Coexpression of two anthocyanin modification genes elicits blue flower coloration through interaction with colorless flavonoids.
Abstract
Various colored cultivars of ornamental flowers have been bred by hybridization and mutation breeding; however, the generation of blue flowers for major cut flower plants, such as roses, chrysanthemums, and carnations, has not been achieved by conventional breeding or genetic engineering. Most blue-hued flowers contain delphinidin-based anthocyanins; therefore, delphinidin-producing carnation, rose, and chrysanthemum flowers have been generated by overexpression of the gene encoding flavonoid 3′,5′-hydroxylase (F3′5′H), the key enzyme for delphinidin biosynthesis. Even so, the flowers are purple/violet rather than blue. To generate true blue flowers, blue pigments, such as polyacylated anthocyanins and metal complexes, must be introduced by metabolic engineering; however, introducing and controlling multiple transgenes in plants are complicated processes. We succeeded in generating blue chrysanthemum flowers by introduction of butterfly pea UDP (uridine diphosphate)–glucose:anthocyanin 3′,5′-O-glucosyltransferase gene, in addition to the expression of the Canterbury bells F3′5′H. Newly synthesized 3′,5′-diglucosylated delphinidin-based anthocyanins exhibited a violet color under the weakly acidic pH conditions of flower petal juice and showed a blue color only through intermolecular association, termed “copigmentation,” with flavone glucosides in planta. Thus, we achieved the development of blue color by a two-step modification of the anthocyanin structure. This simple method is a promising approach to generate blue flowers in various ornamental plants by metabolic engineering.
INTRODUCTION
Flower color is one of the most important breeding targets for ornamental plants because it has a great influence on customer choice in buying potted flowers, garden flowers, and cut flowers. Rose, chrysanthemum, carnation, lily, and gerbera are major ornamental plants in terms of both value and production yields in the global cut flower market. Although white, yellow, orange, red, magenta, and green variants of these flowers have been domesticated from wild species and bred by mutation breeding and interspecific hybridization, violet or blue flowers have not been produced by conventional breeding methods. Hence, this goal is of particular interest both to the floricultural industry and in the field of horticultural and plant sciences.
In general, anthocyanin pigments contribute to the development of red, purple, and blue flower colors. The B-ring hydroxylation and methylation patterns of anthocyanins contribute to the development of flower color. The abovementioned cut flower plants have cyanidin-based anthocyanins with two hydroxyl groups at the 3′ and 4′ positions on the B-ring, which are responsible for reddish coloration. Because these flowers lack flavonoid 3′,5′-hydroxylase (F3′5′H) activity, they do not synthesize delphinidin-based anthocyanins, which have an additional hydroxyl group at the 5′ position and are responsible for the bluish coloration in many plants (1).
The cytochrome P450 enzyme F3′5′H controls the biosynthesis of delphinidin-based anthocyanins (1). Recently, the F3′5′H was introduced into rose and carnation plants, and the resulting genetic transformants produced novel violet flowers that accumulate delphinidin-based anthocyanins (2–5). We also introduced Canterbury bells (Campanula medium) F3′5′H into chrysanthemum. The coloration of these transgenic chrysanthemum plants became bluer, but the flowers were still limited to purple/violet (6, 7). An accumulation of delphinidin-based anthocyanins is necessary to develop purple/violet color but not sufficient to produce a true blue color.
The blue color development of flowers has been well studied, and several mechanisms have been described. These include intramolecular associations within anthocyanin via attached glycosyl and aromatic acyl groups (8, 9), intermolecular associations of anthocyanin with copigment compounds such as aromatic acid derivatives and flavonoids (8, 10), metal complex formation with anthocyanin and flavonoid (8, 11, 12), and modification of vacuolar pH to higher values (8, 13, 14). Because modification of vacuolar pH and metal ion transportation might disturb cellular homeostasis, we initially proceeded by modification of anthocyanin with sugar and aromatic acyl groups to facilitate intramolecular associations.
The major anthocyanins of natural chrysanthemums are cyanidin 3-O-(6″-O-malonyl)glucoside (A1) (15) and cyanidin 3-O-(3″,6″-di-O-malonyl)glucoside (A2) (16), whereas those of purple/violet transgenic chrysanthemums are their delphinidin analogs (A5 and A6) (Fig. 1 and fig. S1) (6). Delphinidin 3-O-(6″-O-malonyl)glucoside-3′,5′-di-O-(6‴,6‴′-di-O-p-coumaroyl)glucoside (ternatin D3) is one of the simplest polyacylated anthocyanins responsible for the blue coloration of Clitoria ternatea (butterfly pea) (fig. S1) (17). In transgenic chrysanthemums, ternatin D3 is likely to be the least-modified blue anthocyanin. Therefore, we tried to synthesize ternatin D3 in chrysanthemums by hydroxylation at the 3′ and 5′ positions and by attachment of p-coumaroyl-glucosyl moieties at these hydroxyl groups. In the process of examining this transformation, we unexpectedly generated blue flower plants by means of glucosylation without aromatic acylation. We then studied the mechanism of blue coloration by analyzing newly synthesized anthocyanins and their intermolecular associations in chrysanthemum petals.
Fig. 1. Scheme for introducing 3′,5′-hydroxylation and 3′,5′-glucosylation into the anthocyanin biosynthetic pathway in chrysanthemum petals.
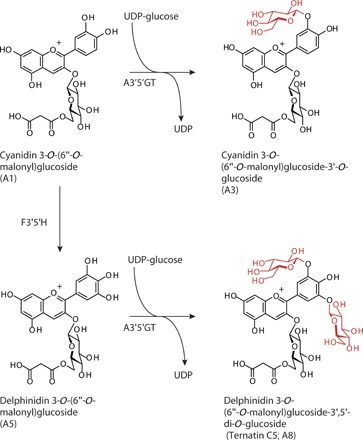
The major anthocyanin of ‘Taihei’ wild-type chrysanthemum was cyanidin-based anthocyanin (A1). F3'5'H, flavonoid 3',5-hydroxylase; A3′5′GT, UDP-glucose:anthocyanin 3′,5′-O-glucosyltransferase.
RESULTS
Blue coloration in transgenic chrysanthemums
First, we introduced the butterfly pea gene CtA3′5′GT encoding UDP (uridine diphosphate)–glucose:anthocyanin 3′,5′-glucosyltransferase (18) into chrysanthemums by using binary vector pB249 (Fig. 1 and fig. S2). Although transgenic plants expressing CtA3′5′GT were obtained, their flowers did not show a color change to bluish hue (table S1). Next, to achieve hydroxylation and glucosylation of the B-ring of anthocyanin in chrysanthemums, we used the binary vector pB423 to coexpress the Canterbury bells gene F3′5′H (CamF3′5′H) and the gene CtA3′5′GT (Fig. 1 and fig. S2). Both transgenes were expressed under the control of the chrysanthemum F3H promoter. We also designed vector pB428 containing a chrysanthemum F3′H RNA interference (RNAi) construct (7, 19) to reduce the accumulation of red cyanidin derivatives, in addition to the CamF3′5′H and CtA3′5′GT cassettes (fig. S2 and table S1), and used this vector for transformation. As a result, 19 of 32 (59%) transgenic ‘Taihei’ chrysanthemum lines harboring the simpler pB423 construct changed their flower color to a blue hue within the blue or violet-blue color groups of the Royal Horticultural Society (RHS) Colour Charts (Fig. 2A and table S1).
Fig. 2. Anthocyanins derived from ray florets of blue-colored transgenic chrysanthemums.
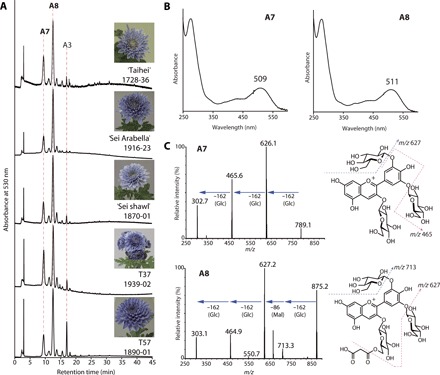
(A) High-performance LC (HPLC) chromatograms of anthocyanins in blue petal extracts detected at 530 nm. (B) Ultraviolet (UV)–visible absorption spectra of A7 and A8 recorded during online HPLC under acidic conditions. An additional spectral maximum at around 320 nm was not detected in either, indicating a lack of aromatic acyl groups. (C) Chemical structures and MS/MS fragmentation spectra of A7 and A8. The product ions of A7 at m/z = 789 [M]+ were 627 (−Glc), 465 (−2 × Glc), and 303 (−3 × Glc). The product ions of A8 at m/z = 875 [M]+ were 713 (−Glc), 627 (−Glc, −Mal), 465 (−2 × Glc, −Mal), and 303 (−3 × Glc, −Mal; delphinidin aglycone). Dotted arrows show the proposed fragmentation scheme. Glc, glucosyl; Mal, malonyl; A7, delphinidin 3,3′,5′-tri-O-glucoside (preternatin C5); A8, delphinidin 3-O-(6″-O-malonyl)glucoside-3′,5′-di-O-glucoside (ternatin C5).
Anthocyanin structures in blue chrysanthemums
The blue ray florets of transgenic chrysanthemums mainly contained anthocyanins A7 and A8 and minor anthocyanin A3 (Fig. 2A and fig. S1). The absorbance spectra indicated the absence of an aromatic acyl moiety in these anthocyanin structures (Fig. 2B). We analyzed A8 in the extracts of blue ray florets by liquid chromatography–photodiode array–mass spectrometry (LC-PDA-MS) and detected a molecular ion at mass/charge ratio (m/z) = 875 [M]+ corresponding to malonylated delphinidin triglucoside (Fig. 2C). We identified the structure of A8 as delphinidin 3-O-(6″-O-malonyl)glucoside-3′,5′-di-O-glucoside (ternatin C5) (Fig. 2) (17) by further analyses of LC-MS, 1H nuclear magnetic resonance (NMR), and nuclear Overhauser effect spectroscopy (NOESY) spectra and high-resolution MS (Supplementary Materials and Methods). Partial hydrolysis of A8 produced A7, which was confirmed by coelution with authentic standard in HPLC. LC-MS analysis yielded the molecular ion of A7 at m/z = 789 [M]+ and product ions corresponding to the loss of the glucosyl moiety from A7. These results suggested that the A7 structure was delphinidin 3,3′,5′-tri-O-glucoside (preternatin C5) (17, 18). From the spectral data of LC-MS (m/z = 697 [M]+), high-resolution MS, and 1H NMR, we determined the minor anthocyanin A3 structure to be cyanidin 3-O-(6″-O-malonyl)glucoside-3′-O-glucoside (Fig. 1). All these anthocyanins were glucosylated at the 3′- and/or 5′-hydroxyl groups (Supplementary Materials and Methods).
Coloration, anthocyanin composition, and transgene expression
We obtained various colored transgenic ‘Sei Arabella’ chrysanthemums carrying CamF3′5′H and CtA3′5′GT. Using these plants, we analyzed the relationship among coloration, anthocyanin composition, and transgene expression (Figs. 3 and 4). Most transgenic lines with flowers in the blue/violet-blue group were distributed below a hue angle of 290°. Almost all lines had shifted from the red-purple group of wild type to the blue/violet-blue group (Fig. 3).
Fig. 3. Flower color distribution of transgenic ‘Sei Arabella’.
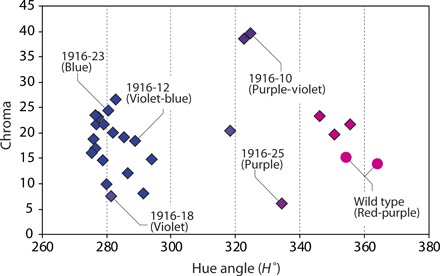
Hue of flower color was evaluated by hue angle (H°), where H° near 270 shows blue color, H° from 300 to 340 shows violet to purple color, and H° over 340 shows red color. Chroma value describes the color strength. The color names given in parentheses are color groups according to the RHS Colour Charts. Circles, wild-type ‘Sei Arabella’; diamonds, transgenic lines.
Fig. 4. Flower coloration, anthocyanin composition, and transgene expression in transgenic ‘Sei Arabella’.
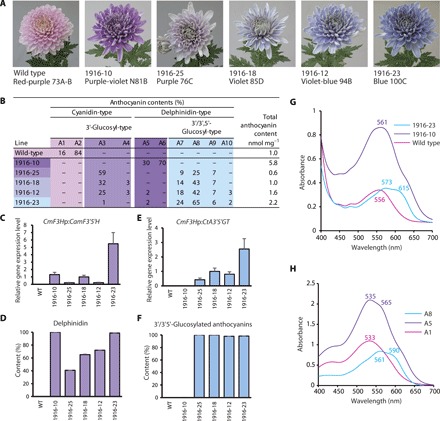
(A) Pictures of representative flower colors of wild-type and transgenic lines. Each line presents the color group and number of RHS Colour Charts. (B) Anthocyanin composition. (C and D) Relative expression of CamF3′5′H (C) and the resultant delphinidin-based anthocyanins (D). Contents of delphinidin-based anthocyanins corresponded well with the expression of CamF3′5′H. (E and F) Relative expression of CtA3′5′GT (E) and the resultant 3′/3′,5′-glucosylated anthocyanins (F). Contents of 3′/3′,5′-glucosylated anthocyanins corresponded well with the expression of CtA3′5′GT. (G) Visible absorption spectra of fresh petals of wild-type (pink), 1916-10 (purple), and 1916-23 (blue) lines. (H) Visible absorption spectra of major anthocyanins derived from pink (A1; wild type), purple-violet (A5), and blue/violet-blue (A8) chrysanthemums in acetate buffer (pH 5.6).
The flowers of wild-type plants accumulated non–3′-glucosylated cyanidin-based anthocyanins (A1 and A2) and exhibited a pink color (Fig. 4, A and B, and fig. S3). The flower color of transgenic 1916-10 plants, which accumulated non–3′/3′,5′-glucosylated delphinidin-based anthocyanins (A5 and A6), exhibited a purple color. On the other hand, the purplish pink flowers of transgenic 1916-25 plants mainly accumulated 3′-glucosylated cyanidin-based anthocyanin (A3). In contrast, the flowers of 1916-18, 1916-12, and 1916-23 lines, which accumulated 3′,5′-glucosylated delphinidin-based anthocyanins (A7 to A9), were blue (Fig. 4, A and B, and fig. S3). We also detected flavone glucosides and caffeoylquinates in ray florets of wild-type and transgenic lines. The composition of these analytes was almost the same in the different flower colored lines (fig. S4). The blue coloration was dependent on the composition ratio of the 3′,5′-glucosylated delphinidin-based anthocyanins (Fig. 4, A and B).
We verified the expression of the transgenes in ray florets by quantitative real-time polymerase chain reaction (qRT-PCR) assay. In purple flower colored lines, transgenic 1916-10 plants expressed only CamF3′5′H, whereas transgenic 1916-25 plants expressed CtA3′5′GT with a low level of CamF3′5′H. The violet/blue transgenic lines 1916-18, 1916-12, and 1916-23 expressed both CamF3′5′H and CtA3′5′GT. The content of the delphinidin-based anthocyanins correlated with the relative expression of CamF3′5′H, whereas the content of the 3′- and/or 5′-glucosylated anthocyanins correlated with the relative expression of CtA3′5′GT (Fig. 4, C to F). These results indicated that CtA3′5′GT efficiently glycosylated both cyanidin- and delphinidin-based anthocyanins in transgenic chrysanthemums. As mentioned above, the content of flavone glucosides and caffeoylquinates, which are candidates for copigments (8, 10), was almost the same among the wild-type and transgenic lines (fig. S4). In addition, expression of CamF3′5′H led to the production of some new flavonoids, such as C2 (fig. S4, B to E).
Spectra of major anthocyanins and intact ray florets
The intact pink-colored petals of host ‘Sei Arabella’ exhibited a visible absorbance spectrum, with the peak of the maximum wavelength of visible absorption (λvismax) at 556 nm. The spectrum of the purple-violet petals of transgenic chrysanthemums was shifted to longer wavelengths, with the λvismax at 561 nm. The blue petals showed a characteristic spectrum with a longer wavelength, with the λvismax at 573 nm, and a second absorption maximum called a shoulder at around 615 nm (Fig. 4G).
The major anthocyanins of pink-colored wild-type plants, purple-violet–colored transgenic lines, and blue-colored lines were A1 and A2, A5 and A6, and A7 and A8, respectively (Fig. 4B and table S2). In the acidic solution of the HPLC solvent, A1 had a λvismax at 518 nm. The λvismax of A5 shifted toward a longer wavelength at 527 nm, whereas that of A8 shifted toward a shorter wavelength at 511 nm (Fig. 2B). In acetate buffer solution at pH 5.6 corresponding to the pH of the petal juice, the λvismax of the A1, A5, and A8 absorption spectra was 533 nm, 535 nm (with a shoulder at 565 nm), and 561 nm (with a shoulder at 590 nm), respectively (Fig. 4H), with successively longer wavelength shifts. Even in solution at the pH of the petals, the λvismax of each major anthocyanin was shorter than its corresponding value for intact petals (Fig. 4, G and H). These results suggested that the flower color of chrysanthemums arises from intermolecular copigmentation of the major anthocyanin with another compound in the petals.
Identification of copigments for blue coloration
We attempted to identify the copigment counterpart of major anthocyanin A8 responsible for generating the blue flower coloration of transgenic chrysanthemums by analyzing an extract from the blue petals of the ‘Taihei’ transgenic chrysanthemum using a cross–thin-layer chromatography (cross-TLC) method (20). After the development of the TLC plate, red, violet, and blue regions were visible in the anthocyanin development lines (Fig. 5A). Observation under UV light irradiation showed that the blue and violet regions crossed with a yellow fluorescent band (X) and an invisible band (Y), respectively (Fig. 5B). These results suggested that bands X and Y contained copigments that shifted the red coloration of anthocyanin A8 to a more bluish coloration.
Fig. 5. Copigments responsible for blue coloration associated with anthocyanin.
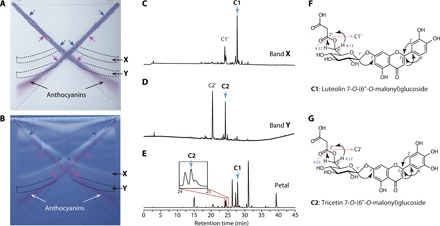
(A and B) Developed TLC plate observed under fluorescent light (A) and UV light (365 nm) (B). Blue arrows indicate blue color parts on the anthocyanin bands overlapping with band X, which showed yellow fluorescence under UV light. Purple arrows indicate violet color parts overlapping with band Y, which was invisible under fluorescent and UV light. Broken lines indicate regions recovered for LC-MS analyses. (C to E) HPLC chromatograms of extracts from the cellulose TLC plate of band X (C) and band Y (D) and of the blue petal extract (E) monitored by absorption at 360 nm for flavonoids. C1 and C1′ were major compounds detected in band X. C2 and C2′ were major compounds detected in band Y. (F and G) Structures of C1 (F) and C2 (G). Arrows indicate heteronuclear multiple-bond coherence. C1′ and C2′ were identified as demalonyl derivatives of C1 and C2, respectively.
We extracted compounds contained in bands X and Y from the cellulose TLC plate. The two major peaks detected in the band X extract were assigned as C1 and C1′, and those in band Y were assigned as C2 and C2′ (Fig. 5, C and D). We analyzed these compounds by LC–PDA–MS analysis: λmax peaks in the UV-visible spectrum were observed at 345, 268, and 236 nm for C1 and at 351, 269, and 235 nm for C2, and their molecular ions ([M + H]+) were determined as m/z = 535 for C1 and 551 for C2 (Fig. 5, C to E, and figs. S4 and S5). We purified these compounds from blue ray florets (Fig. 5E) and identified their structures. C1 and C2 were identified as luteolin 7-O-(6″-O-malonyl)glucoside (21, 22) and tricetin 7-O-(6″-O-malonyl)glucoside, respectively, by high-resolution MS analysis and a series of NMR experiments [1H NMR, 13C NMR, heteronuclear multiple-bond correlation (HMBC), and heteronuclear multiple-quantum coherence (HMQC)] (see Fig. 5, F and G, and Supplementary Materials and Methods). The molecular ions of C1′ and C2′ corresponded to the demalonylated derivatives of C1 and C2, respectively, and thus were identified as luteolin 7-O-glucoside and tricetin 7-O-glucoside, respectively (fig. S5).
In vitro reconstruction of blue flower color
The molar ratio of sum of the 3′,5′-diglucosylated anthocyanins (A7, A8, and A9), C1, and C2 in the ray florets of chrysanthemums was 1:1.7:0.4 on average. The ratio of total flavones to total anthocyanins was approximately 5:1 (table S2). On the basis of the compound ratio, we performed copigmentation experiments in acetate buffer (pH 5.6) to approximate the pH of the ‘Taihei’ chrysanthemum petal juice.
Increasing the concentration of C1 in A8 assay solution led to increased absorption (known as a hyperchromic shift) and a longer wavelength shift (known as a bathochromic shift) of λvismax, with a shoulder peak at around 600 nm (Fig. 6A). On the addition of five or more equivalents of C1 to A8, the color of the buffer solution turned from violet to blue owing to the bathochromic shift (Fig. 6, A and D). The λvismax peak shifted from 560 to 570 nm, and the shoulder region shifted from 590 to around 610 nm, yielding an absorption spectrum similar to that of intact blue petals (Figs. 4G and 6, A and H).
Fig. 6. In vitro reconstruction of petal color by mixing isolated components.
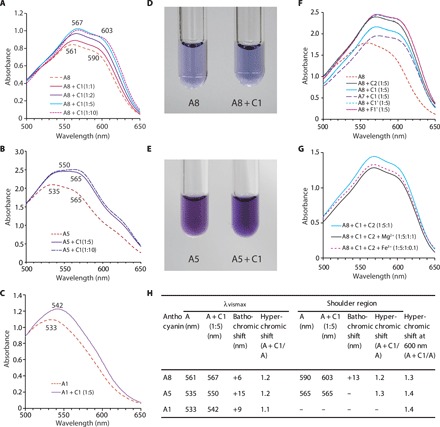
Anthocyanin and copigment were mixed in acetate buffer (pH 5.6). (A) Visible absorption spectra of A8 and its mixture with C1. (B) Visible absorption spectra of A5 and its mixture with C1. (C) Visible absorption spectra of A1 and its mixture with C1. (D) Color of A8 solution (left) and its mixture with C1 (1:10) (right). (E) Color of A5 solution (left) and its mixture with C1 (1:10) (right). (F) Visible absorption spectrum of mixture of A7 or A8 with various flavone glycosides (C1, C2, C1′, and F1′). (G) Visible absorption spectra of a mixture of A8, C1 and C2 solution and its mixture with Fe3+ or Mg2+ ion. (H) Summary of bathochromic shifts and hyperchromic shifts by A8, A5, and A1 solutions by mixing with C1 (1:5). A8, ternatin C5; A5, delphinidin 3-O-(6′′-O-malonyl)glucoside; A1, cyanidin 3-O-(6″-O-malonyl)glucoside; C1, luteolin 7-O-(6″-O-malonyl)glucoside; C2, tricetin 7-O-(6″-O-malonyl)glucoside; C1′, flavone 7-O-glucoside; F1′, apigenin 7-O-glucoside.
Addition of C1 to A5 led to an increase in absorbance at around 565 nm and resulted in the same absorption spectrum as that of intact petals of purple-violet chrysanthemums (Figs. 4G and 6, B and E). Moreover, addition of C1 to A1 also led to an increase in absorbance at around 540 to 550 nm and a longer wavelength shift of the λvismax (Fig. 6C). Overall, we observed longer wavelength shifts of the λvismax and shoulder peaks because of copigmentation following the addition of C1 to A1, A5, and A8. In addition, we confirmed the copigmentation effect of C2, C1′, and apigenin 7-O-glucoside (F1′) on A8 (Fig. 6F).
Because metal ions such as Mg2+ or Fe3+ are often involved in blue flower color development (8, 11, 12), we investigated whether these ions are involved in the blue chrysanthemum coloration. However, we did not observe spectral changes on the addition of Mg2+ or Fe3+ ions to the A8, C1, and C2 mixtures (Fig. 6G).
We observed a bathochromic shift of λvismax and shoulder peaks due to copigmentation with C1 for all tested anthocyanins, whereas the largest shift of λvismax and the shoulder was Δ15 and Δ13 nm for A5 and A8, respectively (Fig. 6H). We observed the effect of copigmentation with C1 for three anthocyanins that have differences in the B-ring structure. Whereas the hyperchromic shift was observed equally among the anthocyanins for the λvismax and shoulder peaks, the bathochromic shift showed a differential effect. The bathochromic shift of λvismax was stronger for A5 than for A8. Thus, the blue coloration of A8 is associated with the bathochromic shift of the shoulder peak of absorption at 590 nm rather than the λvismax peak.
DISCUSSION
Here, we have generated true blue-colored chrysanthemums by introducing only two genes, CamF3′5′H and CtA3′5′GT. Although we also introduced an F3′H RNAi construct to down-regulate the production of red cyanidin derivatives (7, 19), this construct was not essential for the generation of blue flowers. Among all genetically engineered flowers reported to date (9), these transgenic chrysanthemums have produced the most blue-shifted flowers. The blue coloration was dependent on both the coexpression of CamF3′5′H and CtA3′5′GT and the resulting accumulation of 3′,5′-glucosylated delphinidin-based anthocyanins, which do not have aromatic acyl moieties (Fig. 4, A and B). This finding has not been previously reported.
This result was unexpected because it has been previously understood that glycosylation on the 3′- and 5′-hydroxyl groups of anthocyanins is responsible for red coloration in flowers (23–26). For example, 3′,5′-diglucosylated delphinidin galactosides, obtained from berries of the Siberian dogwood Cornus alba ‘Sibirica,’ have absorption spectra with a λvismax peak at a shorter wavelength than their non–3′,5′-glucosylated analogs under acidic conditions (26, 27). Similar results were obtained in this study; under acidic conditions, 3′,5′-glucosylated delphinidin 3-malonylglucoside (A8) had an absorption spectrum having a λvismax peak at a shorter wavelength than both its non–3′,5′-glucosylated analog (A5) and the cyanidin-based analog (A1). Hence, this finding prompted us to examine the mechanisms of blue coloration in the transgenic chrysanthemums.
The coloration of anthocyanins changes under various pH conditions. We therefore analyzed the absorption spectra of the principal accumulated anthocyanins under a weakly acidic condition (pH 5.6), regarded as the pH of the juice from ray floret tissues containing blue-pigmented vacuoles. Under this condition, A8 solution had a λvismax peak at a longer wavelength and showed a bluer coloration than the A5 solution (Figs. 4H and 6). This result indicates that 3′,5′-glucosylation of delphinidin-based anthocyanins produces a more bluish color in planta. Nevertheless, the absorption spectrum of the blue ray florets had λvismax and shoulder peaks at a much longer wavelength than the A8 solution (Fig. 4, G and H). This suggests that, besides the violet coloration under weakly acidic conditions, there is an additional mechanism concerned with blue color development. One assumed mechanism of blue coloration is the intramolecular association of polyacylated anthocyanins, in which a hydrophobic stacking structure is formed between aromatic acyl moieties and the anthocyanidin chromophore (8, 9). However, anthocyanins without aromatic acyl moieties, such as A8, cannot form any such interaction by themselves. We therefore considered the possibility of intermolecular associations of the 3′,5′-diglucosylated anthocyanins with copigments (8, 10).
Shimizu-Yumoto et al. (20) developed the cross-TLC method, which can comprehensively detect copigments in a sample loaded on the TLC plate based on coloration changes in the development line of anthocyanins caused by overlap with development lines of copigments. We therefore applied the cross-TLC method to detect copigments in the extract of the blue ray florets of transgenic chrysanthemums. Consequently, C1 and C2 compounds were detected as copigment candidates (Fig. 5). These candidate copigment compounds were determined to be the major flavone luteolin 7-O-(6″-O-malonyl)glucoside (C1) and the minor tricetin 7-O-(6″-O-malonyl)glucoside (C2). When A8 was added to the C1 solution at pH 5.6, the λvismax and shoulder peaks shifted to a longer wavelength and showed a bluer color than A8 solution without C1. A spectral pattern similar to that of blue ray florets was regenerated when C1 was mixed with A8 at a ratio of 5:1. C1 had a similar copigmentation effect on A7 (Fig. 6F). C2 also had the same effect on A8.
We also detected other major flavones, apigenin 7-O-(6″-O-malonyl)glucoside (F1) and acacetin 7-O-(6″-O-malonyl)glucoside (F2) in the petals (table S2). The demalonyl analog of F1, apigenin 7-O-glucoside, and the demalonyl analog of C1, luteolin 7-O-glucoside, also had a copigmentation effect in vitro (Fig. 6F). Because a malonyl moiety is not involved in copigmentation, F1 is likely to contribute to the blue coloration as a copigment in planta. However, we did not detect a clear copigmentation effect of F1 by the cross-TLC method. In this case, we assumed that the crossing positions of the anthocyanin and F1 overlapped with those of C1 and C2 and showed a violet or blue color (Fig. 5).
The molar ratio of flavones to anthocyanins in ray florets of blue chrysanthemum lines was distributed between 2.2 and 10.4. The content of flavone glucosides as copigments is thought to be sufficient for blue coloration by association with anthocyanin molecules in the ray florets of transgenic chrysanthemums. We believe that caffeoylquinates are not copigments involved in blue pigmentation in planta because they accumulate separately from anthocyanins, which are present in epidermal cells (fig. S6). Metal ions are involved in intermolecular associations leading to blue flower color development in some blue flowers (10–12). We therefore investigated whether the spectrum of the 3′,5′-diglucosylated anthocyanin and flavone mixture was altered by the addition of Fe3+ or Mg2+ ions. However, neither a spectral change nor an effect on blue color development was observed (Fig. 6G). We conclude that the blue flower color of chrysanthemum is mainly due to an intermolecular association between 3′,5′-diglucosylated delphinidins and flavone 7-glucoside derivatives.
Flavone 7-glycoside derivatives have been reported as copigments associated with delphinidin-based anthocyanins or its 3′- and/or 5′-methoxylated derivatives to produce blue or violet coloration in the flower petals of lupin (Lupinus sp.) (28) and torenia (Torenia fournieri) (29). The 3′- and/or 4′-hydroxyflavone 7-glycosides including C1 are major flavonoids in wild-type chrysanthemums, whereas C2, which is further hydroxylated at the 5′ position of the B-ring, is undetectable. C2 is synthesized owing to the expression of the introduced CamF3′5′H gene. We also reconstructed the UV-visible absorption spectrum of purple/violet-colored chrysanthemums and that of the pink-colored chrysanthemums in vitro by mixing C1 and A5 and by mixing C1 and A1, respectively (Figs. 4G and 6, B and C). Flavone 7-glycoside derivatives are also involved as copigments in both the purple/violet and pink coloration of transgenic and wild-type chrysanthemum flowers, respectively.
The bathochromic shift for blue coloration is caused both by the hydroxylation and glucosylation of the anthocyanin B-ring and by copigmentation with flavone glucoside (Fig. 6H). An anthocyanin molecule can alter its structure from a flavylium ion structure showing a red color to a quinonoidal base structure showing a purple color and successively to an anionic quinonoidal base structure showing a blue color (8, 10). Copigments can alter this structural equilibrium through hydrophobic stacking interactions with the anthocyanin molecule. A5 seems to be present as the flavylium ion (λvismax = ca. 530 nm) and the quinonoidal base (λvismax = ca. 570 nm) structures in solution at pH 5.6, whereas A8 seems to be present as the quinonoidal base and/or the anionic quinonoidal base (λvismax = ca. 570 and ca. 610 nm) (Fig. 6). We consider that the coloration and spectral change caused by the addition of copigment C1 are due to a change in this equilibrium: The proportion of the quinonoidal base structure increased for A5, whereas that of the anionic quinonoidal base structure increased for A8.
Previously reported structures of blue flower pigments, such as polyacylated anthocyanins and metalloanthocyanins (8, 9), are too complex to be realized by metabolic engineering because the fine manipulation of multiple transgenes would be necessary. In our case, only two transgenes, namely, CamF3′5′H and CtA3′5′GT, were enough to generate blue chrysanthemums: The 3′,5′-diglucosylated delphinidin exhibits a blue color by intermolecular association with flavone glucosides under the weakly acidic pH conditions of general flower petals. The flavone glycosides are naturally occurring flavonoids in a wide variety of plant species; hence, this two-step modification of anthocyanin structure by B-ring hydroxylation and glucosylation is a highly promising approach to generating blue-colored flowers, which overcomes the problems arising from the introduction and regulation of multiple genes.
MATERIALS AND METHODS
Plant materials
Chrysanthemum morifolium ‘Taihei’ (disbud type), ‘Sei Arabella’ (decorative type), ‘Sei Shawl’ (decorative type), 940-0765 (formerly 94-765; disbud type), T27 (small-flowered pompons type), T37 (small-flowered pompons type), and T57 (decorative type) were used for the host in transformation experiments (table S1). They were cultivated in vitro and maintained as previously described (6).
Plant transformation and cultivation
Binary vectors were introduced into the Agrobacterium tumefaciens strain EHA105 (30), and chrysanthemum leaf explants from in vitro propagation were used for transformation, as previously described (6). Transgenic plants were transferred to soil and grown for 1 month under long-day treatment via exposure to 4-hour night-break illuminations; short-day flowering was then induced by exposure to 11-hour day length in a glasshouse for genetically modified plants. After collection, petals were frozen in liquid nitrogen and stored at −80°C until further analysis.
Construction of transgenes
We constructed the three binary vectors (pB423, pB428, and pB249) in fig. S2 and table S1 in accordance with a previously reported protocol (6, 31), with partial modification as described in Supplementary Materials and Methods. All gene expression cassettes were constructed in the pBI121 vector. Canterbury bells (C. medium) F3′5′H (CamF3′5′H; GenBank accession number FW570877) and butterfly pea (C. ternatea) A3′5′GT (CtA3′5′GT, UGT78K8; GenBank accession number AB115560) were expressed by using the CmF3H promoter (GenBank accession number FW570860), Arabidopsis heat shock protein 18.2 gene terminator (32), Agrobacterium NOS terminator, and NtADH-5′ untranslated region (5′UTR) as a translational enhancer (33). The RNAi cassette for down-regulation of CmF3′H was expressed under the control of the CmF3H promoter and Agrobacterium NOS terminator in pB428. We obtained the RNAi trigger sequence of CmF3′H by PCR amplification and the Gateway LR reaction with the pANDA35K vector (34). The primer sets used in vector construction are listed in table S3.
qRT-PCR analysis
Total RNA was extracted from petals of chrysanthemums by using TRIzol Reagent (Thermo Fisher Scientific) and the PureLink RNA Mini Kit (Thermo Fisher Scientific) and then treated with the PureLink DNase Set (Thermo Fisher Scientific), in accordance with the manufacturer’s instructions. First-strand complementary DNAs (cDNAs) were synthesized by the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) from 1 μg of total RNA in 20 μl of reaction mixture. The resulting cDNA (50 ng) in 20 μl of qRT-PCR mixture with SYBR Premix Ex Taq II (Tli RNase H Plus) (Takara Bio) was subjected to qRT-PCR on Takara Thermal Cycler Dice (TP800), in accordance with the manufacturer’s protocol (Takara Bio). The primers BGT49F (5′-caccttccacctctcttgaacc-3′) and BGT133R (5′-aagtgtgtgtgccgatgaatg-3′) were used for CtA3′5′GT expression analyses. Expression of CamF3′5′H mRNA and actin mRNA as a control was analyzed as previously described (6). Levels of transgene mRNAs were normalized to actin mRNA and were presented relative to transformant 1916-18, which contained 65% of delphinidin-based anthocyanins and the same total anthocyanin content as wild type.
Flower color measurement
The ray floret colors of chrysanthemum flowers were evaluated under fluorescent light with reference to the RHS Colour Charts (5th edition). The L* value for lightness, a* value for redness/greenness, and b* value for yellowness/blueness of CIEL*a*b* color space were measured in at least three replicates by a CD100 spectrum colorimeter (Yokogawa Meters and Instruments). The color hue angle (H°) and C* value indicating chroma were calculated based on the L*a*b* values. We calculated H° values and C* values by the arctangent of b*/a* and the square root of the sum of (a*)2 and (b*)2, respectively. Absorbance spectra of fresh petals of chrysanthemums were analyzed with an integrated sphere attachment ISR-2200 on a Shimadzu UV-2450 spectrophotometer (Shimadzu).
LC-MS analyses
LC-MS analysis was performed by an ACQUITY UPLC system with an ACQUITY TQD tandem mass spectrometer (Waters Japan) using an ACQUITY UPLC BEH C18 column [1.7 μm particle diameter, 2.1 mm inner diameter (i.d.) × 100 mm; Waters] at 35°C. The mobile phase was composed of 1% formic acid containing water (solvent A) and 1% formic acid containing acetonitrile (MeCN) (solvent B). A solvent gradient of 0 to 35% B (0 to 40 min) and 35% B (40 to 45 min) at a flow rate of 0.1 ml min−1 was used. Chromatograms were obtained at 360 nm for acylquinates and flavonoids and 530 nm for anthocyanins; photodiode array spectra were recorded from 200 to 800 nm. The measurement conditions were as follows: positive ion mode; capillary voltage, 3.5 kV; cone voltage, 10 V; source block temperature, 150°C; desolvation temperature, 350°C; flow rate of desolvation gas, 500 liters hour−1; flow rate of cone gas, 50 liters hour−1; collision energy, 20 to 50 V; scan range, 180 to 1080 m/z; detector voltage, 1.7 kV.
Cross-TLC method for copigment detection
A 10% acetic acid extract from blue petals of transgenic chrysanthemums was loaded on a cellulose TLC glass plate (100 × 100 mm; Merck Millipore), as previously described (20). The loaded TLC plate was developed with n-butyl alcohol/acetic acid/water (4:1:2, v/v) at room temperature. After the developed plate had been air-dried, the coloration change and associated compounds were observed on fluorescent light (light box New5000 inverter, Fujicolor) and under UV light (254/365 nm; CSN-15 AC, Cosmo-Bio). The Rf values of anthocyanins on cellulose TLC were as follows: ternatin C5 (A8), 0.15; preternatin C5 (A7), 0.11; cyanidin 3-O-(6″-O-malonyl)glucoside-3′-O-glucoside (A3), 0.30. We recovered cellulose powders containing copigment candidates from the TLC plate by spatula and soaked them with 300 μl of 10% acetic acid containing 40% MeCN solution to reextract the copigments. The extracts were centrifuged and filtered for analysis by LC-MS.
Isolation of anthocyanins and flavones
Frozen petals from blue-colored chrysanthemums (fresh weight, ~2.5 kg) were soaked in ~5.9 liters of 10% formic acid and filtered through a 100-mesh filter. This operation was repeated with 6 liters of 10% formic acid. The combined extract containing flavonoids was centrifuged at 3000 rpm for 10 min, and the supernatant was passed through and adsorbed on a column containing 3 liters of Diaion HP-20 resin (Mitsubishi Kasei). The column was washed with 9 liters of 0.1% formic acid, and anthocyanins and flavonoids were eluted with 0.1% formic acid in methyl alcohol (MeOH). After evaporation and lyophilization, the residue (14.2 g) was dissolved in 0.1% formic acid in 20% MeCN and separated by chromatography on YMC-Pack ODS A (S-15 μm, 50 mm i.d. × 250 mm) in the same solvent, at a flow rate of 30 ml min−1 and with detection at 360 nm. We collected 11 consecutive fractions: Delphinidin 3,3′,5′-tri-O-glucoside (A7; preternatin C5), delphinidin 3-O-(6″-O-malonyl)glucoside-3′,5′-di-O-glucoside (A8; ternatin C5), and cyanidin 3-O-(6″-O-malonyl)glucoside-3′-O-glucoside (A3; Cy3MG3′G) were collected in fraction 1; tricetin 7-O-(6″-O-malonyl)glucoside (C2; Tr7MG) was collected in fraction 2; and luteolin 7-O-(6″-O-malonyl)glucoside (C1; Lt7MG) was collected in fraction 3. Each fraction was evaporated and lyophilized and then dissolved in 0.1% formic acid in 10% MeCN. Fraction 1 was treated by preparative HPLC for rough separation of A3 and A8 using 0.1% formic acid in 35% MeOH at a flow rate of 30 ml min−1. A3 and A8 were finally purified and isolated from these fractions by preparative HPLC using 0.5% formic acid in 20% MeOH, at a flow rate of 28 ml min−1 and with detection at 530 nm. C1 and C2 were isolated from fractions 2 and 3, respectively, by preparative HPLC using 0.1% formic acid in 35% MeOH, at a flow rate of 30 ml min−1 and with detection at 280 nm. The two isolated anthocyanins and two flavone fractions were evaporated and lyophilized; ternatin C5 (A8; 52 mg), Cy3MG3′G (A3; 9.8 mg), Lt7MG (C1; 201.5 mg), and Tr7MG (C2; 52 mg) were obtained as dark reddish or yellowish powders.
High-resolution MS analysis
High-resolution mass spectra of purified anthocyanins and flavones were obtained by an Agilent 1200 LC instrument equipped with electrospray ionization–time-of-flight–MS (JMS-T100LP, JEOL). Samples were dissolved in MeOH (0.01 mg ml−1), and 10- to 20-μl aliquots were analyzed. The measurement conditions were as follows: solvent, MeOH; needle voltage, 2.2 kV; orifice 1 voltage, 85 V; orifice 2 voltage, 10 V; ring lens voltage, 25 V; peaks voltage, 2 kV; flow rate of drying gas, 1 liter min−1; flow rate of nebulizing gas, 0.5 liter min−1; desolvating temperature, 250°C; orifice 1 temperature, 80°C. The high-resolution MS data are provided in Supplementary Materials and Methods.
NMR experiments of anthocyanins and copigments
Anthocyanins A8 and A3 were purified from petals of blue-colored transgenic chrysanthemums. A8 and A3 were dissolved in 10% (v/v) trifluoroacetic acid (TFA)–CD3OD mixture and TFA–dimethyl sulfoxide (DMSO)–d6 mixture, respectively. The purified copigments C1 and C2 from petals of blue-colored transgenic chrysanthemums were dissolved in DMSO-d6. 1H NMR and NOESY spectra of anthocyanins and 1H NMR, 13C NMR, HMBC, HMQC, and homonuclear correlation spectroscopy spectra of copigments were obtained on a JNM-ECZ600R/S1 spectrometer (JEOL). The observation frequency for 1H and 13C measurement was 600.17 and 150.91 MHz, respectively. The residual proton peak of CD3OD was used as the internal standard (3.3 parts per million). The NMR data are provided in Supplementary Materials and Methods.
Petal color reconstruction experiments
The petal color reconstruction experiments were conducted in acetate buffer at pH 5.6, the average pH value of petals in homogenate measurements. Petals (~5 g) of chrysanthemums were ground in liquid nitrogen and soaked in 15 ml of distilled water. The petal juice was measured immediately with a D-71 pH meter equipped with a 9611-10D pH electrode (Horiba). The concentrations (nanomoles per milligram of petals) of anthocyanins and flavones in petals were calculated, with reference to standard graphs of delphinidin 3-glucoside (Funakoshi) and luteolin (Funakoshi), from the HPLC peak area at 530 and 360 nm, respectively. The flavones—luteolin 7-O-glucoside (C1′), apigenin 7-O-glucoside (F1′), C1, and C2—were dissolved in DMSO. C1′ and F1′ were purchased from Funakoshi. The anthocyanins—preternatin C5 (A7), ternatin C5 (A8), A3, delphinidin 3-O-(6″-O-malonyl)glucoside (Dp3MG; A5) (35), and cyanidin 3-O-(6″-O-malonyl)glucoside (Cy3MG, A1) (15, 36)—were dissolved in distilled water. A5 was purified from petals of a butterfly pea mauve line (35). A1 was synthesized and purified from the reaction product of cyanidin 3-glucoside (Funakoshi) and malonyl coenzyme A (Sigma-Aldrich) by using crude enzyme extract from petals of butterfly pea (36).
For petal color reconstruction in vitro, buffer solution [96 to 78 μl of 100 mM acetate buffer (pH 5.6)] and flavone (2 to 20 μl of 10 mM flavone in DMSO) were added to a 1.5-ml microcentrifuge tube. Next, anthocyanin (2 μl of 10 mM anthocyanin aqueous solution) was added to the mixture in a total volume of 100 μl, and the absorption spectrum was recorded in a super-micro black cell (path length, 10 mm; Shimadzu) by using a UV-2450 spectrophotometer (Shimadzu). Spectra were also recorded in the presence of metal ions. Divalent magnesium salt, Mg(OAc)2, was dissolved at 10 mM in distilled water, and 2 μl were added to the mixture. Trivalent ion salt, FeNH4(SO4)2, was dissolved at 1 mM in distilled water, and 2 μl were added to the mixture.
Supplementary Material
Acknowledgments
We thank Inochio Seikoen Ltd. for providing breeding lines and cultivars of chrysanthemums, Aomori Prefectural Industrial Technology Research Center for plasmids containing CmF3Hp and CtA3′5′GT, K. Kato [Nara Institute of Science and Technology (NAIST)] for providing NtADH-5′UTR and AtHSPt, K. Kazuma (Toyama University) for providing anthocyanins (Dp3MG, Cy3MG, and preternatin C5), K. Shimamoto (NAIST) for providing pANDA vectors for RNAi, Institute of Fruit Tree and Tea Science, National Agriculture and Food Research Organization (NARO) for letting us use the glasshouse with insect-proof netting for cultivation of genetically modified plants, and N. Tanikawa (Institute of Vegetable and Floriculture Science, NARO) for discussion on anthocyanin identification experiments. We also thank K. Shimizu, M. Takahashi, Y. Taniji, and Y. Kashiwagi for technical assistance with producing transgenic chrysanthemums and analysis. Funding: This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (Grants-in-Aid for Scientific Research) grant numbers JP21780033 (Grant-in-Aid for Young Scientists B) and JP24658035 (Grant-in-Aid for Challenging Exploratory Research) (to N.N.). Author contributions: N.N., M.N., and R.A. contributed to the planning and writing of the manuscript. N.N. and Y.T. carried out gene isolation and constructed binary vectors. S.Y., R.A., S.K., and N.N. conducted tissue culture and transformation experiments. N.N., M.D., S.Y., and S.K. cultivated the transgenic plants. N.N. conducted flavonoid and molecular biological analyses of transgenic plants. N.N., M.D., and M.N. purified flavonoids and carried out the copigmentation experiment. N.N., Y.T., and R.A. designed and supervised experiments collaboratively with all co-authors. Competing interests: N.N., S.Y., S.K., M.N., M.D., and R.A. are employees of NARO, a nonprofit research organization, and performed this research mainly from a scientific perspective. Y.T. is an employee of a group company of Suntory Holdings Ltd. for scientific research and technology development and performed this research mainly from a commercial perspective. N.N., S.Y., S.K., Y.T., and R.A. are inventors on a patent related to this work held by NARO and Suntory Holdings Ltd. (international publication no. WO/2017/002945; published on 5 January 2017). N.N., M.N., M.D., S.Y., and R.A. are inventors on a separate patent application related to this work filed by NARO and Suntory Holdings Ltd. (application no. PCT/JP2017/010036; filed on 13 March 2017). Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The sequences for all the genes described in this paper have been submitted to the National Center for Biotechnology Information database with the following accession numbers: FW570860 (chrysanthemum F3H promoter), AB115560 (butterfly pea CtA3′5′GT), LC222467 (sequence of CamF3′5′H cassette and CtA3′5′GT cassette in pB423), and LC222468 (sequence of CmF3′H RNAi cassette in pB428).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/7/e1602785/DC1
Supplementary Materials and Methods
fig. S1. Structures of anthocyanins.
fig. S2. Schematic representation of binary vectors used for chrysanthemum transformation.
fig. S3. Anthocyanin profiles of representative ‘Sei Arabella’ transgenic lines.
fig. S4. Flavonoids and caffeoylquinates profiles of representative ‘Sei Arabella’ transgenic lines.
fig. S5. LC-MS analysis of copigment extract from a cellulose TLC plate.
fig. S6. Flavonoids and caffeoylquinates profiles of whole and epidermal petal tissue from a blue transgenic line.
table S1. Flower color changes due to expression of CamF3′5′H and/or CtA3′5′GT.
table S2. Anthocyanin and flavone content and their ratios in petals of ‘Sei Arabella’ and blue/violet-blue–colored transgenic chrysanthemums.
table S3. Primer sets used in binary vector construction.
REFERENCES AND NOTES
- 1.Holton T. A., Brugliera F., Lester D. R., Tanaka Y., Hyland C. D., Menting J. G. T., Lu C.-Y., Farcy E., Stevenson T. W., Cornish E. C., Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366, 276–279 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Fukui Y., Tanaka Y., Kusumi T., Iwashita T., Nomoto K., A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′,5′-hydroxylase gene. Phytochemistry 63, 15–23 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Y. Tanaka, F. Brugliera, in Flowering and its Manipulation, vol. 20 of Annual Plant Reviews, C. Ainsworth, Ed. (Blackwell, 2006), chap. 9. [Google Scholar]
- 4.Katsumoto Y., Fukuchi-Mizutani M., Fukui Y., Brugliera F., Holton T. A., Karan M., Nakamura N., Yonekura-Sakakibara K., Togami J., Pigeaire A., Tao G.-Q., Nehra N. S., Lu C.-Y., Dyson B. K., Tsuda S., Ashikari T., Kusumi T., Mason J. G., Tanaka Y., Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48, 1589–1600 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Tanaka Y., Brugliera F., Flower colour and cytochromes P450. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noda N., Aida R., Kishimoto S., Ishiguro K., Fukuchi-Mizutani M., Tanaka Y., Ohmiya A., Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol. 54, 1684–1695 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Brugliera F., Tao G.-Q., Tems U., Kalc G., Mouradova E., Price K., Stevenson K., Nakamura N., Stacey I., Katsumoto Y., Tanaka Y., Mason J. G., Violet/blue chrysanthemums—Metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors. Plant Cell Physiol. 54, 1696–1710 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Yoshida K., Mori M., Kondo T., Blue flower color development by anthocyanins: From chemical structure to cell physiology. Nat. Prod. Rep. 26, 884–915 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Sasaki N., Nakayama T., Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration. Plant Cell Physiol. 56, 28–40 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Trouillas P., Sancho-García J. C., De Freitas V., Gierschner J., Otyepka M., Dangles O., Stabilizing and modulating color by copigmentation: Insights from theory and experiment. Chem. Rev. 116, 4937–4982 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Kondo T., Yoshida K., Nakagawa A., Kawai T., Tamura H., Goto T., Structural basis of blue-colour development in flower petals from Commelina communis. Nature 358, 515–518 (1992). [Google Scholar]
- 12.Shiono M., Matsugaki N., Takeda K., Phytochemistry: Structure of the blue cornflower pigment. Nature 436, 791 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K., Kondo T., Okazaki Y., Katou K., Cause of blue petal colour. Nature 373, 291 (1995). [Google Scholar]
- 14.Fukuda-Tanaka S., Inagaki Y., Yamaguchi T., Saito N., Iida S., Colour-enhancing protein in blue petals. Nature 407, 581 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Saito N., Toki K., Honda T., Kawase K., Cyanidin 3-malonylglucuronylglucoside in Bellis and cyanidin 3-malonylglucoside in Dendranthema. Phytochemistry 27, 2963–2966 (1988). [Google Scholar]
- 16.Nakayama M., Koshioka M., Shibata M., Hiradate S., Sugie H., Yamaguchi M.-a., Identification of cyanidin 3-O-(3″,6″-O-dimalonyl-β-glucopyranoside) as a flower pigment of Chrysanthemum (Dendranthema grandiflorum). Biosci. Biotechnol. Biochem. 61, 1607–1608 (1997). [Google Scholar]
- 17.Terahara N., Toki K., Saito N., Honda T., Matsui T., Osajima Y., Eight new anthocyanins, ternatins C1–C5 and D3 and preternatins A3 and C4 from young Clitoria ternatea flowers. J. Nat. Prod. 61, 1361–1367 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Kogawa K., Kato N., Kazuma K., Noda N., Suzuki M., Purification and characterization of UDP-glucose: Anthocyanin 3′,5′-O-glucosyltransferase from Clitoria ternatea. Planta 226, 1501–1509 (2007). [DOI] [PubMed] [Google Scholar]
- 19.He H., Ke H., Keting H., Qiaoyan X., Silan D., Flower colour modification of chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLOS ONE 8, e74395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu-Yumoto H., Hayashi N., Ichimura K., Nakayama M., Slantingly cross loading sample system enables simultaneous performance of separation and mixture to detect molecular interactions on thin-layer chromatography. J. Chromatogr. A 1245, 183–189 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Schwinn K. E., Markham K. R., Giveno N. K., Floral flavonoids and the potential for pelargonidin biosynthesis in commercial chrysanthemum cultivars. Phytochemistry 35, 145–150 (1993). [Google Scholar]
- 22.Sugawara T., Igarashi K., Identification of major flavonoids in petals of edible chrysanthemum flowers and their suppressive effect on carbon tetrachloride-induced liver injury in mice. Food Sci Technol. Res. 15, 499–506 (2009). [Google Scholar]
- 23.Saito N., Harborne J. B., A cyanidin glycoside giving scarlet coloration in plants of the Bromeliaceae. Phytochemistry 22, 1735–1740 (1983). [Google Scholar]
- 24.J. B. Harborne, R. J. Grayer, in The Flavonoids: Advances in Research Since 1980, J. B. Harborne, Ed. (Springer, 1988), chap. 1. [Google Scholar]
- 25.Ø. M. Andersen, M. Jordheim, in Flavonoids: Chemistry, Biochemistry and Applications, Ø. M. Andersen, K. R. Markham, Eds. (Taylor & Francis, 2006), chap. 10. [Google Scholar]
- 26.Ø. M. Andersen, M. Jordheim, in Comprehensive Natural Products II: Chemistry and Biology, L. Mander, H.-W. Liu, Eds. (Elsevier Science, 2010), chap. 3.16. [Google Scholar]
- 27.Bjorøy Ø., Fossen T., Andersen Ø. M., Anthocyanin 3-galactosides from Cornus alba ‘Sibirica’ with glucosidation of the B-ring. Phytochemistry 68, 640–645 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Takeda K., Harborne J. B., Waterman P. G., Malonylated flavonoids and blue flower colour in lupin. Phytochemistry 34, 421–423 (1993). [Google Scholar]
- 29.Aida R., Yoshida K., Kondo T., Kishimoto S., Shibata M., Copigmentation gives bluer flowers on transgenic torenia plants with the antisense dihydroflavonol-4-reductase gene. Plant Sci. 160, 49–56 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Hood E. E., Gelvin S. B., Melchers L. S., Hoekema A., New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218 (1993). [Google Scholar]
- 31.Kuroda M., Kimizu M., Mikami C., A simple set of plasmids for the production of transgenic plants. Biosci. Biotechnol. Biochem. 74, 2348–2351 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Nagaya S., Kawamura K., Shinmyo A., Kato K., The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51, 328–332 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Satoh J., Kato K., Shinmyo A., The 5′-untranslated region of the tobacco alcohol dehydrogenase gene functions as an effective translational enhancer in plant. J. Biosci. Bioeng. 98, 1–8 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Miki D., Shimamoto K., Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Kazuma K., Noda N., Suzuki M., Flavonoid composition related to petal color in different lines of Clitoria ternatea. Phytochemistry 64, 1133–1139 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Kogawa K., Kazuma K., Koto N., Noda N., Suzuki M., Biosynthesis of malonylated flavonoid glycosides on the basis of malonyltransferase activity in the petals of Clitoria ternatea. J. Plant Physiol. 164, 886–894 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/7/e1602785/DC1
Supplementary Materials and Methods
fig. S1. Structures of anthocyanins.
fig. S2. Schematic representation of binary vectors used for chrysanthemum transformation.
fig. S3. Anthocyanin profiles of representative ‘Sei Arabella’ transgenic lines.
fig. S4. Flavonoids and caffeoylquinates profiles of representative ‘Sei Arabella’ transgenic lines.
fig. S5. LC-MS analysis of copigment extract from a cellulose TLC plate.
fig. S6. Flavonoids and caffeoylquinates profiles of whole and epidermal petal tissue from a blue transgenic line.
table S1. Flower color changes due to expression of CamF3′5′H and/or CtA3′5′GT.
table S2. Anthocyanin and flavone content and their ratios in petals of ‘Sei Arabella’ and blue/violet-blue–colored transgenic chrysanthemums.
table S3. Primer sets used in binary vector construction.


