Imagine a paleontologist confronted with of a fossil of a single footprint. She could probably risk some conjectures about the imprinting creature, such as whether it possessed paws, claws, or feet, and estimate its weight, height, and other attributes. But the information contained in a single static print remains limited. The situation improves if a multitude of footprints from a population of creatures becomes available. Now, stride lengths can be calculated, variability between individuals can be assessed, and speeds can be estimated. Furthermore, correlating footprints from different geographies could lead to clues about the population's migration patterns. With appropriate assumptions, our paleontologist could begin to map how the population evolved over time.
As this example illustrates, even though a footprint is a static entity, the sum of many footprints could in principle provide clues about the dynamics of a population. In this vein, the work of Middendorf, Ziv, and Wiggins in this issue of PNAS (1) seeks to understand the broad strokes of the evolutionary dynamics that shaped a species from a static network of protein–protein interactions. By way of “footprints,” Middendorf et al. (1) use substructures in the protein–protein network called network motifs whose count provides a window into the dynamics that left the record. These counts are the markers that the authors use to “learn” which motifs differentiate between competing hypothetical evolutionary schemes. The outcome of the study suggests a dominant evolutionary mechanism that shaped Drosophila melanogaster.
Evolution Revealed in Manmade Networks
Networks of all sorts evolve, and it is often possible to form hypotheses about such evolution by direct examination of the structure of the network (2–4). Consider the U.S. telephone network, which evolved over the past 100 years in response to changes in traffic load and advances in technology. The telephone network was originally engineered to carry voice traffic. The addition of data traffic, which eventually dominated voice, was an evolutionary pressure to this network that forced radical changes in its topology. In the 1960s, as the volume of data traffic carried on the network occupied a significant amount of bandwidth, the facilities of many large cities became saturated, and it was necessary to relieve the congestion by rerouting interstate traffic. These changes led to a network topology that resembled that of the interstate highway network, where “beltways” surround the major cities to keep interstate congestion off local roads. These beltways can be observed directly as motifs in today's telephone network topology. Today's topology is the result of evolutionary forces that could be deduced by modeling changes in topology necessary to overcome the rise in data traffic. On a larger scale, there was a shift in topology that resulted from the need to add major capacity in the southern half of the U.S. accompanying the population shift to warmer climates. Notice that the topology changes necessary to respond to local and global pressures are different.
Characterizing Biological Networks
In contrast with the telephone network example, where design and control are explicitly engineered, our understanding of biological network design principles and of mechanisms that control the traffic of biological information is very poor. Part of today's challenge, therefore, is to elucidate the principles on which biological networks evolve.
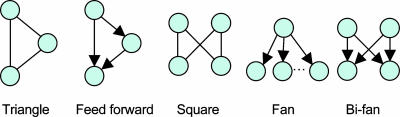
In recent years, biological network architectures have been characterized by properties such as sparseness (5), small-world (6, 7), and scale-free (8). These network characterizations are global in that a single number, such as the average connectivity or the radius (average number of hops between any two nodes), describes a property of the whole network. An alternative approach to characterize a network is via topological motifs. If the number of occurrences of a motif is large compared with what is expected by chance, then a case can be made that such a motif represents a reusable functional module or is the consequence of evolutionary mechanisms. Thus, motifs such as those shown in Fig. 1 have been discovered from a number of complex networks (9, 10). The feed-forward triangle and the bi-fan square (see Fig. 1), for example, occurred with a Z score of >10, both in the Escherichia coli and Saccharomyces cerevisiae gene regulatory networks (10).
Fig. 1.
Some of the most common motifs found in biological networks. Nodes indicate cellular components, such as genes or proteins. Edges represent associations between nodes, such as binding (undirected edges) or influence (directed edges).
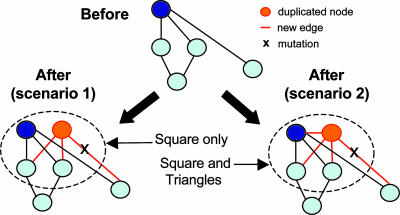
A high Z score does not necessarily imply biological relevance for a motif. Efforts are underway to find biological interpretations to what statistical significance can only suggest. Alon and coworkers (11) have associated the feed-forward motif with a rapid-response filter of noisy inputs. Cycles, both positive and negative, can be associated with feedback loops leading to responses over an extended time frame. The relatively large number of squares and triangles observed in the protein interaction network of Drosophila (1) directly points to possible mechanisms at work in its evolutionary history (see Fig. 2).
Fig. 2.
Two examples of network evolution through the DMC mechanism. Before the iteration, a node is chosen at random (blue node) and replicated (orange node) along with its connections (orange edges). The edges linking the original or replicated node and its neighbors can be mutated and rendered unfunctional (shown with an X in scenarios 1 and 2). The original and duplicated nodes can be conjoined (only scenario 2) with some finite probability. Scenario 1 leads to the creation of square subgraphs, whereas scenario 2 leads to the creation of square and triangle subgraphs.
Protein–Protein Interaction Networks
The two-hybrid studies that produced the protein interaction map for D. melanogaster (12) provide a valuable genome-wide view of protein interactions but have a number of shortcomings (13). Even if the protein–protein interactions were determined with high accuracy, the resulting network would still require careful interpretation to extract its underlying biological meaning. Specifically, the map is a representation of all possible interactions, but one would only expect some fraction to be operating at any given time. Hence, the map is a static imprint of all possible interactions that clearly lacks dynamic information. Combine this feature with the considerable sources of noise and artifacts known to protein–protein networks and one can see the difficulties in using interaction data. However, as in our footprint analogy, interpreting a multitude of static records may give clues to dynamic interactions, even if some records are faulty (i.e., not every footprint needs to be intact).
Machine-Learning Approaches
Rather than discovering all motifs in a network (a task that would result in a combinatorial explosion of subgraphs), previous studies (10) have used statistical criteria to select significant subgraphs from a family of prescribed subnetworks, such as all n-node subgraphs (n = 3 and 4). Similar statistical criteria were used to cluster disparate networks into superfamilies that share the same (normalized) vector of Z scores over all three- or four-node subgraphs (14). However, problems may arise with these approaches if the underlying null hypothesis is not posed carefully (15). Middendorf et al. (1) avoided the use of statistical considerations in the selection or normalization of subgraph counts. They explored all subgraphs that verified two criteria: (i) subgraphs produced by walks of up to eight hops and (ii) subgraphs with up to seven edges. Although such criteria keep the search computationally tractable, other important motifs could be missed. The raw counts of these motifs in the Drosophila network were used as inputs into a learning machine previously trained to recognize networks grown by using seven different putative evolutionary trends: duplication–mutation–complementation (DMC) (Fig. 2), duplication–mutation followed by random attachment, linear preferential attachment, small-world networks, random static networks, random growing networks, and aging vertex networks. As a machine-learning algorithm, the authors chose the alternating decision tree (ADT) (16), which allows for a reasonably straightforward identification of the main subgraphs used in the decisions. With the Drosophila network, the ADT algorithm chose the DMC mechanism as the best choice to explain the subgraph counts in the input network. Indeed, two of the most discriminative subgraphs chosen by the ADT to make its decision were triangles and squares (table 6 in ref. 1), which can be easily interpreted in the context of the DMC mechanism (Fig. 2).
Future Directions
The study by Middendorf et al. (1) is a laudable step toward extracting evolutionary information from static biological networks, but certainly much remains unanswered. Most likely, the evolution of protein–protein maps involves complexities that we are yet to fully grasp. For example, whereas the small-scale features contained in the network motifs are well captured by the DMC mechanism, some large-scale features of the Drosophila interaction map, such as the size of the giant component (table 1, SM, in ref. 1), do not seem to be recapitulated by DMC. This inconsistency should not be too surprising because different aspects of the interaction map may respond to different evolutionary pressures. As with the U.S. phone system, different pressures shaped the network at the city and country levels. Such a notion is compatible with theoretical work that characterized small-world graphs as a superposition of multiple graphs rather than a homogenous entity (3). One might expect analogous superpositions in biological networks. For example, a hierarchical arrangement of connected clusters is described for E. coli metabolic networks (17). Refinements in techniques as well as larger data sets from more species will help us to better trace the evolutionary past and the logical relations that make the networks functional in the present (18).
See companion article on page 3192.
References
- 1.Middendorf, M., Ziv, E. & Wiggins, C. H. (2005) Proc. Natl. Acad. Sci. USA 102, 3192-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorogovtsev, S. N. & Mendes, J. F. F. (2003) Evolution of Networks: From Biological Nets to the Internet and WWW (Oxford Univ. Press, Oxford).
- 3.Watts, D. J. & Strogatz, S. H. (1998) Nature 393, 440-442. [DOI] [PubMed] [Google Scholar]
- 4.Albert, R. & Barabasi, A. L. (2000) Phys. Rev. Lett. 85, 5234-5237. [DOI] [PubMed] [Google Scholar]
- 5.Wagner, A. (2002) Genome Res. 12, 309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner, A. & Fell, D. A. (2001) Proc. R. Soc. London B 268, 1803-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arita, M. (2004) Proc. Natl. Acad. Sci. USA 101, 1543-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabasi, A. L. (2000) Nature 407, 651-654. [DOI] [PubMed] [Google Scholar]
- 9.Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. (2002) Nat. Genet. 31, 64-68. [DOI] [PubMed] [Google Scholar]
- 10.Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D. & Alon, U. (2002) Science 298, 824-827. [DOI] [PubMed] [Google Scholar]
- 11.Mangan, S., Zaslaver, A. & Alon, U. (2003) J. Mol. Biol. 334, 197-204. [DOI] [PubMed] [Google Scholar]
- 12.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., et al. (2003) Science 302, 1727-1736. [DOI] [PubMed] [Google Scholar]
- 13.Jansen, R. & Gerstein, M. (2004) Curr. Opin. Microbiol. 7, 535-545. [DOI] [PubMed] [Google Scholar]
- 14.Milo, R., Itzkovitz, S., Kashtan, N., Levitt, R., Shen-Orr, S., Ayzenshtat, I., Sheffer, M. & Alon, U. (2004) Science 303, 1538-1542. [DOI] [PubMed] [Google Scholar]
- 15.Artzy-Randrup, Y., Fleishman, S. J., Ben-Tal, N. & Stone, L. (2004) Science 305, 1107. [DOI] [PubMed] [Google Scholar]
- 16.Middendorf, M., Kundaje, A., Wiggins, C., Freund, Y. & Leslie, C. (2004) Bioinformatics 20, Suppl. 1, I232-I240. [DOI] [PubMed] [Google Scholar]
- 17.Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N. & Barabasi, A. L. (2002) Science 297, 1551-1555. [DOI] [PubMed] [Google Scholar]
- 18.Bowers, P. M., Cokus, S. J., Eisenberg, D. & Yeates, T. O. (2004) Science 306, 2246-2249. [DOI] [PubMed] [Google Scholar]