Abstract
Protein modeling and molecular dynamics hold a unique toolset to aide in the characterization of clinical variants which may result in disease. Not only do these techniques offer the ability to study under characterized proteins, but they do this with the speed that is needed for time sensitive clinical cases. In this paper we retrospectively study a clinical variant in the XIAP protein, C203Y, while addressing additional variants seen in patients with similar GI phenotypes as the C203Y mutation. In agreement with the clinical tests performed on the C203Y patient, protein modeling and molecular dynamics suggest that direct interactions with RIPK2 and Caspase3 are altered by the C203Y mutation and subsequent loss of Zn coordination in the second BIR domain of XIAP. Interestingly, the variant does not appear to alter interactions with SMAC, resulting in further damage to the caspase and NOD2 pathways. Additionally, for the first time we suggest an amino acid target, F228, of XIAP in drug design to increase specificity of activating Caspase signaling while not interring with NOD2 signaling or other BIR domain protein pathways. This paper shows the exciting applications of molecular modeling in the classification and characterization of genetic variants identified in next generation genome sequencing.
Keywords: XIAP, Caspase3, SMAC, RIPK2, Clinical variants, protein modeling
Introduction
The completion of the human genome was one of the biggest breakthroughs in human knowledge. Following its completion, technology has continued to advance, which has now allowed for sequencing of a genome for only a few thousand dollars. These prices will make genomic sequencing common practice, cheaper than many clinical tests currently being used. The human sequencing so far has identified millions of variants, with characterization of these variants only in its infancy. The influx of this new genomic data is likely to continue transforming fields such as biochemistry and molecular biology. Genomes are currently being employed in understanding common diseases such as cardiovascular (Tennessen et al., 2012) or cancer (Forbes et al., 2011); with the results challenging at both the individual genome and at an epidemiological level. For rare diseases, however, the use of genomic sequencing is becoming a viable option in clinical identification (Jacob et al., 2013). With the cost of sequencing a genome at several thousand dollars, utilization of whole genome sequencing could supplement or supersede some expensive clinical tests.
One of the first cases in which whole exome sequencing was used to treat a patient, was in a male child suffering from a Crohn’s like inflammatory bowel disease (Worthey et al., 2011). Sequencing revealed a single amino acid missense mutation (C203Y) in a protein, XIAP. This case reached wide audiences, gaining a voice in popular media through stories such as the Pulitzer prize winning, “One in a billion: a boy’s life, a medical mystery.” Variants in XIAP were not previously associated with gastrointestinal (GI) disease, but more commonly seen in X-linked lymphoproliferative syndrome (Rigaud et al., 2006). XIAP is located on the X-chromosome, and therefore the single copy of XIAP in a male child with a loss of function mutation (hemizygous) can result in disease. Following the identification of this variant in the XIAP protein, subsequent variations of XIAP have been discovered in patients with inflammatory bowel disease (IBD), celiac-like disease, Crohn’s-like disease, severe infectious mononucleosis, and hemophagocytic lymphohistiocytosis (HLH) (Speckmann and Ehl, 2013; Speckmann et al., 2013; Veillette et al., 2013).
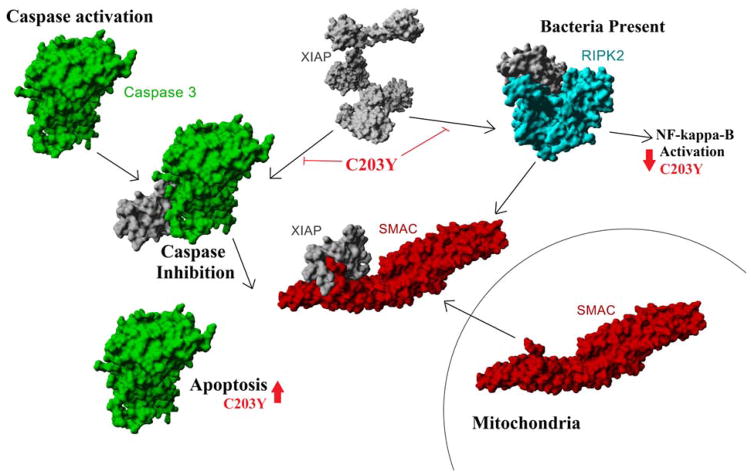
XIAP has several characterized functions, most notably the inhibition of activated Caspases (Eckelman et al., 2006) and regulation of NOD1/2 signaling (Damgaard et al., 2013) through interaction with RIPK2 (Krieg et al., 2009). Both Caspase inhibition and NOD signaling of XIAP can be blocked by the release of the SMAC/Diablo protein from the mitochondria (Chai et al., 2000; Krieg et al., 2009; Shi, 2002) (Figure 1). In many cancer phenotypes, activation of Caspase proteins has the potential to result in apoptotic cell death (Lin et al., 2013). Therefore, many groups have developed SMAC mimetics in hopes of blocking XIAP inhibition of Caspase and autophagy, allowing for selective cell death in cancer cells (Huang et al., 2013; Lin et al., 2013; Mahadevan et al., 2013; Saleem et al., 2013). However, some SMAC mimetics additionally block XIAP activation of the NOD signally pathway (Damgaard et al., 2013; Krieg et al., 2009), suggesting some potential immunocompetence in treating cancer patients with SMAC mimetics. Additionally, the role of XIAP somatic mutations in caner have not been assessed for their ability to endogenously alter apoptosis of cancer.
Figure 1. XIAP in Caspase 3 and NOD2 signaling pathways.
Following Caspase 3 (green) activation by initiator caspase proteins, Caspase 3 is inhibited by the binding of XIAP (gray). SMAC (red), also known as Diablo, is released from the mitochondria as a result of apoptotic signaling, which then displaces XIAP-Caspase 3 interaction allowing for induction of apoptosis. In addition to Caspase regulation, XIAP is known to interact and regulate RIPK2 (cyan) in the NOD pathway of bacterial activation.
Continual increase in genomic sequencing has shown practical clinical use (Jacob et al., 2013); however, suggests the development of new tools in the analysis and characterization of variants in perturbing molecular pathways. From a variant stand point, there exist many tools to address the impact of a variant relative to species’ evolution, thousands of additionally sequenced human exomes/genomes, and known variants seen in diseases. Yet, these tools currently identify a large fraction of variants as Variants of Uncertain Significance (VUS) or Genes of Uncertain Significance (GUS), in which no clinical decision can be made often due to the lack of knowledge for the particular gene. An area that is ideal for use in characterizing VUS and GUS, but currently underutilized, is molecular modeling and dynamics. We hypothesize that molecular modeling and dynamics will serve as a valuable tool in understanding VUS and GUS of genomic sequencing. The XIAP variant previously discovered by our group, C203Y, was initially a VUS that required months of benchtop characterization, delaying treatment for a patient in extreme need of treatment options. Using knowledge of XIAP modeling and dynamics we hope to gain insights into XIAP function while additionally showing the practical application of molecular modeling and dynamics in characterization of VUS and GUS from genomic sequencing.
The XIAP protein has several known structures and well characterized pathways, making it a strong candidate to begin utilizing the tools of molecular modeling and dynamics in understanding genetic variation. Much like the Enterprise served as the tool for Jean-Luc Picard and crew to understand space in Star Trek: The Next Generation; molecular modeling will serve as an enterprise for researchers to understand the space in which variants of next generation sequencing result in disease. Using molecular modeling, sequence analysis, and structures we have addressed known mutations for XIAP as to their potential perturbation in molecular interactions, ultimately suggesting potential sites in XIAP that may be novel to design inhibitors to and suggest novel drug development for treatment options in the future.
Methods
Structural analysis of XIAP
Structures of XIAP were identified by BLAST analysis with the human XIAP sequence against the protein data bank (pdb). A full atomic model was created by joining the BIR1 (pdb file 2pop, amino acids 10–100), BIR2 (1i3o, amino acids 124–240), BIR3 (1f9x, amino acids 241–356), UBA (2kna, amino acids 357–449) and the RING-type (4ic2, amino acids 429–497) domains with an ab initio model prediction using QUARK (Xu and Zhang, 2012) for amino acids 101–123. The domains were placed together so that interactions between XIAP with TAB1 (pdb file 2pop), Caspase3/7 (1i3o and 1i4o), Caspase9/SMAC (1g73 and 1nw9), and the RING-type dimers (4ic2) did not result in steric clash between the binding partners. Domain alignments with protein interaction partners was performed using the MUSTANG algorithm (Konagurthu et al., 2006). Using the compiled model, homology modeling was performed using the sequence of human (Uniprot P98170, amino acids1–497), mouse (Q60989, 1–496), and rat (Q9R0I6, 1–496) XIAP with YASARA. Amino acids that were identified to vary in XIAP in disease patients were then identified on the structure of the full XIAP, allowing for rapid identification of how the variants could perturb interaction with various proteins. To determine SMAC binding with the BIR2 or BIR3 domain, the known structure of BIR3 interaction with SMAC (pdb file 1g73) had the BIR3 homology modeled to the sequence of the BIR2 domain using YASARA, followed by energy minimizations of either BIR3/SMAC or BIR2/SMAC using the Amber03 (Duan et al., 2003) force field with 0.997 g/mL of water. Binding energy was then calculated using YASARA (Krieger et al., 2002).
Sequence analysis of BIR domain proteins
Sequences were obtained for human (h) Survivin (Uniprot O15392), Bruce (Q9NR09), cIAP1 (Q13490), cIAP2 (Q13489), XIAP (P98170), ILP2 (Q96P09), MLIAP (Q96CA5), and NAIP (Q13075) with the individual BIR domains parsed. XIAP sequences for Drosophila (d, I6L9G1), Gallus (c, F6TJT0), Xenopus (x, A5D8Q0), Mus (m, Q60989), and Rattus (r, Q9R0I6) were additionally used. Sequence alignments were performed using Clustal Omega (Sievers et al., 2011) with default settings. A phylogenetic analysis of the domains was performed using maximum likelihood with the JTT matrix (Jones et al., 1992) and a consensus of 1000 bootstrap replicates (Felsenstein, 1985) using MEGA5 (Tamura et al., 2011). Initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analysis involved 31 amino acid sequences for 66 amino acids. Variants of XIAP associated with disease were identified in published literature, utilizing only single amino acid changes (including mutations to stop codons (X) and not mutations that result in a frameshift.
Molecular dynamic simulations
Molecular dynamic (md) simulations were performed on two protein complexes, XIAP interaction with Caspase-3 or XIAP interaction with the top docking prediction of RIPK2. For the XIAP-Caspase-3 md simulation, pdb structure 1i3o was cleaned in YASARA, removing all water and adding hydrogen atoms to all molecules. The structure was placed in a simulation box of 118,70,79Å (x,y,z axes) and cell neutralization was performed using 0.9% mass fraction of cations and anions, pH of 7.4, and water density of 0.997g/mL. The structure was energy minimized using the Amber03 (Duan et al., 2003) force field to reduce crystal packing forces of the structure. Individual molecules of the file were then separated into individual objects joining the two subunits of Caspase-3 into a single object and the BIR2 domain of XIAP with its respective Zn ion. The Zn ion was held in place during the simulation using constrained distances of the Zn from its four connecting residues. The md simulations were then performed for 12.5 nanoseconds (ns) using the default md_run macro in YASARA. Trajectories were analyzed using the md_analyze and md_analyzeres macros (www.yasara.org/macros). In addition to the wild type md simulations, three other simulations were performed. First, the cysteine 203 (Cys,C) of XIAP was substituted with a tyrosine (Tyr,Y) to create XIAP C203Y with the three additional contact points with the Zn ion left constrained. Second, the Zn was removed and all four contacts no longer restrained, but the cysteine was left at amino acid 203 (XIAP –Zn). Lastly the C203Y mutation was made in addition to removal of the Zn ion (XIAP C203Y-Zn). All simulations were performed for 12.5 ns.
Docking of XIAP to RIPK2 was performed using the pdb structure 4c8b of human RIPK2. The structure was parsed down to a single domain of RIPK2 and had missing loops modeled using YASARA. The remaining structure was placed in a 60Å simulation square and rigid body docking of the XIAP BIR2 domain was performed using AutoDock VINA (Trott and Olson, 2010) with 10 dockings performed on 5 separate rotamer ensembles of RIPK2. Following calculation of binding energy, the top docking conformation was inserted into a simulation square extending 5Å from all atoms, and the cell was neutralized with the same conditions used for Caspase-3 analysis. The protein complex was then energy minimized and md simulations performed for 15 ns. The complex was simulated as the wild type or as the XIAP C203Y mutation with the Zn ion removed (XIAP C203Y-Zn).
Results
Structural model of the complete XIAP protein
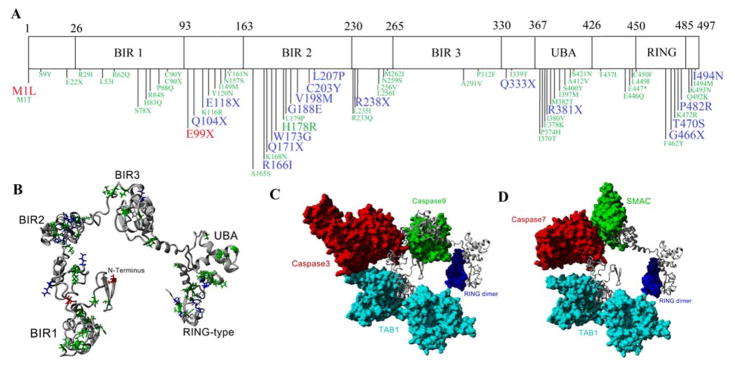
Single amino acid mutations that have been associated with various disease phenotypes (red and blue) or in the Catalog of Somatic Mutations in Cancer (COSMIC) database (green) were identified on the domains of XIAP (Figure 2A). XIAP contains a high density of disease variants in the BIR2 domain, while a high number of variants from COSMIC are found in the UBA domain. To understand where the variants are located on the structure, a complete model was built for human (Figure 2B), mouse and rat XIAP based on known structures (Table 1) such that the interactions with TAB1, Caspase3/7, Caspase 9/SMAC, and the RING dimer were maintained (Figure 2C–D). The structures between human, mouse, and rat aligned with an average of 91.53% homology with only a 0.42Å carbon alpha root-mean squared deviation (RMSD) between the structures. In addition the structural Z-scores were −0.68, −0.76, and −0.83 relative to other PDB structures suggesting accurate predictions of the global structure, with anything >−2 considered a good model. The disease variants were then identified on the full structure (Figure 2B), allowing for rapid identification of how all variants may alter protein packing or protein-protein interaction.
Figure 2. Domains of the XIAP protein.
A) Schematic breakdown of XIAP into the BIR1, BIR2, BIR3, UBA, and RING domains. Single amino acid variants are shown below the schematic with those in red from Speckmann et al 2013, blue from Damgaard et al 2013, and green from the Catalogue of Somatic Mutations in Cancer (COSMIC). Larger sized green text represents reoccurring variants seen in COSMIC. B) Compiled model for the entire XIAP protein with the variants from A mapped onto the structure. C–D) Using the XIAP protein as a scaffold, known XIAP-protein interactions are shown for TAB1 (cyan) with the BIR1 domain of XIAP, Caspase 3/7 (red) with the BIR2 domain, Caspase9/SMAC (green) with the BIR3 domain, and the RING domain dimer (blue).
Table 1. Known structures of XIAP.
Structures highlighted in yellow were used for modeling the complete XIAP structure (Figure 2B) and those in red used to model XIAP-protein interactions (Figure 2C–D).
| Entry | Method | Resolution (Å) | Chain | Position | Domain | Binding partner | Used for full model |
|---|---|---|---|---|---|---|---|
| 1–9 | |||||||
| 2POI | X-ray | 1.8 | A | 10–99 | BIR1 | native (I222 space) | |
| 2QRA | X-ray | 2.5 | A/B/C/D | 10–99 | BIR1 | native (P21 space) | |
| 2POP | X-ray | 3.1 | B/D | 10–100 | BIR1 | TAB1 | x |
| 1C9Q | NMR | - | A | 124–240 | BIR2 | native | |
| 4j3y | X-ray | 1.45 | A/C | 152–236 | BIR2 | native | |
| 1I3O | X-ray | 2.7 | E/F | 124–240 | BIR2 | Caspase 3 | x |
| 1I4O | X-ray | 2.4 | C/D | 120–260 | BIR2 | Caspase 7 | x |
| 1I51 | X-ray | 2.45 | E/F | 124–240 | BIR2 | Caspase 7 | |
| 1KMC | X-ray | 2.9 | C/D | 124–242 | BIR2 | Caspase 7 | |
| 4j45 | X-ray | 1.48 | A/C | 152–236 | BIR2 | ATAA (peptide) | |
| 4j44 | X-ray | 1.3 | A/C | 152–236 | BIR2 | AIAV (peptide) | |
| 4j46 | X-ray | 1.42 | A/C | 152–236 | BIR2 | AVPI (peptide) | |
| 4j47 | X-ray | 1.35 | A/C | 152–236 | BIR2 | SVPI (peptide) | |
| 4j48 | X-ray | 2.1 | A/C | 152–236 | BIR2 | AMRV (peptide) | |
| 1F9X | NMR | - | A | 241–356 | BIR3 | native | x |
| 1G73 | X-ray | 2 | C/D | 238–358 | BIR3 | SMAC | x |
| 1NW9 | X-ray | 2.4 | A | 253–350 | BIR3 | Caspase-9 | x |
| 4HY0 | X-ray | 2.84 | A/B/C/D/E/F/G/H | 238–357 | BIR3 | T3256336 (antagonist) | |
| 1G3F | NMR | - | A | 241–356 | BIR3 | SMAC peptide (9AA) | |
| 1TFQ | NMR | - | A | 241–356 | BIR3 | Antagonist (998) | |
| 1TFT | NMR | - | A | 241–356 | BIR3 | Antagonist (997) | |
| 2JK7 | X-ray | 2.82 | A | 241–356 | BIR3 | SMAC MIMETIC (BI6) | |
| 3CLX | X-ray | 2.7 | A/B/C/D | 241–356 | BIR3 | SMAC MIMETIC (X22) | |
| 3CM2 | X-ray | 2.5 | A/B/C/D/E/F/G/H/I/J | 241–356 | BIR3 | SMAC MIMETIC (X23) | |
| 3CM7 | X-ray | 3.1 | A/B/C/D | 241–356 | BIR3 | SMAC MIMETIC (X22) | |
| 3EYL | X-ray | 3 | A/B | 241–356 | BIR3 | SMAC MIMETIC (SMK) | |
| 3G76 | X-ray | 3 | A/B/C/D/E/F/G/H | 241–356 | BIR3 | bivalent compound (CZ3) | |
| 4EC4 | X-ray | 3.3 | A/B/C/D/E/F/G/J/K/L | 241–356 | BIR3 | SMAC MIMETIC (0O6) | |
| 2OPY | X-ray | 2.8 | A | 249–354 | BIR3 | SMAC MIMETIC (CO9) | |
| 2OPZ | X-ray | 3 | A/B/C/D | 249–357 | BIR3 | SMAC homolog AVPF | |
| 2VSL | X-ray | 2.1 | A | 250–345 | BIR3 | SMAC MIMETIC (15P) | |
| 3HL5 | X-ray | 1.8 | A/B | 256–346 | BIR3 | Antagonist (CS3) | |
| 3UW5 | X-ray | 1.71 | A/B | 296–348 | BIR3 | Antagonist (GDC0152) | |
| 3UW4 | X-ray | 1.79 | A | 338–348 | BIR3 | Antagonist (GDC0152) | |
| 2KNA | NMR | - | A | 357–449 | UBA | native-dimer | x |
| 4IC2 | X-ray | 2.2 | A/B | 429–497 | RING-type | native | x |
| 4IC3 | X-ray | 1.78 | A/B | 429–497 | RING-type | native (F495L) | |
| 2ECG | NMR | - | A | 430–497 | RING-type | native |
Evolution of the BIR domain
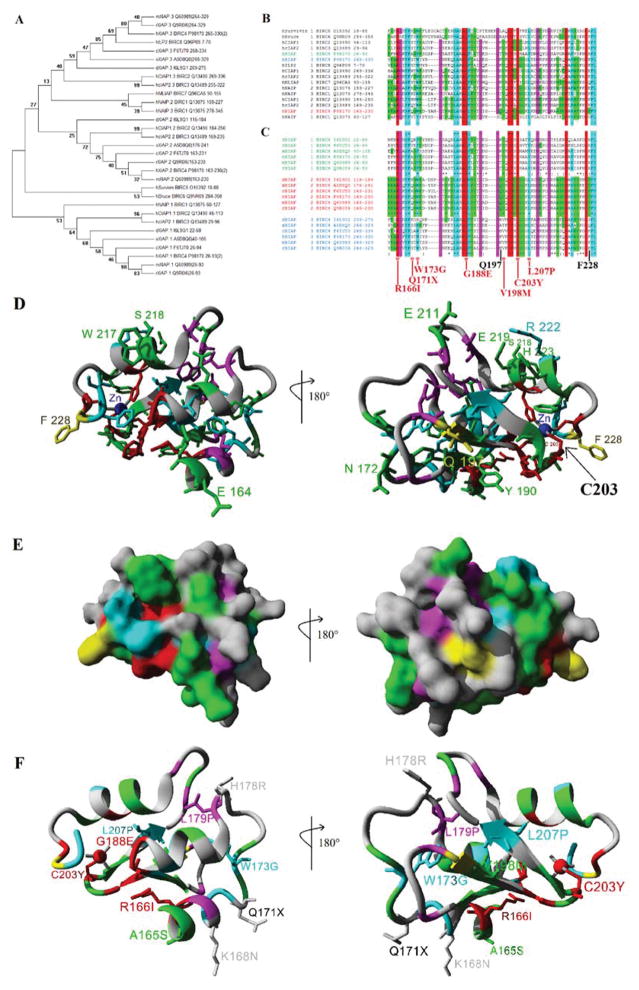
The XIAP protein contains three separate BIR domains. In the human a total of 8 BIR containing proteins have been identified with a total of 16 separate BIR domains. Each of these BIR domains were parsed, aligned, and a phylogenetic tree created (Figure 3A). Across species the BIR1 and BIR3 domain of XIAP cluster separately with the Bir2 domain clustering close to the human cIAP1 and cIAP2 BIR2 domains. Sequence analysis revealed eight amino acids that are 100% conserved (red) and several additionally functionally conserved (cyan) in all human BIR domains (Figure 3B). To identify if there are any amino acids that are specific and conserved in the second BIR domain of XIAP, sequence alignments from multiple species for XIAP were performed (Figure 3C). 18 amino acids that are conserved in the BIR2 domain of XIAP are also conserved in other human BIR domains (red and cyan), while 6 amino acids are conserved in all three of the XIAP BIR domains but not all human BIR containing proteins (magenta). The genetic variant first identified in the child with the Crohn’s like disease was a C203Y mutation (Worthey et al., 2011). This amino acid is 100% conserved in all BIR domains and is known to contribute to the four coordinated Zn binding, of which are four of the eight 100% conserved amino acids (red) of the BIR domain. Amino acids 166 and 188 that have variants associated with disease are also 100% conserved in all BIR domains. Amino acids 173 and 207, which contain disease associated variants, are functionally conserved in all BIR domains. 21 amino acids were found conserved in the BIR2 domain of XIAP but not in the other XIAP BIR domains (green).
Figure 3. Sequence analysis of BIR domains.
A) Phylogenetic analysis using maximum likelihood for human (h), mouse (m), rat (r), chicken (c), Xenopus (x), and Drosophila (d) proteins containing 31 total BIR domains. The names for each are shown with the first letter representing the species, followed by the protein name, sometimes containing a .x where x is the number of the BIR domain for that protein, then the Uniprot accession number and finally the amino acids from the protein used for the analysis. Values at each node of the tree represent the percent of 1000 bootstrap analyses matching the tree shown. B–C) Sequence alignments of the human Bir domains (B), with the Bir domains listed for human (h), mouse (m), rat (r), chicken (c), Xenopus (x), and Drosophila (d) for each of the three Bir domains of XIAP (C). Amino acids highlighted in red are 100% conserved in all Bir domains, those in cyan are functionally conserved in the Bir domains, Magenta are conserved in all three of the Bir domains of XIAP and those in green are conserved only in the XIAP Bir2 domain across the species. The location of the C203Y is 100% conserved in all Bir domains. D) Coloring of the sequence alignment in B–C shown on the structure of Bir2 from XIAP. Amino acids that are surface exposed and specific to the XIAP Bir 2 domain are labeled. Those shown in yellow (Q197 and F228) were 100% conserved in multiple species of Bir2 domains but were not seen in other XIAP Bir domains (1/3). E) The molecular surface of the Bir2 domain of XIAP showing high probability of protein interaction sites. F) Disease associated variants in the Bir2 domain from Figure 2A shown on the structure with conservation color. Those variants that fall at highly conserved sites are shown as side chains and labeled.
To understand the specificity of the XIAP BIR2 domain relative to other BIR domain proteins, a BIR domain structure was used to map the conservation from the sequence alignment above. Several variants that were highly conserved in the BIR2 domain (green) map to the surface of the protein, suggesting potential protein interaction (Figure 3D–E). Two amino acids, Q197 and F228, both map to the surface of the BIR2 domain and are conserved in all species XIAP BIR2 domains and not in any other human BIR domain, suggesting critical sites to target with future drug design. Using the same protein structure, the genetic variants in the BIR2 domain of XIAP associated with disease were also identified to determine their contribution to protein folding or surface exposure (Figure 3F).
XIAP and Caspase
Previously, the XIAP BIR2 domain was crystalized with Caspase-3 (Figure 4A). Molecular dynamic simulations were performed either on the wild type BIR2 domain, with the C203Y mutation, removal of the Zn ion or C203Y without Zn (Figure 4B–C). The global movement of XIAP was most strongly perturbed in the C203Y mutation (red, Figure 4B), not altering any of the protein dynamics of Caspase3, but altering a stretch of amino acids in XIAP around the C203Y mutation (Figure 4C). These results suggest that the removal of the Zn did not perturb interaction of the two proteins as a result of the high binding energy between XIAP and Caspase3, with some perturbation following the C203Y mutation only as a result of the insertion of a Tyr that is restrained in global positioning due to the Zn. Thus modeling approaches independent of protein-protein interacts were needed to determine the outcomes of the C203Y mutation, and suggest that when assessing single point mutations using dynamics, considerations need to be placed first on large changes to structural conformation.
Figure 4. XIAP interaction with Caspase 3.
A) Structure (pdb file 1i3o) showing the interaction of two Bir2 domains of XIAP (gray) interacting with two molecules of Caspase 3 (cyan and blue) with a zoomed in view of the Zn ion (magenta) and location of amino acid C203Y. B–C) 12.5 ns of md simulation for Caspase 3 interacting with the wild type XIAP (blue), XIAP with C203Y with Zn coordinating the three remaining sites (red), wild type XIAP without the Zn ion (green), or XIAP with the C203Y mutation and the removal of Zn (purple). The carbon alpha RMSD over the simulation shows only the C203Y mutation (red) to perturb dynamics (B). Movement of individual amino acids over the simulation for Caspase 3 shows perturbations to XIAP in the C203Y mutation (red) through changes at amino acids 210–220 dynamics (C). As molecular dynamics suggested that C203Y mutation of XIAP post protein interaction results in negligible change, we utilized ab initio protein modeling without Zn coordination (red) that results in a protein with similar fold space as the wt Zn coordinated structure (gray) as seen by structural alignments.
Unlike most Zn finger proteins and other Zn binding domains that contain few hydrophobic amino acids, the BIR domain contains strong hydrophobic packing with the Zn ion coordinating a hydrophilic region resulting in the surface exposure of several hydrophobic amino acids for protein interactions. Ab initio modeling independent of the known BIR2 structures confer the hydrophobic collapse independent of the Zn coordination (Figure 4D). Contrary to the possibility that the C203Y and resulting loss of Zn coordination leads to complete unfolding of the molecule, it appears that the surface exposed hydrophobic amino acids internally collapse resulting in a loss of surface exposed contacts. Utilizing this ab initio model we next sought to address changes in the binding properties to several other XIAP-interactions (below).
XIAP and RIPK2
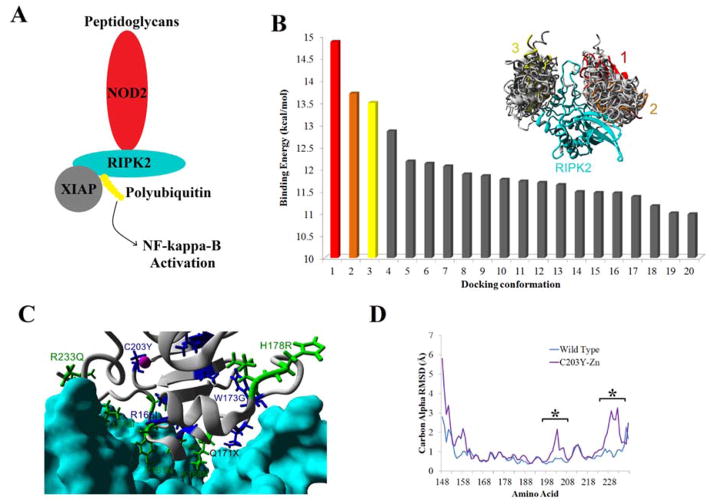
One area current structural work cannot help in understanding XIAP genetic variants, is for regulation of the NOD2 pathway through interaction with RIPK2 (Figure 5A). To begin predictions to allow for analysis, in silico rigid body AutoDock experiments were used to predict the most likely conformation of XIAP interaction with RIPK2. Two clustering regions were found based on the docking with the highest and second highest binding energy conformations in the same cluster (Figure 5B). This binding conformation when energy minimized provides numerous contact points between the two proteins with several variants found at the interface (Figure 5C). The C203Y mutation and loss of Zn binding results in the loss of several amino acid contacts, with molecular dynamic simulations (Figure 5D) confirming perturbations to two regions of XIAP (Figure 8G) following the mutation.
Figure 5. XIAP interaction with RIPK2 in NOD signaling.
A) Depiction of the NOD2 signaling pathway, where binding of a peptidoglycan to NOD2 results in RIPK2 recruitment, binding of XIAP and subsequent ubiquitination of RIPK2 resulting in NF-kappa-B activation. B) To predict the site of interaction of XIAP with RIRK2, rigid body docking was performed with the top twenty docking results shown. Most of these top 20 dockings clustered together into two sites, however the top two docking energies can be seen in only one of the two conformations. C) Based on the top docking results from B, the location of disease association (blue) or COSMIC (green) variants in the Bir2 domain of XIAP are shown for interaction with RIPK2 (cyan). D) Molecular dynamic simulations of the wild type docked complex (cyan) or on the XIAP C203Y mutation without a Zn ion (purple) showing the carbon alpha RMSD average movement of each amino acid for XIAP. Results show sites with differential trajectories represented with a *.
XIAP and SMAC
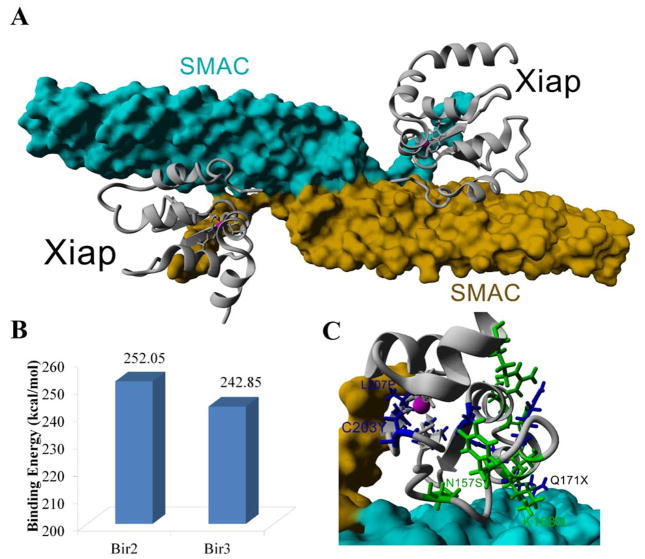
The known interaction of XIAP to SMAC occurs through a dimer of the SMAC protein with two separate XIAP domains (Figure 6A). One of the two BIR3 domains of the known structure was mutated to the BIR2 sequence and energy minimized. The physical constraints between the two domains of XIAP in this structure places the N and C-termini 47Å apart. Having a 180° rotation on the Omega, Phi, and, Psi bonds of a peptide, maximizing the distance covered by a peptide chain, only yields a distance of 3.6Å between nitrogen atoms. A gap of 11 amino acids is seen between the two BIR domains interacting with SMAC. Therefore it is physically impossible (maximum of 39.6Å) to cover the minimum distance (47Å) between the domains. This is strong evidence that a single molecule of XIAP could not simultaneously have the dimer of SMAC bind to both the BIR2 and BIR3 domains without having significant structural alteration to the BIR domains. Therefore, we utilized the individual domains to calculate binding energy with SMAC for both BIR2 and BIR3 (Figure 6B). Interestingly, four COSMIC variants (L256I, L256V, N259S, and M262I) in the BIR3 domain map directly to the interface of binding with SMAC. For the BIR2 domain, the C203Y mutation would alter only slight contacts with SMAC (Figure 6C).
Figure 6. XIAP interaction with SMAC.
A) The known structure of a dimer of SMAC (cyan and yellow) bound to two molecules of Bir3 XIAP (gray). B) Utilizing the Bir3 bound structure, the Bir domain was mutated to that of Bir2 and following energy minimizations the theoretical binding energy determined, confirming that both Bir domains have a high probability of binding. C) Mapping the Bir2 variants of disease association (blue) or seen in COSMIC (green) shows several variants close to the interaction sites.
Altered protein interactions
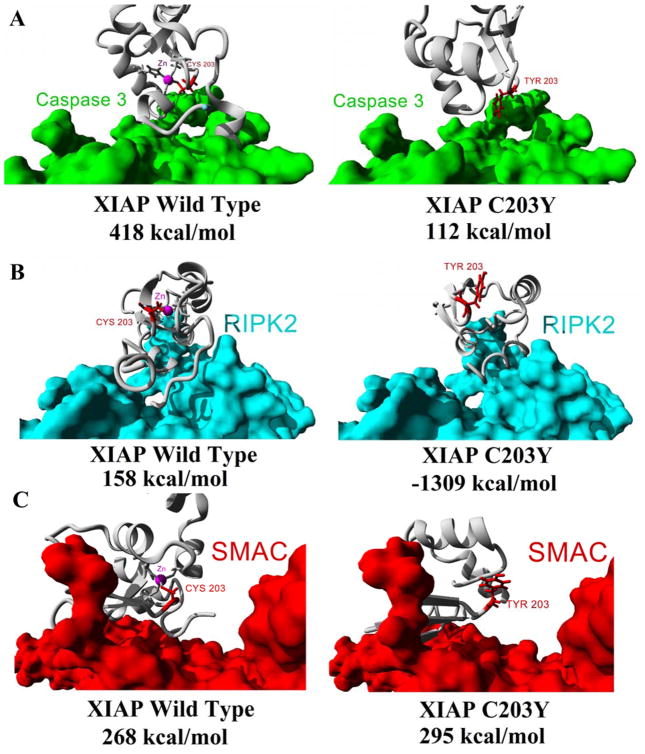
Utilizing the ab initio model of XIAP C203Y protein above (Figure 4D) we assessed the theoretical binding energies for wt XIAP (left) or the C203Y variant (right) to Caspase 3 (Figure 7A), RIPK2 (Figure 7B), or SMAC (Figure 7C). The binding energies are greatly altered for XIAP C203Y for binding to Caspase 3 (wt=418 kcal/mol, C203Y=112 kcal/mol) and RIPK2 (wt=158 kcal/mol, C203Y=−1309 kcal/mol) but not for binding to SMAC (wt=268 kcal/mol, C203Y=295 kcal/mol) suggesting the mutation seen in the patient to alter interactions with two of the three macromolecules. Maintaining interaction with SMAC suggests further perturbation in the decrease of NF-kappa-B activation and an increase in Caspase3 apoptotic signaling (Figure 1). A representative video can be seen on our YouTube channel showing all models and interactions (https://www.youtube.com/watch?v=FPQPXByeUGs).
Figure 7. Binding energy calculations for interaction of wt or C203Y XIAP with several proteins.
Based on the structures of figures 4–6, the ab initio modeled C203Y XIAP protein structure was docked to either Caspase 3 (A), RIPK2 (B), or SMAC (C) and the binding energies calculated for either the wt (left) or C203Y ab initio model (right).
Discussion
Genomic sequencing is becoming a powerful tool in understanding how genetic variation can result in disease. However, to go from simply detection of a variant, to detailed atomic understanding of the mechanisms altered by such a variant, requires additional tools. In this study we have taken a highly publicized clinical genomic sequencing case from our group, XIAP C203Y, and assessed the applicably of molecular modeling to understanding how atomic mechanisms may be altered in the disease progression. Analysis of clinically relevant XIAP BIR2 domain variants show several to fall in highly conserved sites that contribute to either the Zn coordinated structure (C203Y) or the folding of the domain (R166I, W173G, G188E, V198M, and L207P). Several additional early stop codon (E99X, Q104X, E118X, and Q171X) or frame shift mutations have been identified that result in the complete removal of the BIR2 and subsequence domains resulting in disease. In this paper we also analyzed all known variants in XIAP seen in the genome sequencing for somatic mutations in cancer (from COSMIC) in hopes of identifying if a subset of these genomes may contain any type of variants that result in blockade of the Caspase pathway. Several COSMIC variants were identified to be clustered in the UBA domain of XIAP. COSMIC variants were also seen that could alter the interaction of XIAP with Caspase3, Caspase7, and SMAC proteins based on the known structures of XIAP. None of the variants identified in the disease patients were seen in the COSMIC database.
One worry of using the designed SMAC mimetics in treating cancer is the risk of inducing the GI and immune secondary phenotypes associated with variants in XIAP that were first identified with the C203Y variant in our patient. Additionally these SMAC mimetics may block the non-caspase dependent pathways of other BIR domain proteins, most of which are not currently characterized. Analysis of all human BIR domains revealed amino acids Q197 and F228 of XIAP to be interesting sites for driving drug specificity to only the BIR2 domain of XIAP and no other BIR containing proteins. Using the known protein interaction sites, amino acid Q197 interacts with the SMAC N-terminus while F228 does not interact with any part of SMAC. On the contrary, amino acid F228 directly contacts both Caspase-3 and Caspase7, while Q197 does not. Based on our top docking conformation of XIAP BIR2 with RIPK2, neither of these amino acids contributes to binding. Therefore, we suggest that targeting XIAP amino acid F228 and surrounding amino acids with a new class of inhibitors may allow for activation of Caspase while not perturbing the XIAP activation of the NOD2 pathway or other BIR containing protein pathways.
Following sequencing of a human genome, the time it takes to interpret and propose experiments to validate hypotheses of how the variant results in disease far exceeds that of any conventional clinical test on the market. Many scientists are continuing to push the genomic sequencing as a possible clinical based test, especially with the wet lab cost of sequencing reaching several thousand dollars. This opens up the exciting possibility for scientists in the molecular modeling community to help in the interpretation of clinical variants, but will require our community to learn from existing variants and applying new approaches. The research community has numerous tools and resources that can significantly reduce the time it takes to discover the mechanisms in which these clinical variants result in disease phenotype.
References
- Chai JJ, Du CY, Wu JW, Kyin S, Wang XD, Shi YG. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- Damgaard RB, Fiil BK, Speckmann C, Yabal M, zur Stadt U, Bekker-Jensen S, Jost PJ, Ehl S, Mailand N, Gyrd-Hansen M. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol Med. 2013;5:1278–1295. doi: 10.1002/emmm.201303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wu Z, Mei Y, Wu M. XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J. 2013;32:2204–2216. doi: 10.1038/emboj.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HJ, Abrams K, Bick DP, Brodie K, Dimmock DP, Farrell M, Geurts J, Harris J, Helbling D, Joers BJ, et al. Genomics in Clinical Practice: Lessons from the Front Lines. Sci Transl Med. 2013;5:194cm5–194cm5. doi: 10.1126/scitranslmed.3006468. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: A multiple structural alignment algorithm. Proteins Struct Funct Bioinforma. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Knoefel WT, Reed JC. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci U S A. 2009;106:14524–14529. doi: 10.1073/pnas.0907131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Lin YF, Lai TC, Chang CK, Chen CL, Huang MS, Yang CJ, Liu HG, Dong JJ, Chou YA, Teng KH, et al. Targeting the XIAP/caspase-7 complex selectively kills caspase-3-deficient malignancies. J Clin Invest. 2013;123:3861–3875. doi: 10.1172/JCI67951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D, Chalasani P, Rensvold D, Kurtin S, Pretzinger C, Jolivet J, Ramanathan RK, Von Hoff DD, Weiss GJ. Phase I trial of AEG35156 an antisense oligonucleotide to XIAP plus gemcitabine in patients with metastatic pancreatic ductal adenocarcinoma. Am J Clin Oncol. 2013;36:239–243. doi: 10.1097/COC.0b013e3182467a13. [DOI] [PubMed] [Google Scholar]
- Rigaud S, Fondanèche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, Ahmad B. Inhibitors of apoptotic proteins: new targets for anticancer therapy. Chem Biol Drug Des. 2013;82:243–251. doi: 10.1111/cbdd.12176. [DOI] [PubMed] [Google Scholar]
- Shi YG. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann C, Ehl S. XIAP deficiency is a mendelian cause of late-onset IBD. Gut. 2013 doi: 10.1136/gutjnl-2013-306474. [DOI] [PubMed] [Google Scholar]
- Speckmann C, Lehmberg K, Albert MH, Damgaard RB, Fritsch M, Gyrd-Hansen M, Rensing-Ehl A, Vraetz T, Grimbacher B, Salzer U, et al. X-linked inhibitor of apoptosis (XIAP) deficiency: the spectrum of presenting manifestations beyond hemophagocytic lymphohistiocytosis. Clin Immunol Orlando Fla. 2013;149:133–141. doi: 10.1016/j.clim.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A, Pérez-Quintero LA, Latour S. X-linked lymphoproliferative syndromes and related autosomal recessive disorders. Curr Opin Allergy Clin Immunol. 2013;13:614–622. doi: 10.1097/ACI.0000000000000008. [DOI] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, et al. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012 doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]