Abstract
Background
Electronic health records (EHRs) can provide insights into diagnoses, treatment patterns, and clinical outcomes. Acquired hemophilia (AH) is an ultrarare bleeding disorder characterized by factor VIII inhibiting autoantibodies.
Aim
To identify patients with AH using an EHR database.
Methods
Records were accessed from a large EHR database (Humedica) between January 1, 2007 and July 31, 2013. Broad selection criteria were applied using the International Classification of Diseases, Ninth Revision, clinical modification (ICD-9-CM) code for intrinsic circulating anticoagulants (286.5 and all subcodes) and confirmation of records 6 months before and 12 months after the first diagnosis. Additional selection criteria included mention of “bleeding” within physician notes identified via natural language processing output and a normal prothrombin time and prolonged activated partial thromboplastin time.
Results
Of 6,348 patients with a diagnosis code of 286.5 or any subcodes, 16 males and 15 females met the selection criteria. The most common bleeding locations reported was gastrointestinal (23%), vaginal (16%), and endocrine (13%). A wide range of comorbidities was reported. Natural language processing identified chart note mention of “hemophilia” in 3 patients (10%), “bruise” in 15 patients (48%), and “pain” in all 31 patients. No patients received a prescription for approved/recommended AH treatments. Four patient cases were reviewed to validate whether the identified cohort had AH; each patient had bleeding symptoms and a normal prothrombin time and prolonged activated partial thromboplastin time, although none received hemostatic treatments.
Conclusion
In ultrarare disorders, ICD-9-CM coding alone may be insufficient to identify patient cohorts; multimodal analysis combined with in-depth reviews of physician notes may be more effective.
Keywords: acquired hemophilia, electronic health record, database, big data
Introduction
Acquired hemophilia A (AH) is an ultrarare, but potentially life-threatening autoimmune bleeding disorder characterized by the development of inhibitory autoantibodies (inhibitors) directed against plasma clotting factor VIII (FVIII).1 The incidence of AH is estimated at approximately 1 case per million per year, although the true incidence may be higher due to frequent under-reporting.2–4 Because of this low incidence, AH is not well studied and qualifies as an ultrarare disorder.5,6
In contrast to congenital hemophilia, AH affects both men and women and is most common in the elderly population (median age has been reported to be as high as 78 years7) and in postpartum females (median age in postpartum females, 28 years8).1 Diagnosis usually follows an unexpected bleeding event, surgery, or trauma and is often associated with an underlying medical condition (i.e., cancer, pregnancy, certain drugs);9 however, approximately half of all cases have no known cause (idiopathic).1 Unlike congenital hemophilia, the most common bleeding symptoms of AH include extensive bruising and cutaneous purpura, soft tissue bleeding (i.e., deep muscle bleeding), internal hemorrhage (including retroperitoneal), and persistent vaginal bleeding in postpartum women.1,10 Although often misdiagnosed as a different acquired bleeding disorder, such as disseminated intravascular coagulation, acquired von Willebrand syndrome, or acquired factor XIII deficiency,1,11 or complicated by potential anticoagulant treatment, AH can be differentiated by the characteristic coagulation test results of a normal prothrombin time (PT) and a prolonged activated partial thromboplastin time (aPTT) that does not correct after mixing with normal plasma.1
In the context of rare diseases, such as AH, where the feasibility of conducting randomized clinical trials is limited due to small patient numbers, electronic health record (EHR) databases are a potentially valuable tool for obtaining patient data.12 Furthermore, these databases can be analyzed using software applications in a relatively inexpensive and rapid manner, as compared to clinical trial studies. The organized format of information contained in EHRs enables researchers to study sequential data on symptoms and diagnosis to assess the presence of comorbidities,13 which may provide practical clinical information for improving disease management strategies and outcomes.14 One mechanism for organizing data from medical records involves the International Classification of Diseases, Ninth Revision (ICD-9) codes or its clinical modification (ICD-9-CM), which are used by physicians for epidemiologic and billing purposes to report patient health status and resource use; these codes may serve as inclusion and exclusion criteria to identify specific patient populations within a database.13 AH has historically been identified based on an ICD-9-CM code of 286.5 (hemorrhagic disorder due to intrinsic circulating anticoagulants), although the specific code 286.52 (AH) was created in 2012 to separate AH from diagnoses such as heparin overdose and lupus anticoagulant, and has a mapped code in ICD-10 (D68.311).
Although ICD-9-CM coding has the potential to organize the reporting of medical information, many clinicians document detailed patient information as free-text chart notes because of the greater ability to express patient case nuances and uncertainties, and in some instances, the requirements for medical documentation in standardized EHR platforms.12,14,15 Therefore, studying such unstructured (free-text) clinical narratives may provide contextual information about procedures, protocols, and patient characteristics that are not captured in the structured/coded EHR data (such as ICD-9-CM coding and laboratory results) and may be useful for identifying patients with rare or commonly undiagnosed conditions during retrospective data analysis. To more systematically access data contained in the chart notes, natural language processing (NLP) applications have been developed to scan EHR notes using an algorithm that recognizes relevant terms and clinical concepts (or “attributes”).12 To increase the sensitivity of this computational analysis process, the context in which a key term appears in the chart notes can also be processed for qualifying attributes to derive clinical meaning (or “sentiment”).16 The extracted content is then collated into a database for further analysis based on signs, diseases, and symptoms associated with the study population.
A careful consideration of the inclusion and exclusion criteria when analyzing EHR data is critical for obtaining an analysis population that reflects the patient population of interest. Here we report data from a study designed to define a population of patients with AH. The patient identification strategy used both ICD-9-CM–based inclusion criteria and NLP-based keyword screening, as well as laboratory test results and prescription information. We present the challenges encountered with obtaining accurate EHR data on an ultrarare disease, which support the need for improved coding processes and identification techniques.
Materials and methods
This study is a retrospective analysis of data from a large EHR database, and it was designed to identify patients with AH. The patient identification strategy combines standard identification techniques used in secondary database analysis with clinical information from the EHR, including a text search of physician notes using NLP. Patient records were accessed from Humedica, a service offering access to a large (~13 million patients) national, de-identified, longitudinal EHR database, between January 1, 2007 and July 31, 2013. Because the records are de-identified and no individually identifiable data were collected or analyzed, Institutional Review Board approval to conduct the study was not required. These EHR data were acquired directly from providers across the USA, spanning both the inpatient and outpatient care settings. Given the rare nature of the disease, broad selection criteria were initially applied by using an ICD-9-CM code of 286.5 (hemorrhagic disorder due to intrinsic circulating anticoagulants) and all 286.5 subcodes (implemented in 2012: 286.52 [AH], 286.53 [antiphospholipid antibody with hemorrhagic disorder], and 286.59 [other hemorrhagic disorder due to intrinsic circulating anticoagulants, antibodies, or inhibitors]). Additional inclusion criteria were activity in the database 6 months prior to and 12 months after the first diagnosis code of interest identified in the database for AH, no prescription for an anticoagulant (warfarin, heparin, Lovenox® [sanofi-aventis US, LLC, Bridgewater, NJ, USA], Fragmin® [Pfizer Inc, New York, NY, USA], Eliquis® [Bristol-Myers Squibb Co, Princeton, NJ, USA], Pradaxa® [Boehringer Ingelheim Pharmaceuticals, Inc, Ridgefield, CT, USA], or Xarelto® [Janssen Pharmaceuticals Inc, Titusville, NJ, USA), no diagnosis for systemic lupus (ICD-9-CM code 695.4) or lupus erythematosus (ICD-9-CM code 710.0), inclusion in the integrated delivery network, and mention of NLP terms for “blood” or “bleed” in the chart notes. (Full NLP list had 53 terms for blood/bleeding; some of the more common terms included black or bloody stools, bleeding, bleeding after intercourse, bleeding between periods, bleeding diathesis, bleeding disorder, bleeding duodenal ulcer, bleeding per rectum, bleeding risk, bleeding tendency, bleeding ulcer, blood clot, blood clotting disorder, blood disease, blood disorder, blood dyscrasia, blood issues, blood loss.)1 Cases were also required to have a record of a normal PT (defined as <16 seconds) and a clearly prolonged aPTT (defined as >50 seconds). Humedica’s NLP software was able to derive clinical meaning from the key terms using associated attributes, which are relevant descriptive features that further characterize the terms, from the surrounding chart notes. This information was then used to create semistructured variables to represent concepts, such as signs, diseases, and symptoms associated with the study population, for further analysis.
Results
Of ~13 million patients with usable records in the EHR database, 6,348 (~0.049%) had a diagnosis code of 286.5 or one of the 286.5 subcodes (286.52, 286.53, and 286.59). After applying the full set of inclusion criteria, an analysis population of 31 (16 males and 15 females) was obtained (Figure 1). The mean (SD) age of patients at the time of obtaining access to the database (2014) was 78 (20.3) years, and the median (interquartile range) age was 79 (30) years. Of the patients having any 286.5* diagnosis code (umbrella code indicating the broad diagnostic category “coagulation defects”),2 most patients had a diagnosis code of 286.5 (23 patients; 74%); other 286* codes were also identified (Table 1). Three patients (10%) also had a diagnosis code for congenital hemophilia A (286.0); however, only 1 patient (3%) was coded using the specific diagnosis code for AH (286.52). Other relevant diagnoses included 1 patient with liver disease (3%). No patients received a prescription for US Food and Drug Administration-approved AH treatment (the bypassing agent recombinant activated factor VII [NovoSeven® RT; Novo Nordisk A/S, Bagsvaerd, Denmark17]) or other treatments that have been reported for congenital hemophilia or AH (FVIII, factor IX, or desmopressin acetate [Stimate®; CSL Behring LLC, King of Prussia, PA, USA]).
Figure 1.
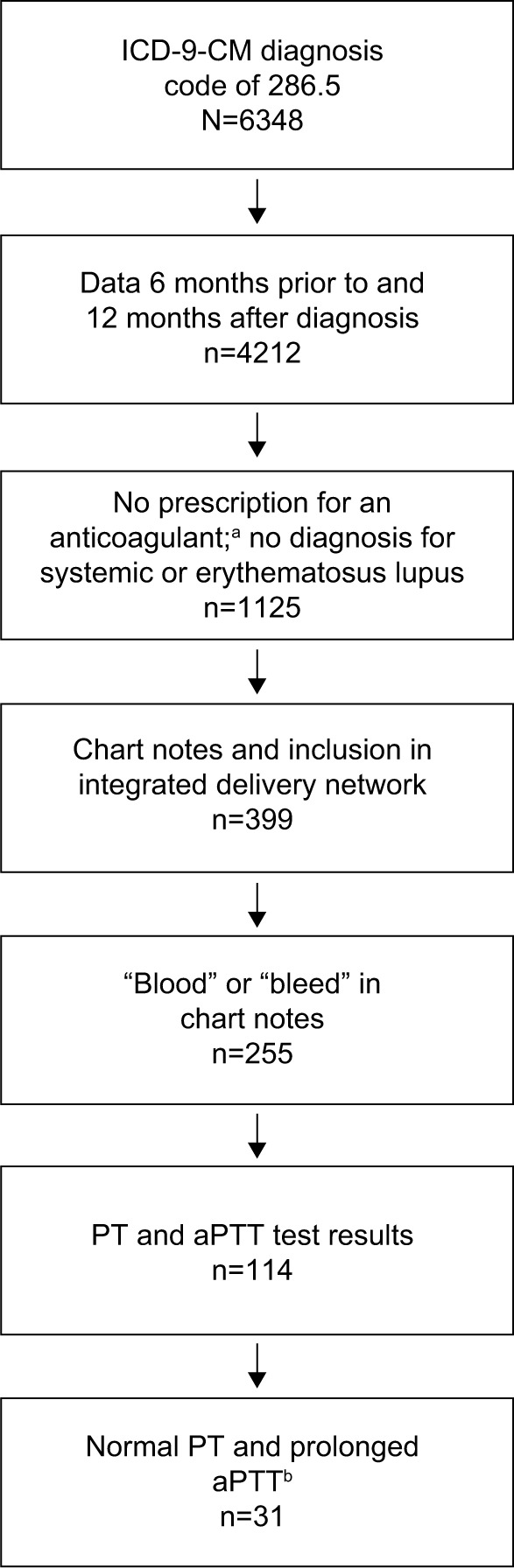
Patient population.
Notes: aAnticoagulants included warfarin, heparin, Lovenox, Fragmin, Eliquis, Pradaxa, and Xarelto. bNormal PT, <16 seconds; prolonged aPTT, >50 seconds.
Abbreviations: aPTT, activated partial thromboplastin time; ICD-9-CM, International Classification of Diseases, Ninth Revision, clinical modification; PT, prothrombin time.
Table 1.
Diagnosis codes
| Diagnosis code, n (%)a | Patients, n=31 |
|---|---|
| 286 – Coagulation defects | |
| 286.0 – Congenital factor VIII disorder | 3 (10) |
| 286.1 – Congenital factor IX disorder | 0 (0) |
| 286.2 – Congenital factor XI deficiency | 0 (0) |
| 286.3 – Congenital deficiency of other clotting factors | 0 (0) |
| 286.4 – von Willebrand’s disease | 0 (0) |
| 286.5 – Hemorrhagic disorder due to intrinsic circulating anticoagulants | 23 (74) |
| 286.52b – Acquired hemophilia | 1 (3) |
| 286.53b – Antiphospholipid antibody with hemorrhagic disorder | 0 (0) |
| 286.59b – Other hemorrhagic disorder due to intrinsic circulating anticoagulants, antibodies, or inhibitors | 4 (13) |
| 286.6 – Defibrination syndrome | 0 (0) |
| 286.7 – Acquired coagulation factor deficiency | 3 (10) |
| 286.9 – Other and unspecified coagulation defects | 4 (13) |
Notes:
Multiple selections allowed. Study selection criteria included an ICD-9-CM code of 286.5 or any 286.5 subcode.
286.5 subcodes established in 2012.
Abbreviation: ICD-9-CM, International Classification of Diseases, Ninth Revision, clinical modification.
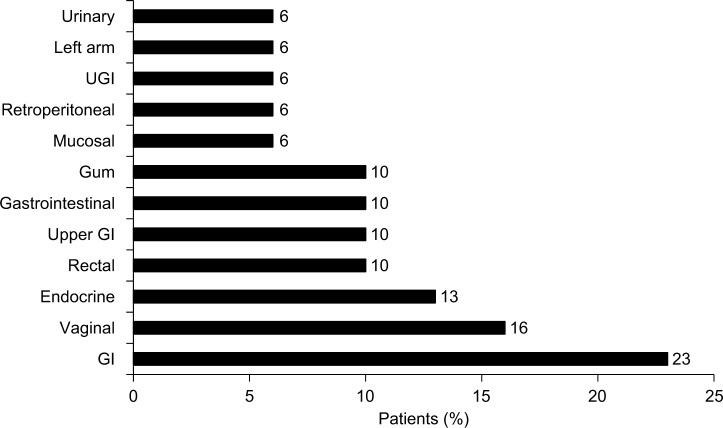
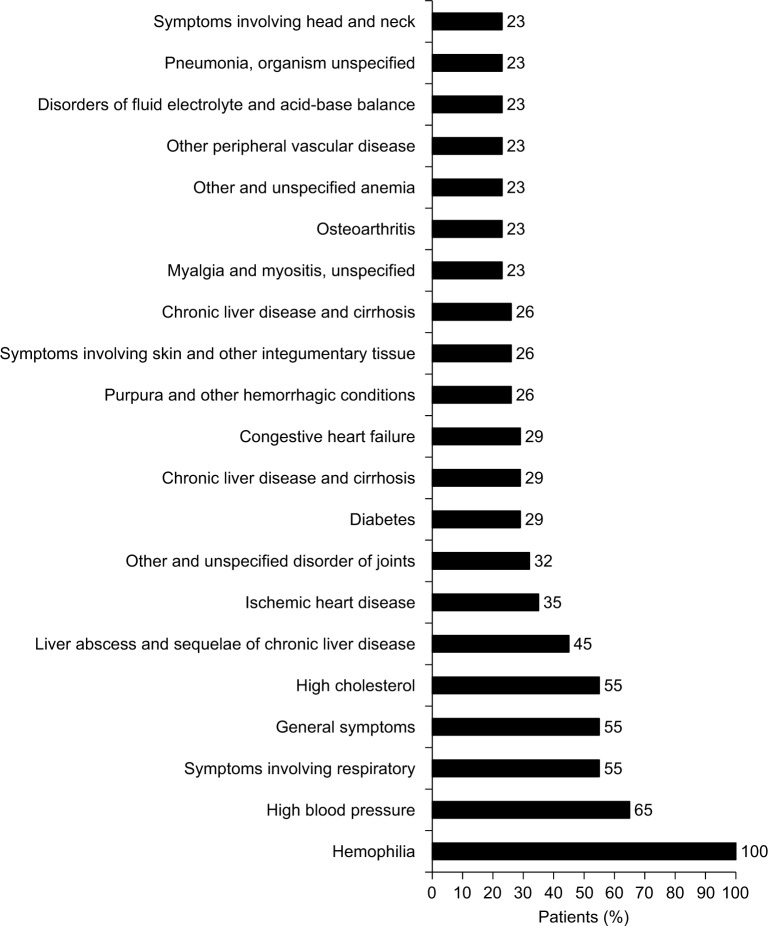
The most common bleeding locations associated with chart note mention of NLP terms for “blood” or “bleed” were gastrointestinal (23%), vaginal (16%), and endocrine (13%), as shown in Figure 2; however, there were several patient records that included documentation of bleeds that lacked information on bleed location. Patients exhibited a wide range of comorbidities, many of which may be related to advanced age, and all exhibited hemophilia as a comorbidity (Figure 3). The next most common comorbidities were high blood pressure (65%), symptoms involving the respiratory system (55%), general symptoms (55%), and high cholesterol (55%). Approximately one-third of the patients (32%) had “other and unspecified disorder of joints.” Using NLP, chart note mentions of “hemophilia” were identified in 3 patients (10%), “bruise” in 15 patients (48%), and “pain” in all 31 patients. For patients who had a location specified, the most common sites of pain were the chest (94%), abdomen (90%), back (68%), and neck (65%), as shown in Figure 4.
Figure 2.
Reported bleeding locations.
Figure 3.
Comorbidities.
Figure 4.
Sites of pain.
To evaluate whether patients had been accurately identified as having AH, 4 de-identified patients were selected at random for individual review of all available data by 2 clinicians; of these, 2 patients had also received an ICD-9-CM diagnosis code for congenital hemophilia A (286.0) and 1 patient had received ICD-9-CM codes for both congenital hemophilia A and AH (286.52). Although these individuals may not represent the entire study population, the patient cases summarized below exemplify some of the inconsistencies and challenges encountered with the AH patients extracted from the database using the inclusion and exclusion criteria in this study.
Case examples
Patient 1 was a male aged older than 89 years with a record of bruising and ecchymoses for approximately 6 months prior to the 286.5 diagnosis. Although the identifiable NLP terms in his medical record included “suspect” mention of “clotting factor deficiency”, no AH diagnosis was reported. His record included results from 13 aPTT tests, with the time ranging from 27.5 to 60.2 seconds (normal range ~28–40 seconds).
Patient 2 was a 70-year-old female with breast cancer and diagnosis codes of 286.59 (other hemorrhagic disorder due to intrinsic circulating anticoagulants, antibodies, or inhibitors) and 286.0 (congenital FVIII disorder). Her chart notes included isolated mention of hematemesis from a Mallory–Weiss tear, multiple mentions of hematuria, and, paradoxically, hemorrhagic cystitis and a renal pelvis blood clot on the same date. Her record included results from 12 aPTT tests, with the time ranging from 48.7 to 73.8 seconds.
Patient 3 was a 32-year-old pregnant female with a record of bruising and irregular menses and diagnosis codes of 286.5 (hemorrhagic disorder due to intrinsic circulating anticoagulants) and 286.0. Her chart notes included conflicting mention of venous thrombosis and blood clots, as well as FVIII deficiency, congenital hemophilia, and FVIII inhibitors. Her record included results from 17 aPTT tests, with the time ranging from 32.2 to 62.3 seconds.
Patient 4 was a 72-year-old female with diagnosis codes of 286.52 and 286.0 and approximately 3 years of note data. Her chart notes included recurrent mentions of melena and bloody stools throughout the first 2 years and frequent hematuria throughout the last year. The only mention of coagulopathy occurred concurrently with stroke and pneumonia; a note mentioning “improved” coagulopathy occurred on the same day. Her record included one aPTT result, which was 53 seconds.
Discussion
The objective of this study was to identify a population of patients with AH, an ultrarare bleeding disorder, using a strategy combining ICD-9-CM diagnoses, laboratory results, prescriptions, and NLP. The data analysis criteria specifying absence of prescription for an anticoagulant, no diagnosis of systemic lupus or lupus erythematosus, mention of NLP terms for “blood” or “bleed” in the chart notes, and evidence of a normal PT and prolonged aPTT were intended to increase the likelihood that all identified patients were accurately diagnosed with AH. Only 31 patients, from an initial 13 million included in the database, met all of the inclusion criteria (0.00029%). Additionally, only 1 patient in the identified cohort received the ICD-9-CM code specific for AH (286.52), which was established during the period from which patient records are referenced. However, given the incidence of AH,2–4 we anticipated ~13 cases. Although the average age of the study cohort was 78 years and many age-related comorbidities were identified, consistent with the known demographics of AH,7,9,18,19 none of the patients received a prescription for any typical known or effective AH treatment. Additionally, whereas for 15 patients (48%) NLP identified chart note mention of the term “bruise”, widespread ecchymosis was notably absent from the most common bleeding locations identified from the notes, despite being a common symptom of AH.20 Furthermore, among the 4 patients for whom a comprehensive data review was performed, none exhibited convincing evidence of having AH.
Whereas information documented in a structured manner, such as that provided by EHRs, has been considered to be useful for clinical research,21 various coding practices may affect the quality of EHR data entry. ICD-9 data are typically entered by a coder who assigns a code based on the information provided by the clinician as well as the available data in the patient’s medical record, such as pharmacy and prescription records. At some institutions, where coding is required to be complete before discharging a patient in order to meet insurance and billing requirements, diagnostic codes may be entered based on incomplete information, perhaps before the clinician has assigned a diagnosis, thereby creating inaccurate diagnoses.15,22 In the USA where diagnostic codes are integral for billing purposes, the level of reimbursement may be an influential factor during the coding process, which may potentially impact the utility of ICD-9-CM codes as an accurate source of clinical information in EHR research.14,23 Furthermore, in the context of ultrarare disorders such as AH, the accuracy of data entry may be limited by a coder’s experience and familiarity with coding for rare disorders, including potential misinterpretations of ICD-9-CM criteria. Additional factors affecting data entry include the use of variable terminology among clinicians or patients to describe a condition or symptom, the legibility of handwritten notes during transcription, dictation errors associated with voice recognition software, and facility coding guidelines that can affect the accuracy of data entry.22,24 In some cases, coders may enter individual codes for each condition instead of a combination code, which can further complicate database analyses due to the excess information required to sift through.22 Some of these coding practices may, therefore, have contributed to inaccurate ICD-9-CM coding in this study, resulting in an overidentification or misidentification of AH.
Inconsistent use of specialized codes may also hinder analyses of EHRs. A Canadian study of ICD-9-CM coding in the Alberta claims database found that only 43.5% of claims contained a detailed ICD-9-CM code (>3 digits), with the lowest proportion of use being among family physicians.25 Investigators proposed that family physicians may be less likely to use 4- or 5-digit codes compared with specialists, because many of their entries are initially investigative rather than diagnostic and may be limited in specificity for the purpose of referral to other specialists. Clinicians also demonstrated unique patterns of ICD-9-CM code use, with some codes being used more commonly than others in certain specialties and practices.26
In light of such challenges in obtaining accurate patient data from EHRs, patient chart notes may be used as an additional source of clinical information in EHR-based studies. Data from chart notes, however, are also subject to certain data limitations. Inaccuracies in chart notes may be caused by outdated information contained in replicated text that is carried forward, resulting in documentation errors.27,28 The volume and quality of data entered into the medical records may be influenced by physicians’ resistance to using EHR, which may be associated with a belief that data entry reduces the amount of time spent interacting with patients.28,29 In a survey of 1 hospital emergency department, Hill et al found that physicians reported spending 44% of their time at the computer and only 28% in direct patient care.29 To reduce the time associated with data entry, physicians may, therefore, copy and paste text to quickly populate patient notes;27,28 studies of medical records have identified large sections of copied text that contain outdated or incorrect information.27,30 In one such study, 82% of residents’ progress notes and 74% of attending physicians’ notes contained 20% or more copied text.27,30 Oftentimes, physicians may overlook the implications of such recording practices on research; while 71% of physicians acknowledged that inconsistencies and outdated information were more common in copied and pasted text, only 19% felt that it could have a negative impact on documentation.31
NLP is an important tool to extract useful information from chart notes; however, analyzing free text using NLP applications can be difficult. Redundancies from copied notes, typographic errors, and variability in grammar, abbreviations, and acronyms create complications. Additionally, mention of excluded diagnoses or attributes may complicate the accurate interpretation of text. It is likely that a potentially long list of symptoms, including multiple keywords (e.g., review of systems), may be associated with a common negative attribute indicated only at the beginning of the list (e.g., “the patient had no complaints of…” or “the examination revealed no evidence of…”).
Together, these issues highlight the complexity of information and the potential for error in EHR databases that pose significant challenges to investigators searching for relevant, contextual data, especially in ultrarare disorders.32 These issues are also consistent with the coding and chart note discrepancies observed in our AH cohort, supporting the need for more sophisticated and sensitive EHR analysis strategies. For ultrarare disorders, in order to effectively identify patient cohorts of interest, ICD-9 coding should be combined with a multimodal analysis strategy that includes in-depth reviews of physician notes.33 EHR databases have been shown to yield more accurate patient cohorts when multiple sources are used for screening.33 Future studies should be designed to account for the potential limitations of missing or inaccurately entered data, which can skew results32 and reduce the value of data, particularly in studies of rare conditions with highly limited numbers of patients.27 Additionally, guiding principles established by the American Medical Informatics Association may be useful in ensuring efficient, reliable, and high-quality information reporting that can be used for policymaking, education, reimbursement, and research purposes.23 In the context of diseases such as AH, which are relatively more common among older individuals,20 use of a Medicare database may be useful in providing access to patient records of interest.
Study limitations
Our ability to identify a population of patients with AH may have been limited by multiple factors. Inherent limitations in using an EHR database include the potential for ICD-9 coding errors, variable coding practices, and data inconsistencies. The use of chart notes and NLP to inform inclusion criteria may also be limited by variable chart use among physicians and potential errors in NLP translation. Our ability to characterize the analysis population was limited by the small patient numbers obtained, due to the extreme rarity of AH. Additionally, numbers of AH patients identified in this analysis may have been limited by the inclusion and exclusion criteria (i.e., requiring activity in the database at least 6 months prior to and 12 months after the first diagnosis code of interest and no prescription for an anticoagulant or diagnosis of systemic or erythematosus lupus); however, these criteria were considered to be important in ensuring that patient database activity could be tracked and for excluding patients with diagnoses other than AH.
Conclusion
NLP approaches to analysis of EHRs hold promise and have demonstrated utility in population-based studies for commonly occurring disorders. In this study, ICD-9-CM codes, laboratory test results, NLP output, and treatments were not consistently aligned. This study highlights that in ultrarare disorders, ICD-9-CM coding alone may not be sufficient to identify cohorts, and multimodal analysis combined with in-depth reviews of physician notes may be more effective. Furthermore, improving the quality of EHR records may be critical in supporting “big data” initiatives aimed at analyzing trends in medical care.
Acknowledgments
Writing assistance was provided by Anna Abt, PhD, and Shawn Keogan, PhD, of ETHOS Health Communications in Yardley, Pennsylvania, and was supported financially by Novo Nordisk Inc., Plainsboro, New Jersey, in compliance with international Good Publication Practice guidelines. We thank Ekaterine Bakhtadze Bagci, Christina Stentoft Hoxer, Anders Krabbe, Snejana Krassova, and Lars Wilkinson for their review and input to the manuscript.
The abstract of this paper was presented, in part, at the 57th Annual Meeting of the American Society of Hematology as a poster presentation. The poster’s abstract was published in “Poster Abstracts” in Blood. 2015;126:3271. http://www.bloodjournal.org/content/126/23/3271.
Footnotes
Authors’ contribution
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
M Wang is a consultant for Novo Nordisk Inc. A Cyhaniuk is a principal consultant of AC Analytical Solutions, LLC. DL Cooper and NN Iyer are employees of Novo Nordisk Inc., the sponsor of the study. The authors report no other conflicts of interest in this work.
References
- 1.Giangrande P. Acquired Hemophilia, Revised Edition. Montréal, Québec, Canada: World Federation of Hemophilia; 2012. [Accessed June 15, 2017]. (Treatment of Hemophilia No. 38). http://www1.wfh.org/publication/files/pdf-1186.pdf. [Google Scholar]
- 2.Franchini M, Mannucci PM. Acquired haemophilia A: a 2013 update. Thromb Haemost. 2013;110(6):1114–1120. doi: 10.1160/TH13-05-0363. [DOI] [PubMed] [Google Scholar]
- 3.Franchini M, Gandini G, Di Paolantonio T, Mariani G. Acquired hemophilia A: a concise review. Am J Hematol. 2005;80(1):55–63. doi: 10.1002/ajh.20390. [DOI] [PubMed] [Google Scholar]
- 4.Collins P, Macartney N, Davies R, Lees S, Giddings J, Majer R. A population based, unselected, consecutive cohort of patients with acquired haemophilia A. Br J Haematol. 2004;124(1):86–90. doi: 10.1046/j.1365-2141.2003.04731.x. [DOI] [PubMed] [Google Scholar]
- 5.National Institute for Clinical Excellence . Citizens Council Report: Ultra Orphan Drugs. London, UK: National Institute for Clinical Excellence; 2004. [Accessed June 15, 2017]. Available from: https://www.nice.org.uk/Media/Default/Get-involved/Citizens-Council/Reports/CCReport04UltraOrphanDrugs.pdf. [Google Scholar]
- 6.European Union Committee of Experts on Rare Diseases (EUCERD) 2012 Report on the State of the Art of Rare Disease Activities in Europe. Part V: Activities of Member States and Other European Countries in the Field of Rare Diseases. [Accessed June 15, 2017]. Available from: http://ec.europa.eu/health//sites/health/files/rare_diseases/docs/eucerd2012_report_state_of_art_rare_diseases_activities_5.pdf.
- 7.Collins PW, Hirsch S, Baglin TP, et al. Acquired hemophilia A in the United Kingdom: a 2-year national surveillance study by the United Kingdom Haemophilia Centre Doctors’ Organisation. Blood. 2007;109(5):1870–1877. doi: 10.1182/blood-2006-06-029850. [DOI] [PubMed] [Google Scholar]
- 8.Hauser I, Schneider B, Lechner K. Post-partum factor VIII inhibitors. A review of the literature with special reference to the value of steroid and immunosuppressive treatment. Thromb Haemost. 1995;73(1):1–5. [PubMed] [Google Scholar]
- 9.Knoebl P, Marco P, Baudo F, et al. Demographic and clinical data in acquired hemophilia A: results from the European Acquired Haemophilia registry (EACH2) J Thromb Haemost. 2012;10(4):622–631. doi: 10.1111/j.1538-7836.2012.04654.x. [DOI] [PubMed] [Google Scholar]
- 10.Franchini M, Targher G, Montagnana M, Lippi G. Laboratory, clinical and therapeutic aspects of acquired hemophilia A. Clin Chim Acta. 2008;395(1–2):14–18. doi: 10.1016/j.cca.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Sakurai Y, Takeda T. Acquired hemophilia A: a frequently overlooked autoimmune hemorrhagic disorder. J Immunol Res. 2014;2014:320674. doi: 10.1155/2014/320674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sujansky WV. The benefits and challenges of an electronic medical record: much more than a “word-processed” patient chart. West J Med. 1998;169(3):176–183. [PMC free article] [PubMed] [Google Scholar]
- 13.Tomines A, Readhead H, Readhead A, Teutsch S. Applications of electronic health information in public health: uses, opportunities & barriers. EGEMS (Wash DC) 2013;1(2):1019. doi: 10.13063/2327-9214.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan N, Hanauer D, Keller B. Mining electronic health records. Computer. 2010;43(10):77–81. [Google Scholar]
- 16.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NovoSeven RT Coagulation Factor VIIa (Recombinant) Room Temperature Stable [prescribing information] Plainsboro, NJ: Novo Nordisk; 2014. [Google Scholar]
- 18.Borg JY, Guillet B, Le Cam-Duchez V, Goudemand J, Levesque H, SACHA Study Group Outcome of acquired haemophilia in France: the prospective SACHA (Surveillance des Auto antiCorps au cours de l’Hemophilie Acquise) registry. Haemophilia. 2013;19(4):564–570. doi: 10.1111/hae.12138. [DOI] [PubMed] [Google Scholar]
- 19.Tiede A, Klamroth R, Scharf RE, et al. Prognostic factors for remission of and survival in acquired hemophilia A (AHA): results from the GTH-AH 01/2010 study. Blood. 2015;125(7):1091–1097. doi: 10.1182/blood-2014-07-587089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lak M, Sharifian RA, Karimi K, Mansouritorghabeh H. Acquired hemophilia A: clinical features, surgery and treatment of 34 cases, and experience of using recombinant factor VIIa. Clin Appl Thromb Hemost. 2010;16(3):294–300. doi: 10.1177/1076029608331227. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–186. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health Serv Res. 2005;40(5 Pt 2):1620–1639. doi: 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cusack CM, Hripcsak G, Bloomrosen M, et al. The future state of clinical data capture and documentation: a report from AMIA’s 2011 policy meeting. J Am Med Inform Assoc. 2013;20(1):134–140. doi: 10.1136/amiajnl-2012-001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.American Health Information Management Association AHIMA e-HIM work group on speech recognition in the EHR. 2003. [Accessed August 26, 2015]. http://library.ahima.org/xpedio/groups/public/documents/ahima/bok1_022107.hcsp?dDocName=bok1_022107. Published.
- 25.Cunningham CT, Cai P, Topps D, Svenson LW, Jette N, Quan H. Mining rich health data from Canadian physician claims: features and face validity. BMC Rese Notes. 2014;7:682. doi: 10.1186/1756-0500-7-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royer JA, Hardin JW, McDermott S, et al. Use of state administrative data sources to study adolescents and young adults with rare conditions. J Gen Intern Med. 2014;29(Suppl 3):S732–S738. doi: 10.1007/s11606-014-2925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowman S. Impact of electronic health record systems on information integrity: quality and safety implications. Perspect Health Inf Manag. 2013;10:1c. [PMC free article] [PubMed] [Google Scholar]
- 28.Pollack R. Doctors, hospitals rethinking electronic medical records mandated by 2009 law. Washington Examiner. Oct 10, 2014. [Accessed June 15, 2017]. Available from: http://www.washingtonexaminer.com/doctors-hospitals-rethinking-electronic-medical-records-mandated-by-2009-law/article/2554622.
- 29.Hill RG, Jr, Sears LM, Melanson SW. 4000 clicks: a productivity analysis of electronic medical records in a community hospital ED. Am J Emerg Med. 2013;31(11):1591–1594. doi: 10.1016/j.ajem.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 30.Thornton JD, Schold JD, Venkateshaiah L, Lander B. Prevalence of copied information by attendings and residents in critical care progress notes. Crit Care Med. 2013;41(2):382–388. doi: 10.1097/CCM.0b013e3182711a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrenn JO, Stein DM, Bakken S, Stetson PD. Quantifying clinical narrative redundancy in an electronic health record. J Am Med Inform Assoc. 2010;17(1):49–53. doi: 10.1197/jamia.M3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders CM, Saltzstein SL, Schultzel MM, Nguyen DH, Stafford HS, Sadler GR. Understanding the limits of large datasets. J Cancer Educ. 2012;27(4):664–669. doi: 10.1007/s13187-012-0383-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21(2):221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]