Abstract
Circulating monocytes mediate inflammation in atherosclerosis and may serve as easily accessible reporters of disease. To search for markers of atherosclerosis, we compared the in vivo transcriptomes of monocytes purified from patients undergoing carotid endarterectomy and normal subjects by using the serial analysis of gene expression technique. We selected a subset of differentially expressed monocyte-specific genes and confirmed their expression levels. The Finkel–Biskis–Jinkins osteosarcoma (FOS) gene was significantly increased in patients, and the highest levels of FOS associated with patients who had previously undergone coronary revascularization. The correlation between coronary revascularization and FOS was higher than that compared with the cardiac risk marker high sensitivity C-reactive protein. In vitro inhibition of FOS using small interfering RNA and 3-hydroxy-3-methyl-glutaryl CoA reductase inhibitor simvastatin (statin) affected monocyte activation and suggested an important role in pathogenesis. Given the prominent role of FOS in inflammation and calcification, its association with atherosclerosis severity has clear pathophysiologic bases as well as clinical implications as a marker. Our results suggest that analysis of gene expression in circulating cells may provide biological and clinical insights into human atherosclerosis, and that this type of approach may be applicable for studying other types of diseases.
Keywords: monocytes, serial analysis of gene expression, Finkel–Biskis–Jinkins osteosarcoma, 3-hydroxy-3-methyl-glutaryl CoA reductase inhibitor, high sensitivity C-reactive protein
The importance of inflammation in atherosclerosis has been well established, and inflammatory markers such as high-sensitivity C-reactive protein (hsCRP) are being used for cardiac risk stratification (1, 2). With increasing prevalence of atherosclerosis risk factors such as aging, obesity, and the metabolic syndrome, the discovery of new biomarkers and therapeutic targets may assist in the management of this common disease (3–5).
Cardiovascular investigators have been limited by a number of factors such as difficulty in obtaining diseased tissue, functional complexity of the system, and lack of in vitro human disease models. We reasoned that the variety of blood cells that circulate throughout the body present an ideal tissue for atherosclerosis studies for four reasons. (i) They are easily accessible and include inflammatory cells such as monocytes that are critical elements in the atherosclerotic process. (ii) Circulating blood cells are in contact with the diseased endovascular lumen and as such may serve as reporters. (iii) The variety of blood cells have defined cell surface markers facilitating their purification to homogeneity for gene expression analysis. (iv) There are immortalized human monocytic cell lines, which retain differentiated phenotypes, and can thus support in vitro studies.
To identify disease markers and genes involved in atherosclerosis, we quantified gene expression in circulating monocytes from a limited set of patients with atherosclerosis and normal subjects by using the serial analysis of gene expression (SAGE) technique (6–8). This comparison revealed higher levels of various stress response and inflammatory gene transcripts in the monocytes of patients compared with normal controls, and one in particular, the Finkel–Biskis–Jinkins osteosarcoma (FOS) gene, was strongly expressed in the circulating monocytes of patients. FOS was first identified as an osteosarcoma oncogene, and its importance in inflammation and calcification fits with the known pathogenetic changes in atherosclerosis (9–15). In comparison with plasma hsCRP, measurement of FOS transcript levels showed that it was more significantly associated with severe atherosclerosis. Although FOS has recently been localized to smooth muscle cells and macrophages in plaques, its pathophysiologic role and clinical utility in atherosclerosis remain unexplored (16). We present complementary clinical and basic experimental data showing that FOS is a marker and mediator of atherosclerosis and that similar approaches examining the circulating transcriptome in other conditions may be equally fruitful.
Materials and Methods
Human Subjects. All patients and normal volunteers were recruited after informed consent in accordance with the National Institutes of Health Internal Review Board. The patients were selected from those scheduled to undergo carotid endarterectomy (CEA) for atherosclerotic disease. A significant fraction of the patients also had concomitant coronary heart disease. The normal control subjects were screened to ensure absence of significant atherosclerosis based on history and physical examination, electrocardiogram, echocardiogram, exercise stress testing, and carotid artery ultra-sonogram with intima-media thickness (IMT) measurements. The mean IMT measurements of the right and left common carotid arteries for the controls, 0.84 ± 0.12 mm and 0.85 ± 0.18 mm, respectively, were within the low cardiovascular risk category. The exclusions criteria for all subjects were: chronic infections, vasculitis or any other inflammatory disease, neoplastic disease, immunosuppressive therapy, and chemotherapy.
Blood Purification. Blood samples were collected in citratecontaining Vacutainer CPT tubes (Becton Dickinson) from controls and from patients intraoperatively and processed within 1 h to obtain mononuclear cells (MNC) (17). MNCs were washed at 4°C and resuspended in RNA lysis/binding buffer (Dynal, Great Neck, NY). For monocytes purification, CD14 MicroBeads and Fc Blocking reagent were used per protocol (Miltenyi Biotec, Auburn, CA). Cell viability was >95% by Trypan blue exclusion with high purity as determined by flow-cytometry (>95% CD14+) and RT-PCR (Fig. 4 A and B, which is published as supporting information on the PNAS web site).
Macrophage Purification. Within 1 h of surgical resection, human carotid artery plaques were processed as described with modifications (18, 19). The tissue was cubed (0.5 mm) and digested in Hanks' balanced salt solution (HBSS), Hepes (4.8 mg/ml), containing collagenase type IV (450 units/ml), DNase I (500 units/ml), and trypsin inhibitor (1 mg/ml) (Worthington) for 30 min to 1 h at 37°C. The cell suspension was sequentially filtered through 600- to 40-μm nylon filters (Spectrum Laboratories, Houston) and macrophages isolated by using CD14 Microbeads. Cell viability and macrophage purity were similar to that described above for monocytes (Fig. 4B).
Cells and Tissue Culture. All human monocytic cell lines were obtained from the American Type Culture Collection. The Mono-Mac6 cell line was a generous gift of H. W. Ziegler-Heitbrock (20). Untouched primary monocytes were isolated from blood of healthy donors by using a negative selection kit (Miltenyi Biotec) and cultured as described (21).
SAGE. We used the LongSAGE protocol that increases the specificity of this sequencing-based gene expression technique (7). A SpectruMedix 192-capillary automated sequencer (SpectruMedix, State College, PA) was used for sequencing 50,000–100,000 tags per library. SAGE tags were obtained by using the sage2000 software (www.sagenet.org), normalized to 100,000 tags per library and identified by using the Unigene/SAGEmap database (22). Tags matching a single Unigene cluster were summed and fold-change/total tag queries were performed by using Microsoft access.
Quantitative Real-Time RT-PCR. mRNA from lysates were purified by binding to poly(dT) magnetic beads (Dynal) and reverse transcribed by using SuperScript II (Invitrogen). Primer sequences for all of the various genes are provided in Table 3, which is published as supporting information on the PNAS web site. Standard quantitative RT-PCR was performed in duplicates at least two to three times by using SYBR Green (Molecular Probes) and TaqMan protocols on the 7900HT (Applied Biosystems) (23). RT-PCR data were normalized by measuring average cycle threshold (Ct) ratios between candidate genes and two different control genes, eukaryotic translation initiation factor (EIF3S5 or TIF) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The formula 2Ct(Candidate)/2Ct(Control) was used to calculate normalized ratios. Color-coded normalized fold changes were generated from log-transformed control-normalized ratios (normalized Ct ratio divided by the average Ct ratio of all control samples) by using cluster 2.2 and treeview software (http://rana.lbl.gov/EisenSoftware.htm) (23).
Immunohistochemistry and Western Blotting. The following antibodies were used: rabbit polyclonal anti-FOS (Santa Cruz Biotechnology), mouse monoclonal anti-human CD14 (Immunotech, Westbrook, ME), mouse monoclonal anti-GAPDH (Ambion), and negative control mouse IgG (Biocare, Walnut Creek, CA). Serial cryosections (8–10 μm) of carotid and coronary plaques were immunostained with Vector blue substrate (Vector Laboratories) and developed by secondary antibody conjugated to alkaline phosphatase (24). Western blotting was done as described (25).
Plasma hsCRP Measurements. Plasma hsCRP levels were determined by using an immunoassay per protocol (BioCheck, Foster City, CA). To ensure accuracy, all samples were remeasured and validated by an external laboratory (Quest Diagnostics, Teterboro, NJ).
FOS Inhibition by Small Interfering RNA (siRNA). Nonspecific and FOS siRNA duplexes were purchased from Dharmacon Research (Lafayette, CO) and transfected according to standard protocol (Amaxa, Gaithersburg, MD).
Monocyte Function. Cells were pretreated with 10 μM simvastatin and/or 1 mM mevalonate for 20 h and then stimulated with phorbol 12-myristate 13-acetate (PMA, Sigma). Unattached cells were gently washed twice and pooled. The adherent cells were released with trypsin-EDTA, and viable cells were counted by using Trypan blue. Twenty-four-hour cumulative monocyte chemoattractant protein 1 (MCP-1) release into the medium was determined by immunoassay (R & D Systems).
Statistical Analysis. P values for SAGE tag counts were calculated accounting for sample size differences between libraries as described (26). Data are expressed as mean ± SE. P values were calculated with the use of a two-tailed Student's t test. The significance of paired t test results between normal subjects and patients was confirmed with the Mann–Whitney test. In light of the number of comparisons performed in this exploratory study, statistical significance was ascribed to P values <0.01. The receiver operating characteristic plots were generated by obtaining sensitivity and specificity at multiple threshold values for FOS and hsCRP. (analyse-it Software, Leeds, England).
Results
SAGE. We used the strategy of creating a limited number of SAGE libraries by using purified monocytes to screen for monocyte-specific candidate genes, followed by quantitative RT-PCR using MNCs to more rapidly confirm candidate genes in larger groups of subjects. Briefly, monocytes were purified from the whole MNC fraction by using a CD14 antibody conjugated to magnetic beads, and the remaining fraction was considered the monocyte-depleted MNC (non-monocyte) fraction. We made a total of seven SAGE libraries, five by using CD14+ monocytes and two by using non-monocyte MNCs. The five monocytes libraries were made as follows: two from patients (P1 and P2) with atherosclerosis undergoing CEA surgery; one from an age-matched control subject (C1) without clinical evidence of atherosclerosis; and two from younger age subjects (A1 and A2) to serve as additional controls and to exclude age-related changes (Table 1). The patients P1 and P2 were selected because both had a history of coronary heart disease. Two non-monocyte libraries were made from subjects P1 and A1 for screening out potential candidate genes also expressed in non-monocytes.
Table 1. Subjects.
| SAGE subjects
|
RT-PCR confirmation subjects
|
||||||
|---|---|---|---|---|---|---|---|
| P1 | P2 | C1 | A1 | A2 | Controls (n = 19) | Patients (n = 25) | |
| Age, yr | 71 | 72 | 68 | 45 | 39 | 70 ± 5 | 74 ± 8 |
| Gender | M | F | F | M | M | 42% male | 60% male |
| Systolic blood pressure, mm Hg | 134 | 120 | 145 | 130 | 115 | 141 ± 18 | 143 ± 20 |
| LDL, mg/dl | 101 | 100 | 65 | 87 | 124 | 110 ± 33 | 103 ± 28 |
| HDL, mg/dl | 44 | 66 | 48 | 55 | 47 | 56 ± 21 | 53 ± 16 |
| Diabetes mellitus | – | – | – | – | – | 5% | 8% |
| Current smoker | – | – | – | – | – | 0% | 4% |
| Family history of CHD | – | + | – | – | – | 36% | 48% |
| History of CHD | + | + | – | – | – | 0% | 56% |
| Body mass index, kg/m2 | 29.2 | 27.7 | 28.0 | 25.6 | 23.0 | 26.3 ± 3.8 | 26.4 ± 4.9 |
| Framingham 10-yr CHD risk, % | 18 | 8 | 9 | 3 | 3 | 13 ± 9 | 15 ± 10 |
Individual profiles of the subjects used for SAGE library construction and group profiles of normal subjects (Controls) and CEA patients with atherosclerosis (Patients) used for quantitative RT-PCR. Subjects with atherosclerosis: P1, patient 1; P2, patient 2. Subjects without clinically significant atherosclerosis: C1, age-matched control 1; A1, younger age control 1; A2, younger age control 2. CHD, coronary heart disease.
A total of 460,012 SAGE tags, or an estimated 2- to 3-fold redundant coverage of the transcriptome, from the five monocyte SAGE libraries (P1, P2, C1, A1, A2) were sequenced and matched to 13,154 genes in the Unigene database. Based on known tags expressed on average at least two tags per library, the pairwise correlation coefficients between monocyte libraries were very high, 0.9992 ± 0.0004. As expected of monocytes purified by using the CD14 surface antigen, CD14 transcripts were greatly enriched as were other monocyte markers such as CD163 (Table 2; SAGE libraries available at http://cgap.nci.nih.gov/SAGE/). In contrast, the non-monocyte SAGE libraries were enriched in T and B lymphocyte markers such as CD3E and CD79A, respectively (Table 2).
Table 2. SAGE libraries.
| Normalized SAGE tag counts
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Monocyte
|
Non-monocyte
|
||||||||
| Gene description | SAGE tag sequence | Unigene ID | P1 | P2 | C1 | A1 | A2 | P1 | A1 |
| Hematopoietic markers | |||||||||
| Monocyte | |||||||||
| CD14 antigen | TGGTCCAGCGCCCTGAA | 163867 | 47 | 64 | 100 | 106 | 78 | 4 | 0 |
| CD163 antigen | GAGGTTCCTGGGGGACA | 504641 | 23 | 20 | 21 | 11 | 20 | 0 | 0 |
| Non-monocyte | |||||||||
| CD3E antigen, epsilon (TiT3 complex) | TAAGTTGTCCCCCATCC | 3003 | 0 | 0 | 0 | 0 | 5 | 28 | 54 |
| CD79A antigen (Ig-associated alpha) | TATGAGGACATCTCCCG | 79630 | 0 | 2 | 2 | 2 | 2 | 30 | 20 |
| Pan-leukocyte | |||||||||
| CD99 antigen | GGATGTGAAAGGCTGGC | 495605 | 26 | 48 | 32 | 29 | 39 | 66 | 54 |
| Monocyte candidate genes | |||||||||
| FOS, osteosarcoma viral oncogene homolog | TGGAAAGTGAATTTGAA | 25647 | 111 (5.8) | 138 (7.3) | 19 | 27 | 11 | 8 | 4 |
| DUSP1, dual specificity phosphatase 1 | CTTGACATACCTACCAG | 171695 | 71 (3.0) | 92 (3.8) | 24 | 17 | 14 | 12 | 2 |
| NFKBIA, NFK gene in B cell inhibitor, alpha | TAACAGCCAGGAGTGCT | 81328 | 40 (2.0) | 56 (2.8) | 20 | 10 | 15 | 16 | 16 |
| ID2, inhibitor of DNA binding 2 | CTAAACTTTTTATAAAA | 180919 | 32 (1.7) | 42 (2.2) | 19 | 7 | 5 | 16 | 12 |
| PER1, period homolog 1 | GAGTCCCTGGTGCTGCC | 445534 | 30 (1.8) | 60 (3.5) | 17 | 1 | 1 | 0 | 2 |
| SAP30, sin3-associated polypeptide, 30 kDa | TAGAAATGTTCTTTGTG | 512813 | 10 (1.7) | 30 (5.0) | 6 | 3 | 4 | 2 | 0 |
Hematopoietic markers and monocyte candidate gene tag counts are tabulated under the various SAGE libraries, along with their associated sequences and gene identification numbers. A total of seven SAGE libraries are shown, five CD14+ monocyte (Monocyte) and two monocyte-depleted (Non-monocyte) libraries. The tag counts shown are normalized to 100,000 tags per library. Parentheses represent patient-to-control C1 tag ratio. P1, patient 1; P2, patient 2; C1, age-matched control 1; A1, younger age control 1; A2, younger age control 2.
Evaluation of Candidate Genes. SAGE tag comparisons were made between the two patients P1 and P2 and the control C1 monocyte SAGE libraries (Table 2). To raise the stringency and reproducibility of our screen, we considered only tags increased at least 1.5-fold in both P1 and P2 monocyte libraries to obtain a list of 297 candidates (with total tag counts in all libraries ≥25 tags) (Table 4, which is published as supporting information on the PNAS web site). To each tag from this preliminary list, the following additional criteria were applied: (i) low tag counts in both control A1 and A2 monocyte libraries to rule out age-related differences; and (ii) low tag counts in non-monocyte libraries for selecting relatively monocyte-specific genes. We also observed 267 genes repressed by 1.5-fold in P1 and P2 monocytes libraries but were not evaluated further in this study (Table 5, which is published as supporting information on the PNAS web site).
Using the above criteria, we selected six candidate genes [FOS, dual specificity phosphatase 1 (DUSP1), nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor-α (NFKBIA), inhibitor of DNA binding 2 (ID2), period homolog 1 (PER1), and sin3-associated polypeptide (SAP30)] all associated with regulatory or transcriptional functions (Table 2). The two most differentially expressed candidates were FOS, a protooncogene involved in proliferation and differentiation, and DUSP1, a stress response phosphatase important for mitogen-activated protein kinase (MAPK) regulation (P < 0.001, Table 4) (14, 27). A few differentially expressed SAGE tags were without gene assignment, but follow-up revealed that most were polymorphic tags from highly expressed known genes. We did not observe strong differentially expressed candidates in the non-monocyte SAGE libraries P1 and A1 containing mixed populations of cells.
To minimize sample processing and purification requirements, we examined the feasibility of using the whole MNC fraction for measuring monocyte-specific gene expression. This concept seemed possible because the monocyte content of patient and control MNC samples was similar, 20 ± 9% and 22 ± 9%, respectively. The fold change ratios of FOS and DUSP1 mRNA levels (patients versus controls) in the MNC and monocyte fractions were identical, indicating that we could use MNCs to accurately and rapidly detect monocyte-specific gene expression (Fig. 5, which is published as supporting information on the PNAS web site). Additionally, it confirmed that the monocyte purification step had not altered their expression levels.
Quantitative RT-PCR of Subject Samples. To confirm our SAGE data, a total of 25 patients scheduled for CEA and 19 age-matched normal control subjects were selected (Table 1). Although the patient and control subjects were closely matched by age, notable differences could be seen due to the inherent risk factors associated with atherosclerosis such as male gender, family history, and prior history of coronary artery disease. Treatment for hypertension and hyperlipidemia were more prevalent among the patients compared with controls, 92% vs. 32% and 80% vs. 37%, respectively, but the measured blood pressure and low-density lipoprotein (LDL) cholesterol levels were comparable at the time of the study.
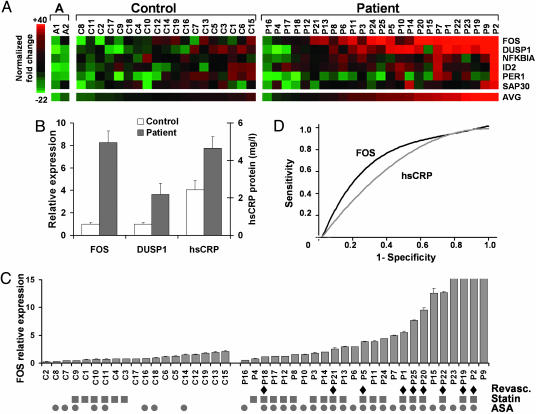
The relative expression levels of the six candidate genes were color-coded and ordered by their average values (AVG) in the control and patient groups (Fig. 1A). The high levels of FOS and DUSP1 in patients confirmed SAGE data and largely determined the patient ordering. The mean FOS and DUSP1 RT-PCR fold increases in patients over controls were 8.3 ± 2.2 (P = 0.003) and 3.6 ± 0.9 (P = 0.009), respectively (Fig. 1B). The statistical significance of the fold difference in FOS expression levels was reconfirmed by using the two-tailed Mann–Whitney test (P < 0.0001). In subsequent studies, FOS also gave the most consistent difference between patients and controls, leading us to focus on this gene.
Fig. 1.
Analysis and significance of candidate gene mRNA levels in control subjects and CEA patients. (A) Normalized fold-change expression levels of the candidate genes are color-coded (red, induced; green, repressed). The subjects are ordered by the average expression values of the six genes (AVG). The three groups are composed of: A, younger controls A1 and A2; Control, C1–C19; and Patient, P1–P25. (B) Relative expression levels of FOS and DUSP1 mRNAs, and plasma hsCRP protein levels in Control (n = 19) versus Patient (n = 25). Values shown as mean ± SE. (C) Controls and patients ordered by the relative level of FOS expression within each group. Diamonds, indicate history of coronary revascularization either by angioplasty or coronary artery bypass graft surgery (Revasc.); squares, current 3-hydroxy-3-methyl-glutaryl (HMG) CoA reductase inhibitor treatment (Statin); circles, current aspirin treatment (ASA). All RT-PCR measurements were done in duplicate and repeated at least twice. For presentation purposes, the FOS relative expression values were truncate at 15; the actual values for P23, P19, P2, and P9 were 19.5 ± 0.2, 27.3 ± 2.6, 34.7 ± 3.5, and 39.6 ± 2.2, respectively. (D) Receiver operating characteristic curves for the utility of FOS (black) and hsCRP (gray) at identifying coronary revascularization history among all patients in the study.
Clinical Significance of Increased FOS Level. Because plasma hsCRP has been shown to be a clinically useful indicator of inflammation and predictor of future cardiovascular events, we tested whether FOS might be similarly diagnostic. In comparison with FOS, hsCRP was not as significantly elevated in patients versus control subjects, 1.9 ± 0.2-fold (P = 0.22), and the correlation between hsCRP and FOS levels was low (correlation coefficient <0.6) (Fig. 1B). We measured another inflammatory plasma marker IL-6, but it too did not show as marked a difference as FOS (data not shown).
We next wondered whether there were any patient characteristics that could account for the variations in FOS levels. We examined all available information such as CEA surgical outcome (3 months to >1 year follow-up), cardiac risk factors, and associated medical conditions and medications, as well as quantitative measures such as body mass index (BMI) and 10-year Framingham cardiac risk (Table 1). The large number of variables in a limited patient population did not allow us to do a controlled multivariate analysis. Surprisingly, although all patients had peripheral vascular disease, we observed that history of coronary revascularization (coronary artery bypass graft surgery or angioplasty) seemed to associate with higher FOS level (Fig. 1C). Empirically taking the highest control subject's FOS level as the threshold for a positive test, eight of the nine coronary revascularization patients were detected (89% sensitivity). Combining the average (AVG) RT-PCR values of the top six candidates did not improve the sensitivity. The receiver operating characteristic (ROC) at identifying coronary revascularization history among all patients in the study revealed higher sensitivities and specificities for FOS compared with hsCRP (Fig. 1D).
Expression of FOS in Plaques. We reasoned that candidate genes involved in pathogenesis should be expressed and up-regulated in atherosclerotic plaque macrophages. As a first step, we performed immunohistochemistry on serial sections of CEA plaques and observed specific colocalization of FOS to CD14+ cells (Fig. 2A). We also examined atherosclerotic plaques in coronary arteries and observed strong colocalization of FOS and CD14 immunoreactivity (Cor-CD14, Cor-FOS, Fig. 2A). Notably, the normal appearing segment of the coronary artery without CD14+ macrophage infiltration (arrowhead) did not stain for FOS. To ascertain FOS expression in macrophages, we first purified CD14+ cells from a number of carotid plaques and verified macrophage markers (Fig. 4B). Using RT-PCR, we observed progressive enrichment of the candidate genes (particularly FOS) in MNCs, monocytes (Mono), and plaque macrophages (Mac), respectively (Fig. 2B).
Fig. 2.
Expression of FOS in human plaque macrophages and in activated human monocytic cells. (A) Fresh frozen sections of human carotid and coronary (Cor) artery plaques stained with hematoxylin/eosin (H&E), negative control Ig (Control Ig), and antibodies against CD14 or FOS. Shown is CD14+ staining of macrophages colocalized with FOS immunoreactivity. Similar colocalization of CD14 and FOS can be seen in the coronary artery plaque (Cor-CD14 and Cor-FOS) whereas, in the more “normal” appearing segment (black arrowhead), there is virtual absence of staining. Note that the CD14 staining gives a more diffuse appearance consistent with cell membrane staining whereas the FOS pattern is more punctate, consistent with nuclear localization (carotid plaque, ×25 magnification; coronary plaque, ×50 magnification). (B) From four patients (labeled 1–4), the normalized RT-PCR expression levels of the candidate genes are shown for the corresponding MNC, monocyte (Mono), and carotid plaque purified macrophage (Mac) fractions. Note the consistent pattern of increased mRNA levels in the purified macrophages. (C) Five different human monocytic cell lines were evaluated by RT-PCR after stimulation with 20 nM PMA at the indicated time points (h).
To further establish the biological significance of these candidate genes, we examined their expression in several different monocytic cell lines in response to PMA, commonly used for activating monocytes into macrophage-like cells (28). As early as 3 h after PMA treatment, there was induction of some candidates. DUSP1 was repressed in two of the cell lines, but FOS was uniformly induced in all, validating it as our best indicator of monocyte activation (Fig. 2C).
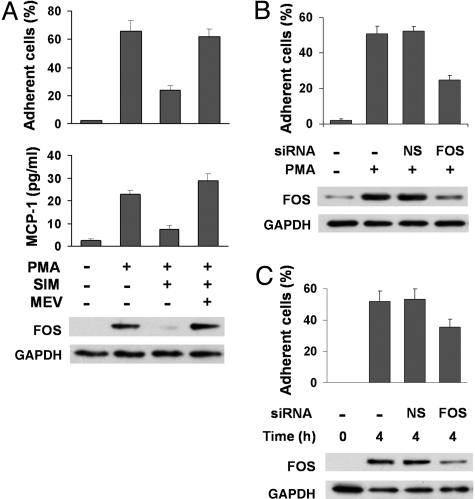
Modulation of FOS Affects Monocyte Function. When the human monocyte leukemia THP1 cells are activated by PMA, a prominent phenotype is attachment of the suspension cells to a number of different substrata including fibronectin, polylysine, and uncoated plastic (data not shown). Because statins have been shown to have potent antiatherogenic properties, we examined its effect on FOS expression, cell adhesion, and release of MCP-1, important for atherosclerotic plaque formation (5, 29). Pretreatment with statin inhibited PMA induction of FOS protein in parallel with monocyte adhesion and MCP-1 release (Fig. 3A). These effects were reversed by the addition of mevalonate, the product of statin-inhibited 3-hydroxy-3-methyl-glutaryl (HMG)-CoA reductase, demonstrating pharmacologic specificity.
Fig. 3.
Functional effects of statin and FOS siRNA inhibition on monocyte activation by PMA. (A) THP1 cells were pretreated with simvastatin and/or mevalonate for 20 h before stimulation with 2 nM PMA. Cell adhesion and cumulative MCP-1 release into medium were determined 4 h and 24 h after PMA stimulation, respectively. Western blot shows FOS protein levels after 4 h of PMA stimulation relative to control protein GAPDH. P values for the difference in cell adhesion and MCP-1 release after statin treatment were 0.004 and 0.04, respectively. (B) Thirty minutes after siRNA transfection, THP1 cells were stimulated with 2 nM PMA for 4 h. Negative control (-) cells were mock transfected without siRNA. The percentage adhesion difference between the nonspecific sequence (NS) and FOS-specific (FOS) siRNAs was significant, P = 0.006. Data shown are representative of experiments repeated at least three times in duplicate or triplicate. (C) Primary monocytes express FOS protein within 4 h of plating. FOS siRNA decreased FOS protein level and adhesion compared with mock transfection (-) and nonspecific siRNA (NS).
We also examined the genetic inhibition of FOS transcripts using siRNA molecules. FOS-specific siRNA transfection partially inhibited the induction of FOS protein and cell adhesion 4 h after PMA treatment (Fig. 3B). In contrast, mock transfection or siRNA directed toward nonspecific sequences (NS) had no inhibitory effects on FOS protein level and adhesion. We also tried some other inflammatory stimuli such as IL-6 and LPS. LPS increased THP1 adhesion with an associated increase in FOS protein level, whereas IL-6 alone or in combination with LPS had negligible effect (data not shown). The transformed nature of the THP1 cells may explain their lack of response to IL-6; thus, we investigated whether our observations were applicable to primary human monocytes isolated from normal donors. In contrast to THP-1 cells, which require a stimulus to adhere, primary monocytes spontaneously attached within 4 h of plating in association with strong FOS protein induction (Fig. 3C) (21). The partial inhibition of FOS protein using siRNA specifically inhibited adhesion as observed in THP1 cells.
Discussion
Our findings of highly expressed regulatory genes in the monocytes of patients with atherosclerosis demonstrate the utility of focusing on the in vivo transcriptome of readily available cells. The previously described involvement of some of our candidate genes in various stress response pathways is consistent with an activated state of monocytes in atherosclerosis. FOS stands out as the most differentially expressed marker, followed by another stress response gene DUSP1. Although not much is known about DUSP-1 in atherosclerosis, it has recently been linked to aortic lesion formation in mouse models, validating our unbiased screen of the circulating transcriptome (30). Also, increasing interest in genes such as JUN kinase 1 (JNK1) for treating diabetes, the strongest risk factor for coronary artery disease, points to the importance of FOS-associated pathways in artherosclerosis (31). It should be noted that the SAGE transcriptomes of cultured human monocytes have been reported, but their correlation to our data was low (data not shown), indicating that the context and techniques used in gene expression studies are critical determinants (32).
In comparison with control subjects, FOS transcript levels are increased >8-fold in patients requiring CEA and are more sensitive and specific to disease severity versus the plasma hsCRP test. The only coronary revascularization patient missed by FOS levels was 1 of 3 patients on maximum statin doses, all of whom also had low FOS levels. Of the 25 patients enrolled in our study, 1 patient had an ischemic event on follow up. Nine months after the CEA surgery, the patient (P9) with the highest FOS level, who was not on statin treatment, had subsequent thrombosis of a femoral artery bypass graft requiring emergent revascularization. Finally, it is noteworthy that control subjects on statin treatment clustered together with lower levels of FOS whereas this pattern is difficult to evaluate in the patient group because of its widespread use (Fig. 1C). It is tempting to speculate that, without statin treatment, the difference in FOS levels between patients and controls may have been more marked. Our data showing relatively acute functional inhibition of monocyte activation by statin treatment correlate well with clinical observations such as the benefits of preoperative or early high dose statin treatment in acute coronary syndromes (33, 34). The rapid expression of FOS in primary human monocytes as they activate and adhere and the specific inhibition of this property by FOS siRNA demonstrate its importance in nontransformed cells as well.
Although our observations have potential clinical utility, a number of issues need to be addressed in the future. We have simplified the RT-PCR test using whole mononuclear cell fractions, which permits studies such as the one presented. However if a more simple, sensitive, and specific FOS assay can be developed, larger prospective clinical trials could be performed to determine the clinical utility of FOS levels. FOS is a reactive transcriptional regulator, and this functional property may make it useful as a monitor of disease activity or treatment efficacy (10). However, like hsCRP, FOS may be elevated in other inflammatory conditions such as rheumatoid arthritis and will need to be tested in the appropriate patient populations (35, 36).
From a public health perspective, better tests are needed to identify patients with subclinical atherosclerosis or with disease in multiple vascular territories for risk stratification and treatment (4). FOS expression may be the molecular equivalent of coronary artery calcium scores currently used for coronary artery disease screening (37). Targeted patient management based, in part, on sensitive molecular tests may become reality with more genetic insights into atherosclerosis. The quantitative and digital nature of our human monocyte SAGE database will make it easily accessible online to all and serve as a foundation for future studies examining the circulating human transcriptome.
Supplementary Material
Acknowledgments
We thank Zambia Mcleod, Betsy Framer, and Jane Yates for administrative assistance, Thomas H. Shawker and the ultrasound staff of the National Institutes of Health (NIH) Diagnostic Radiology Department, and Amy Rapkiewicz of the NIH Laboratory of Pathology. We thank Toren Finkel and Elizabeth G. Nabel for support and guidance, and Victor Velculescu, Kenneth Kinzler, Michael N. Sack, Neal Epstein, Fred Bunz, Carlo Rago, and Myron Waclawiw for helpful discussions. This research was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institutes, NIH.
Author contributions: W.D.P., J.-G.K., S.M., and P.M.H. designed research; W.D.P., O.Y.M., J.-G.K., S.M., L.D.B., B.H., H.H.T., L.K., and P.M.H. performed research; W.D.P., O.Y.M., J.-G.K., S.M., and P.M.H. analyzed data; and W.D.P. and P.M.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SAGE, serial analysis of gene expression; hsCRP, high sensitivity C-reactive protein; statin, 3-hydroxy-3-methyl-glutaryl CoA reductase inhibitor; MNC, mononuclear cells; siRNA, small interfering RNA; CEA, carotid endarterectomy; FOS, Finkel–Biskis–Jinkins osteosarcoma; Ct, cycle threshold; PMA, phorbol 12-myristate 13-acetate; MCP-1, monocyte chemoattractant protein 1; DUSP1, dual specificity phosphatase 1.
References
- 1.Ross, R. (1999) N. Engl. J. Med. 340, 115-126. [DOI] [PubMed] [Google Scholar]
- 2.Libby, P., Ridker, P. M. & Maseri, A. (2002) Circulation 105, 1135-1143. [DOI] [PubMed] [Google Scholar]
- 3.Beckman, J. A., Creager, M. A. & Libby, P. (2002) J. Am. Med. Assoc. 287, 2570-2581. [DOI] [PubMed] [Google Scholar]
- 4.Chaves, P. H., Kuller, L. H., O'Leary, D. H., Manolio, T. A. & Newman, A. B. (2004) Am. J. Geriatr. Cardiol. 13, 137-151. [DOI] [PubMed] [Google Scholar]
- 5.Willerson, J. T. & Ridker, P. M. (2004) Circulation 109, II2-II10. [DOI] [PubMed] [Google Scholar]
- 6.Polyak, K. & Riggins, G. J. (2001) J. Clin. Oncol. 19, 2948-2958. [DOI] [PubMed] [Google Scholar]
- 7.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508-512. [DOI] [PubMed] [Google Scholar]
- 8.Patino, W. D., Mian, O. Y. & Hwang, P. M. (2002) Circ. Res. 91, 565-569. [DOI] [PubMed] [Google Scholar]
- 9.Finkel, M. P., Biskis, B. O. & Jinkins, P. B. (1966) Science 151, 698-701. [DOI] [PubMed] [Google Scholar]
- 10.Ransone, L. J. & Verma, I. M. (1990) Annu. Rev. Cell Biol. 6, 539-557. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, R. S., Spiegelman, B. M. & Papaioannou, V. (1992) Cell 71, 577-586. [DOI] [PubMed] [Google Scholar]
- 12.Wang, Z. Q., Ovitt, C., Grigoriadis, A. E., Mohle-Steinlein, U., Ruther, U. & Wagner, E. F. (1992) Nature 360, 741-745. [DOI] [PubMed] [Google Scholar]
- 13.Liebermann, D. A., Gregory, B. & Hoffman, B. (1998) Int. J. Oncol. 12, 685-700. [DOI] [PubMed] [Google Scholar]
- 14.Shaulian, E. & Karin, M. (2002) Nat. Cell Biol. 4, E131-E136. [DOI] [PubMed] [Google Scholar]
- 15.Doherty, T. M., Fitzpatrick, L. A., Shaheen, A., Rajavashisth, T. B. & Detrano, R. C. (2004) Mayo Clin. Proc. 79, 197-210. [DOI] [PubMed] [Google Scholar]
- 16.Lavezzi, A. M., Milei, J., Grana, D. R., Flenda, F., Basellini, A. & Matturri, L. (2003) Int. J. Cardiol. 92, 59-63. [DOI] [PubMed] [Google Scholar]
- 17.Holodniy, M., Mole, L., Yen-Lieberman, B., Margolis, D., Starkey, C., Carroll, R., Spahlinger, T., Todd, J. & Jackson, J. B. (1995) J. Clin. Microbiol. 33, 1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.St. Croix, B., Rago, C., Velculescu, V., Traverso, G., Romans, K. E., Montgomery, E., Lal, A., Riggins, G. J., Lengauer, C., Vogelstein, B. & Kinzler, K. W. (2000) Science 289, 1197-1202. [DOI] [PubMed] [Google Scholar]
- 19.Liu-Wu, Y., Svenningsson, A., Stemme, S., Holm, J. & Wiklund, O. (1997) Cytometry 29, 155-164. [PubMed] [Google Scholar]
- 20.Ziegler-Heitbrock, H. W., Thiel, E., Futterer, A., Herzog, V., Wirtz, A. & Riethmuller, G. (1988) Int. J. Cancer 41, 456-461. [DOI] [PubMed] [Google Scholar]
- 21.Seager Danciger, J., Lutz, M., Hama, S., Cruz, D., Castrillo, A., Lazaro, J., Phillips, R., Premack, B. & Berliner, J. (2004) J. Immunol. Methods 288, 123-134. [DOI] [PubMed] [Google Scholar]
- 22.Lash, A. E., Tolstoshev, C. M., Wagner, L., Schuler, G. D., Strausberg, R. L., Riggins, G. J. & Altschul, S. F. (2000) Genome Res. 10, 1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerutti, J. M., Delcelo, R., Amadei, M. J., Nakabashi, C., Maciel, R. M., Peterson, B., Shoemaker, J. & Riggins, G. J. (2004) J. Clin. Invest. 113, 1234-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, J., Zhang, L., Hwang, P. M., Kinzler, K. W. & Vogelstein, B. (2001) Mol. Cell 7, 673-682. [DOI] [PubMed] [Google Scholar]
- 25.Chan, T. A., Hermeking, H., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1999) Nature 401, 616-620. [DOI] [PubMed] [Google Scholar]
- 26.Audic, S. & Claverie, J. M. (1997) Genome Res. 7, 986-995. [DOI] [PubMed] [Google Scholar]
- 27.Farooq, A. & Zhou, M. M. (2004) Cell Signal 16, 769-779. [DOI] [PubMed] [Google Scholar]
- 28.Auwerx, J. (1991) Experientia 47, 22-31. [DOI] [PubMed] [Google Scholar]
- 29.Gu, L., Okada, Y., Clinton, S. K., Gerard, C., Sukhova, G. K., Libby, P. & Rollins, B. J. (1998) Mol. Cell 2, 275-281. [DOI] [PubMed] [Google Scholar]
- 30.Reddy, S. T., Nguyen, J. T., Grijalva, V., Hough, G., Hama, S., Navab, M. & Fogelman, A. M. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1676-1681. [DOI] [PubMed] [Google Scholar]
- 31.Kaneto, H., Nakatani, Y., Miyatsuka, T., Kawamori, D., Matsuoka, T. A., Matsuhisa, M., Kajimoto, Y., Ichijo, H., Yamasaki, Y. & Hori, M. (2004) Nat. Med. 10, 1128-1132. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto, S., Suzuki, T., Dong, H. Y., Yamazaki, N. & Matsushima, K. (1999) Blood 94, 837-844. [PubMed] [Google Scholar]
- 33.Dotani, M. I., Morise, A. P., Haque, R., Jain, A. C., Gupta, N. & Gibson, C. M. (2003) Am. J. Cardiol. 91, 1107-1109. [DOI] [PubMed] [Google Scholar]
- 34.Cannon, C. P., Braunwald, E., McCabe, C. H., Rader, D. J., Rouleau, J. L., Belder, R., Joyal, S. V., Hill, K. A., Pfeffer, M. A. & Skene, A. M. (2004) N. Engl. J. Med. 350, 1495-1504. [DOI] [PubMed] [Google Scholar]
- 35.Ridker, P. M. (2003) Circulation 107, 363-369. [DOI] [PubMed] [Google Scholar]
- 36.Onda, K., Rimbara, E., Hirano, T., Oka, K., Abe, H., Tahara, K., Takanashi, H., Tsuboi, N., Niitsuma, T. & Hayashi, T. (2004) J. Rheumatol. 31, 464-469. [PubMed] [Google Scholar]
- 37.Greenland, P., LaBree, L., Azen, S. P., Doherty, T. M. & Detrano, R. C. (2004) J. Am. Med. Assoc. 291, 210-215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.