Abstract
Soluble P loop NTPases represent a large protein family and are involved in diverse cellular functions. Here, we functionally characterized the first member of the Mrp/Nbp35 subbranch of this family, the essential Nbp35p of Saccharomyces cerevisiae. The protein resides in the cytosol and nucleus and carries an Fe/S cluster at its N terminus. Assembly of the Fe/S cluster requires the mitochondrial Fe/S cluster (ISC)-assembly and -export machineries. Depletion of Nbp35p strongly impairs the activity of the cytosolic Fe/S protein, isopropylmalate isomerase (Leu1p), whereas mitochondrial Fe/S enzymes are unaffected. Moreover, defects in the de novo maturation of various cytosolic and nuclear Fe/S proteins were observed in the absence of Nbp35p, demonstrating the functional involvement of Nbp35p in the biogenesis of extramitochondrial Fe/S proteins. Furthermore, Nbp35p genetically interacts with the closely similar P loop NTPase, Cfd1p, and the hydrogenase-like Nar1p, both of which were recently shown to perform a crucial function in cytosolic and nuclear Fe/S protein biogenesis. Hence, our study suggests that eukaryotic Nbp35 NTPases function in Fe/S protein maturation. The findings provide strong evidence for the existence of a highly conserved and essential machinery dedicated to assembling cytosolic and nuclear Fe/S proteins.
Keywords: iron–sulfur cluster (ISC), ISC-assembly machinery, mitochondria, biogenesis
In eukaryotes, iron–sulfur (Fe/S) proteins are located in mitochondria, the cytosol, and the nucleus. Investigations of the past few years have provided insights into the pathways leading to the synthesis of Fe/S clusters and their insertion into apoproteins. The best knowledge by far is available for maturation of mitochondrial Fe/S proteins, which is accomplished by the so-called Fe/S cluster (ISC)-assembly machinery located in the matrix space (1–3). This complex machinery has been inherited during evolution from the bacterial ISC machinery and consists of some 10 proteins, many of which are essential for cell viability in yeast. ISC components include the cysteine desulfurase Nfs1p as a sulfur donor, the Isu proteins serving as a scaffold for synthesis of the cluster, the electron transfer chain NADH/Arh1p/Yah1p, and the chaperone proteins Ssq1p, Jac1p, and Mge1p, which are thought to assist steps after synthesis of the Fe/S cluster on the Isu proteins (4–9).
In contrast, the maturation of Fe/S proteins that reside outside of mitochondria has been poorly characterized. Recently, two proteins with a specific function in the maturation of extramitochondrial Fe/S proteins have been identified: the P loop NTPase Cfd1p and the iron-only hydrogenase-like protein Nar1p (10, 11). Virtually nothing is known concerning the precise mode of action of these proteins. Remarkably, the mitochondrial ISC-assembly machinery also performs a crucial task in the biogenesis of extramitochondrial Fe/S proteins in that it cooperates with two other mitochondrial proteins, namely the mitochondrial inner-membrane ATP-binding cassette (ABC) transporter Atm1p and the sulfhydryl oxidase Erv1p of the intermembrane space (5, 12). In light of the complexity of Fe/S protein assembly in eukaryotes and bacteria (13), it is reasonable to expect that further components participate in this poorly defined process. In this work, we report the identification of a cytosolic component of this essential process, the P loop NTPase Nbp35p from the yeast Saccharomyces cerevisiae.
Soluble P loop NTPases comprise one of the largest protein families with members present in all three kingdoms of life (14). Numerous subgroups of the family can be distinguished based on primary sequence information. Individual subgroup members perform a wide variety of cellular functions, for example, in translation, signal transduction, signal-sequence recognition, protein transport and localization, chromosome partitioning, metal insertion, and membrane transport. The function of quite a number of family subgroups is still unknown. One such example is the protein Nbp35 (nucleotide-binding protein of 35 kDa), which appears to execute a critical task in the eukaryotic cell, as indicated by the essential character of the NBP35 gene in yeast (15). The protein belongs to the Mrp/Nbp35 subgroup of the P loop NTPases that shares highest sequence similarity with members of other subgroups involved in nitrogen fixation, metal insertion, and bacterial cell division (14, 16). Close relatives of full-length Nbp35 are present in all eukaryotes thus far sequenced, but are not found in Bacteria and Archaea. A speciality of the Nbp35 protein is an N-terminal extension of ≈50 residues with four conserved cysteine residues, which were predicted to represent a binding motif for either a metal [e.g., zinc (14)] or an Fe/S cluster. The Mrp members of the Mrp/Nbp35 subgroup characteristically lack this N-terminal cysteine-rich motif and are found in Eukarya, Bacteria, and Archaea. To date, two Mrp members have been analyzed in some detail. The yeast P loop NTPase Cfd1p is required for maintenance of Fe/S protein activities in the cytosol (10). Likewise, the Salmonella protein ApbC was reported to perform an auxiliary role in Fe/S protein maturation, cooperating with ApbE (17).
Here, we report that Nbp35p from the yeast Saccharomyces cerevisiae is an Fe/S protein with a dual localization in both the cytosol and nucleus. In a combination of in vivo and biochemical experiments, we show that Nbp35p is required for maintenance of extramitochondrial Fe/S protein activities. Iron radiolabeling experiments suggest that Nbp35p plays a direct role in the biogenesis of both cytosolic and nuclear Fe/S proteins. Hence, our results identify Nbp35p as a central component of the emerging cytosolic Fe/S protein-assembly apparatus that is essential for eukaryotes.
Materials and Methods
Yeast Strains and Cell Growth. The following strains of S. cerevisiae were used: W303-1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3,112), which served as the wild type, Gal-ATM1 (5), Gal-YAH1 (7), Gal-NFS1 (9), and Gal-NBP35 (this work). In the latter strain, region -69 to -1 of the NBP35 promoter was exchanged for the GAL1-10 promoter by PCR-mediated DNA replacement as described in ref. 18. To deplete the amount of Nbp35p to physiologically critical levels, Gal-NBP35 cells were precultivated for 3 d on glucose-containing solid minimal medium. Cells were grown in rich yeast extract/peptone (YP), minimal synthetic complete (SC) lactate media, or “iron-poor” minimal medium lacking added iron chloride (19). Carbon sources were added at a concentration of 2% (wt/vol).
55Fe Incorporation into Nbp35p and Other Fe/S Cluster Apoproteins in Vivo. In vivo labeling of yeast cells with radioactive iron (55FeCl3) and measurement of 55Fe-incorporation into Fe/S proteins was carried out as described in refs. 5 and 18. The following Fe/S reporter proteins were used: mitochondrial biotin synthase (Bio2p), hemagglutinin (HA)-tagged versions of cytosolic Rli1p and Nar1p, and nuclear Ntg2p. Bio2p, Rli1p, and Ntg2p were overproduced from the 2μ-plasmid p426GPD under the control of the TDH3 promoter (20), whereas vector p416MET25 was used for expression of NAR1 (11). A tandem-affinity purification (TAP)-tagged version of NBP35 was amplified by PCR from a yeast strain that carries the NBP35 gene fused to a TAP-tag coding sequence (Euroscarf accession no. SC0234). A Streptagged version of Nbp35p was generated by using a PCR strategy with an oligonucleotide containing the coding sequence of Strep-tag II (IBA, Göttingen, Germany) fused to the DNA part corresponding to the C terminus of Nbp35p. For deletion of the first 52 residues of Nbp35p, the residual gene with a TAP-tag coding sequence was synthesized by PCR. For overexpression, the respective PCR products were inserted into p426GPD. Nbp35p-TAP and Nbp35p-TAPΔ1-52 were immunoprecipitated with IgG Sepharose (Amersham Pharmacia).
Purification of Recombinant His-Tagged Nbp35p. Nbp35p was overproduced in Escherichia coli in the presence of the bacterial isc operon (21) employing a pET-15b (Novagen) and purified anaerobically by Ni-NTA affinity chromatography. The protein was eluted from the agarose column by 150 mM histidine in 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 1 mM DTT, and 10% (vol/vol) glycerol and appeared as a homogeneous peak in size-exclusion chromatography (Fig. 5, which is published as supporting information on the PNAS web site). The eluate was immediately desalted on a PD10 column (Pharmacia).
Miscellaneous Methods. The following published methods were used: Manipulation of DNA and PCR (22); transformation of yeast cells (23); preparation of whole cell lysates by mechanical cell disruption with glass beads and subcellular fractionation (24); immunostaining and immunoprecipitation (25); in situ immunofluorescence (11); enzyme activities of malate dehydrogenase, aconitase, succinate dehydrogenase, isopropylmalate isomerase, and alcohol dehydrogenase (5); and iron and sulfide determination and EPR experiments (11). Enzyme activities of isopropylmalate isomerase (Leu1p) and alcohol dehydrogenase were measured in freshly prepared postmitochondrial supernatants from cells grown at 30°C in minimal medium. Mitochondria were prepared from cells grown in minimal media containing galactose or lactate/glucose (24). Mitochondrial iron contents were determined as described in ref. 26. Error bars represent the SEM.
Results
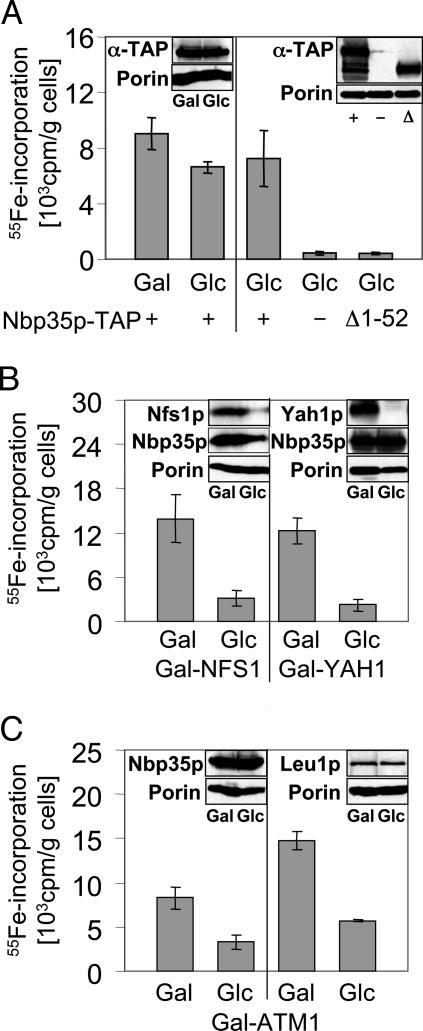
Nbp35p Binds an Fe/S Cluster in Vivo. The soluble P loop NTPase Nbp35p contains, at its N terminus, four conserved cysteine residues that may form a metal-binding site. To investigate the binding of iron to Nbp35p in vivo, the NBP35 gene was fused with a C-terminal TAP-tag and inserted into the high-copy yeast vector p426GPD under the control of the constitutive promoter TDH3. Yeast strains harboring the resulting plasmid p426-NBP35-TAP produced the fusion protein as shown by immunostaining (Fig. 1 Insets). For determination of iron binding, cells containing either p426-NBP35-TAP or the empty vector p426GPD were grown in iron-poor minimal medium and radiolabeled with radioactive 55Fe in the presence of ascorbate for 4 h. Subsequently, a cell lysate was prepared, and Nbp35p-TAP was immunoprecipitated with IgG-Sepharose beads. The amount of radioactivity coimmunoprecipitated with the immunobeads was quantified by liquid-scintillation counting. A significant amount of radioactive iron was coimmunoprecipitated with the IgG beads (Fig. 1A Left). In contrast, background levels of 55Fe were immunoprecipitated from cells containing the empty vector, even though the uptake of iron was similar in both strains (Fig. 1A Right and data not shown). We therefore conclude that Nbp35p binds iron in vivo.
Fig. 1.
Fe/S cluster association at the N terminus of Nbp35p requires a functional mitochondrial ISC machinery. (A) Wild-type cells overproducing TAP-tagged versions of Nbp35p (+) or Nbp35pΔ1-52 (Δ1-52) from vector p426-NBP35-TAP and control cells carrying plasmid p426GPD (-) were grown in iron-poor minimal medium supplemented with either galactose (Gal) or glucose (Glc) for 16 h. Cells were radiolabeled with 55Fe for 4 h, and cell lysates were prepared. TAP-tagged Nbp35p proteins were immunoprecipitated from the lysates with IgG, and the amount of coimmunoprecipitated 55Fe was quantified by liquid scintillation counting. (B) Gal-NFS1 and Gal-YAH1 cells overproducing Nbp35p-TAP were grown as in A to allow or repress, respectively, NFS1 and YAH1 expression. Subsequently, 55Fe binding to Nbp35p-TAP was investigated as described above. (C) Gal-ATM1 cells overproducing Nbp35p-TAP were treated as cells in A, and 55Fe binding to Nbp35p-TAP and the endogenous Leu1p was determined. Insets show immunostaining of the indicated proteins in the cell extracts.
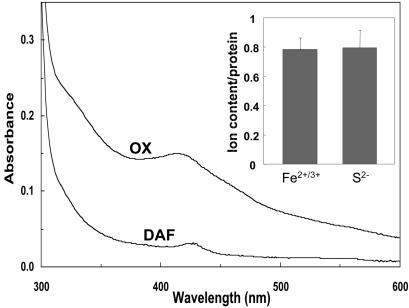
Iron binding to Nbp35p was stable because it was not removed by treatment with EDTA, Triton X-100, or oxygen (data not shown). These characteristics support the idea that bound iron is part of an Fe/S cluster (9). To substantiate this speculation, Nbp35p was overproduced in E. coli and purified under anaerobic conditions. The protein exhibited a structured UV-visible (UV-vis) spectrum that compares well with that of typical 2Fe–2S proteins (absorption peaks at 330 and 420 nm) (Fig. 2). The preparations contained 0.8 acid-labile sulfide and 0.8 iron per polypeptide chain (Inset). Even though Fe/S cluster assembly on the recombinant protein seemed to be incomplete, these data are fully consistent with Nbp35p being an Fe/S protein. This interpretation was confirmed by low-temperature EPR spectroscopy of reduced Nbp35p showing a signal with a gav value of 1.94 (data not shown). However, the biophysical data so far has not allowed us to decide which type of cluster (2Fe–2S or 4Fe–4S) may be associated with this protein. In summary, these data document that Nbp35p contains an Fe/S cluster.
Fig. 2.
Purified Nbp35p is an Fe/S protein. UV-vis spectrum of N-terminally His-tagged Nbp35p (1.9 mg/ml/50 mM Tris·HCl, pH 8/1 μM 5′-deazaflavin) before (OX) and after (DAF) photoreduction by irradiation for 3 min with a commercial slide projector. The contribution of oxidized 5′-deazaflavin to the spectrum before photoreduction has been subtracted. Inset shows the results of colorimetric iron and acid-labile sulfide determinations for five independent samples.
The most likely motif for binding of an Fe/S cluster on Nbp35p is an N-terminal extension containing four conserved cysteine residues (Fig. 6, which is published as supporting information on the PNAS web site). To verify this hypothesis, this region was deleted, and the N-terminally truncated, TAP-tagged protein was overproduced in wild-type yeast cells. 55Fe labeling and immunoprecipitation of Nbp35p-TAPΔ1-52 showed that no radioactivity was associated with this protein (Fig. 1 A Right). This finding indicates that the N-terminal region of Nbp35p was responsible for binding of the Fe/S cluster.
Fe/S Cluster Assembly on Nbp35p Requires the Mitochondrial ISC Apparatus. To further support the notion that Nbp35p binds an Fe/S cluster, we investigated whether 55Fe binding to Nbp35p depends on components of the mitochondrial ISC-assembly and -export machineries. To this end, the reporter plasmid p426-NBP35-TAP was introduced into the conditional yeast ISC-assembly mutants Gal-NFS1 and Gal-YAH1 and the ISC-export mutant Gal-ATM1. These cells harbor the respective genes under the control of the galactose-inducible GAL1-10 promoter (5, 7). To induce or repress the three genes, cells were grown in iron-poor minimal medium containing galactose or glucose, respectively, before radiolabeling with 55Fe. In wild-type cells, the choice of the carbon source did not significantly alter the amount of iron associated with immunoisolated Nbp35p-TAP (Fig. 1A). In extracts of Gal-YAH1, Gal-NFS1, or Gal-ATM1 cells grown under inducing conditions in the presence of galactose, a significant amount of 55Fe radioactivity was coimmunoprecipitated with IgG Sepharose (Fig. 1 B and C). In contrast, 55Fe-incorporation into Nbp35p-TAP was reduced 3- to 5-fold when Nfs1p, Yah1p, or Atm1p were depleted by growth of the respective Gal strains in the presence of glucose. As a control, the cytosolic Fe/S protein Leu1p was immunoprecipitated from the same cell extract. A similar, 3-fold decrease in iron association to Leu1p was observed upon depletion of Atm1p (Fig. 1C) (5). The depletion of Nfs1p and Yah1p under these conditions was verified by immunostaining (Fig. 1B Insets). Because of the similar molecular masses of Atm1p and Nbp35p-TAP (containing a protein-A domain, which binds to IgG during the immunostaining procedure), Atm1p depletion could not be documented in this case but was routinely seen (5). Depletion of Atm1p or Yah1p did not significantly change the amount of Nbp35p (Fig. 1B Insets). However, in depleted Gal-NFS1 cells, Nbp35p levels were somewhat reduced. This observation, occasionally made with our Gal-NFS1 strain under repressive conditions, may be attributed to the proteolytic sensitivity of the Fe/S apoproteins (cf. 11). Hence, we conclude that the apoform of Nbp35p is rather susceptible to proteolysis. Taken together, the dependence of 55Fe binding to Nbp35p on the function of the mitochondrial ISC and ISC-export components further demonstrates that Nbp35p is an Fe/S protein.
Nbp35p Is Localized in both the Nucleus and Cytosol. Nbp35p had first been described as a nuclear protein but, in subsequent systematic localization studies, was also found in the cytosol (15, 27). To clarify this issue, the localization of Nbp35p was analyzed by immunofluorescence microscopy of wild-type and Gal-NBP35 cells by using antibodies directed against Nbp35p. The labeled wild-type cells revealed a uniform fluorescence of the entire yeast cell (Fig. 7A, which is published as supporting information on the PNAS web site). The fluorescence signal was specific for Nbp35p because no labeling was obtained in Gal-NBP35 cells in which Nbp35p was depleted by growth in the presence of glucose. A signal stronger than that in wild-type cells was seen in Gal-NBP35 cells after growth in galactose, consistent with the overproduction of Nbp35p. Together, these results demonstrated that Nbp35p was predominantly located in the cytosol. However, immunofluorescence was also visible in regions that represented the nucleus, as judged by counterstaining of nuclear DNA with DAPI. Hence, a fraction of Nbp35p is located in the nucleus. Biochemical cell fractionation by ultracentrifugation revealed that an overproduced Strep-tagged version of Nbp35p was detected exclusively in the postmitochondrial supernatant and in the cytosol but not in mitochondria, similar to observations made for the cytosolic marker protein Pgk1p (Fig. 7B). In summary, these data demonstrate that Nbp35p is located as a soluble protein in both the cytosol and the nucleus but is excluded from mitochondria.
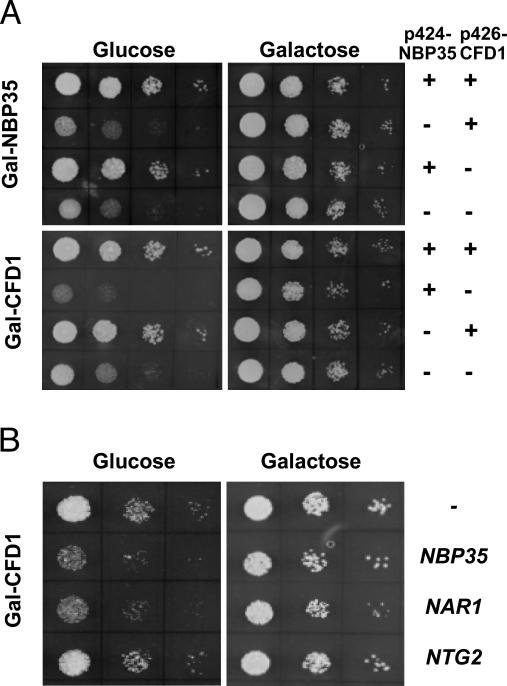
Nbp35p Is Involved in the Maturation of Cytosolic and Nuclear Fe/S Proteins. To initiate the functional investigation of Nbp35p, we first investigated whether the highly similar NBP35 and CFD1 can functionally replace each other. The conditional yeast strains Gal-NBP35 and Gal-CFD1 were constructed, in which the endogenous promoters were replaced by the galactose-inducible GAL1-10 promoter. Under permissive conditions in the presence of galactose, Gal-NBP35 and Gal-CFD1 cells grew at wild-type rates (Fig. 3A). Depletion of Nbp35p or Cfd1p by cultivation in the presence of glucose resulted in substantial growth reduction, which could be complemented to wild-type growth by expression of either NBP35 or CFD1, respectively. However, no reciprocal complementation was observed. This clearly showed that, despite the sequence similarity, the two proteins cannot functionally replace each other. Interestingly, we noted that Cfd1p-depleted cells grew even more slowly when Nbp35p was overproduced. A similar effect was detected when Nar1p was overproduced under these conditions (Fig. 3B). This finding was not due to the potential deleterious effects of overproducing these Fe/S proteins, because increased amounts of Nar1p or Nbp35p did not affect the growth of wild-type cells (data not shown), nor did the Fe/S protein Ntg2p impair growth of Cfd1p-depleted cells. Apparently, a fairly balanced synthesis of Nbp35, Cfd1p, and Nar1p appears to be important for sustained cell growth, suggesting that Nbp35p might play a role in Fe/S protein biogenesis.
Fig. 3.
Overexpression of NBP35 and NAR1 inhibits growth of Cfd1p-depleted Gal-CFD1 cells. (A) Gal-NBP35 and Gal-CFD1 cells were transformed with the high-copy plasmids p424-NBP35 and p426-CFD1-HA or empty vectors (-) to overexpress NBP35 and CFD1 under control of the TDH3 promoter as indicated. Tenfold serial dilutions of cells were cultivated on solid minimal media supplemented with galactose or glucose at 30°C twice for 3 days. (B) Gal-CFD1 cells transformed with high copy vectors encoding the indicated genes were grown as in A.
To experimentally test a potential function of Nbp35p in Fe/S protein maturation, we first determined the enzyme activity of the cytosolic Fe/S protein Leu1p in cell extracts isolated from the conditional mutant Gal-NBP35 after growth in the presence of galactose or glucose. No Leu1p enzyme activity was detectable upon depletion of Nbp35p, whereas wild-type activities were observed for extracts derived from cells grown in the presence of galactose (Table 1). In contrast, the enzyme activity of alcohol dehydrogenase remained similar in Gal-NBP35 cells under both conditions. This result suggests that Nbp35p is required for the maintenance and/or maturation of Leu1p.
Table 1. Enzymatic activities upon depletion of Nbp35p in Gal-NBP35 cells.
| Wild type
|
Gal-NBP35
|
|||
|---|---|---|---|---|
| Compartment | Gal | Glc | Gal | Glc |
| Cytosol | ||||
| Isopropyl malate isomerase | 0.30 ± 0.01 | 0.31 ± 0.05 | 0.40 ± 0.09 | <0.04 |
| Alcohol dehydrogenase | 1.8 ± 0.3 | 2.0 ± 0.3 | 1.6 ± 0.2 | 2.4 ± 0.6 |
| Mitochondria | ||||
| Aconitase | 3.5 ± 0.9 | 3.2 ± 0.4 | 2.9 ± 0.5 | 3.7 ± 0.6 |
| Succinate dehydrogenase | 0.30 ± 0.03 | 0.20 ± 0.02 | 0.20 ± 0.01 | 0.30 ± 0.01 |
| Malate dehydrogenase | 2.0 ± 0.1 | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.5 ± 0.1 |
| Citrate synthase | 2.8 ± 0.4 | 2.9 ± 0.3 | 3.1 ± 0.6 | 3.7 ± 0.5 |
Enzyme activities of cytosolic and mitochondrial proteins were determined using postmitochondrial supernatants and mitochondria, respectively, isolated from wild-type and Gal-NBP35 cells grown in minimal media containing galactose (Gal) or glucose (Glc). Activities are given in units per mg of protein.
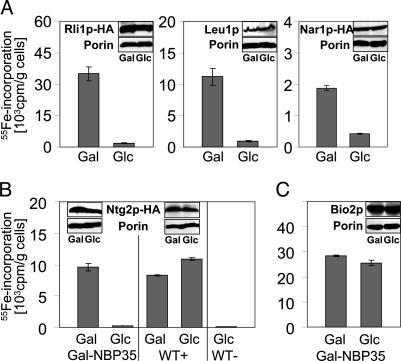
To further investigate the role of Nbp35p, the de novo formation of cytosolic Fe/S proteins was analyzed by an in vivo radiolabeling assay by using 55Fe (5). We used Leu1p, Rli1p, and Nar1p as cytosolic Fe/S-marker proteins. The Rli1p and Nar1p proteins were synthesized with a C-terminal HA tag. With all three reporter proteins, a high amount of 55Fe could be coimmunoprecipitated with Leu1p- or HA-specific antibodies by using Gal-NBP35 cells grown in the presence of galactose (Fig. 4A). The amounts were similar to those obtained with wild-type cells (data not shown). Strikingly, the amount of coimmunoprecipitated 55Fe was 5- to 20-fold lower upon depletion of Nbp35p in the presence of glucose (Fig. 4A). As estimated by immunostaining, the expression levels of Leu1p, Rli1p, and Nar1p remained unchanged upon depletion of Nbp35p, indicating a general defect in the assembly of extramitochondrial Fe/S proteins in the absence of Nbp35p (Fig. 4A Insets). Taken together, these data clearly demonstrate that Nbp35p is required for the maturation of Fe/S proteins located in the cytosol.
Fig. 4.
Nbp35p is required for maturation of Fe/S proteins in the cytosol and nucleus but not in mitochondria. (A) Gal-NBP35 cells overproducing HA-tagged (HA) versions of Rli1p and Nar1p were cultivated in iron-poor minimal media containing galactose (Gal) or glucose (Glc). Cells were labeled with 55Fe and lysed, and the radiolabeled proteins were immunoprecipitated by using anti-HA or anti-Leu1p antibodies. Coimmunoprecipitated 55Fe was quantified by liquid scintillation counting. (B and C) Wild-type (WT+) and Gal-NBP35 cells overproducing a HA-tagged (HA) version of Ntg2p (B) or mitochondrial Bio2p (C), and wild-type cells carrying vector p426GPD (WT-; B) were radiolabeled with 55Fe under permissive (Gal) and repressive (Glc) conditions. Iron binding to Ntg2p and Bio2p was determined by immunoprecipitation as described above. Insets show the immunostaining of indicated proteins in the cell extracts.
Next, we investigated the de novo maturation of the nuclear Fe/S protein Ntg2p after overexpression of an HA-tagged version from plasmid p426GPD. In wild-type cells or in Gal-NBP35 cells grown in the presence of galactose, similarly high amounts of 55Fe could be immunoprecipitated by using HA-specific antibodies, whereas only background signals were obtained in wild-type cells that did not harbor overproduced Ntg2p-HA (Fig. 4B). Upon growth in the presence of glucose, the amount of 55Fe associated with Ntg2p-HA in Gal-NBP35 cells declined to background levels, an observation that was not made in similarly treated wild-type cells. The levels of Ntg2p-HA were unchanged upon depletion of Npb35p, indicating that this protein is also required for the assembly of nuclear Fe/S proteins.
No Influence on Mitochondrial Fe/S Proteins or Iron Content upon Depletion of Nbp35p. To analyze the specificity of the defects in cytosolic and nuclear Fe/S proteins upon depletion of Nbp35, we determined the activity and biogenesis of mitochondrial Fe/S proteins. First, mitochondrial Fe/S proteins aconitase and succinate dehydrogenase were measured in mitochondria from Gal-NBP35 cells grown with either galactose or glucose. Both enzymes, as well as two proteins that do not harbor an Fe/S cofactor (citrate synthase and malate dehydrogenase), displayed wild-type activities in Gal-NBP35 cells under both growth conditions (Table 1). This result was in striking contrast to the enzyme activity of Leu1p (see above) but was consistent with the extramitochondrial localization of Nbp35p. Second, the de novo synthesis of Fe/S clusters on mitochondrial Bio2p was estimated by the 55Fe radiolabeling of Gal-NBP35 cells that overproduced this enzyme. Independently of the carbon source, high amounts of 55Fe were incorporated into Bio2p in Gal-NBP35 cells, indicating that Nbp35p was not responsible for maturation of this mitochondrial Fe/S protein (Fig. 4C). Immunostaining confirmed that Bio2p levels were similar in both the presence and the absence of Nbp35p (Fig. 4C Insets). Taken together, our data demonstrate that Nbp35p is specifically involved in the maturation of cytosolic and nuclear Fe/S proteins but is not required for Fe/S protein biogenesis within mitochondria.
Finally, to examine whether the lack of Nbp35p elicited a similar mitochondrial iron accumulation as did defects in the mitochondrial ISC components (1), we determined the iron content of mitochondria isolated from Nbp35p-depleted Gal-NBP35 cells. The mitochondrial iron content remained at low, wild-type levels upon depletion of Nbp35p (data not shown). Furthermore, the cellular 55Fe uptake was not dramatically altered. Taken together, these data show that Nbp35p is required exclusively for maturation of cytosolic and nuclear proteins.
Discussion
We have characterized the essential P loop NTPase Nbp35p as a component of the machinery responsible for maturation of cytosolic and nuclear Fe/S proteins. The protein carries an Fe/S cluster at the N terminus containing four conserved cysteine residues. Cluster integration depends strictly on the mitochondrial ISC-assembly and -export machineries. Thus, maturation of Nbp35p occurs as does that of other cytosolic Fe/S proteins. However, Nbp35p differs from the similar NTPases, Cfd1p of yeast and ApbC of bacteria, in that the latter do not stably bind iron or an Fe/S cluster (10, 17) (data not shown). These two proteins belong to the Mrp branch of the Mrp/Nbp35 subgroup of P loop NTPases and perform a function in Fe/S protein maturation. Our data suggest that the Nbp35 proteins which are present only in eukaryotes also execute a crucial role in the process of Fe/S protein biogenesis. We therefore propose that the entire Mrp/Nbp35 subgroup of P loop NTPases is dedicated to the generation of Fe/S proteins. Despite the striking structural similarities between Nbp35p and Cfd1p, the two proteins cannot functionally replace each other, even after overproduction from a high-copy expression vector. This may be because of the structural differences at the N terminus. Nevertheless, both proteins show a genetic interaction, in that high levels of Nbp35p exacerbate the growth defects of Cfd1p-depleted cells. A similar observation was made for high levels of Nar1p, indicating that imbalances in the three known components of the extramitochondrial Fe/S protein-assembly machinery are not tolerated by the yeast cells. Moreover, mutants in NBP35, CFD1, and NAR1 show synthetic lethality (31). These genetic data and the functional studies presented herein suggest a critical role of Nbp35p, Cfd1p, and Nar1p in the generation of cytosolic and nuclear Fe/S proteins (10, 11).
The function of Nbp35p, Cfd1p, and Nar1p in the same biosynthetic pathway is reflected in strikingly similar features. The three proteins are present in all eukaryotes sequenced to date (mammals, plants, fungi, and parasites), are located in both the cytosol and nucleus, and are expressed at comparatively low levels. Cells depleted of these proteins show strong growth defects and a specific impairment in the maturation of cytosolic and nuclear but not of mitochondrial Fe/S proteins (10, 11) (data not shown). These data suggest that the same machinery is used for the maturation of cytosolic and nuclear Fe/S proteins. Finally, depletion of these proteins does not lead to increased mitochondrial iron levels or to changes in cellular iron metabolism, because these effects are observed for depletion of the mitochondrial ISC-assembly and -export components. These criteria may, therefore, be useful to distinguish these components.
Despite the fact that we still know little about the molecular mechanisms of Fe/S protein formation in the cytosol and nucleus, a clear picture is emerging from this and previous studies (10, 11). The assembly of extramitochondrial Fe/S proteins requires a specific machinery that is distinct from the well characterized ISC components of mitochondria. We therefore propose to designate this functional unit as cytosolic iron–sulfur protein-assembly (CIA) machinery (3). Because all known cytosolic and nuclear Fe/S proteins strictly depend on the function of the mitochondrial ISC machineries for their conversion to holoproteins, the CIA machinery appears to function downstream of, and not independently of, mitochondria. The molecular basis of this functional cooperation is unclear, and further insights may require the reconstitution of cytosolic Fe/S protein maturation in vitro. Nevertheless, one might speculate on possible scenarios to explain the available data. First, mitochondria may produce a precursor of an Fe/S cluster, export it to the cytosol via the ATP-binding cassette transporter Atm1p, and, subsequently, the CIA machinery may use this moiety for generating the Fe/S cluster for its insertion into apoproteins. Alternatively, and equally likely, mitochondria may produce a compound that is an essential cofactor of the CIA machinery. Synthesis of this compound would occur specifically in mitochondria and would strictly depend on the ISC-assembly machinery. Thus, the compound may either represent a direct product of the ISC pathway or may be produced by a mitochondrial Fe/S protein. Other, more complex scenarios to explain the function of mitochondria in cytosolic and nuclear Fe/S protein maturation are not excluded.
The detection of small amounts of some ISC components in the cytosol and nucleus (see, e.g., ref. 28) has raised the issue of whether they might function in Fe/S protein maturation in these compartments. Two recent studies render this possibility unlikely (29, 30). First, both the cysteine desulfurase Nfs1p and the scaffold proteins Isu1p and Isu2p have been shown to be necessary inside mitochondria for successful participation in Fe/S protein maturation in the cytosol. Second, no synthesis of an Fe/S cluster on an Isu protein that has been mislocalized to the yeast cytosol was observed, indicating that this key step of Fe/S protein biogenesis has to be performed inside mitochondria. Finally, the human Nfs1 and Isu proteins identified in the cytosolic compartment lack essential N-terminal residues that are important for proper function of these proteins. Together, the findings do not support a role for these ISC components outside mitochondria. Rather, the CIA components appear to represent the cytosolic machinery that is responsible for the generation of cytosolic (and nuclear) Fe/S proteins.
The essential character of all three genes encoding the CIA components emphasizes the importance of cytosolic/nuclear Fe/S protein biogenesis for cell viability. This notion requires that at least one essential extramitochondrial Fe/S protein must exist. In principle, Nbp35p and Nar1p, both being Fe/S proteins per se, would fulfil this criterion. However, this “chicken-and-egg” situation is reminiscent of the maturation of mitochondrial 2Fe–2S ferredoxin Yah1p, which is a central ISC component (7) and therefore does not provide a satisfactory explanation for the essential nature of the CIA machinery. Currently, just one cytosolic Fe/S protein with an essential character and no function in Fe/S protein maturation is known to us. This protein, termed Rli1p, was recently identified to play a central role in a step of the complex process of ribosome biogenesis and function (31, 32). Depletion of Rli1p and, not surprisingly, also of the CIA components, results in nuclear accumulation of ribosomal subunits, a defect in ribosomal RNA processing, and, eventually, translational arrest. These findings impressively document the intimate connection between two central cellular processes, which both must have developed early in the evolution of life.
Further insights into the function of Fe/S protein biogenesis in the cytosol and nucleus depend crucially on the availability of an in vitro assay to monitor Fe/S cluster insertion into apoproteins and on the future identification of more components participating in this important biosynthetic pathway. Given the complexity of Fe/S protein maturation in mitochondria and in bacteria (13, 3), it is almost certain that the CIA machinery consists of more than just the three known proteins. The availability of Nbp35p, Cfd1p, and Nar1p has opened the gate for biochemical and genetic investigations aimed at the understanding of how Fe/S clusters destined for cytosolic and nuclear proteins are assembled and inserted into the apoproteins.
Supplementary Material
Acknowledgments
We thank R. Thauer and W. Buckel for access to EPR facilities and N. Richhardt for expert technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Gottfried Wilhelm Leibniz Program), Sonderforschungsbereich 593, by European Commission Grant QLG1-CT-2001-00966, by the Marie Curie European Fellowship HPMF-CT-2002-01750 (to J.B.), by the Deutsches Humangenomprojekt, and by the Fonds der Chemischen Industrie.
Author contributions: U.M. and R.L. designed research; A.H., D.J.A.N., J.B., A.J.P., and U.M. performed research; A.J.P. analyzed data; and R.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ISC, Fe/S cluster; Leu1p, isopropylmalate isomerase; Bio2p, biotin synthase; HA, hemagglutinin; UV-vis, UV-visible; TAP, tandem-affinity purification; CIA, cytosolic iron–sulfur protein assembly.
References
- 1.Lill, R. & Kispal, G. (2000) Trends Biochem. Sci. 25, 352-356. [DOI] [PubMed] [Google Scholar]
- 2.Craig, E. A. & Marszalek, J. (2002) Cell. Mol. Life Sci. 59, 1658-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lill, R. & Mühlenhoff, U. (2005) Trends Biochem. Sci., in press. [DOI] [PubMed]
- 4.Strain, J., Lorenz, C. R., Bode, J., Garland, S., Smolen, G. A., Ta, D. T., Vickery, L. E. & Culotta, V. C. (1998) J. Biol. Chem. 273, 31138-31144. [DOI] [PubMed] [Google Scholar]
- 5.Kispal, G., Csere, P., Prohl, C. & Lill, R. (1999) EMBO J. 18, 3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schilke, B., Voisine, C., Beinert, H. & Craig, E. (1999) Proc. Natl. Acad. Sci. USA 96, 10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange, H., Kispal, G., Kaut, A. & Lill, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, J., Saxena, S., Pain, D. & Dancis, A. (2001) J. Biol. Chem. 276, 1503-1509. [DOI] [PubMed] [Google Scholar]
- 9.Mühlenhoff, U., Gerber, J., Richhardt, N. & Lill, R. (2003) EMBO J. 22, 4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy, A., Solodovnikova, N., Nicholson, T., Antholine, W. & Walden, W. E. (2003) EMBO J. 22, 4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balk, J., Pierik, A. J., Aguilar Netz, D., Mühlenhoff, U. & Lill, R. (2004) EMBO J. 23, 2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange, H., Lisowsky, T., Gerber, J., Mühlenhoff, U., Kispal, G. & Lill, R. (2001) EMBO Rep. 2, 715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frazzon, J. & Dean, D. R. (2003) Curr. Opin. Chem. Biol. 7, 166-173. [DOI] [PubMed] [Google Scholar]
- 14.Leipe, D. D., Wolf, Y. I., Koonin, E. V. & Aravind, L. (2002) J. Mol. Biol. 317, 41-72. [DOI] [PubMed] [Google Scholar]
- 15.Vitale, G., Fabre, E. & Hurt, E. C. (1996) Gene 178, 97-106. [DOI] [PubMed] [Google Scholar]
- 16.Lutkenhaus, J. & Sundaramoorthy, M. (2003) Mol. Microbiol. 48, 295-303. [DOI] [PubMed] [Google Scholar]
- 17.Skovran, E. & Downs, D. M. (2003) J. Bacteriol. 185, 98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mühlenhoff, U., Richhardt, N., Ristow, M., Kispal, G. & Lill, R. (2002) Hum. Mol. Genet. 11, 2025-2036. [DOI] [PubMed] [Google Scholar]
- 19.Sherman, F. (1991) Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- 20.Mumberg, D., Müller, R. & Funk, M. (1995) Gene 156, 119-122. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, M., Saeki, K. & Takahashi, Y. (1999) J. Biochem. 126, 10-18. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 23.Gietz, D., St. Jean, A., Woods, R. A. & Schiestl, R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diekert, K., deKroon, A. I. P. M., Kispal, G. & Lill, R. (2001) Methods Cell Biol. 65, 37-51. [DOI] [PubMed] [Google Scholar]
- 25.Harlow, E. & Lane, D. (1998) Using Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 26.Li, J., Kogan, M., Knight, S. A., Pain, D. & Dancis, A. (1999) J. Biol. Chem. 274, 33025-33034. [DOI] [PubMed] [Google Scholar]
- 27.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- 28.Tong, W. H., Jameson, G. N., Huynh, B. H. & Rouault, T. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9762-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerber, J., Neumann, K., Prohl, C., Mühlenhoff, U. & Lill, R. (2004) Mol. Cell. Biol. 24, 4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mühlenhoff, U., Balk, J., Richhardt, N., Kaiser, J. T., Sipos, K., Kispal, G. & Lill, R. (2004) J. Biol. Chem. 279, 36906-36915. [DOI] [PubMed] [Google Scholar]
- 31.Yarunin, A., Panse, V., Petfalski, E., Tollervey, D. & Hurt, E. (2005) EMBO J. 24, 589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kispal, G., Sipos, K., Lange, H., Fekete, Z., Bedekovics, T., Janaky, T., Bassler, J., Aguilar Netz, D. J., Balk, J., Rotte, C. & Lill, R. (2005) EMBO J. 24, 580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.