 5′-GCGU... and ended with... GCGCAACA-3′ (Fig. 1), demonstrating that complementarity of both vRNA and cRNA is incomplete. Both RNA strands of BDV thus seemed to lack four nucleotides at their respective 5′ ends.
Fig. 1.
Nucleotide sequences at the extreme termini of vRNA and cRNA from authentic BDV strain He/80. Sequences at the 3′ termini were determined by C-tailing of viral RNA, followed by RT-PCR amplification of tailed viral RNA. Sequences at the 5′ termini were determined by ligating a synthetic RNA oligonucleotide to viral RNA, followed by RT-PCR amplification. Shown are sequences from near the 3′ termini (including part of the C tail) and from near the 5′ termini (including part of the synthetic RNA oligonucleotide). Deduced structures of vRNA and cRNA are shown schematically.
Rescue of BDV from cDNA. Genetic manipulation of BDV previously has not been possible. To recover recombinant BDV from cDNA, we constructed plasmids from which viral cRNA could be synthesized under the control of the T7 RNA polymerase promoter (Fig. 2A). To enable efficient expression of plasmid-encoded RNA in transfected cells, we inserted three G nucleotides directly downstream of the T7 promoter. Correct processing of the 5′ and 3′ termini of the artificial BDV cRNA was achieved by inserting a hammerhead (H) and a hepatitis delta virus (δ) ribozyme upstream and downstream of the viral sequence, respectively. For later discrimination between parental BDV strain He/80 (BDVwt) and recombinant virus, we introduced a silent point mutation that created a diagnostic XhoI restriction site (Fig. 2 A). To account for the unusual terminal structure of the BDV genome observed above, we constructed two versions of viral full-length cDNA that differed only in the presence of four additional nucleotides at the 3′ terminus of the cRNA product (Fig. 2B). The two constructs encode viral cRNAs with either noncomplete (nc) or complete (c) ITR.
BSR-T7 cells stably expressing T7 RNA polymerase were transfected with either of these two plasmids and with expression vectors pCA-N, pCA-P, and pCA-L encoding the essential components of the BDV polymerase complex. Three days after transfection, the cells were seeded into new dishes containing Vero cells that are highly susceptible to BDV. Cells were analyzed regularly by immunofluorescence for accumulation of the BDV nucleoprotein in nuclear dots, a reliable visual sign of productive BDV infection. In approximately half of all rescue attempts, a dotted appearance of the BDV nucleoprotein was first detected ≈15–20 days after transfection. The number of cells showing this particular staining pattern steadily increased thereafter, indicating that the virus was spreading. The two rescued viruses were designated rBDVnc and rBDVc, respectively.
Virus stocks prepared from these cultures were used to infect fresh Vero cells. Remarkably, establishment of persistence by rBDVnc was delayed by several months compared with rBDVc (data not shown). RNA from cells was reverse-transcribed and amplified by PCR using BDV-specific primers that anneal 411 bp downstream and 213 bp upstream of the diagnostic XhoI site (Fig. 2 A). As expected, the RT-PCR fragment amplified from BDVwt-infected Vero cells remained intact after incubation with restriction enzyme XhoI (Fig. 2C). In contrast, fragments amplified from rBDVnc- and rBDVc-infected Vero cells were cut by XhoI into two fragments of expected size (Fig. 2C), confirming that these viruses originated from plasmid DNA. Overlapping cDNA fragments of the complete genomes of rBDVnc and rBDVc were amplified by RT-PCR and analyzed by sequencing. The genomes of both recombinant viruses perfectly matched the sequence of the plasmids used for virus rescue (data not shown).
The 3′ terminus of rBDVc-derived cRNA was identical to that of parental virus (Fig. 2D). Based on the structure of the cDNA template, we expected to find the 3′-terminal sequence... GCGC-3′ in rBDVnc, followed by a C-tail. However, we detected... GCGCA-3′ in cRNA molecules of rBDVnc (Fig. 2E). Minor fractions of cRNA molecules contained up to five terminal A residues (see Fig. 7, which is published as supporting information on the PNAS web site). These data indicated that the BDV replicase complex prefers to terminate RNA synthesis by incorporating one or more nontemplated A residues.
Interestingly, the 5′ terminus of vRNA from rBDVc was 5′-GCGC... (Fig. 2D). Because rBDVc was generated from a cDNA template that should yield vRNA with the sequence 5′-UGUUGCGC... at the extreme 5′ terminus, we concluded that the recombinant virus is able to specifically trim its genome like parental BDV. In accordance with the cDNA used for the rescue, the 5′ terminus of vRNA derived from rBDVnc was also 5′-GCGC... (Fig. 2E). For both recombinant viruses we detected complete 3′ vRNA termini and trimmed 5′ cRNA termini (Fig. 2 D and E), respectively. To estimate the frequency of vRNA molecules with complete 5′ termini in rBDVc particles, we cloned the RT-PCR product into a plasmid vector and sequenced 150 individual clones. None represented a full-length viral genome (data not shown), indicating that the frequency of particles with complete vRNAs is low.
Biochemical Analysis of Cells Infected with rBDVnc and rBDVc. To characterize viral gene expression, we extracted RNA and proteins from Vero cells that were either uninfected or persistently infected with BDVwt, rBDVnc, or rBDVc. Viral transcripts were analyzed by Northern blotting using two different cDNA hybridization probes. Although Vero cells infected with rBDVnc and rBDVc had slightly lower levels of viral RNA than cells infected with BDVwt, the overall viral RNA expression patterns were similar for all three viruses (Fig. 3). We also noted no significant differences in the intensities of the signals at 8.9 kb, indicating that the steady-state levels of genome-length viral RNAs were similar. We further detected no obvious difference in the viral protein expression patterns between rBDVnc and rBDVc in persistently infected cells (Fig. 3).
Fig. 3.
Viral RNA and protein expression patterns in Vero cells persistently infected with either authentic or recombinant BDV. (A) Northern blot analysis of RNA from uninfected cells (uninf.) or cells persistently infected with BDVwt, rBDVnc, or rBDVc. Blots were hybridized to cDNA probes comprising nucleotides 976–1749 (Left) or 4014–4729 (Right) of the BDV antigenome. Rehybridization of the membranes with a GAPDH probe confirmed that similar amounts of RNA were loaded into each lane. The identity of prominent bands is indicated. (B) Western blot analysis of lysates from parallel cultures of uninfected and infected Vero cells. Proteins were separated by 10% (for detection of N, P, gp43, and gp84) or 15% PAGE (for detection of M and X). Rabbit antisera specific for the indicated BDV proteins were used. Bound antibodies were visualized by using the ECL detection system.
Impaired Propagation of rBDVnc. C6 rat astroglioma cells were infected with a low dose of BDVwt, rBDVnc, or rBDVc, and the cells were split every third day. Samples were seeded onto coverslips, fixed the same day, and analyzed by indirect immunofluorescence for expression of BDV-N. Propagation of rBDVnc was severely hampered, whereas rBDVc and parental virus replicated comparably well (Fig. 4; see also Fig. 8, which is published as supporting information on the PNAS web site). Thus, despite similar viral RNA and protein levels in persistently infected cells, propagation of rBDVnc in freshly infected cells was severely impaired.
Fig. 4.
Spreading kinetics of authentic and recombinant BDV in C6 cell cultures infected with 102 focus-forming units of BDVwt (▪), rBDVnc (○), or rBDVc (•).
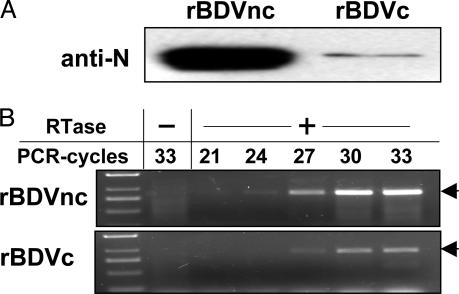
Titers of virus stocks prepared from cells persistently infected with rBDVnc were consistently 10- to 50-fold lower than titers of stocks with rBDVc or wt virus (data not shown). To distinguish between the possibilities that fewer particles were present or that the particles were less infectious, we compared the viral protein contents of purified stocks with equivalent infectivity. If the infectivity of rBDVnc particles is reduced compared with rBDVc, the number of physical particles in the former sample should be significantly higher, and enhanced levels of viral proteins should be present. Indeed, we observed strong signals for BDV nucleoprotein in the rBDVnc sample and only faint signals in the rBDVc sample (Fig. 5A). From the average signal intensity differences observed in three independent experiments, we estimated that the infectivity of rBDVnc particles was ≈10-fold lower than that of rBDVc particles.
Fig. 5.
Reduced infectivity of viral particles from rBDVnc-infected Vero cells. Samples corresponding to 103 focus-forming units of rBDVc and rBDVnc were analyzed by Western blotting using antiserum to the viral nucleoprotein (anti-N) (A) or subjected to reverse transcription in the presence (+) or absence (-) of reverse transcriptase (RTase) (B). PCR primers were selected to specifically amplify a fragment of ≈900 bp (arrow) corresponding to the 5′ terminus of BDV vRNA. Samples were removed from the PCR tubes after completion of the indicated number of cycles. DNA size markers are shown in the left lane.
We used RT-PCR to determine whether particles derived from rBDVnc- and rBDVc-infected cells contained comparable levels of vRNA. Because primer 8908(-) corresponding to the nontranscribed 5′ terminus of the BDV vRNA was used for PCR amplification, the intensities of the RT-PCR signals should directly reflect the abundance of viral genome equivalents in the various samples. When virus stocks with equivalent infectivity were analyzed, we observed that PCR product already became detectable after 24 cycles of amplification in samples from the rBDVnc stock, whereas 27 cycles were required to obtain signals of comparable intensity in samples from rBDVc stocks (Fig. 5B). The rBDVnc stock thus contained ≈8-fold more vRNA than the rBDVc stock, indicating that reduced infectivity of rBDVnc particles cannot be explained by a packaging defect.
Discussion
We showed here that the structure of the BDV genome is unusual in that the vast majority of vRNA and cRNA molecules have recessed 5′ termini. To clarify whether this unique genome structure might be of functional relevance for noncytolytic viral persistence, we established a technology for the generation of recombinant BDV from cDNA-derived viral genomes. Several unusual properties of the BDV polymerase complex complicated the viral rescue. In our successful approach, we acknowledged the results of previous studies with a viral minireplicon (12, 13) that demonstrated that the subunit composition of the BDV polymerase complex requires an ≈10-fold molar excess of N over P for maximal activity and that the viral X protein is a negative regulatory factor. We further used plasmid constructs encoding viral genomes carrying perfect and imperfect ITR, although the existence of the former (seemingly complete) virus genome had not been well documented previously (4, 20, 21). To minimize technical problems that might arise from these special features of BDV, we modified the frequently used protocol for rescue of negative-strand RNA viruses from cDNA (25) in several ways. For example, for efficient synthesis of BDV cRNA with correct 5′ ends by T7 RNA polymerase, we used a hammerhead ribozyme specifically designed for sequence-specific processing of artificial viral transcripts at the 5′ terminus. Our work represents, to our knowledge, the first successful genetic manipulation of a negative-strand RNA virus that predominantly establishes long-term persistence.
An interesting result of our study was that replication-competent BDV could be rescued from complete viral cRNA as well as from incomplete template with truncated 3′ terminus. Because the 3′ end of viral cRNA contains the promoter that directs synthesis of progeny viral genomes, it initially appeared likely that the terminal truncation introduced in rBDVnc would eliminate infectivity altogether. This assumption was clearly not correct. Our analysis showed that rBDVnc was viable but severely attenuated. Interestingly, the genome truncation in rBDVnc did not strongly affect the steady-state levels of viral RNAs and proteins in persistently infected cells. It rather affected the efficacy by which the virus was able to infect new cells. Our data indicate that rBDVnc particles exhibited reduced infectivity, which cannot be explained by excessive production of empty virus particles or inefficient packaging of viral RNA. Rather, it appeared that the artificial truncation of the viral genome in rBDVnc reduced the activity of the viral replicase promoter, which delays production of new templates for viral mRNA synthesis. Inefficient genome amplification at early times after de novo infection of host cells might frequently result in abortive infections.
Analysis of RNA derived from rBDVnc yielded a surprising result. The 3′ terminus of the viral cRNA contained one or more nontemplated A residues in the vast majority of molecules. It is of interest to note that the RNA polymerase of bacteriophage Qβ was shown to add nontemplated A residues to the 3′ terminus of newly synthesized transcripts that are not used for initiation of complementary strand synthesis (26). Thus, the 3′ terminal A nucleotides present in BDV vRNA and cRNA (Fig. 2) most likely are not truly encoded by the viral genome. Rather, they appear to be added by the viral polymerase during the termination process.
Maintenance of the genetic information requires perfect complementarity between genome and antigenome. The unusual terminal structure of the BDV genome implies that the majority of vRNA and cRNA molecules found in BDV particles represent 5′ terminally truncated subgenomic RNA species rather than full-length genomes. Interestingly, the analysis of the terminal genome sequences of rBDVc showed that the majority of vRNA and cRNA molecules also were trimmed like in authentic BDV. Because the rBDVc genome originates from a cDNA molecule that encodes a full-length cRNA, it appears that the subgenomic viral RNA molecules are being produced by programmed terminal trimming during the genome replication process (see model in Fig. 6). It is presently not understood how 5′-terminal trimming of the viral genome is achieved. The specificity of the truncations argues against RNA degradation, although it is conceivable that a replication complex-associated endonuclease activity is specifically removing four nucleotides from the 5′ end of the majority of nascent viral transcripts. For several reasons, we favor the alternative possibility that the BDV replicase complex might frequently initiate RNA synthesis with G as first base on the C residue at template position 5 (Fig. 2B), which would result in 5′-recessed cRNA molecules (see model in Fig. 6). Rarely, the BDV replicase complex also might initiate at the penultimate C residue, resulting in the synthesis of completely replication-competent (full-length) cRNA. It is possible that the CGC sequence context of the C at position 5 (Fig. 2B) favors initiation at this site. If we assume that the second replication step (synthesis of vRNA from cRNA templates) follows the same basic rules, the vast majority of vRNA and cRNA molecules in infected cells are expected to be replication-defective but, nevertheless, transcription-competent. A scenario in which only full-length genomes efficiently transmit infectivity could readily explain the observation that BDV titers obtained from persistently infected tissue culture or from brains of infected animals remain low, although high levels of viral RNA and proteins can be detected (27). A detailed mutational analysis of the BDV ITR in the context of the minireplicon as well as in the context of recombinant viruses will be required to resolve the mechanism of the observed BDV genome-trimming process.
Fig. 6.
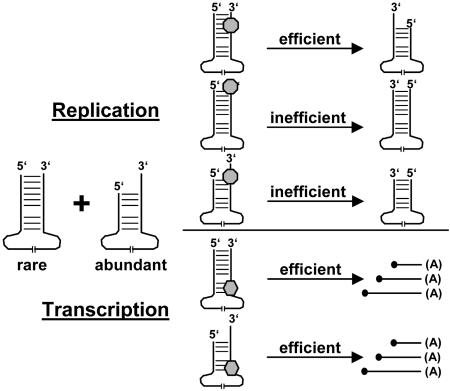
Schematic drawing depicting the proposed preferential replication of full-length BDV genomes. Our model predicts that genomes with 5′-terminal deletions are efficiently transcribed, although they are not replicated well. Note that panhandle presentation of BDV genomes is used for clarity. The formation of panhandles during BDV replication has not been formally proven.
The terminal sequences of rBDVnc were almost identical to those of rBDVc and parental BDV, except that the cRNA molecules of rBDVnc lacked a correct 3′ terminus. The fact that rBDVnc was unable to reconstitute a cRNA with correct 3′ terminus supports the view that rare full-length RNA molecules indeed serve as templates for cRNA and probably also vRNA synthesis. If alternative mechanisms generated the 3′ overhangs, rBDVnc should possess cRNA with correct 3′ terminus because the 5′ termini of vRNA derived from rBDVnc are indistinguishable from the 5′ termini of the majority of vRNA molecules of rBDVc or authentic BDV. Based on these considerations, it is clear that full-length vRNA and cRNA molecules must exist in rBDVc. However, the frequency of these full-length molecules appears to be low because we did not detect any full-length molecules among 150 individual plasmid clones derived from PCR fragments representing the genomic 5′ end of rBDVc. We cannot exclude a technical bias in our experimental system that might affect the frequency of detectable full-length molecules. However, it should be noted that previous attempts in which other strategies were used to determine the genomic 5′ ends yielded identical result (4, 20, 21). Because different techniques exclusively identified subgenomic vRNA and cRNA molecules and because we observed clear phenotypic differences between the two recombinant viruses, it appears unlikely that our results are due to a technical bias in our experimental system.
It is important to note that the majority of vRNA molecules have complete 3′ termini. This finding indicates that initiation of replication occurs almost exclusively at full-length templates. Efficient initiation of BDV replication thus seems to require complementarity of the 5′ and 3′ termini (see model in Fig. 6). It is difficult to envisage how the 5′ terminus should influence the efficiency of replication initiation at the 3′ terminus of the same RNA molecules, unless one assumes a direct interaction of the two termini. Our data thus support the view that the terminal sequences of the vRNA and cRNA molecules of BDV interact to form a panhandle structure that facilitates initiation of replication.
From the fact that high levels of viral proteins are present in cells persistently infected with rBDVnc, it follows that 5′-recessed vRNAs are excellent templates for mRNA synthesis (see model in Fig. 6). Because the BDV transcriptase complex initiates at template position 44, it is conceivable that viral mRNA synthesis is not strongly influenced by the presence or absence of a perfect ITR. To account for the attenuated phenotype of rBDVnc, we speculate that high replicase activity is required to provide sufficient templates for viral mRNA synthesis at early times after infection of new host cells and that this requirement is probably less stringent in persistently infected cells.
It is of interest to note that the genome termini of viruses of the genus Hantavirus also look different from most other negative-strand RNA viruses. For example, like BDV, Hantaan virus features a U residue at the 5′ terminus of genomic RNA, which in fact is not the first base to be incorporated into the growing polynucleotide chain (28). In Hantaan virus, the viral replicase complex starts RNA synthesis with G at an internal C template. By employing a unique prime-and-realign mechanism, it then moves the newly synthesized primer a few nucleotides toward the 3′ terminus of the template before it continues RNA synthesis. Like Hantaan virus, BDV most likely starts replication by incorporating a G residue templated at internal C residues. However, in the case of BDV, we do not need to postulate a prime-and-realign mechanism to account for the observed structures of the viral replication products (see model in Fig. 6). In Seoul virus, another member of the Hantavirus genus, the 3′ and 5′ termini of vRNA and cRNA exhibit terminal deletions if the virus is derived from persistently infected cells (29). It was speculated that accumulation of terminally deleted RNAs may play a role in Seoul virus persistence. However, unlike in the case of BDV, 5′-terminal deletions of the viral RNAs were comparatively rare in Seoul virus, the deletions had no uniform size, and the biological significance of the truncations could not be analyzed due to the lack of a reverse genetics system for this virus.
What might be the benefit of a viral multiplication strategy that includes genome trimming and thus programmed deletion of genetic information? BDV is highly neurotropic in natural and experimental hosts. It replicates in neurons and astrocytes without inducing cytopathic effects. Long-term persistence in the CNS is presumably best achieved if viral replicase activity is low. Our results indicate that programmed genome truncation, which limits viral genome amplification, represents a suitable strategy for maintaining noncytolytic viral persistence.
Supplementary Material
Acknowledgments
We thank Annette Ohnemus for excellent technical assistance, Matthias Görlach for help with the design of the hammerhead ribozyme, Amiya Banerjee for helpful discussion, Veronika von Messling (Institut National de la Recherche Scientifique, Institut Armand-Frappier, Université du Québec, Québec) for providing plasmid pBRT7-MV(+), and Daniel Gonzalez-Dunia and Adolfo Garcia-Sastre for critically reading the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant SCHN 765/1-3. U.S. was supported by a grant from the Swiss National Science Foundation.
Author contributions: U.S., M.S., and P.S. designed research; U.S. and M.S. performed research; U.S. and M.S. analyzed data; and U.S. and P.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BDV, Borna disease virus; ITR, inverted terminal repeats; rBDVc, BDV rescued from cDNA encoding complete ITR; rBDVnc, BDV rescued from cDNA encoding a viral genome with noncomplete ITR; BDVwt, wild-type BDV; vRNA, negative-sense RNA molecules; cRNA, positive-sense RNA molecules; L, polymerase; N, nucleoprotein; P, phosphoprotein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY705791 and AY705792).
References
- 1.Staeheli, P., Sauder, C., Hausmann, J., Ehrensperger, F. & Schwemmle, M. (2000) J. Gen. Virol. 81, 2123-2135. [DOI] [PubMed] [Google Scholar]
- 2.Rott, R. & Becht, H. (1995) Curr. Top. Microbiol. Immunol. 190, 17-30. [DOI] [PubMed] [Google Scholar]
- 3.de la Torre, J. C. (2002) Front. Biosci. 7, d569-d579. [DOI] [PubMed] [Google Scholar]
- 4.Briese, T., Schneemann, A., Lewis, A. J., Park, Y. S., Kim, S., Ludwig, H. & Lipkin, W. I. (1994) Proc. Natl. Acad. Sci. USA 91, 4362-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briese, T., de la Torre, J. C., Lewis, A., Ludwig, H. & Lipkin, W. I. (1992) Proc. Natl. Acad. Sci. USA 89, 11486-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubitt, B. & de la Torre, J. C. (1994) J. Virol. 68, 1371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneemann, A., Schneider, P. A., Kim, S. & Lipkin, W. I. (1994) J. Virol. 68, 6514-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneemann, A., Schneider, P. A., Lamb, R. A. & Lipkin, W. I. (1995) Virology 210, 1-8. [DOI] [PubMed] [Google Scholar]
- 9.Billich, C., Sauder, C., Frank, R., Herzog, S., Bechter, K., Takahashi, K., Peters, H., Staeheli, P. & Schwemmle, M. (2002) Biol. Psychiatry 51, 979-987. [DOI] [PubMed] [Google Scholar]
- 10.Pletnikov, M. V., Moran, T. H. & Carbone, K. M. (2002) Front. Biosci. 7, d593-607. [DOI] [PubMed] [Google Scholar]
- 11.de la Torre, J. C. (2002) J. Infect. Dis. 186, Suppl. 2, S241-S247. [DOI] [PubMed] [Google Scholar]
- 12.Perez, M., Sanchez, A., Cubitt, B., Rosario, D. & de la Torre, J. C. (2003) J. Gen. Virol. 84, 3099-3104. [DOI] [PubMed] [Google Scholar]
- 13.Schneider, U., Naegele, M., Staeheli, P. & Schwemmle, M. (2003) J. Virol. 77, 11781-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong, H. K., Cheong, C., Lee, Y. S., Seong, B. L. & Choi, B. S. (1999) Nucleic Acids Res. 27, 1392-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, M. T., Parvin, J. D., Gupta, S., Krystal, M. & Palese, P. (1987) Proc. Natl. Acad. Sci. USA 84, 8140-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronnholm, R. & Pettersson, R. F. (1987) Virology 160, 191-202. [DOI] [PubMed] [Google Scholar]
- 17.Barr, J. N. & Wertz, G. W. (2004) J. Virol. 78, 1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wertz, G. W., Whelan, S., LeGrone, A. & Ball, L. A. (1994) Proc. Natl. Acad. Sci. USA 91, 8587-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapparel, C. & Roux, L. (1996) Virology 225, 163-171. [DOI] [PubMed] [Google Scholar]
- 20.Pleschka, S., Staeheli, P., Kolodziejek, J., Richt, J. A., Nowotny, N. & Schwemmle, M. (2001) J. Gen. Virol. 82, 2681-2690. [DOI] [PubMed] [Google Scholar]
- 21.Cubitt, B., Oldstone, C. & de la Torre, J. C. (1994) J. Virol. 68, 1382-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Messling, V., Zimmer, G., Herrler, G., Haas, L. & Cattaneo, R. (2001) J. Virol. 75, 6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richt, J. A., Furbringer, T., Koch, A., Pfeuffer, I., Herden, C., Bause-Niedrig, I. & Garten, W. (1998) J. Virol. 72, 4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauder, C., Muller, A., Cubitt, B., Mayer, J., Steinmetz, J., Trabert, W., Ziegler, B., Wanke, K., Mueller-Lantzsch, N., de la Torre, J. C. & Grasser, F. A. (1996) J. Virol. 70, 7713-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conzelmann, K. K. (1998) Annu. Rev. Genet. 32, 123-162. [DOI] [PubMed] [Google Scholar]
- 26.Goodman, H. M., Billeter, M. A., Hindley, J. & Weissmann, C. (1970) Proc. Natl. Acad. Sci. USA 67, 921-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauli, G. & Ludwig, H. (1985) Virus Res. 2, 29-33. [DOI] [PubMed] [Google Scholar]
- 28.Garcin, D., Lezzi, M., Dobbs, M., Elliott, R. M., Schmaljohn, C., Kang, C. Y. & Kolakofsky, D. (1995) J. Virol. 69, 5754-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, B. J. & Schmaljohn, C. (2000) J. Virol. 74, 1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.