Abstract
Testicular necrosis is a sensitive endpoint for cadmium (Cd2+, Cd) toxicity across all species tested. Resistance to Cd-induced testicular damage is a recessive trait assigned to the Cdm locus on mouse chromosome 3. We first narrowed the Cdm-gene-containing region to 880 kb. SNP analysis of this region from two sensitive and two resistant inbred strains demonstrated a 400-kb haplotype block consistent with the Cd-induced toxicity phenotype; in this region is the Slc39a8 gene encoding a member of the solute-carrier superfamily. Slc39a8 encodes SLC39A8 (ZIP8), whose homologs in plant and yeast are putative zinc transporters. We show here that ZRT-, IRT-like protein (ZIP)8 expression in cultured mouse fetal fibroblasts leads to a >10-fold increase in the rate of intracellular Cd influx and accumulation and 30-fold increase in sensitivity to Cd-induced cell death. The complete ZIP8 mRNA and intron-exon splice junctions have no nucleotide differences between two sensitive and two resistant strains of mice; by using situ hybridization, we found that ZIP8 mRNA is prominent in the vascular endothelial cells of the testis of the sensitive strains of mice but absent in these cells of resistant strains. Slc39a8 is therefore the Cdm gene, defining sensitivity to Cd toxicity specifically in vascular endothelial cells of the testis.
Keywords: metal influx, vascular endothelial cells, solute carrier gene superfamily, in situ hybridization
Cd is a toxic and carcinogenic nonessential metal (1), which can enter the body through the intestine, skin, and lung and accumulates in the kidney (1–3). The level of Cd in the environment has risen with advances in industrialization, and the role of Cd in human disease is of increasing concern. The mechanisms of Cd toxicity are poorly understood, although it is known that Cd exerts its effects intracellularly, and there are polypeptides such as metallothionein (4) and reduced glutathione (5) that bind Cd and afford protection. The subcellular events by which Cd is taken up by cells or removed from cells remain obscure, although such knowledge could provide potential therapeutic targets for protection or intervention against Cd toxicity. Several proteins transport Cd into bacteria, yeast, plants, and mammalian cells in culture (6–11), but their specific roles in causing toxicity are unclear; these studies underscore the difficulties in extrapolating from observations in cell culture to the intact animal.
Nature has provided a fascinating genetic system as a foothold into identifying a gene involved in Cd toxicity. It is known that Cd-induced testicular necrosis is common across all animal species having testes: rodents, opossum, armadillos, frogs, pigeons, roosters, and fish (12–17). Cellular events that precede Cd-induced testicular toxicity indicate that vascular endothelial cell injury is the earliest and, perhaps, the causative event (16, 18–24).
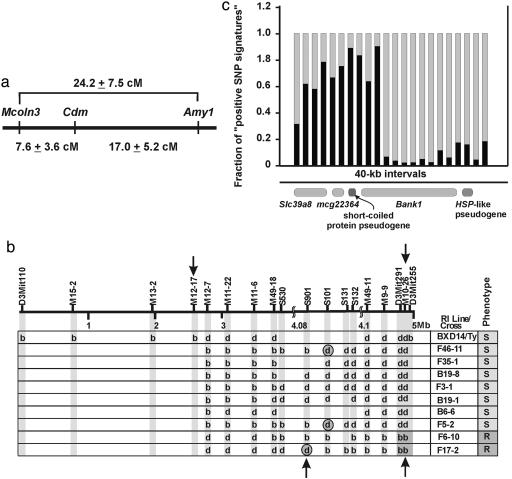
Some inbred mouse strains are resistant to Cd-induced testicular toxicity (25). The resistance phenotype segregates largely as an autosomal-recessive Mendelian trait, and the gene responsible for the trait was named Cdm (26). The wild-type Cdm allele thus confers testicular sensitivity to Cd. The Cdm gene was mapped to a 24-centiMorgan (cM) segment (27) (Fig. 1a) between amylase-1 (Amy1) and varitint-waddler (Va) on mouse Chr 3 (30). Phenotyping 26 BXD/Ty recombinant inbred lines and using quantitative histology to assess testicular necrosis, our lab refined the Cdm gene locus from >24 cM to 0.64 cM (31). In this study, we now have identified the Cdm gene as a member of the solute carrier Slc39 gene family.
Fig. 1.
Scheme showing how the Cdm-gene-containing region was refined from >24 cM on mouse Chr 3 to 4.96 Mb, 2.37 mM, and 880 kb containing three putatively functional genes. (a) Genetic map originally generated by Taylor et al. (26). Varitint waddler (28) has now become the mucolipin 3 gene (Mcoln3) (29). (b) Phenotype-genotype association studies with the recombinant inbred line BXD14/Ty (b, B6; d, D2 allele) showing that a double crossover occurred between M12-7 and M10-26 (arrows above) and then nine recombinants derived from the (B6D2)F1 × B6 backcross further refined the Cdm locus to a region between S901 and M10-26 (arrows below). Gray circled genotypes are recombinants that ultimately define the 880-kb segment containing Cdm. (c) SNP analysis over the 880-kb segment showing “the fraction of positive SNP signatures” occurring in 40-kb intervals. “Positive” denotes those SNPs in D2 and 129S6 (Cd-sensitive strains) that differ from that in B6 and A/J (Cd-resistant strains) divided by the total number of SNPs in 40-kb intervals.
Materials and Methods
Animals. All mouse experiments were conducted in accordance with the National Institute of Health standards for the care and use of experimental animals and the University of Cincinnati Medical Center Institutional Animal Care and Use Committee. C57BL/6J (B6), DBA/2J (D2), 129S6/SvEvTac (129S6), and A/J inbred strains and the BXD14/Ty recombinant inbred line were purchased from The Jackson Laboratory.
Mapping the Cdm Locus. DNA from the above-described mice was prepared by using standard methods. Potential polymorphic microsatellite markers were identified by PCR by choosing primer sets adjacent to d(CA) repeats, generally >20 repeats in length. PCR was conducted, and amplicons from B6 DNA were compared with those from D2 DNA. PCR products showing >5% difference by agarose gel electrophoresis were chosen for analysis. Recently discovered microsatellite markers and SNP sites (Fig. 1b) were amplified by using primer sets shown in Table 1, which is published as supporting information on the PNAS web site.
For fine-mapping the chromosomal crossover positions in B6D2F1 × B6 backcross offspring, we used several SNPs. The positions of these SNPs are shown in the context of 20 bp of contiguous sequence in Table 2, which is published as supporting information on the PNAS web site. SNPs analysis was conducted by using SNPs documented in the Celera Mouse Genome Database (www.celeradiscoverysystem.com).
Treatment of the Mice. Testicular sensitivity to Cd was assessed as described in ref. 31.
Cloning of the Solute-Carrier (SLC)39A8 (ZIP8) cDNA. Oligo-dT-primed reverse transcription was carried out on B6 and D2 mouse total testicular RNA. Primers for amplification began at the start codon and ended at the stop codon; a consensus Kozak sequence at the start site was included for efficient expression. Restriction sites were added at the 5′ (BamHI) and 3′ (ClaI) ends for cloning into the pRevTRE vector (Invitrogen). A mutant ZRT-, IRT-like protein (ZIP)8 (ZIP8m) was generated by using a 5′ primer with a single-base deletion in the third codon of the ZIP8 cDNA. ZIP8 with a C-terminal hemagglutinin (HA) tag was generated (ZIP8ha) by using PCR and a 3′ primer in which the termination codon was replaced with the coding sequence of an HA tag, followed by a termination codon.
Delivery of ZIP8 cDNA into Mouse Fetal Fibroblasts Tet-Off Cells. The cDNAs described below were inserted into the pRevTet-off vector (modified from the Invitrogen vector by replacing G418 resistance with puromycin resistance; ref. 32), which were used to generate retrovirus and infect immortalized mouse fetal fibroblasts (33) that express a Tet-off Tet receptor. Cells were selected for resistance to puromycin (3 μg/ml). Cells were infected with retrovirus (rv)-encoding control luciferase (LUC), ZIP8, ZIP8m, or ZIP8ha to generate the rvLUC, rvZIP8, rvZIP8m, and ZIP8ha cell lines, respectively.
Northern Blot Hybridization. ZIP8 mRNA levels were measured by standard analysis in ref. 34.
Western Immunoblot Analysis. The rvZIP8ha cells and rvLUC control cells were harvested in PBS and homogenized (100 strokes with a tight-fitting Dounce homogenizer) in 10 mM Tris, pH 7.4/10 mM KCl9491 mM EDTA containing phenylmethylsulfonyl fluoride, leupeptin, and aprotinin. Homogenates were centrifuged at 500 × g for 10 min and then 20,000 × g for 20 min. This supernatant was centrifuged at 100,000 × g for 30 min to generate a soluble cytosolic fraction, and the pellet was suspended by pipetting in homogenization buffer to generate a membrane fraction. Proteins were quantified by using the BCA protein assay (Pierce). Proteins were denatured, run on SDS/PAGE gels, transferred to nitrocellulose, and blotted as described in ref. 34. A rabbit affinity-purified polyvalent anti-HA antibody (Bethyl Laboratories; Montgomery, TX) was used at 1/10,000 dilution.
Immunohistochemical Analysis. The rvZIP8ha and rvLUC control cells were grown on fibronectin-coated cover slips, fixed, blocked, and then reacted with the anti-HA antibody (described above; 1/2,000 dilution) and secondary FITC-conjugated goat anti-rabbit antibody (Alexa Fluor 488, Molecular Probes) as described in ref. 35. Images were recorded by confocal microscopy (Zeiss LSM510).
Measuring Cd Uptake. Cd uptake was performed by control cell lines with 109Cd [3.64 mCi/mg (1 Ci = 37 GBq) in 0.5 M HCl; NEZ058; PerkinElmer] as described in ref. 36.
Measuring Cd Toxicity. Cd toxicity was assessed in cell lines after a 32-h treatment with the indicated Cd concentration. Cell viability was determined as the cleavage of 3-(4,5-dimethlythiazol-2-yl)-2,5-diphenyl tetrasodium bromide or lactate dehydrogenase release, both according to the manufacturer's protocol (Promega).
In Situ Hybridization. Templates for cRNA probes included a portion of the ZIP domain unique to ZIP8 and were generated by PCR with the primers: P081, 5′-AATTAACCCTCACTAAAGGGGGATCCGCTATGCCAACCCCGCTG-3′; and P082, 5′-GTAATACGACTCACTATAGGGCATCGATGCAAGATCACAAAGTCCCCT-3′.
PCR products contained a 5′ T3 polymerase promoter and 3′ T7 polymerase promoter for generation of sense and antisense probes, respectively. We prepared the 35S-labeled single-stranded RNA probes (2 × 109 cpm/μg) by using uridine 5′-(α-[35S]thio)triphosphate (800 Ci/mmol) and T3 and T7 polymerases. Tissues were fixed, sectioned, and hybridized as described in ref. 37. After light photomicroscopy, coverslips were removed with xylene; residual emulsion was digested with 1% sodium hydroxide. After stepwise washes (water, Kodac fixer, water, and PBS), indirect immunofluorescence was performed by using anti-CD31 (Pharmingen), a biotinylated secondary antibody, and a fluorophore-tagged label (Alexa Fluor-488 streptavidin; catalog no. S11223).
Statistical Analysis. Statistical significance between groups was determined by way of analysis of variance for means and SD (95% confidence intervals) between each group by statistical linear models and Student's t test. All assays were performed in duplicate or triplicate and repeated at least twice. Statistical analyses were performed with the use of sas statistical software (SAS Institute, Cary, NC). The determinations of Km and Vmax values for ZIP8 and ED50 values for Cd toxicity were determined by using sigma plot (developed by Jandel Scientific; purchased by SPSS, Chicago; and sold by Systat Software, Point Richmond, CA).
Results
Refinement from 4.96 Mb to 880 kb. The 0.64-cM Cdm-gene-containing region was determined (31) to be the 4.96-Mb segment between D3Mit110 and D3Mit255 (Fig. 1b). By using newly discovered polymorphic microsatellite markers, this region was decreased further to a 2.37-Mb segment defined by markers M12–17 and M10–26, in which D2 (sensitive) markers are flanked by B6 (resistant) markers in the Cd-sensitive BXD14/Ty recombinant inbred line. DNA from 1,164 B6D2F1 × B6 backcross offspring was then analyzed by multiplex PCR, and nine informative previously uncharacterized recombinants further narrowed the Cdm-containing region to 880 kb, residing between S901 and M10–26 (Fig. 1b). This 880-kb segment contains two pseudogenes and three putatively functional genes: Bank1, encoding a B cell scaffold protein and signaling molecule with ankyrin repeats (38); mcg22364, having no known function; and Slc39a8, a member of the SLC39 family (39) of the solute-carrier gene superfamily (40). We resequenced all exons (including 5′ and 3′ untranslated regions) and all intron-exon splice junctions (at least 30 bp) of these three genes, comparing two Cd-sensitive inbred strains (D2 and 129S6) with two Cd-resistant inbred strains (B6 and A/J). Although only two nonsynonymous-coding SNPs in the Bank1 gene were detected, neither was in accord with the strain distribution pattern for Cd-induced testicular toxicity and, thus, neither is a candidate for the Cd toxicity trait.
Analysis of all SNPs (from the Celera Mouse Genome Database) in the two Cd-sensitive and two Cd-resistant inbred strains between S901 and M10–26 revealed a ≈400-kb haplotype block predictive of phenotype (Fig. 1c), suggesting an ancestral relationship between the sensitive and resistant strains, and further refined the Cdm-gene-containing region; also, this analysis eliminated Bank1 as a candidate gene.
Two genes thus remained as Cdm candidates. The mcg22364 gene encodes an 87-residue protein, based on its longest ORF. Neither the gene nor its protein shares homology with any other gene or protein in the National Center for Biotechnology Information database. This gene is hypothetical, based on only two ESTs.
Association of Cd Toxicity with the SLC39A8 Transporter. The remaining gene, Slc39a8, is one of 14 members so far identified in the mouse Slc39 family of metal-ion transporters; 15 SLC39 genes exist in the human genome (39). SLC39 genes are members of the ZIP family, best known for ZRT1 and ZRT2, the major Zn2+ uptake transporters in Saccharomyces cerevisiae, and IRT1, the major iron transporter in Arabidopsis thaliana. ZIP proteins have been described in ref. 39 as transporters of Zn2+, Fe2+, and Mn2+. In general, ZIP proteins transport metal ions from outside the cell, or they are transported from intracellular organelles into the cytoplasm.
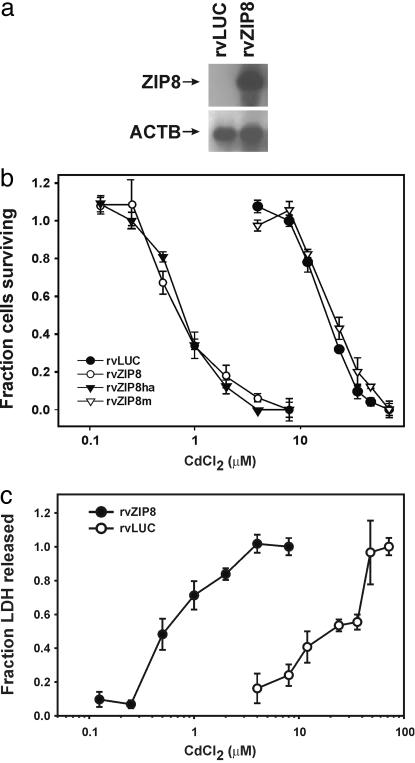
We found that ZIP8 mRNA is expressed in rvZIP8 cells but not in the control rvLUC cells (Fig. 2a). Cd was toxic to the rvZIP8m cells and rvLUC cells, both negative controls, with an ED50 for cell death of ≈22 μM, as seen by both the 3-(4,5-dimethlythiazol-2-yl)-2,5-diphenyl tetrasodium bromide (Fig. 2b) and lactate dehydrogenase (Fig. 2c) assays. The same was true for the parent mouse fetal fibroblast cell line (data not shown). On the other hand, rvZIP8 cells and rvZIP8ha cells were sensitized to Cd, with an ED50 of 0.69 μM. We conclude that ZIP8 is a putative metal transporter whose expression sensitizes cells to Cd toxicity.
Fig. 2.
Cd toxicity in mouse fetal fibroblasts that express ZIP8 cDNA. (a) Northern blot of ZIP8 mRNA in rvZIP8 cells or rvLUC control cells. Total RNA was size-separated, and the blots were hybridized with cRNA probes for the indicated mRNA. ACTB, β-actin mRNA as a control for lane loading. (b and c) Dose–response curves for Cd-induced cell death. Cells were treated for 32 h with the indicated concentration of CdCl2 and cell death monitored by using b, the 3-(4,5-dimethlythiazol-2-yl)-2,5-diphenyl tetrasodium bromide assay, or c, the lactate dehydrogenase release assay. rvZIP8ha cells contain the HA-tagged ZIP8 protein. rvZIP8m cells contain a mutated ZIP8 cDNA. Data represent means ± SD of triplicate determinations.
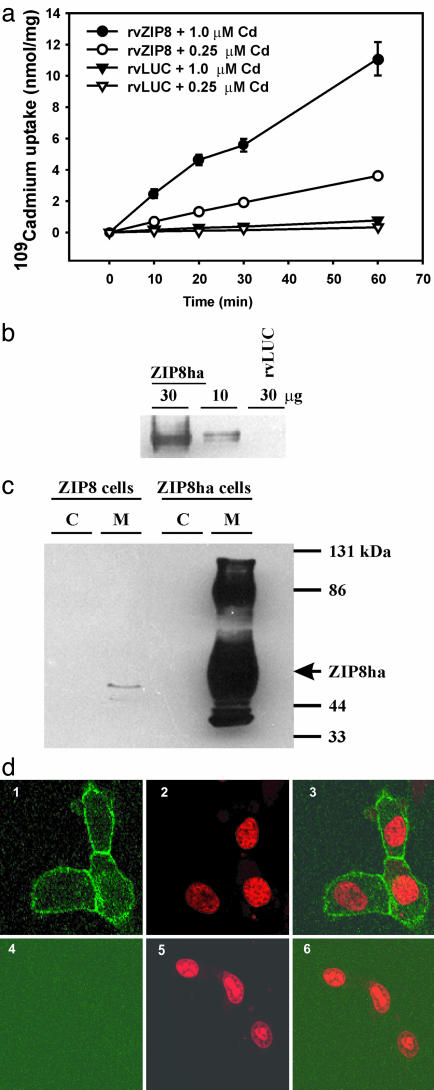
Membrane-Associated ZIP8 Stimulates Cd Uptake. The rate of intracellular Cd uptake was linear for at least 1 h with <25% of Cd internalized. At 0.25 and 1 μM Cd, rvZIP8 cells accumulated Cd at 60 and 180 pmol/min per mg protein, respectively (Fig. 3a), compared with 5.3 and 12.4 pmol/min per mg protein, respectively, in rvLUC control cells. Because of the striking increase in Cd uptake by ZIP8, we determined that Cd transport by ZIP8 is temperature-sensitive and saturable (data not shown). The apparent Km for ZIP8-mediated transport of Cd is 8.4 ± 0.84 μM with a Vmax of 204 ± 6.6 pmol/min per mg protein. These studies were conducted in the presence of 10% FBS, meaning that transport can be directly compared to toxicity studies; serum constituents are known to bind Cd such that its apparent Km is likely to be a considerable overestimation of the actual Km value.
Fig. 3.
Increased Cd uptake caused by membrane-localized ZIP8. (a) Time/dose-dependent 109CdCl2 uptake into rvZIP8 or rvLUC cells. (b) Western blot of ZIP8ha in microsomes (30 vs. 10 μg per lane). (c) Western blot of ZIP8ha in cytosol (C) or microsomes (M) (30 μg per lane) from rvLUC or rvZIP8ha cells. Arrow denotes band at 55 kDa. (d) Localization of the ZIP8ha protein. rvZIP8ha cells (Upper) or rvLUC cells (Lower) were fixed and incubated with a primary anti-HA antibody and a secondary goat anti-rabbit FITC-conjugated antibody. Cells were counterstained with propidium iodide (PI) to visualize nuclei. Confocal fluorescent microscopy detected FITC (Left), PI (Center), or both FITC and PI (Right).
Is ZIP8 located in the plasma membrane? Western blot analysis of rvZIP8ha cells showed that ZIP8 is detected as a tightly running doublet, migrating at ≈55 kDa (Fig. 3b). After overexposure, the ZIP8ha protein was seen in the membranes but not detected in cytosol (Fig. 3c). In addition to the abundant 55-kDa form of ZIP8ha, a slower-migrating 86-kDa form is detected; the nature of this band is not known. We had used rvZIP8ha cells to determine the cellular localization of ZIP8; the rvZIP8ha cells are sensitized to Cd to the same degree as rvZIP8 cells (Fig. 2b), suggesting that addition of the C-terminal HA tag does not affect transporter function. ZIP8ha is strictly associated with the plasma membrane (Fig. 3d). When rvZIP8ha cells were made permeable by using detergents, ZIP8ha was detected in membranes throughout the cell (data not shown).
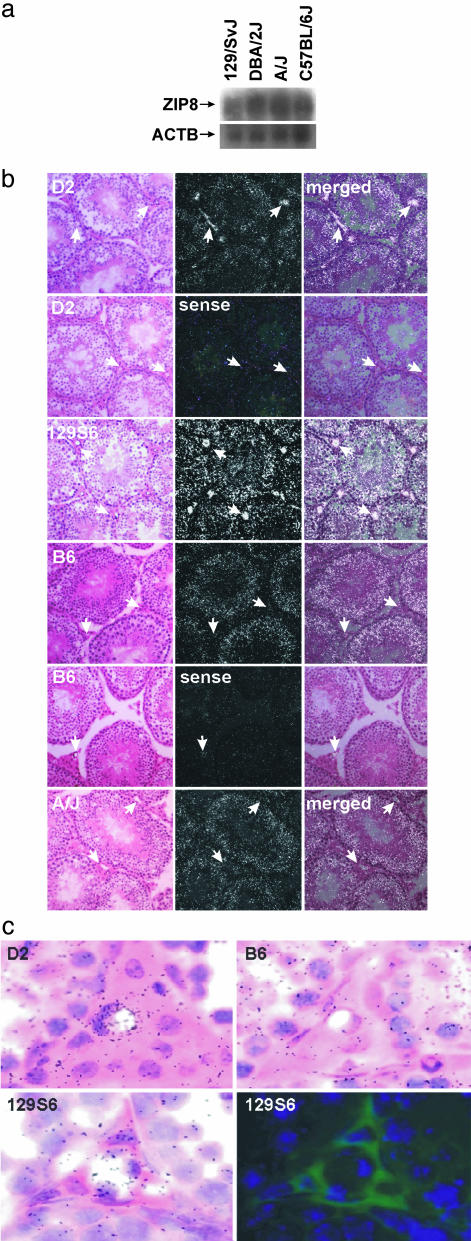
ZIP8 mRNA and in Situ Hybridization Analysis. All Slc39a8 exons and splice junctions revealed no differences between two sensitive (D2 and 129S6) and two resistant (B6 and A/J) mouse strains; this finding suggests that a strain-specific difference exists in the testicular accumulation of ZIP8 mRNA, rather than a mutated or absent ZIP8 protein. Northern blot analysis of poly(A+) RNA from the testis of these four inbred strains showed no difference in the absolute levels of ZIP8 mRNA between the sensitive and resistant mouse strains (Fig. 4a). Real-time PCR of the whole testis also revealed no significant differences in ZIP8 mRNA levels among the two sensitive and two resistant strains (data not shown).
Fig. 4.
Localization of ZIP8 mRNA from sensitive (D2 and 129S6) versus resistant (B6 and A/J) inbred mouse strains. (a) Northern analysis of testicular ZIP8 mRNA by using poly(A+) RNA. ACTB, β-actin mRNA as a control for lane loading. (b) In situ hybridization of ZIP8 mRNA in testis of the four inbred strains. Left show hematoxylin-and-eosin tissue staining (bright-field), Center show signal from in situ hybridization (dark-field), and Right show both images overlaid. Arrows show ZIP8 mRNA localized in the vascular endothelial cells of D2 and 129S6, a feature not detected in B6 or A/J mice. (c) High-magnification bright-field ZIP8 in situ image of testicular capillaries from the indicated mouse strains. (Lower) Photographic emulsion showing capillary in situ pattern (Left) was stripped and section-stained with an anti-CD31 endothelial-specific antibody that was detected by using a biotinylated secondary antibody and Alexa Fluor-488 streptavidin. Antibody reactivity is shown in green. DAPI (blue color) was used as a nuclear counterstain.
In situ hybridization was therefore performed. Analysis of the silver-grain distribution shows that ZIP8 was expressed similarly in the seminiferous tubules of all four inbred strains, with most silver grains over Sertoli cells (Fig. 4b). Sense-strand controls showed a diffuse nonspecific pattern of hybridization. Importantly, the two sensitive strains showed robust ZIP8 hybridization consistent with vascular endothelial cells of the testis; in contrast, this hybridization pattern was absent in the two resistant strains (Fig. 4b). High magnification bright-field photos of the interstitium showed the typical difference in silver-grain density between resistant and sensitive strains (Fig. 4c Upper); this analysis shows a sensitive-strain-specific accumulation of ZIP8 hybridization over testicular vascular endothelial cells. As further proof of endothelial localization, a capillary from the sensitive 129S6 strain with high silver-grain density was photographed, stripped of photographic emulsion, and reacted with an endothelial-cell-specific antibody (Fig. 4c Lower). High silver-grain density can be observed to colocalize with endothelial cell-specific staining. These data are consistent with endothelial cell ZIP8 mRNA accumulation in sensitive strains of mice, consistent with previous studies in refs. 16 and 18–24. In accord with these findings, loss of vascular endothelial ZIP8 mRNA expression in the resistant inbred strains protects the testis from Cd toxicity.
Northern blot analysis of ZIP8 mRNA in total RNA (data not shown) showed an order of concentration of lung > kidney > liver = testis. A similar pattern has been demonstrated for ZIP8 in human tissues (41). ESTs for ZIP8 are ubiquitous, furthermore, found in >30 tissues and cell types (www.ncbi.nlm.nih.gov). Hence, possible participation of ZIP8 in Cd toxicity could be widespread. Examining ZIP8 expression in other tissues by in situ hybridization, we found that ZIP8 mRNA was detected in lung, kidney, liver, lung, and intestine without differences in cell-type-specific expression between the two sensitive and two resistant strains of mice (data not shown).
Slc39a8 Gene Structure and Alternative Splicing. What is the mechanistic basis for the difference in accumulation of ZIP8 mRNA in vascular endothelial cells of the testis between these inbred strains? Based on ESTs from various tissues (“sequence information” entry in NCBI UniGene link of Mm.30239 and Hs.284205), the mouse Slc39a8 and human SLC39A8 genes both have nine exons; introns 2–8 and exons 2–9 span >66 kb and >83 kb in mouse and human, respectively (Table 3, which is published as supporting information on the PNAS web site). Both species have three exons 1 as the result of alternative promoter usage and splicing, and several distinct exons 9 were caused by the use of different poly(A+) sites.
If the mouse Slc39a8 exons 1 (Fig. 5, which is published as supporting information on the PNAS web site) are driven by independent promoters or enhancers, perhaps one of these promoters/enhancers might specify testicular endothelial-specific expression of ZIP8 mRNA. Would the transcript driven by this promoter be specifically absent from testicular RNA of Cd-resistant mice? Using real-time PCR to determine the levels of the transcripts initiated from each promoter, we could not detect the mRNA-containing exon 1c in testis; transcripts initiated at exons 1b and 1a were measurable but did not differ in amount between the sensitive and resistant strains (data not shown). Thus, neither the promoter that initiates at exon 1a nor exon 1b is specific to the testis.
Discussion
Cd is a nonessential metal. Therefore, ZIP8 is not a Cd transporter per se but likely has evolved to transport one or more essential metals. Other members of the SLC39 family have been shown to transport divalent Zn, Fe, and Mn (39, 41–46). During the course of uptake studies, we found that the transport of 0.25 μM CdCl2 was 50% inhibited by divalent Zn and Mn at concentrations of 13 μM and 2.9 μM, respectively (data not shown). These studies suggest that ZIP8 is more selective for transport of Mn than Zn, although the cation transported by ZIP8 in cells cannot be predicted based solely on inhibitor selectivity.
The data in this article strongly suggest that Slc39a8 is the Cdm gene and that ZIP8, the transporter product of the mouse Slc39a8 gene, functions normally for Mn+2, and perhaps Zn+2, ions. Cd+2 presumably participates as an opportunistic hitchhiker, being transported inadvertently into the vascular endothelial cells of the testis, resulting in increased cellular accumulation and toxicity.
There exists a haplotype block, shared in a phenotype-specific manner among at least the four strains studied herein and encompassing Slc39a8 (Fig. 1c). As a result, a majority of SNPs in and around the Slc39a8 gene are shared by sensitive or resistant strains, respectively. This conclusion limits our ability to eliminate nonfunctional SNPs. By alignment of the SLC39A8 gene and surrounding sequences in humans and rats (both presumed to be Cd-sensitive) and Cd-sensitive mouse strains, versus Cd-resistant mouse strains, there are less than a dozen “candidate SNPs” within 10 kb of the 5′ flanking region and the three largest introns (data not shown); however, because the expression difference between sensitive and resistant mouse strains is apparently limited to testicular vascular endothelial cells, determining perhaps a single SNP that results in a change in expression will likely require analysis in transgenic mice. Nonetheless, our data strongly implicate a difference in organ- and cell-type-specific Slc39a8 transcription as the mechanistic basis of resistance to Cd-induced testicular toxicity, but whether this result is due to an altered enhancer, locus-control region, or other DNA element remains to be determined.
Mammalian SLC39A8 is a largely uncharacterized gene. During a screen of innate immune activation of monocytes (41), human ZIP8 mRNA levels increased after treatment of monocytes with live and heat-killed Mycobacterium bovis bacillus Calmette–Guérin cell-wall lipopolysaccaride and inflammatory cytokines such as type-α TNF.
The very ancient SLC superfamily includes genes encoding passive transporters, ion-coupled transporters, and exchangers. Currently, there are 43 families with 298 putatively functional transporter genes (40). Metals are known to be transported by five families: SLC11 proton-coupled metal-ion transporter (47), SLC30 zinc effluxor (48), SLC31 copper transporter (49), SLC39 metal-ion transporter (39), and the SLC41 MgtE-like magnesium transporter (40). Except for the SLC30 family of effluxors (formerly known as the cation diffusion facilitator family), transporters in the other four families pump metal ions into the cell.
Cd uptake and efflux, and the inhibition of metal transport by Cd, are well characterized in plants, yeast, and mollusk (6, 8, 50–59). In mammalian-cultured cells, many studies of Cd transport and inhibition of divalent cation transport by Cd have shown Cd uptake and toxicity (11, 42, 43, 60–61), but there were no studies correlating Cd uptake with toxicity in any organ or specific cell type of any intact vertebrate.
Is ZIP8 important in Cd uptake and toxicity in mammalian organ systems aside from testis? As we have shown, the inbred strains used in this study do not display differences in ZIP8 mRNA accumulation in tissues examined except the endothelial cell of the testis. ZIP8 is present in a variety of tissues, including lungs and kidneys, two important target organs for Cd toxicity. Testicular endothelial cells are a unique population, involved in maintaining a blood–testis barrier with relatively impermeable tight junctions (62); this barrier is structurally different from other vascular endothelial cells, even when compared with the blood–brain barrier. It is our understanding that testicular vascular endothelial cells represent a very special endothelial cell population and that no good in vitro model for these cells exists.
Our observation of vascular endothelial cell toxicity might also give important insights into the molecular mechanisms of other types of heavy metal-associated human diseases. Epidemiological studies suggest Cd exposure can lead to testicular tumors in humans (1, 63, 64); Cd-induced cancer of the rodent testis has been shown experimentally (63, 64). In humans, chronic exposure to Cd leads to renal and pulmonary toxicity, possibly osteoporosis, and “itai-itai” disease (3, 65). Interindividual variations in susceptibility to Cd toxicity might exist among people from the same area, with presumed similar amounts of exposure to Cd from the soil (1, 66); these data suggest that allelic differences in one or more human genes might be involved in resistance to Cd toxicity.
In conclusion, we have shown here that the absence of Slc39a8 expression in vascular endothelial cells of certain inbred mouse strains is associated with resistance to Cd-induced testicular toxicity, a phenotype ascribed to the Cdm gene. Although numerous studies of Cd uptake and toxicity have been performed in bacteria, yeast, plants, invertebrates, and mammalian cell culture, the gene Slc39a8 demonstrates Cd toxicity in an intact vertebrate. Slc39a8 is expressed in a variety of tissues, but its differential expression among inbred mouse strains is apparently specific to the vasculature of the testis.
Supplementary Material
Acknowledgments
We thank our colleagues, especially David P. Witte and Alvaro Puga, for valuable discussions, technical advice, and a critical reading of the manuscript; and Jingchun Luo, Joanna Watson and Ge Zhang (DNA studies), Stacey Andringa (histology), and Meredith Farmer, Chris Woods, and Pam Groen (in situ hybridization) for technical assistance. This work was supported in part by National Institutes of Health Grants R01 ES10416 (to D.W.N.) and P30 ES06096 (to T.P.D., M.L.M., L.J., and D.W.N.).
Author contributions: T.P.D. and D.W.N. designed research; T.P.D., L.H., B.W., and M.L.M. performed research; L.J., K.F.S., and X.C. contributed new reagents/analytic tools; T.P.D., L.H., M.L.M., C.S.B., and D.W.N. analyzed data; and T.P.D. and D.W.N. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: cM, centimorgan; HA, hemagglutinin; LUC, luciferase; rv, retrovirus; SLC, solute-carrier; ZIP, ZRT-, IRT-like protein.
References
- 1.Järup, L. (2003) Br. Med. Bull. 68, 167-182. [DOI] [PubMed] [Google Scholar]
- 2.Swiergosz-Kowalewska, R. (2001) Microsc. Res. Tech. 55, 208-222. [DOI] [PubMed] [Google Scholar]
- 3.Takebayashi, S., Jimi, S., Segawa, M. & Takaki, A. (2003) Clin. Exp. Nephrol. 7, 18-26. [DOI] [PubMed] [Google Scholar]
- 4.Klaassen, C. D. & Liu, J. (1998) Environ. Health Perspect. 106, 297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singhal, R. K., Anderson, M. E. & Meister, A. (1987) FASEB J. 1, 220-223. [DOI] [PubMed] [Google Scholar]
- 6.Williams, L. E., Pittman, J. K. & Hall, J. L. (2000) Biochim. Biophys. Acta 1465, 104-126. [DOI] [PubMed] [Google Scholar]
- 7.Himeno, S., Yanagiya, T., Enomoto, S., Kondo, Y. & Imura, N. (2002) Tohoku J. Exp. Med. 196, 43-50. [DOI] [PubMed] [Google Scholar]
- 8.Hall, J. L. & Williams, L. E. (2003) J. Exp. Bot. 54, 2601-2613. [DOI] [PubMed] [Google Scholar]
- 9.Morgan, D. & DeCoursey, T. E. (2003) Front. Biosci. 8, S1266-S1279. [DOI] [PubMed] [Google Scholar]
- 10.Thevenod, F. (2003) Nephron Physiol. 93, P87-P93. [DOI] [PubMed] [Google Scholar]
- 11.Bressler, J. P., Olivi, L., Cheong, J. H., Kim, Y. & Bannona, D. (2004) Ann. N.Y. Acad. Sci. 1012, 142-152. [DOI] [PubMed] [Google Scholar]
- 12.Alsberg, C. L. & Schwartze, E. W. (1919) J. Pharmacol. Exp. 13, 504-505. [Google Scholar]
- 13.Schwartze, E. W. & Alsberg, C. L. (1923) Exper. Therap. 21, 1-22. [Google Scholar]
- 14.Parizek, J. (1956) Nature 177, 1036-1037. [DOI] [PubMed] [Google Scholar]
- 15.Parizek, J. (1957) J. Endocrinol. 15, 56-63. [DOI] [PubMed] [Google Scholar]
- 16.Chiquoine, A. D. (1964) Anat. Rec. 149, 23-36. [DOI] [PubMed] [Google Scholar]
- 17.Sangalang, G. B. & O'Halloran, M. J. (1972) Nature 240, 470-471. [DOI] [PubMed] [Google Scholar]
- 18.Gunn, S. A., Gould, T. C. & Anderson, W. A. (1963) Am. J. Pathol. 42, 685-702. [PMC free article] [PubMed] [Google Scholar]
- 19.Waites, G. M. & Setchell, B. P. (1966) J. Endocrinol. 34, 329-342. [DOI] [PubMed] [Google Scholar]
- 20.Clegg, E. J. & Carr, I. (1967) J. Pathol. Bacteriol. 94, 317-322. [DOI] [PubMed] [Google Scholar]
- 21.Gupta, R. K., Barnes, G. W. & Skelton, F. R. (1967) Am. J. Pathol. 51, 191-205. [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, M. H. (1969) J. Reprod. Fertil. 19, 551-553. [DOI] [PubMed] [Google Scholar]
- 23.Setchell, B. P. & Waites, G. M. H. (1970) J. Endocrinol. 47, 81-86. [DOI] [PubMed] [Google Scholar]
- 24.Schlaepfer, W. W. (1971) Lab. Invest. 25, 556-564. [PubMed] [Google Scholar]
- 25.Lucis, O. J. & Lucis, R. (1969) Arch. Environ. Health 19, 334-336. [DOI] [PubMed] [Google Scholar]
- 26.Taylor, B. A., Heiniger, H. J. & Meier, H. (1973) Proc. Soc. Exp. Biol. Med. 143, 629-633. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, B. A. (1976) J. Hered. 67, 389-390. [DOI] [PubMed] [Google Scholar]
- 28.Cloudman, A. M. & Bunker, L., Jr. (1945) J. Hered. 36, 258-263. [Google Scholar]
- 29.Zhu, Y., King, B. L., Parvizi, B., Brunk, B. P., Stoeckert, C. J., Jr., Quackenbush, J., Richardson, J. & Bult, C. J. (2003) Genome Biol. 4, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonhomme, F., Benmehdi, F., Britton-Davidian, J. & Martin, S. (1979) C. R. Seances Acad. Sci. D. 289, 545-548. [PubMed] [Google Scholar]
- 31.Dalton, T. P., Miller, M. L., Wu, X., Menon, A., Cianciolo, E., McKinnon, R. A., Smith, P. W., Robinson, L. J. & Nebert, D. W. (2000) Pharmacogenetics 10, 141-151. [DOI] [PubMed] [Google Scholar]
- 32.Bergwitz, C., Wendlandt, T., Potter, E., Glomb, I., Gras, K., von zur Muhlen, A. & Brabant, G. (2000) Histochem. Cell Biol. 113, 145-150. [DOI] [PubMed] [Google Scholar]
- 33.Solis, W. A., Childs, N. L., Weedon, M. N., He, L., Nebert, D. W. & Dalton, T. P. (2002) Toxicol. Appl. Pharmacol. 178, 93-101. [DOI] [PubMed] [Google Scholar]
- 34.Dalton, T. P., Dieter, M. Z., Matlib, R. S., Childs, N. L., Shertzer, H. G., Genter, M. B. & Nebert, D. W. (2000) Biochem. Biophys. Res. Commun. 267, 184-189. [DOI] [PubMed] [Google Scholar]
- 35.Robison, J. G., Elliott, J., Dixon, K. & Oakley, G. G. (2004) J. Biol. Chem. 279, 34802-34810. [DOI] [PubMed] [Google Scholar]
- 36.Jumarie, C., Campbell, P. G., Berteloot, A., Houde, M. & Denizeau, F. (1997) J. Membr. Biol. 158, 31-48. [DOI] [PubMed] [Google Scholar]
- 37.Qian, J., Colbert, M. C., Witte, D., Kuan, C. Y., Gruenstein, E., Osinska, H., Lanske, B., Kronenberg, H. M. & Clemens, T. L. (2003) Endocrinology 144, 1053-1061. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama, K., Su Ih, I. H., Tezuka, T., Yasuda, T., Mikoshiba, K., Tarakhovsky, A. & Yamamoto, T. (2002) EMBO J. 21, 83-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eide, D. J. (2004) Pflügers Arch. 447, 796-800. [DOI] [PubMed] [Google Scholar]
- 40.Hediger, M. A., Romero, M. F., Peng, J. B., Rolfs, A., Takanaga, H. & Bruford, E. A. (2004) Pflügers Arch. 447, 465-468. [DOI] [PubMed] [Google Scholar]
- 41.Begum, N. A., Kobayashi, M., Moriwaki, Y., Matsumoto, M., Toyoshima, K. & Seya, T. (2002) Genomics 80, 630-645. [DOI] [PubMed] [Google Scholar]
- 42.Gaither, L. A. & Eide, D. J. (2000) J. Biol. Chem. 275, 5560-5564. [DOI] [PubMed] [Google Scholar]
- 43.Gaither, L. A. & Eide, D. J. (2001) J. Biol. Chem. 276, 22258-22264. [DOI] [PubMed] [Google Scholar]
- 44.Lasswell, J., Rogg, L. E., Nelson, D. C., Rongey, C. & Bartel, B. (2000) Plant Cell 12, 2395-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dufner-Beattie, J., Wang, F., Kuo, Y. M., Gitschier, J., Eide, D. & Andrews, G. K. (2003) J. Biol. Chem. 278, 33474-33481. [DOI] [PubMed] [Google Scholar]
- 46.Dufner-Beattie, J., Langmade, S. J., Wang, F., Eide, D. & Andrews, G. K. (2003) J. Biol. Chem. 278, 50142-50150. [DOI] [PubMed] [Google Scholar]
- 47.Mackenzie, B. & Hediger, M. A. (2004) Pflügers Arch. 447, 571-579. [DOI] [PubMed] [Google Scholar]
- 48.Palmiter, R. D. & Huang, L. (2004) Pflügers Arch. 447, 744-751. [DOI] [PubMed] [Google Scholar]
- 49.Petris, M. J. (2004) Pflügers Arch. 447, 752-755. [DOI] [PubMed] [Google Scholar]
- 50.Gaxiola, R. A., Fink, G. R. & Hirschi, K. D. (2002) Plant Physiol. 129, 967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guerinot, M. L. (2000) Biochim. Biophys. Acta 1465, 190-198. [DOI] [PubMed] [Google Scholar]
- 52.Ortiz, D. F., Kreppel, L., Speiser, D. M., Scheel, G., McDonald, G. & Ow, D. W. (1992) EMBO J. 11, 3491-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bovet, L., Eggmann, T., Meylan-Bettex, M., Polier, J., Kammer, P., Marin, E., Feller, U. & Martinoia, E. (2003) Plant Cell. Environ. 26, 371-381. [Google Scholar]
- 54.Finkemeier, I., Kluge, C., Metwally, A., Georgi, M., Grotjohann, N. & Dietz, K. J. (2003) Plant Cell. Environ. 26, 821-833. [DOI] [PubMed] [Google Scholar]
- 55.Persans, M. W., Nieman, K. & Salt, D. E. (2001) Proc. Natl. Acad. Sci. USA 98, 9995-10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salt, D. E. & Wagner, G. J. (1993) J. Biol. Chem. 268, 12297-12302. [PubMed] [Google Scholar]
- 57.Hirschi, K. D., Korenkov, V. D., Wilganowski, N. L. & Wagner, G. J. (2000) Plant Physiol. 124, 125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidoumou, Z., Gnassia-Barelli, M. & Romeo, M. (1997) Bull. Environ. Contam. Toxicol. 58, 318-325. [DOI] [PubMed] [Google Scholar]
- 59.Li, L., He, Z., Pandey, G. K., Tsuchiya, T. & Luan, S. (2002) J. Biol. Chem. 277, 5360-5368. [DOI] [PubMed] [Google Scholar]
- 60.Bannon, D. I., Abounader, R., Lees, P. S. & Bressler, J. P. (2003) Am. J. Physiol. Cell. Physiol. 284, C44-C50. [DOI] [PubMed] [Google Scholar]
- 61.Okubo, M., Yamada, K., Hosoyamada, M., Shibasaki, T. & Endou, H. (2003) Toxicol. Appl. Pharmacol. 187, 162-167. [DOI] [PubMed] [Google Scholar]
- 62.Kamimura, Y., Chiba, H., Utsumi, H., Gotoh, T., Tobioka, H. & Sawada, N. (2002) Med. Electron Microsc. 35, 139-145. [DOI] [PubMed] [Google Scholar]
- 63.IARC. (1993) Scand. J. Work Environ. Health 19, 360-363. [DOI] [PubMed] [Google Scholar]
- 64.Waalkes, M. P. (2003) Mutat. Res. 533, 107-120. [DOI] [PubMed] [Google Scholar]
- 65.Emmerson, B. T. (1970) Ann. Intern. Med. 73, 854-855. [DOI] [PubMed] [Google Scholar]
- 66.Elinder, C. G., Edling, C., Lindberg, E., Kagedal, B. & Vesterberg, O. (1985) Am. J. Ind. Med. 8, 553-564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.