Abstract
We demonstrate an experimental approach to structural studies of unfolded and partially folded proteins in which conformational distributions are probed at a site-specific level by 2D solid-state 13C NMR spectroscopy of glassy frozen solutions. Experiments on chemical denaturation of the 35-residue villin headpiece subdomain, a model three-helix-bundle protein with a known folded structure, reveal that 13C-labeled residues in the three helical segments of the folded state have markedly different conformational distributions in the unfolded state. Moreover, the 2D solid-state NMR line shapes near the unfolding midpoint do not fit a simple two-state model, in which the conformational distributions of the unfolded component are assumed to be independent of denaturant concentration. Comparison with solid-state NMR spectra of peptides containing the individual helical segments suggests an alternative two-step description of conformational distributions in partially folded states of the helical villin headpiece subdomain, in which chemical denaturation is viewed as a disruption of tertiary contacts followed by equilibration of local secondary structure according to the intrinsic helical propensities of individual segments.
Keywords: chemical denaturation, magnetic resonance, protein structure, two-state system, villin
Processes involving unfolded or partially folded states of proteins are both fundamentally intriguing and biologically relevant (1), but the structural properties of these states are not well understood. The kinetics and thermodynamics of protein folding have traditionally been studied primarily by optical spectroscopies, calorimetry, and scattering techniques. Although these techniques provide crucial information about global structural transitions, they provide relatively little direct information about site-specific structural features. Although the folded structures of proteins can be determined precisely by x-ray crystallography and multidimensional liquid-state NMR, crystallography is not applicable, and liquid-state NMR provides limited information in studies of unfolded proteins. Simple analyses of protein-folding data treat the unfolded state as a uniform random coil with conformational properties described by statistical models for polymer chains. Evidence for nonstatistical conformational distributions in unfolded proteins has been found in recent experiments (2–8) and simulations (9–12), but a detailed understanding of these distributions has not been achieved yet. A more complete experimental characterization of the structural properties of unfolded and partially folded states of proteins would (i) facilitate the interpretation of kinetic and thermodynamic data on protein folding, (ii) contribute to the development of more accurate and precise descriptions of protein-folding pathways, (iii) permit more stringent tests of computer simulations of these states, and (iv) possibly shed light on protein-aggregation phenomena such as amyloid formation (13). Progress in these areas will depend on the introduction of new experimental approaches.
Detailed structural characterization of unfolded and partially folded states of proteins is inherently difficult both because these states are disordered, implying that each structural parameter (e.g., torsion angle, interatomic distance) takes on a site-specific distribution of values, and rapid exchange among populated conformations in the liquid state tends to produce experimental data that are dynamically averaged functions of the structural parameters. In principle, solid-state NMR measurements on glassy frozen solutions can overcome these difficulties. With appropriate choices of isotopic labels and solid-state NMR techniques, the solid-state NMR data are sensitive to site-specific structural parameters and can differentiate among competing models for distributions of these parameters. In addition, dynamical averaging is suppressed at temperatures well below the solvent glass transition (Tg). The applicability of various solid-state NMR techniques to disordered systems similar to unfolded proteins has been demonstrated in studies of synthetic organic polymers (14, 15) and biopolymers (16–18), including helical peptides in frozen glycerol/water (19). A recent structural study of an HIV-related peptide/antibody complex further illustrates the structural-information content of solid-state NMR data in glassy frozen solutions (20).
In this article, we describe a solid-state NMR study of site-specific conformational distributions in a 35-residue three-helix-bundle protein (HP35) (21, 22) that has been the subject of recent computational (10, 11, 23, 24) and experimental (25–28) studies. We demonstrate that information about these distributions in unfolded and partially folded states, which are induced by chemical denaturation with guanidine hydrochloride (Gdn·HCl), is contained in 2D solid-state 13C NMR spectra of 13C-labeled HP35 in frozen glycerol/water. Specifically, we show that the backbone conformational distributions in unfolded HP35 are reflected in crosspeak line shapes in the 2D spectra and are significantly different at 13C-labeled sites in the three helical segments of the folded state. The crosspeak line shapes further reveal that backbone conformational distributions near the denaturation midpoint of HP35 are not represented accurately by a simple two-state model but can be described by the two-step model presented below.
For the solid-state NMR data in frozen solution to be relevant to our understanding of protein folding under more conventional experimental conditions, it is important that structural aspects of the protein-folding phenomena under study not be altered qualitatively by the use of low temperatures and a glass-forming solvent. Circular dichroism (CD) and NMR data presented below indicate that the solid-state NMR indeed are relevant, at least in the case of HP35.
Materials and Methods
HP35 (amino acid sequence LSDEDFKAVFGMTRSAFANLPLWKQQNLKKEKGLF, representing residues 42–76 of the 76-residue chicken villin headpiece) was synthesized with standard solid-phase methods and purified by high-performance liquid chromatography (water/acetonitrile gradient, C18 reverse-phase column). The folded state of HP35 contains the three helical segments H1, H2, and H3 [residues 44–50, 55–58, and 63–72, respectively (21, 22)]. Uniformly 13C-labeled residues were introduced at V50, A57, and L69 as probes of the conformational distributions in H1, H2, and H3. These particular amino acids were selected to ensure well resolved 2D 13C NMR signals. The three peptides HP42–52,HP53–61, and HP62–76 (HPn–m represents residues n through m) were also synthesized with the same labeled residues.
CD measurements were performed on a Jasco (Easton, MD) 720 instrument with a home-built low-temperature chamber. Samples for CD measurements shown in Fig. 1 were 30 μM HP35, pH 5, with a 2-mm path length. Thermodynamic parameters for HP35 folding in glycerol/water at low temperatures were derived from the CD data as explained in the Fig. 1 legend. CD spectra at -40°C and [Gdn·HCl] = 0, 4, and 7 M were found to be independent of HP35 concentration from 17 μM to 9.2 mM, indicating that significant protein aggregation does not occur in this concentration range and therefore does not affect the data shown in Fig. 2.
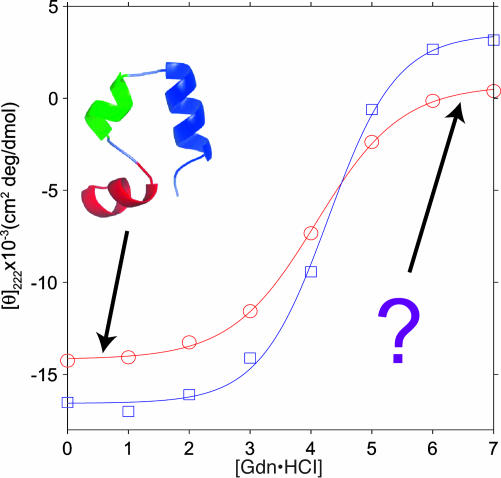
Fig. 1.
Chemical denaturation of HP35 by Gdn·HCl at 20°C (○) and -40°C (□), monitored by the CD signal at 222 nm. The known folded HP35 conformation at low [Gdn·HCl] is shown as a ribbon diagram. Solid-state NMR data shown in Figs. 2, 3, 4 probe the unknown conformational distributions at high [Gdn·HCl]. Solid lines are least-squares fits to a standard, simple, two-state model (40): CD(g) = CDfold - (CDfold - CDunf){exp[(mg - ΔG0)/RT]/{1 + exp[(mg - ΔG0)/RT]}}, where CDfold and CDunf are the CD signals of the folded and unfolded states, g · [Gdn·HCl], and ΔG0 is the free energy of unfolding at g = 0. At 20°C, m = 0.83 ± 0.10 kcal/mol·M (1 cal = 4.18 J), ΔG0 = 3.41 ± 0.40 kcal/mol, CDfold = -14,180 ± 320 cm2·deg/dmol, and CDunf = 710 ± 46 cm2·deg/dmol. At -40°C, m = 0.82 ± 0.21 kcal/mol·M, ΔG0 = 3.51 ± 0.90 kcal/mol, CDfold = -16,570 ± 810 cm2·deg/dmol, and CDunf = 3,510 ± 1,230 cm2·deg/dmol. At 4°C (data not shown), m = 0.79 ± 0.06 kcal/mol·M, and ΔG0 = 3.36 ± 0.25 kcal/mol. At -20°C (data not shown), m = 0.71 ± 0.07 kcal/mol·M, and ΔG0 = 3.17 ± 0.31 kcal/mol. (Errors are 95% confidence limits.)
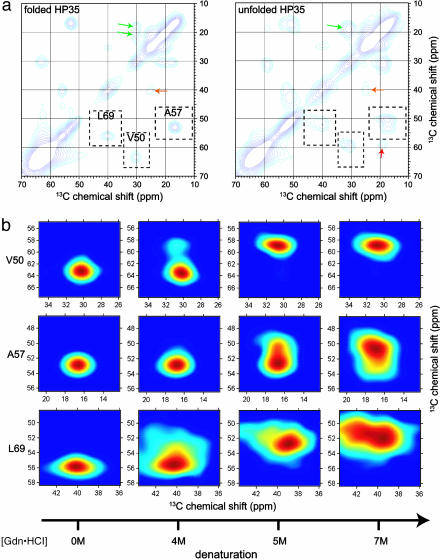
Fig. 2.
Two-dimensional solid-state 13C spectra of HP35 in frozen solution. (a) Aliphatic regions of 2D spectra at [Gdn·HCl] = 0M(Left)and7M(Right). Dashed boxes enclose Cα/Cβ crosspeaks for 13C-labeled V50, A57, and L69. Contour levels increase by successive factors of 1.3. Arrows indicate Cβ/Cγ crosspeaks of V50 (green), Cβ/Cγ crosspeaks of L69 (orange), and Cα/Cγ crosspeaks of V50 (red). (b) Expansions showing the dependence of Cα/Cβ crosspeak shapes on [Gdn·HCl]. Color scales run from the maximum signal intensity in each crosspeak (dark red) to 15% of the maximum intensity (dark blue). Differences in crosspeak shapes at high [Gdn·HCl] reflect site-specific differences in conformational distributions.
Solid-state NMR measurements were carried out at a 13C NMR frequency of 100.8 MHz by using a Varian Infinity console and Varian magic-angle spinning (MAS) probe with 6-mm sample rotors (240-μl sample volume). Solid-state NMR samples were 5–10 mM HP35 in 1:1 vol/vol glycerol/water, 1 mM CH3COONa, 6 mM CuSO4 (to reduce the proton spin-lattice relaxation time to 1 s in the frozen state), pH 5, with the indicated [Gdn·HCl] values. Samples of peptide mixtures were pH 7, with 6 mM CuNa2EDTA in place of CH3COONa and CuSO4. Solid-state NMR signals of HP42–52 alone and HP53–61 alone were identical to those in the peptide mixture, showing that peptide association does not affect the data shown in Fig. 4. Samples were frozen in the MAS rotors by immersion in liquid N2, producing cooling rates of ≈10°C/s. 2D spectra shown in Figs. 2 and 4 were acquired at -120°C sample temperatures, with MAS at 6.7 kHz and two-pulse phase-modulated proton decoupling (29) at 75 kHz. To maximize Cα/Cβ crosspeak intensities, finite-pulse radio frequency-driven recoupling (30) was applied during 1.6-ms mixing periods by using 30-μs 13C π pulses and a carrier frequency centered in the aliphatic region of the 13C NMR spectrum. Data were acquired with active rotor synchronization to minimize distortions arising from MAS side bands of natural-abundance glycerol 13C NMR signals, with coaddition of spectra generated by Herzfeld–Roberts–Griffin and Hagemeyer–Schmidt–Rohr–Spiess processing (31) to maximize the signal-to-noise ratio. Total measurement times were 4–10 days per 2D spectrum depending on the HP35 and Gdn·HCl concentrations. All 13C chemical shifts are referenced to adamantane at 25°C (38.56 ppm).
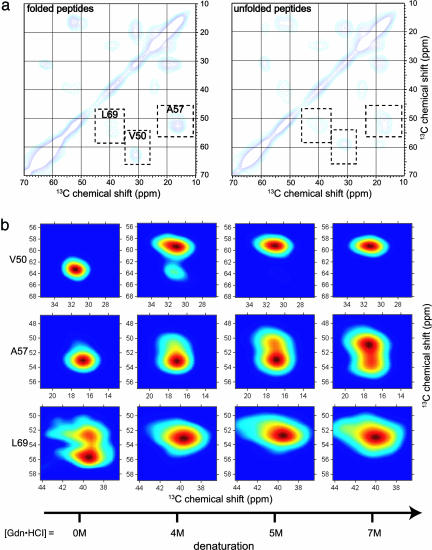
Fig. 4.
Two-dimensional solid-state 13C spectra of HP42–52, HP53–61, and HP62–76 peptides (1:1:1 mixture) in frozen solution. (a) Aliphatic regions of 2D spectra at [Gdn·HCl] = 0M(Left)and7M(Right). (b) Expansions showing the dependence of Cα/Cβ crosspeak shapes on [Gdn·HCl] for V50 (in HP42–52), A57 (in HP53–61), and L69 (in HP62–76).
Results
CD data shown in Fig. 1 are consistent with a helical folded structure and a cooperative, two-state unfolding of HP35 by addition of Gdn·HCl in glycerol/water, with a denaturation midpoint at [Gdn·HCl] ≈ 4.1 M between 20 and -40°C. The denaturation midpoint and thermodynamic parameters extracted from the data shown in Fig. 1 (see legend) are nearly identical to those extracted from CD data in aqueous solution at 4°C by McKnight et al. (21), indicating that the use of glycerol/water and low temperatures has little effect on the Gdn·HCl denaturation of HP35. The 13C NMR chemical shifts discussed below provide additional strong evidence that the folded structure of HP35 is preserved in frozen glycerol/water.
Fig. 2a shows aliphatic regions of 2D solid-state 13C NMR spectra of HP35 in folded ([Gdn·HCl] = 0 M) and unfolded ([Gdn·HCl] = 7 M) states (see Fig. 5, which is published as supporting information on the PNAS web site, for full 2D spectra). As established by previous empirical (32, 33) and ab initio (34) studies, 13C chemical shifts for Cα and Cβ sites in proteins and peptides are determined primarily by backbone conformation. In the folded state, Cα/Cβ crosspeaks for all three 13C-labeled residues are relatively sharp (2.3–3.9 ppm full width at half maximum), consistent with a well defined conformation in a rigid, noncrystalline solid state (20). Cα and Cβ chemical shifts are consistent with the expected helical secondary structure at these sites. All 13C chemical shifts are within 0.2 ppm of those observed in aqueous solution at 25°C, including those of V50 methyl carbons that are sensitive to tertiary contacts through ring-current effects (25). In the unfolded state, significant broadening of NMR lines and changes in chemical shifts are observed, reflecting increased conformational disorder and changes in the predominant conformations. Cα/Cβ crosspeak line shapes for the three labeled sites are qualitatively different in the unfolded state. The crosspeak for L69 broadens substantially in both dimensions and loses nearly all intensity at its initial (i.e., [Gdn·HCl] = 0 M) position; the A57 crosspeak broadens and shifts but maintains significant intensity at its initial position; and the V50 crosspeak shifts to a new position but remains relatively sharp. The L69 crosspeak line shape is consistent with nearly complete disorder in H3, i.e., population of all sterically allowed regions of the backbone ϕ,Ψ torsion-angle space, with no preference for helical ϕ,Ψ values. The crosspeak line shape for A57 is consistent with ≈25% α-helix content in H2. The crosspeak position for V50 indicates loss of nearly all α-helix content in H1 and is consistent with (but does not prove) a predominant polyproline II conformation in the unfolded state (35). These data rule out a uniform statistical model for conformational distributions in unfolded HP35.
Differences among Cα/Cβ crosspeak line shapes for V50, A57, and L69 in unfolded HP35 do not result simply from inherent differences in 13C NMR chemical-shift distributions or conformational propensities for valine, alanine, and leucine residues. Crosspeak line shapes in unfolded HP35 are significantly different from crosspeak line shapes in “random-coil” pentapeptides GGXGG (36) in frozen glycerol/water (X = V, A, L; see Fig. 6, which is published as supporting information on the PNAS web site), which reveal the inherent spectroscopic properties of these residues.
Fig. 2b shows the evolution of the Cα/Cβ crosspeak line shapes near the CD denaturation midpoint. Crosspeak line shapes, S(ν1, ν2; g), for each labeled residue can be fit individually to a linear combination of the fully folded and fully unfolded line shapes, i.e., S(ν1, ν2; g) = ffold(g)Sfold(ν1, ν2) + funf(g)Sunf(ν1, ν2), where ν1 and ν2 are the NMR frequencies in the two dimensions, g · [Gdn·HCl], Sfold(ν1, ν2) and Sunf(ν1, ν2) are the crosspeak line shapes for the folded and unfolded states (normalized to unit volume), with ffold(g) + funf(g) = 1. However, as shown in Fig. 3 (first and second columns), good fits cannot be obtained when ffold and funf are required to be the same for all labeled residues. This result shows that a simple two-state model for chemical denaturation of HP35, in which partially denatured states would have conformational distributions describable as the sum of a fraction ffold of folded proteins and a fraction funf of unfolded proteins [with Sunf(ν1, ν2) independent of [Gdn·HCl]], does not describe the solid-state NMR data adequately.
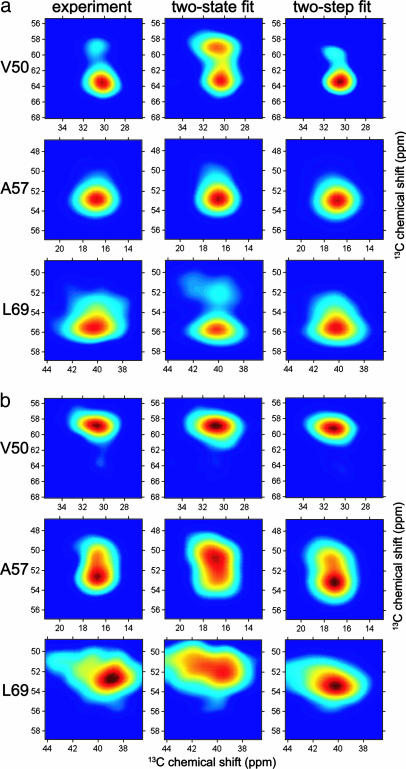
Fig. 3.
Least-squares fits of experimental Cα/Cβ crosspeak shapes to two-state and two-step models (see Results) for the conformational distribution of HP35 near its denaturation midpoint. (a) [Gdn·HCl] = 4 M. Folded fractions ffold derived from the fits are 0.48 ± 0.04 and 0.50 ± 0.05 for the two-state and two-step models, respectively. (b) [Gdn·HCl] = 5 M. Folded fractions are 0.05 ± 0.05 and 0.00 ± 0.05 for the two-state and two-step models, respectively. The two-step model represents the crosspeak shapes accurately, whereas the simple two-state model does not.
Fig. 4 shows 2D solid-state 13C NMR spectra of an equimolar mixture of HP42–52, HP53–61, and HP62–76. These spectra closely resemble spectra of HP35 in Fig. 2 but indicate more conformational disorder in the peptide mixture than in the protein at [Gdn·HCl] = 0 M. Comparison of Figs. 2 and 4 suggests that crosspeak line shapes for HP35 at a particular [Gdn·HCl] near the denaturation midpoint may be modeled as a linear combination of line shapes for fully folded HP35 and line shapes for the peptide mixture at the same [Gdn·HCl], i.e., S(ν1, ν2; g) = ffold(g)Sfold(ν1, ν2) + fpep(g)Spep(ν1, ν2; g), where fpep(g) is the fractional contribution of the [Gdn·HCl]-dependent crosspeak line shape Spep(ν1, ν2; g) for the peptide mixture. As shown in Fig. 3 (first and third columns), good fits are obtained by using the same values of ffold and fpep for all labeled residues. This result supports the following two-step description of chemical denaturation: (i) in the presence of denaturant, the conformational distribution of HP35 consists of a fraction ffold of folded proteins and a fraction fpep of proteins in which tertiary contacts have been disrupted; and (ii) proteins with disrupted tertiary contacts have local conformational distributions dictated by the intrinsic secondary structure propensities of the three segments containing H1, H2, and H3. In other words, at a given [Gdn·HCl], disruption of tertiary contacts by addition of denaturant produces nearly the same local conformational distributions as does disruption of tertiary contacts by division of the HP35 sequence into the three separate peptides. Because the three peptides have different intrinsic dependences of helix content on [Gdn·HCl] and because the solid-state NMR data are sensitive to local conformational distributions, the simple two-state model cannot fit the solid-state NMR data.
Discussion
Data shown in Figs. 2, 3, 4 demonstrate that information about the conformational distributions in unfolded and partially folded proteins can be obtained from crosspeak line shapes in 2D solid-state 13C NMR spectra. To a first approximation, the inhomogeneously broadened Cα/Cβ crosspeak line shapes are maps of backbone ϕ,Ψ torsion-angle distributions at the 13C-labeled residues, because the Cα and Cβ chemical shifts are determined largely by the local ϕ and Ψ values. 2D spectroscopy reveals the correlations between Cα and Cβ chemical shifts, enhancing the information content of the line shapes, and allows the NMR signals of multiple labeled residues to be resolved.
The ϕ,Ψ torsion-angle distributions probed by our measurements in frozen glycerol/water are approximately the equilibrium distributions at an effective temperature Teff at which local structural relaxation becomes too slow to keep up with temperature-dependent changes in the equilibrium distributions as the samples are cooled. At the relatively low cooling rates of the experiments described above (≈10 °C/s), we expect Teff to be several degrees above the solvent glass transition (Tg ≈ -75°C). At [Gdn·HCl] = 5 M, Cα/Cβ crosspeak line shapes are essentially identical when the cooling rate is increased to ≈1,000°C/s (see Fig. 7, which is published as supporting information on the PNAS web site), consistent with rapid structural relaxation in HP35 (26, 27) and the relatively weak dependence of the equilibrium conformational distributions on temperature indicated by our CD (Fig. 1) and liquid-state NMR (see Fig. 8, which is published as supporting information on the PNAS web site) data. The absence of a significant dependence on cooling rate also shows that our solid-state NMR data are not affected by protein aggregation, formation of microcrystalline ice, precipitation of buffer salts, or other extraneous factors.
In principle, the Cα/Cβ crosspeak line shapes could serve as quantitative constraints on backbone conformational distributions, provided that the dependences of chemical shifts on ϕ and Ψ were known accurately (see Fig. 9, which is published as supporting information on the PNAS web site, for ab initio chemical-shift calculations and line-shape simulations). However, even a qualitative and empirical analysis of the solid-state NMR data yields important information. Specifically, the evolution of crosspeak line shapes with increasing [Gdn·HCl] (Fig. 2) shows that 13C-labeled residues in the three helical segments of folded HP35 have markedly different conformational distributions in unfolded HP35. V50 in H1 adopts a relatively well ordered but nonhelical conformation; A57 in H2 is more conformationally disordered, but retains significant helix content; and L69 in H3 is the most disordered of the three labeled residues in unfolded HP35. Moreover, conformational distributions near the denaturation midpoint are not described accurately by a simple two-state model. As described above, comparison of crosspeak line shapes for intact HP35 with those for peptides containing H1, H2, and H3 (Fig. 4) suggests an alternative, two-step description of conformational distributions near the denaturation midpoint, which accurately fits the solid-state NMR data (Fig. 3).
The good fit of CD data to a simple two-state model (Fig. 1) may be explained by the fact that CD contributions from individual helices are not resolved. The two-step model indicates that helical CD signal at [Gdn·HCl] ≥ 5 M, where fits to the solid-state NMR data yield ffold ≈ 0 (see Fig. 3 legend), does not arise from a nonzero population of fully folded HP35 molecules, as would be inferred from a simple two-state analysis of the CD data. Rather, the helical CD signal arises from the partial helical content of HP35 molecules with disrupted tertiary contacts (especially in H2). Thus, in the case of HP35, the solid-state NMR data reveal a significantly different physical picture of chemical denaturation than would be indicated by a conventional treatment of CD data, including a more rapid loss of fully folded molecules with increasing denaturant concentration and a larger secondary structure content in molecules with disrupted tertiary structure.
The two-step model for conformational distributions in HP35 is reminiscent of the diffusion-collision model for protein-folding kinetics (37). Although recent diffusion-collision calculations by Islam et al. (24) were unable to explain the rapid folding (26–28) of HP35, this discrepancy is likely caused by an underestimation of the helical propensities of the HP35 segments, as discussed by Islam et al. and supported by the data shown in Fig. 4. Although CD and proton NMR data for HP42–53, HP52–61, and HP62–76 have been reported by Tang et al. (25) as evidence that these peptides are “basically unstructured” in aqueous solution at 25°C, their data seem consistent with helix contents >20%. One might expect the helix contents of the three peptides (and denatured HP35) to be enhanced by glycerol and low temperatures, but liquid-state 13C NMR chemical shifts for the peptide mixture show only minor effects at <0°C (see Fig. 8).
Relatively simple solid-state NMR techniques were used in the experiments described above, and the structural information was extracted solely from isotropic 13C NMR chemical shifts. This work can be extended by quantitative measurements of interatomic distance and torsion-angle distributions by using appropriate solid-state NMR techniques to measure and correlate nuclear magnetic dipole–dipole couplings and chemical-shift anisotropies (14–16, 18–20, 31, 38). Other likely extensions include kinetic measurements based on rapid-mixing and freeze-quench techniques (39) and applications to proteins with nonhelical secondary structures.
Supplementary Material
Author contributions: R.H.H. designed research; R.H.H. performed research; R.H.H. and R.T. analyzed data; and R.H.H. and R.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HP35, 35-residue three-helix-bundle protein; Gdn·HCl, guanidine hydrochloride; HPn–m, residues n through m of villin headpiece protein.
References
- 1.Dobson, C. M. (2003) Nature 426, 884-890. [DOI] [PubMed] [Google Scholar]
- 2.Mayor, U., Grossmann, J. G., Foster, N. W., Freund, S. M. V. & Fersht, A. R. (2003) J. Mol. Biol. 333, 977-991. [DOI] [PubMed] [Google Scholar]
- 3.Arcus, V. L., Vuilleumier, S., Freund, S. M. V., Bycroft, M. & Fersht, A. R. (1995) J. Mol. Biol. 254, 305-321. [DOI] [PubMed] [Google Scholar]
- 4.Shortle, D. & Ackerman, M. S. (2001) Science 293, 487-489. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie, J. R. & Shortle, D. (1997) J. Mol. Biol. 268, 170-184. [DOI] [PubMed] [Google Scholar]
- 6.Cavagnero, S., Nishimura, C., Schwarzinger, S., Dyson, H. J. & Wright, P. E. (2001) Biochemistry 40, 14459-14467. [DOI] [PubMed] [Google Scholar]
- 7.Kortemme, T., Kelly, M. J. S., Kay, L. E., Forman-Kay, J. & Serrano, L. (2000) J. Mol. Biol. 297, 1217-1229. [DOI] [PubMed] [Google Scholar]
- 8.Segel, D. J., Fink, A. L., Hodgson, K. O. & Doniach, S. (1998) Biochemistry 37, 12443-12451. [DOI] [PubMed] [Google Scholar]
- 9.Zagrovic, B., Snow, C. D., Khaliq, S., Shirts, M. R. & Pande, V. S. (2002) J. Mol. Biol. 323, 153-164. [DOI] [PubMed] [Google Scholar]
- 10.Duan, Y., Wang, L. & Kollman, P. A. (1998) Proc. Natl. Acad. Sci. USA 95, 9897-9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen, M. Y. & Freed, K. F. (2002) Proteins 49, 439-445. [DOI] [PubMed] [Google Scholar]
- 12.Wong, K. B., Clarke, J., Bond, C. J., Neira, J. L., Freund, S. M. V., Fersht, A. R. & Daggett, V. (2000) J. Mol. Biol. 296, 1257-1282. [DOI] [PubMed] [Google Scholar]
- 13.Tycko, R. (2004) Curr. Opin. Struct. Biol. 14, 96-103. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt-Rohr, K., Hu, W. & Zumbulyadis, N. (1998) Science 280, 714-717. [DOI] [PubMed] [Google Scholar]
- 15.Dabbagh, G., Weliky, D. P. & Tycko, R. (1994) Macromolecules 27, 6183-6191. [Google Scholar]
- 16.van Beek, J. D., Hess, S., Vollrath, F. & Meier, B. H. (2002) Proc. Natl. Acad. Sci. USA 99, 10266-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, J., Parkanzky, P. D., Khunte, B. A., Canlas, C. G., Yang, R., Gabrys, C. M. & Weliky, D. P. (2001) J. Mol. Graph. Model. 19, 129-135. [DOI] [PubMed] [Google Scholar]
- 18.Yao, X. L. & Hong, M. (2004) J. Am. Chem. Soc. 126, 4199-4210. [DOI] [PubMed] [Google Scholar]
- 19.Long, H. W. & Tycko, R. (1998) J. Am. Chem. Soc. 120, 7039-7048. [Google Scholar]
- 20.Sharpe, S., Kessler, N., Anglister, J. A., Yau, W. M. & Tycko, R. (2004) J. Am. Chem. Soc. 126, 4979-4990. [DOI] [PubMed] [Google Scholar]
- 21.McKnight, C. J., Doering, D. S., Matsudaira, P. T. & Kim, P. S. (1996) J. Mol. Biol. 260, 126-134. [DOI] [PubMed] [Google Scholar]
- 22.McKnight, C. J., Matsudaira, P. T. & Kim, P. S. (1997) Nat. Struct. Biol. 4, 180-184. [DOI] [PubMed] [Google Scholar]
- 23.Zagrovic, B., Snow, C. D., Shirts, M. R. & Pande, V. S. (2002) J. Mol. Biol. 323, 927-937. [DOI] [PubMed] [Google Scholar]
- 24.Islam, S. A., Karplus, M. & Weaver, D. L. (2002) J. Mol. Biol. 318, 199-215. [DOI] [PubMed] [Google Scholar]
- 25.Tang, Y. F., Rigotti, D. J., Fairman, R. & Raleigh, D. P. (2004) Biochemistry 43, 3264-3272. [DOI] [PubMed] [Google Scholar]
- 26.Wang, M. H., Tang, Y. F., Sato, S. S., Vugmeyster, L., McKnight, C. J. & Raleigh, D. P. (2003) J. Am. Chem. Soc. 125, 6032-6033. [DOI] [PubMed] [Google Scholar]
- 27.Kubelka, J., Eaton, W. A. & Hofrichter, J. (2003) J. Mol. Biol. 329, 625-630. [DOI] [PubMed] [Google Scholar]
- 28.Hansen, K. C., Rock, R. S., Larsen, R. W. & Chan, S. I. (2000) J. Am. Chem. Soc. 122, 11567-11568. [Google Scholar]
- 29.Bennett, A. E., Rienstra, C. M., Auger, M., Lakshmi, K. V. & Griffin, R. G. (1995) J. Chem. Phys. 103, 6951-6958. [Google Scholar]
- 30.Ishii, Y. (2001) J. Chem. Phys. 114, 8473-8483. [Google Scholar]
- 31.Tycko, R. & Berger, A. E. (1999) J. Magn. Reson. 141, 141-147. [DOI] [PubMed] [Google Scholar]
- 32.Spera, S. & Bax, A. (1991) J. Am. Chem. Soc. 113, 5490-5492. [Google Scholar]
- 33.Wishart, D. S. & Sykes, B. D. (1994) J. Biomol. NMR 4, 171-180. [DOI] [PubMed] [Google Scholar]
- 34.Oldfield, E. (2002) Annu. Rev. Phys. Chem. 53, 349-378. [DOI] [PubMed] [Google Scholar]
- 35.Chellgren, B. W. & Creamer, T. P. (2004) Biochemistry 43, 5864-5869. [DOI] [PubMed] [Google Scholar]
- 36.Merutka, G., Dyson, H. J. & Wright, P. E. (1995) J. Biomol. NMR 5, 14-24. [DOI] [PubMed] [Google Scholar]
- 37.Karplus, M. & Weaver, D. L. (1979) Biopolymers 18, 1421-1437. [Google Scholar]
- 38.Tycko, R. (2001) Annu. Rev. Phys. Chem. 52, 575-606. [DOI] [PubMed] [Google Scholar]
- 39.Lin, Y., Gerfen, G. J., Rousseau, D. L. & Yeh, S. R. (2003) Anal. Chem. 75, 5381-5386. [DOI] [PubMed] [Google Scholar]
- 40.Jackson, S. E. & Fersht, A. R. (1991) Biochemistry 30, 10428-10435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.