Abstract
Transmission ratio distortion (TRD) and sterility are male-specific quantitative trait phenomena associated with the mouse t haplotype. TRD occurs in t haplotype-heterozygous males and is caused by the deleterious action of distorter products on sperm bearing a wild-type responder locus. It has been proposed that t-mediated male sterility is a severe manifestation of TRD caused by homozygosity for distorter loci; thus, the distorter and sterility loci would be identical. In this, study a transgenic approach was used to identify the proximal sterility locus, tcs1 (S1), and test its role in TRD. Mice transgenic for a wild-type bacterial artificial chromosome (BAC) derived from the S1-critical region were bred onto t haplotype backgrounds. Mating results conclusively showed that the BAC is sufficient to restore fertility in otherwise sterile males. Multiple mutations were identified in the t alleles of Synj2 and Serac1, two genes in the BAC; thus, they are candidates for S1. In addition, whereas the BAC transgene rescued sterility, it had no effect on TRD. These results uncouple the proximal t haplotype sterility locus, S1, from TRD, demonstrating that S1 and the proximal distorter locus, D1, are not the same gene.
Keywords: synaptojanin 2, t complex
The t complex spans ≈20 cM of mouse chromosome 17 and exists in two forms: a variant known as the t haplotype (t) present in 20% of the feral house mouse population, and the wild-type form (+) (Fig. 1). The t haplotype differs from + by the presence of four nonoverlapping inversions (1), Inv (17)1-Inv (17)4, that effectively suppress recombination between t and +; thus, the t haplotype is inherited en bloc. Two male-specific phenomena occur in t haplotype mice: transmission ratio distortion (TRD) and sterility (reviewed in ref. 2). TRD, or non-Mendelian segregation, occurs in t/+ males and is characterized by the transmission of the t haplotype to up to 99% of offspring (reviewed in ref. 3). Several recessive lethal and semilethal t alleles of multiple loci affecting embryonic development have been discovered. These alleles are subdivided into complementation groups such that mice homozygous for two complementing t haplotypes are viable, yet the males are invariably sterile because of homozygosity of sterility loci. Females are equally affected by the lethal loci but do not exhibit TRD or sterility.
Fig. 1.
The t complex and the S1-critical region. (A) Structure and organization of the t complex. The complex exists as two variants, + and t haplotype, distinguished by four nonoverlapping inversions (arrows). Markers defining the inversions are shown. TRD and sterility loci are shown below the t haplotype (S1/D1, etc.) (B) BAC tile of the S1-critical region. The RPCI-23 BACs are 335F22 (335), 387F2 (387), 456P8 (456), 60J11 (60), 52A7 (52), and 22M9 (22). The structure and organization of 335, 387, 456, 60, and 52 are published elsewhere (13). BAC 22M9 is 225 kb and overlaps 387 and 456 (tick marks). (C) The following are in 22M9. Genes: Snx9 (3′ half only), Synj2, Serac1, and D17Wsu155e. Pseudogenes: Fibrillarin-like and Ftl1-rs9. The transcriptional orientations are shown.
The current model proposes that in t/+ males, t distorters expressed during spermatogenesis interact deleteriously with the + allele of the cis-acting responder (Tcr; R); thus, R+ sperm are incapacitated. However, Rt is refractory to distorter effects, and Rt sperm are normal and capable of fertilization (4). Distorter and sterility factors are genetically inseparable, leading to the proposal that distortion and sterility are caused by the same mutations, and the end effect is distorter-dosage-dependent. Accordingly, a double dose of distorters overcomes the refractory nature of Rt and causes sterility in viable t/t males (5). Evidence exists for three distorter/sterility loci (6–8): Distorter 1/Sterility 1 (D1/S1), D2/S2, and D3/S3; and two distortion-only loci: D4 and D5 (Fig. 1). Only Rt has been cloned and shown to encode a mutant kinase (SMOK1Tcr), consisting of the C terminus of SMOK1, a sperm motility kinase, fused to the N terminus of RSK6, a ribosomal kinase (9).
Distortion and sterility effects attributable to D1/S1 are caused by amorphic mutations because a deletion of the wild-type genomic region harboring D1/S1 recapitulates TRD and sterility when in trans to distal partial t haplotypes or complete t haplotypes, respectively (10). Various combinations and dosages of S1, D2/S2, and D3/S3 result in sterility or reduced fertility. Whereas homozygosity of S1 alone does not cause infertility, mice homozygous for S1 and heterozygous for D2/S2 (abbreviated as S1 S2/S1 +) are sterile (5). A similar dosage effect has been observed for TRD (8). For example, males heterozygous for partial t haplotypes typically transmit the t haplotype chromosome at a lower frequency than do their complete t haplotype counterparts, albeit at a higher rate than expected. These observations underscore the interpretation of these phenomena as classical quantitative traits and the effecting loci as quantitative trait loci.
The assumption that a single gene encodes each distorter/sterility pair was questioned when data obtained from a panel of deletion-bearing mice implied that the D1/S1 locus is actually two loci (11, 12). Subsequently, a 1-Mbp high-resolution physical map of the S1 region uncovered several genes (Fig. 1), including Synj2 (13), and Tctex1, a candidate for D1/S1 (14).
In the present study, we report that (i) a bacterial artificial chromosome (BAC) derived from the S1-critical region contains two genes, Synj2 and Serac1, both of which are mutated in t haplotypes; (ii) the BAC is sufficient to restore fertility in sterile males; and (iii) the BAC has no effect on TRD. Our results confirm our hypothesis that D1 and S1 are different genes.
Materials and Methods
Mice. Mice were maintained at The Jackson Laboratory or at Bates College in accordance with Institutional Animal Care and Use Committee protocols. The chromosome 17 variants used in this study were D17Aus9df5J (abbreviated as 5J), a deletion spanning D1 and S1 (15), the complete t haplotype tw2 (16), and the proximal partial t haplotypes th49 (4) and tw82 (17). Mice had mixed genetic backgrounds, including contributions from FVB/NJ, C3H/HeJ, and C57BL/6J.
BAC Manipulation. BAC RPCI-23-22M9 (22M9) containing a complete copy of Synj2 was identified by PCR amplification of DNA pools (Research Genetics, Huntsville, AL) with Synj2-specific primers. BAC DNA was purified by CsCl density gradient centrifugation (18).
BAC Analysis. Gene composition of 22M9 was determined by the use of restriction enzyme fingerprinting tables to identify overlapping, sequenced BACs (19); the public human and mouse genomic databases; the high-throughput genomic sequence database at the National Center for Biotechnology Information; and genscan (20).
Generation of BAC Transgenics (Tgs). BAC Tg animals were made at The Jackson Laboratory, using standard techniques (21). Briefly, pronuclei of 472 FVB/NJ zygotes were microinjected with 1 pl of 2 or 4 ng/μl BAC DNA. Embryos were surgically implanted into recipient CByB6F1 pseudopregnant females, and 51 pups were obtained. Tg pups were identified by PCR typing with chloramphenicol acetyltransferase gene-specific primers or Sp6/T7-specific primers. Of 51 pups, 3 founders were obtained. Only two, Tg18 and Tg26, contained both BAC ends, and presumably, the intervening genomic DNA. These two founders were mated to C57BL/6J, and Tg offspring were identified by PCR as described.
RT-PCR. Total RNA was isolated from mouse testis with TRIzol (Invitrogen) according to the manufacturer's protocol. Approximately 10 μg were used for polydT-primed first-strand cDNA synthesis with retroscript (Ambion, Austin, TX). PCR was performed with gene-specific primers and high-fidelity Taq polymerase from various suppliers. The amplicons were sequenced directly by the dideoxy chain termination method on 16-capillary ABI 3100 DNA sequencers or subcloned into the pCR II vector (Invitrogen) before sequencing. Sequence contigs were assembled with vectornti (Invitrogen) or sequencher (Gene Codes). Primer sequences used in this study are available on request.
Mouse Genotyping. Tg-bearing mice were identified as described. The th49, tw82, or tw2 haplotypes were detected by PCR amplification of polymorphic markers, including D17Jcs115 (22) and t haplotype-specific alleles of Vil2 (Mouse Genome Informatics accession ID no. 3033301) and Hbaps4 (23). 5J/+ mice always exhibit a short tail, whereas 5J/t mice are tailless, abrogating the need for molecular genotyping of carriers. The genetic profiles of 5J and of the t haplotypes used in this study have been described (11).
Western Blot Analysis. Analysis of β-fertilin (ADAM2) maturation in testicular and epididymis/vas deferens-derived sperm was performed as described in ref. 24 with antibody 9D2.2 (25)(Chemicon). Sperm proteins were resolved on SDS/9% PAGE gels (26).
Statistical Analysis. TRD was analyzed by χ2 using Fisher's Exact Test. For sterility analysis, male fertility was expressed as offspring per female per month (OFM) as described (5).
Results
Analysis of S1 Candidate Genes. RT-PCR of testis total RNA. A 1-Mbp genomic segment spanning the S1-critical region was known to contain Synj2 (13), encoding a phosphatidylinositol polyphosphate phosphatase (27) with two distinct catalytic sites (27, 28).
Synj2 expression was observed in wild-type testicular germ-cell-enriched fractions (data not shown) and in whole testis of wild-type, tw2/tw2, and th49/tw2 mice by RT-PCR (Fig. 2). Synj2 amplicons from various t haplotypes and wild-type inbred strains were sequenced, and a C → T transition at nucleotide 1436 was observed in all t haplotypes tested (tw2, tw32, tw82, tTuw32, and th49) but not in wild-type strains (C57BL/6J, 129/SvJ, BALB/cByJ, C3H/HeOuJ, and CAST/Ei). The mutation is in exon 11 and replaces Ser-385 with Phe (Table 1). By using only tw2, other t haplotype mutations were discovered (Table 1).
Fig. 2.
RT-PCR expression analysis of Serac1 (A–E) and Synj2 (F). Lanes are with (+) or without (-) RT enzyme. Wild-type (+) and t haplotype (t) analysis is indicated below each lane. Mutations are shown as arrows. Residue numbers are derived from NP_796285 (SERAC1) and NP_035653 (SYNJ2). Start (arrowhead) and stop (asterisk) sites of translation are indicated for Serac1.
Table 1. Analysis of S1 candidate genes within BAC RPCI-23–22M9.
| Gene symbol | Function | Human homolog | Mutation | Coordinates* | RT-PCR | |
|---|---|---|---|---|---|---|
| RPCI-23-22M9 | DNA | Protein | 3110767–3368420 | |||
| Synj2 | Phosphoinositide 5-phosphatase | SYNJ2 | A562→G | N94D | 3216322–3318403 | Y |
| C1436→T | S385F | |||||
| A1603→G | NC | |||||
| C3884→T | NC | |||||
| A3905→C | Q1208P | |||||
| A4016→G | 3′ UTR | |||||
| Serac1 | Unknown | SERAC1 | C74→A, C98→A | 5′ UTR | 3180874–3216545 | Y |
| T112→C | V4A | |||||
| T118→A, T119→C | F6Y | |||||
| T121→G | F7C | |||||
| T141→C | NC | |||||
| C185→G | T25M | |||||
| A301→C, C302→A | T64Q | |||||
| T1215→C | NC | |||||
| A1574→G | N488S | |||||
| T1866→C | NC | |||||
| T1976→G | NC | |||||
| G1994→A | 3′ UTR | |||||
| D17Wsul55e | Unknown | BC060317 | 3175122–3180761 | Y | ||
| Fibrillarin | Pseudogene | 3138435–3139391 | N | |||
| Ftl2 | Pseudogene | 3127986–3128906 | N | |||
Synj2 is one of two paralogous genes found in mammals. It encodes a three-domain protein: an N-terminal SacI-like domain with 3′ and 4′ phosphatase activity specific for lipid-bound inositol, a central 5-phosphatase domain-specific lipid-bound and soluble inositol protein, and a proline-rich, heterogeneous C terminus derived from alternative splicing (27–30). The S385F mutation occurs within the region joining the N-terminal domain to the central 5-phosphatase domain. S385 is conserved in human and rat synaptojanin 2 (SYNJ2), whereas the homologous residue in SYNJ1 (the paralog) is Asn in mouse, human, rat, cow, zebrafish, Fugu rubripes, Drosophila melanogaster, Anopheles gambiae, and Caenorhabditis elegans. Inositol polyphosphate 5-phosphatases are important in sperm function, as demonstrated by the sterility observed in males homozygous for a Inpp5b null allele (24); thus, we proceeded to generate BAC Tgs to test the hypothesis that S1 is encoded by the t allele of Synj2.
Shortly afterward, the sequence from RPCI-23 BACs 387F2, 456P8, and 60J11 became available, closing the S1 critical region (Fig. 1). genscan (20) analysis of the BAC sequences predicted a gene distal to Synj2 which has been named Serac1. Further analysis confirmed that Serac1 is contained within 22M9.
The function of serine active site containing 1 (SERAC1) is unknown. interpro analysis of the ORF predicts an esterase/lipase/thioesterase domain (accession no. IPR000379) and an ARM repeat fold (accession no. IPR008938). RT-PCR analysis confirmed that the gene is expressed in the testis (Fig. 2). A comparison of wild-type and t alleles revealed a cluster of t-specific mutations (Table 1), located within the first 25 codons of the Serac1 ORF, and several other mutations dispersed throughout the transcript. Several mutations are nonsynonymous and introduce nonconservative amino acid replacements (Phe7Cys and Thr25Met), whereas others are less drastic (Val4Ala, Phe6Tyr, and Asn488Ser). One mutation in particular, Thr64Gln, represents one of two alleles found in wild-type strains, because this site is Gln in t and BALB/cByJ, but is Thr in (NMRI/GSF × CD1) F1 mice (GenBank accession no. NM_011523) (30). In addition, several mutations were found within UTRs.
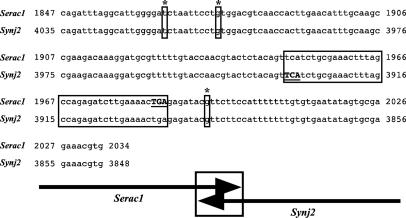
Synj2 and Serac1 overlap tail to tail such that the same genomic segment encodes a fragment of the ORFs of both genes (Fig. 3). At the mRNA level, nucleotides 1847–2034 of Serac1 are the complements of nucleotides 3848–4035 of Synj2, and a subset of the mutations in Serac1 correlates with mutations in Synj2: mutations 1866, 1976, and 1994 in Serac1 correspond to mutations 4016, 3905, and 3884, respectively, in Synj2 (Table 1).
Fig. 3.
Serac1 and Synj2 overlap. Mutated nucleotides (small boxes; see Table 1), and coding-coding overlap (large boxes) are shown. Translation stop sites (all caps) are shown. The transcriptional orientations are indicated below the alignment.
Mapping of BAC ends onto the mouse assembly (Ensembl Build 31) using global survey sequences (National Center for Biotechnology Information accession IDs AZ083735 and AZ083736) identified three more genes: Fibrillarin-like, Ftl-rs9, and D17Wsu155e. Fibrillarin-like and Ftl-rs9 are inactive pseudogenes, whereas D17Wsu155e is a small ORF encoding a hypothetical 71-aa protein of unknown function that is 38% identical to the 78-aa protein Rex1-S of Chlamydomonas reinhardtii, required in DNA repair (31). Testis expression of D17Wsu155e was confirmed by RT-PCR (data not shown), and a comparison of the genomic sequence of the t and wild-type alleles yielded no differences that would lead to amino acid changes, thus D17Wsu155e was excluded as an S1 candidate.
BAC Effect on Fertility Rescue and TRD Experiments. To test the hypotheses that S1 is encoded in 22M9, and that D1 and S1 are distinct genetic elements, we generated male mice with various combinations of complete and partial t haplotypes with or without the BAC. Fecundity and transmission ratios were tested with the following results.
Sterility Rescue Test. Three assays were performed. The first assay tested th49/tw2 males, the second assay tested 5J males, and the third assay tested tw82/tw2 males. In each case, sibling males with or without the Tg were tested, and in all three assays, 22M9 provided strong evidence of sterility rescue.
th49/tw2. The first test used a partial t haplotype, th49, in trans to tw2, a complete t haplotype. th49 harbors S1 but not the stronger sterility locus, S2 (4). Thus, the genotype of non-Tg males is (S1++/S1, S3, S2) and they are expected to be sterile. Control and experimental males were mated with two females each for several months. The number of offspring sired by each male was normalized (5) and compared with the fecundity of non-Tg control male siblings. The fertility of Tg males was remarkably similar to wild-type mice (Tg18: 7.72 OFM and Tg26: 6.64 OFM), whereas the non-Tg sibling controls were infertile (0.02 OFM), with the exception of one male (Table 2 and Fig. 4). The fecundity of the fertile non-Tg th49/tw2 male was indistinguishable from wild-type males and is likely due to complex genetic background effects or an undetected change in th49 or tw2. These results unequivocally demonstrate that 22M9 has a significant effect on fertility.
Table 2. Results of fertility assay by using Tg and non-Tg males.
| Mating test results
|
||||||
|---|---|---|---|---|---|---|
| Genotype | Fertile | Semisterile | Sterile | OFM range | OFM average | SD |
| tw2/Aus9df5J;Tg18/+ | 5 | 2 | 5 | 0–8.66 | 2.52 | 2.88 |
| tw2/Aus9df5J;Tg26/+ | 1 | 1 | 2 | 0–12.40 | 3.62 | 5.94 |
| tw2/Aus9df5J;+/+ | 0 | 1 | 14 | 0–1.09 | 0.08 | 0.28 |
| tw2/tw82;Tg18/+ | 9 | 0 | 0 | 4.71–11.54 | 7.81 | 2.38 |
| tw2/tw82;Tg26/+ | 2 | 0 | 0 | — | 10.0 | 0.86 |
| tw2/tw82;+/+ | 2 | 1 | 3 | 0–6.05 | 2.12 | 2.65 |
| tw2/th49;Tg18/+ | 9 | 1 | 1 | 0–11.33 | 7.72 | 3.78 |
| tw2/th49;Tg26/+ | 3 | 0 | 0 | 3.88–8.09 | 6.64 | 2.40 |
| tw2/th49;+/+ | 1 | 0 | 11 | 0–0.18 | 0.02 | 0.06 |
Fig. 4.
Results of Tg rescue analysis. The results are expressed as a mean of all experiments for each cluster of genotypes (described in Materials and Methods) plus the SE of the mean, for each experiment.
5J/tw2. The second test used 5J, a deletion that spans the S1/D1 critical region, in trans to tw2; therefore, non-Tg males are (null+ +/S1, S3, S2) and considered homozygous for the S1 amorphic mutation, whereas BAC Tgs would be (+ + +/S1, S3, S2) if the wild-type allele of S1 were in 22M9. Twelve 5J/tw2;Tg18/+ and 4 5J/tw2;Tg26/+ males were tested, and their fecundity was compared with that of 15 non-Tg siblings. Non-Tg male fecundity was extremely low (0.08 OFM), whereas Tg male fecundity was significantly increased (Tg18: 2.52 OFM and Tg26: 3.62 OFM). These results are consistent with the first assay and confirm that 22M9 is capable of rescuing S1 infertile males. However, a minority of Tg males remained sterile, a phenomenon that may be due to the health effects of harboring a large deletion and not to factors affecting fertility per se (11).
tw82/tw2. The last assay used male mice heterozygous for a partial proximal t haplotype (tw82) harboring S1 but not the stronger sterility locus, S2, and a complete t haplotype (tw2). Accordingly, non-Tg males are (S1++/S1, S3, S2) and are expected to be sterile. Interestingly, non-Tg males were semisterile (2.12 OFM); however, the Tg counterparts were considerably more fecund (Tg18: 7.72 OFM and Tg26: 6.64), which is consistent with the previous assays. tw82 is derived from tw32, a complete t haplotype known to exhibit variability in fertility assays, sometimes resulting in unexpected fertility in carrier males (10). Although it is possible that a hypomorphic tw82 allele of S1 with residual wild-type function caused the semisterility of non-Tg males, it is more likely that background genetic modifiers affected the results because none of the mice were inbred.
In conclusion, the results of all three assays demonstrate that 22M9 rescues S1-induced male sterility. Of a total of 41 Tg males tested, 33 were fertile (average OFM = 6.4) whereas only 5 of 33 non-Tg control siblings exhibited any fertility (average OFM = 0.74; Table 2).
TRD Test. ES cell-derived deletions in chromosome 17 segregate into two groups when introduced in male mice: (i) deletions that cause sterility when in trans to complete t haplotypes, or TRD when in trans to partial t haplotypes; and (ii) deletions that cause sterility when in trans to complete t haplotypes but not TRD when in trans to partial t haplotypes (11). If D1 and S1 are the same, then tw2 in the presence of the Tg is functionally equivalent to a long distal partial t haplotype (+ R D3, D2) such as t6 (4), because 22M9 would supply the D1/S1 wild-type allele.
t6/+ males transmit t6 to ≈65% of their offspring on average (4, 6, 10, 11), which is significantly lower than the 95% observed for tw2 (16, 32). Therefore, we tested the transmission of tw2 from tw2/+ males with or without Tg18. If the wild-type allele of D1 is in 22M9, we would observe a significant decrease in the transmission of tw2 from tw2/+;Tg18/+ males. The results clearly showed that Tg18 had no effect on tw2 because tw2/+;Tg18/+ males transmitted tw2 to 94% of offspring compared with 97% transmission by tw2/+ males (χ2 = 0.10, P = 0.75).
Western Blot Analysis of ADAM2. Male mice homozygous for the Inpp5b knock-out are sterile and exhibit defects in the proteolytic maturation of ADAM2 (24). ADAM2 and ADAM1 form a heterodimer necessary for sperm–egg binding (33) and maturation of ADAM2, which occurs in the epididymis, is affected by inositol metabolism (24). Although the maturation protease is unknown, it is thought to be a serine protease (34).
The prediction that SERAC1 contains a serine active site characteristic of serine proteases, prompted us to test ADAM2 maturation in fertile Tg animals and their non-Tg siblings. The results show that ADAM2 is processed normally in rescued and sterile males (Fig. 5). Consequently, the t alleles of Serac1 and Synj2 do not affect ADAM2 maturation.
Fig. 5.
Western Blot analysis of ADAM2. The th49/tw2 males (described in Materials and Methods) are sterile in the absence of the transgene (Tg18-), and fertile in its presence (Tg18+). ADAM2 was derived from testis sperm (T) or vas deferens sperm (V). Molecular mass markers (kDa) are shown.
Discussion
Genetic studies have uncovered five distorter (D1-D5) and three sterility (S1-S3) loci, although the genes remain unknown (2, 3). D1/S1, D2/S2, and D3/S3 comprise distorter/sterility loci; it is hypothesized that each is encoded by a single-mutant allele. However, the results of our Tg rescue analysis, and deletion (12) and high-resolution mapping studies (11), do not support this hypothesis for D1/S1; instead, we propose that D1 and S1 are different mutations.
TRD is caused by a deleterious interaction between distorters and SMOK1, resulting in the inactivation of + gametes. SMOK1Tcr, however, is resistant to distorter action (3). The trans-acting inactivation effect of distorters may be facilitated by diffusion across cytoplasmic bridges connecting synchronously developing sperm (35), whereby they gain access to + gametes. Alternatively, distorter gene expression before meiosis may distribute distorters throughout meiotic partners where they could affect SMOK1. SMOK1 however, is postmeiotically expressed and restricted to the gamete of origin, despite apparently being soluble (9). The resistance of SMOK1Tcr to harmful distorter effects may be due to multiple mutations and reduced kinase activity (9), although the chemistry of distorter/SMOK1 interactions is unknown.
To determine whether BAC 22M9 ameliorates TRD by providing a wild-type copy of D1, we bred it onto the tw2 background. If 22M9 harbors wild-type D1, Tg tw2 males should have the same levels of TRD as t6 males because t6 is a long distal characterized by (+ D3 and D2) alleles. However, the expected decrease of tw2 transmission was not observed. Instead Tg males transmitted tw2 at a rate equal to non-Tg tw2 siblings (≈95%). The persistence of a mutant D1 locus in Tg tw2 males used for TRD analysis was deemed unimportant, because D1 is amorphic and its effects are elicited by deleting the genomic interval that harbors it (10). Thus, D1 encodes a functionless product, or the product is not expressed. We conclude that the wild-type allele of D1 is not encoded in 22M9.
t haplotype sterility is thought to be the consequence of a double dose of distorters, referred to as sterility factors in this context (5). In t/+ males, SMOK1Tcr counteracts the effect of the distorters (4) but this resistance is absent in t/t males; thus, the type of responder makes no difference on the outcome (36), and R+/R+, Rt/R+, or Rt/Rt males are sterile provided that at a minimum, the male is homozygous for S2. If the male is heterozygous for S2, then it must be homozygous for S1 to be infertile (5).
Male sterility can be induced by deleting, in + chromosomes, the genetic interval harboring the S1 locus (10–12), indicating that S1 is an amorphic mutation whose lack of function has deleterious consequences on fertility provided one S2 allele is present. In fact, the fecundity of males carrying S1 only is indistinguishable from wild type (10), suggesting that the + allele of S1 is redundant, except when its absence is compounded by heterozygosity for S2.
We bred 22M9 onto various backgrounds that otherwise produce sterile or semisterile males, expecting that if 22M9 harbors the + allele of S1, Tg mice would be heterozygous for the S1 mutation and exhibit normal fertility. With one exception, this is what we observed (Fig. 4). The exception was when 22M9 was bred onto the 5J/tw2 background; these males are sterile in the absence of the BAC, whereas their Tg counterparts were mildly fecund, averaging three OFM (Table 2). The diminished rescue effect is likely due to adverse health effects brought on by the large size of the 5J deletion, which spans several cM. Taken together, our results demonstrate that 22M9 rescues S1-mediated male sterility, which is in stark contrast to its lack of effect on TRD. Therefore, different genes encode S1 and D1.
Analysis of 22M9 uncovered Synj2, Serac1, and D17Wsu155e, and several pseudogenes. D17Wsu155e was ruled out because the t allele sequence and expression profile is indistinguishable from +. The Synj2 and Serac1 t alleles, however, harbor multiple mutations, making them strong candidates for S1 (Table 1). Interestingly, the t alleles do not qualitatively differ in expression relative to the + alleles, insofar as both genes are expressed in sterile males at levels similar to wild-type and rescued Tg males.
SYNJ2 functions in the inositol-signaling pathway. It has two catalytic motifs: an N-terminal SacI-like domain that dephosphorylates positions 3 and 4 of phosphatidylinositol, and a central domain that dephosphorylates position 5 of lipidic and soluble inositol, including inositol-3,4,5-trisphosphate (IP3) (27, 28). SYNJ2 functions in clathrin-mediated endocytosis (37) and has been localized to various cellular compartments, including the outer mitochondrial membrane (38), the plasma membrane, where it functions as a Rac1 effector (39), and the spermatid manchette (29); its function in spermatogenesis or sperm function is unknown. However, regulation of the inositol signaling pathway is important in spermatogenesis and sperm function as seen by the effect of knocking out Inpp5b (24, 40), and by the expression of Inpp5d (SHIP) in round spermatids (41). Both genes encode proteins with substrate specificity similar to SYNJ2.
SYNJ2 mutations fall in three categories. N94D and S385F are in the SacI domain. Interestingly, residue 94 is D in rat and human SYNJ2, suggesting that the mutation originated in wild-type mice after the divergence of t and + lineages. S385F is in a highly conserved site in mouse, rat, and human SYNJ2, and is drastic. S385 is in the vicinity of R381, a critical Arg residue that when mutated to His, abolishes SacI-phosphatase activity (29). The same effect is observed in the yeast homolog of SAC1 (42). Thus, we suspect that S385F may adversely affect enzyme activity by disrupting the three-dimensional structure in the vicinity of R381.
The C-terminal mutation Q1208P, is in a unique region of SYNJ2 arising from an alternative splicing event not observed in human or rat; therefore, this residue does not have an equivalent in human or rat SYNJ2. The last category is characterized by mutations within the 3′ UTR, none of which appear to affect the splicing or expression of the gene.
If Synj2 is S1, we assume that its phosphatase activity is lost in S1/S1 homozygotes because deletion of the genomic interval harboring it mimics the S1 effect, but only in the presence of the D2/S2 mutation (10–12). The lost activity could be compensated by other testis-expressed phosphatases, including inositol polyphosphate 5-phosphatase (INPP5)B and INPP5D. An interaction between S1 and S2 is suggested because at least one S2 t allele is necessary to elicit sterility in S1/S1 homozygotes (10). Perhaps S2 encodes a general regulatory protein that enhances phosphatase activity in SYNJ2, INPP5B, and INPP5D. In the absence of SYNJ2, up-regulation of other phosphatases by the wild-type product of S2 could be sufficient to restore normal function. However, a single S2 t allele would create a conditional haploinsufficiency, characterized by up-regulation of phosphatase activity below a critical threshold, thus severely compromising all sperm. Regulation of the inositol pathway by modulating IP3 levels is critical to sperm function. IP3 is required for mobilizing intracellular Ca2, an action that affects sperm hypermotility (43) and the acrosomal reaction (44), required at critical steps during fertilization. S2 mutations might affect this regulatory network, allowing IP3 accumulation and misregulation of calcium release. Although levels of IP3 or phosphatase activity in t/t males have not been measured relative to +/+ males, this hypothesis is consistent with the fact that S2 homozygotes are sterile in the absence of other mutant t sterility loci (5). The region harboring S2 is several cM, making candidate gene analysis impractical. Interestingly, there are reports of abnormal hypermotility and flagellar morphology in t/t males as a consequence of improper calcium regulation (45, 46). In addition, the differential expression in developing sperm of calmodulin and other cytoplasmic Ca2+-binding proteins (47), and the localization of IP3 receptors to various sperm organelles, including the acrosome (48), demonstrates that spermatogenesis is affected by Ca2+ regulation. Our hypothesis predicts the presence of abnormally high levels of IP3 in t/t spermatozoa.
Serac1 is ubiquitously expressed and its expression in t haplotypes is not qualitatively different from wild type. Its function remains unknown, yet it appears to belong to the family of serine active-site-containing proteins characteristic of many hydrolases, including serine proteases. Interestingly, in the Inpp5b knock-out males, defects in ADAM2 maturation were observed (24). ADAM2 is essential for normal sperm function (49), and its maturation depends on an as-of-yet identified protease, although it has been suggested to be a serine protease (34). Consequently, we tested the maturation of ADAM2 in sterile t haplotypes and their rescued counterparts and observed normal processing in both, thus ruling out SERAC1 as the ADAM2 maturation protease (Fig. 5).
The tail-to-tail overlap in Serac1 and Synj2 (Fig. 3) suggests that they might be coevolving to a limited extent, because silent mutations in one could be harmful to the other. Interestingly, this overlap is not observed in the human, chimp, or rat orthologs (Ensembl). A recent analysis of overlapping genes within the mouse genome found that 31 of 578 overlapping genes include so-called coding-coding overlap such as that observed with Serac1 and Synj2 (50). A similar analysis of the human genome did not find an instance in which coding-coding overlap in one organism is conserved in the other (50), thus substantiating our finding that the Serac1-Synj2 overlap is unique to mouse. What effect, if any, this overlap has on the expression of these genes is unknown. However, if the genes have the same spatial-temporal expression profiles, the possibility exists that the transcripts associate by means of complementarity and this in turn could affect their expression profiles and/or stability.
We conclude that S1 is encoded either by Synj2 or Serac1. Further studies requiring a knock-out approach could unravel which of the two genes is S1. If the t allele of Synj2 encodes S1, it raises the interesting question of why are multiple inositol-metabolizing enzymes, with overlapping substrate specificity, required for proper sperm function? The answer may lie in nonoverlapping subcellular localization or in temporally regulated differences in enzyme activity. Alternatively, if Serac1 encodes S1, further work will be required to dissect the role of SERAC1 in spermatogenesis, beginning with identifying its function.
Acknowledgments
We thank the Cell Biology and Microinjection Facility at The Jackson Laboratory for making the Tg mice used in this study, Chris Smith (Mount Desert Island Biological Laboratory) for DNA sequencing, and Mary Ann Handel for critical reading of the manuscript. A.P. thanks Mary Hughes (Bates College) for colony work. This work was supported by National Science Foundation Grant MCB-9983161 (to J.C.S.), Biomedical Research Infrastructure Network Program of the National Center for Research Resources Grant P20 RR-016463 (to A.P.), Bates College (A.P.), and National Institutes of Health Cancer Center Support Grant P30-CA034196A (to The Jackson Laboratory).
Author contributions: J.C.S. and A.P. designed research; J.C.S., J.L.R., and A.P. performed research; J.C.S., J.L.R., and A.P. analyzed data; and A.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TRD, transmission ratio distortion; BAC, bacterial artificial chromosome; Tg, transgenic; OFM, offspring per female per month; SERAC1, serine active site containing 1; IP3, inositol-3,4,5-trisphosphate; ADAM2, β-fertilin; INPP5, inositol polyphosphate 5-phosphatase.
References
- 1.Hammer, M. F., Schimenti, J. & Silver, L. M. (1989) Proc. Natl. Acad. Sci. USA 86, 3261-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraser, L. R. & Dudley, K. (1999) BioEssays 21, 304-312. [DOI] [PubMed] [Google Scholar]
- 3.Lyon, M. F. (2003) Annu. Rev. Genet. 37, 393-408. [DOI] [PubMed] [Google Scholar]
- 4.Lyon, M. F. (1984) Cell 37, 621-628. [DOI] [PubMed] [Google Scholar]
- 5.Lyon, M. F. (1986) Cell 44, 357-363. [DOI] [PubMed] [Google Scholar]
- 6.Lyon, M. F. & Mason, I. (1977) Genet. Res. 29, 255-266. [Google Scholar]
- 7.Silver, L. M. & Remis, D. (1987) Genet. Res. 49, 51-56. [DOI] [PubMed] [Google Scholar]
- 8.Silver, L. M. (1989) Genet. Res. 54, 221-225. [DOI] [PubMed] [Google Scholar]
- 9.Herrmann, B. G., Koschorz, B., Wertz, K., McLaughlin, K. J. & Kispert, A. (1999) Nature 402, 141-146. [DOI] [PubMed] [Google Scholar]
- 10.Lyon, M. F. (1992) Genet. Res. 59, 27-33. [DOI] [PubMed] [Google Scholar]
- 11.Planchart, A., You, Y. & Schimenti, J. C. (2000) Genetics 155, 803-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett, D. & Artzt, K. (1990) Genet. Res. 56, 179-183. [DOI] [PubMed] [Google Scholar]
- 13.Planchart, A. & Schimenti, J. C. (2001) Mamm. Genome 12, 668-670. [DOI] [PubMed] [Google Scholar]
- 14.Lader, E., Ha, H. S., O'Neill, M., Artzt, K. & Bennett, D. (1989) Cell 58, 969-979. [DOI] [PubMed] [Google Scholar]
- 15.You, Y., Bergstrom, R., Klemm, M., Lederman, B., Nelson, H., Ticknor, C., Jaenisch, R. & Schimenti, J. (1997) Nat. Genet. 15, 285-288. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, L. C., Bennett, D. & Beasley, A. B. (1962) Genetics 47, 285-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett, D., Dunn, L. C. & Artzt, K. (1976) Genetics 83, 361-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 19.Soderlund, C., Humphray, S., Dunham, A. & French, L. (2000) Genome Res. 10, 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, B. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 22.Bergstrom, D. E., Bergstrom, R. A., Munroe, R. J., Lee, B. K., Browning, V. L., You, Y., Eicher, E. M. & Schimenti, J. C. (2003) Mamm. Genome 14, 817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schimenti, J. & Hammer, M. (1990) Mouse Genome 87, 108. [Google Scholar]
- 24.Hellsten, E., Evans, J. P., Bernard, D. J., Janne, P. A. & Nussbaum, R. L. (2001) Dev. Biol. 240, 641-653. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura, H., Cho, C., Branciforte, D. R., Myles, D. G. & Primakoff, P. (2001) Dev. Biol. 233, 204-213. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 27.Nemoto, Y., Arribas, M., Haffner, C. & DeCamilli, P. (1997) J. Biol. Chem. 272, 30817-30821. [DOI] [PubMed] [Google Scholar]
- 28.Khvotchev, M. & Sudhof, T. C. (1998) J. Biol. Chem. 273, 2306-2311. [DOI] [PubMed] [Google Scholar]
- 29.Nemoto, Y., Wenk, M. R., Watanabe, M., Daniell, L., Murakami, T., Ringstad, N., Yamada, H., Takei, K. & DeCamilli, P. (2001) J. Biol. Chem. 276, 41133-41142. [DOI] [PubMed] [Google Scholar]
- 30.Seet, L. F., Cho, S., Hessel, A. & Dumont, D. J. (1998) Biochem. Biophys. Res. Commun. 247, 116-122. [DOI] [PubMed] [Google Scholar]
- 31.Cenkci, B., Petersen, J. L. & Small, G. D. (2003) J. Biol. Chem. 278, 22574-22577. [DOI] [PubMed] [Google Scholar]
- 32.Dunn, L. C. (1960) Am. Nat. 94, 385-393. [Google Scholar]
- 33.Cho, C., Ge, H., Branciforte, D., Primakoff, P. & Myles, D. G. (2000) Dev. Biol. 222, 289-295. [DOI] [PubMed] [Google Scholar]
- 34.Lum, L. & Blobel, C. P. (1997) Dev. Biol. 191, 131-145. [DOI] [PubMed] [Google Scholar]
- 35.Braun, R. E., Behringer, R. R., Peschon, J. J., Brinster, R. L. & Palmiter, R. D. (1989) Nature 337, 373-376. [DOI] [PubMed] [Google Scholar]
- 36.Lyon, M. F. (1982) Genet. Res. 40, 102-103. [Google Scholar]
- 37.Rusk, N., Le, P. U., Mariggio, S., Guay, G., Lurisci, C., Nabi, I. R., Corda, D. & Symons, M. (2003) Curr. Biol. 13, 659-663. [DOI] [PubMed] [Google Scholar]
- 38.Nemoto, Y. & DeCamilli, P. (1999) EMBO J. 18, 2991-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malecz, N., McCabe, P. C., Spaargaren, C., Qiu, R., Chuang, Y. & Symons, M. (2000) Curr. Biol. 10, 1383-1386. [DOI] [PubMed] [Google Scholar]
- 40.Hellsten, E., Bernard, D. J., Owens, J. W., Eckhaus, M., Suchy, S. F. & Nussbaum, R. L. (2002) Biol. Reprod. 66, 1522-1530. [DOI] [PubMed] [Google Scholar]
- 41.Liu, Q., Shalaby, F., Jones, J., Bouchard, D. & Dumont, D. J. (1998) Blood 91, 2753-2759. [PubMed] [Google Scholar]
- 42.Kearns, B. G., McGee, T. P., Mayinger, P., Gedvilaite, A., Phillips, S. E., Kagiwada, S. & Bankaitis, V. A. (1997) Nature 387, 101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho, H. C. & Suarez, S. S. (2001) Biol. Reprod. 65, 1606-1615. [DOI] [PubMed] [Google Scholar]
- 44.De Blas, G., Michaut, M., Trevino, C. L., Tomes, C. N., Yunes, R., Darszon, A. & Mayorga, L. S. (2002) J. Biol. Chem. 277, 49326-49331. [DOI] [PubMed] [Google Scholar]
- 45.Olds-Clarke, P. (1983) Genet. Res. 42, 151-157. [DOI] [PubMed] [Google Scholar]
- 46.Olds-Clarke, P. & Johnson, L. R. (1993) Dev. Biol. 155, 14-25. [DOI] [PubMed] [Google Scholar]
- 47.Slaughter, G. R., Meistrich, M. L. & Means, A. R. (1989) Biol. Reprod. 40, 395-405. [DOI] [PubMed] [Google Scholar]
- 48.Walensky, L. D. & Snyder, S. H. (1995) J. Cell Biol. 130, 857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho, C., Bunch, D. O., Faure, J. E., Goulding, E. H., Eddy, E. M., Primakoff, P. & Myles, D. G. (1998) Science 281, 1857-1859. [DOI] [PubMed] [Google Scholar]
- 50.Veeramachaneni, V., Makalowski, W., Galdzicki, M., Sood, R. & Makalowska, I. (2004) Genome Res. 14, 280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]