Abstract
We describe opportunities and challenges in RNA structural modeling and design, as recently discussed during the second Telluride Science Research Center workshop organized in June 2016. Topics include fundamental processes of RNA, such as structural assemblies (hierarchical folding, multiple conformational states and their clustering), RNA motifs, and chemical reactivity of RNA, as used for structural prediction and functional inference. We also highlight the software and database issues associated with RNA structures, such as the multiple approaches for motif annotation, the need for frequent database updating, and the importance of quality control of RNA structures. We discuss various modeling approaches for structure prediction, mechanistic analysis of RNA reactions, and RNA design, and the complementary roles that both atomistic and coarse-grained approaches play in such simulations. Collectively, as scientists from varied disciplines become familiar and drawn into these unique challenges, new approaches and collaborative efforts will undoubtedly be catalyzed.
A pessimist sees the difficulty in every opportunity; an optimist sees the opportunity in every difficulty.
— Winston S. Churchill
Main Text
Only decades ago considered a neglected cousin, RNA has become a superstar in its own right. Indeed, from this molecule’s remarkable ability to facilitate genome editing and chemical catalysis to its tantalizing repertoire of structural and functional motifs, it is no wonder that RNA research now attracts many scientists from varied disciplines. Not only have such RNA studies enhanced our understanding of fundamental biological processes; they have also directly impacted medicine, biotechnology, and genome engineering.
Yet as technologies for RNA structure determination and analysis advance, practical issues and deep basic-science questions have emerged. Efforts to confront challenges in RNA structure analysis, prediction, and design have resulted in many innovative interdisciplinary approaches to analyze, classify, predict, simulate, and design RNA molecules. While many successes have been reported, progress in the field has been hampered by limited experimental resolution and an incomplete understanding of RNA tertiary structure, especially for large RNAs. The difficult problem of understanding and predicting RNA tertiary structure from its primary as well as secondary structure remains unsolved in general.
In a unique collaborative atmosphere inspired by the Telluride Science Research Center, a small group of RNA practitioners from the mathematical/physical to the biological sciences gathered in an informal workshop titled “Challenges in RNA Structural Modeling and Design” (https://www.telluridescience.org/meetings/workshop-details?wid=553) organized by us to share recent studies, discuss field advances, debate ongoing challenges, and present innovative solutions to major unsolved problems. From these broad-minded scientists working on both the genomic and molecular levels, using a novel array of experimental, mathematical, statistical, and computational methods to investigate RNA, several interesting challenges and issues emerged.
In this perspective, we highlight those challenges from the workshop presentations and informal discussions, representative of many ongoing efforts in the field. Interesting issues discussed at the first meeting held in June 2014 were reported in Pyle and Schlick (1), along with a collection of RNA articles by workshop participants and other RNA researchers.
In particular, we highlight important issues in structural assemblies that emerge as we grow to appreciate the greater flexibility and functional versatility of RNAs, concerning the order of folding of various elements in the RNAs and the clustering of RNA secondary-structure substates. We discuss the importance and difficulty of annotating RNA motifs, both in biological and practical terms (database conformity, database updating, etc.). Related issues in structural accuracy and quality control also emerge, especially in light of the increasing large RNA structures arising from cryo-electron microscopy. Biochemical mapping is also discussed in important contexts such as structure prediction and functional inference; however, incorporating chemical reactivity data requires a better appreciation of the dependency of these measurements on the cellular environment and on the multiple conformational states, related to the structural assemblies theme above. We also highlight the need to develop both atomic level models for simulations as well as coarse-grained models for RNA simulation, structure prediction, and RNA design. The former approach is necessary for incorporating details such as ion binding and for understanding detailed dynamics and chemical reactivity of RNAs, but the latter can accelerate conformational sampling and suggest modular motifs for RNA design. Finally, we discuss challenges in predicting structures of large RNAs and the working protocols that have been developed, containing both experimental and computational components.
Structural assemblies
Is RNA folding truly hierarchical?
One of the tenets of RNA folding has been the hierarchical nature of RNA tertiary structural assembly, and the concept that structural organization occurs in discrete states or transitions (2, 3). These states involve organization of secondary (2D) structure, or basepairing arrangements, followed by cooperative transitions to the three-dimensional (3D) structure. This paradigm has been important for computational approaches, because many predictions start with the 2D structure and predict the 3D structure consistent with those interactions.
Recent work in the field, such as the structure/function experiments by in Sarah Woodson's lab (4), suggests more intricate folding patterns. While scaffolds presented by 2D structural elements of stems and helix junctions appear to facilitate and guide tertiary structure assembly (4), many functional experiments involving activity assessment and native electrophoresis measurements on mutant ribozymes show that these sequence variants exhibit multiple structures rather than a dominant fold when the tertiary structure is compromised (5). Single-molecule FRET experiments show that these structural assemblies also exhibit frequent fluctuations into transient populations that depend on the concentration of magnesium ions in the solution. Furthermore, the active sites form last in the tertiary folding process. Proteins are observed to guide and link successive stages of RNA folding (6), and various conformational switches, whether internal or external (ions, proteins), help dictate the overall folding pathways.
Thus, secondary and tertiary structural folding appear to be more closely linked, and the hierarchical folding concept may be more fluid than originally conceived. Whether these observations for ribozymes apply more generally remains to be seen, but caution is warranted in applying the hierarchical folding paradigm of RNA to computational prediction.
Clustering challenges emerge for secondary-structure substates
Secondary-structure prediction approaches have been used extensively as a first step in assessing RNA structure from sequence. Basepairing arrangements are deduced by seeking a minimum free energy using nearest-neighbor rules and dynamic programming algorithms (7, 8). However, 2D structure prediction algorithms often produce a large ensemble of candidates, and the minimum free energy state is not necessarily the biologically relevant state. This occurs due to imperfections in the empirical functions used for 2D-structure predictions, the fact that some sequences fold into more than structure, and other biological factors not considered in the calculations. Thus, it is important to examine alternative low-energy states.
Because quantifying these alternative states based on energy ranking alone may not be sufficient, it is necessary to develop qualitative measures to assess the 2D-structure candidates produced. Clustering methods are often used for this purpose, but purely mathematical clustering approaches may only separate the candidates by relative energy rankings. Work from the lab of Christine Heitsch (9) has produced profiling approaches that cluster the candidates by coarser arrangements of the helices and loops to discern similar and dissimilar motifs, and in helical branching patterns of variable lengths and basepair identities. Further automated efforts for improving the underlying energy functions and our analyses of the resulting predictions are needed for large RNAs, where the number of low energy states increases exponentially and cannot be examined by simple visual inspection.
RNA motifs, databases, and structure quality
Extracting and analyzing RNA motifs, automating RNA structure annotation, and updating RNA structural databases define open challenges
One of the recurring issues discussed throughout the meeting involves interpreting and reconciling RNA structural information and annotations produced by different RNA analysis and bioinformatics tools.
For example, it is well known that various 2D structure prediction algorithms often yield different (and multiple) answers, especially for large RNAs. But even simpler tasks such as annotating the 2D structure of RNAs from solved 3D structures (as described in a Protein Data Bank (PDB) file) can be difficult. For example, pseudoknots that are well established in the solved 3D structure are often removed from the 2D structural output, and thus not always identified. Another issue is that when an RNA is divided into different chains, the sequence files often present the RNA as a single file, even though the chains may not be basepaired. This representation requires visualization of the RNA to determine whether the chains are paired, and then dividing the file accordingly before determining the 2D structure.
Thus, practitioners often opt to develop their own analytical programs to remedy issues they have encountered. As a whole, we are benefiting from improved and more analytical tools for viewing, annotating, and manipulating RNA molecules, but are also burdened with reconciling results from the various approaches.
Some of these issues were highlighted in talks by Xiang-Jun Lu and Blake Sweeney.
Lu et al. (10) presented the program Dissecting the Spatial Structure of RNA (DSSR) (http://x3dna.org) that aims to automate the analysis and annotation of nucleic acids and their complexes including RNA, RNA/RNA, RNA/DNA, and DNA quadruplex complexes. This modern software package has its roots in 3DNA originally developed for DNA. A modern programming design now makes it robust and efficient, with a large forum for user comments and requests.
DSSR identifies a range of canonical and noncanonical basepairs, including those with modified nucleotides, in various tautomeric or protonation states. It detects and classifies higher-order coplanar base associations, stacked pairs, RNA stems, various loop types (hairpin, bulge, internal, and junction loops), and various query motifs (like k-turns and reverse k-turns). Pseudoknots are handled in a special way with the introduction of junction loops and thus are not removed from the 2D structure. DSSR’s integration with widely used molecular visualization programs (Jmol, www.jmol.org; and Pmol, www.pmol.org) also make it easy to use.
Sweeney from the Zirbel/Leontis labs presented the latest enhancements in extensive annotation efforts for RNAs (11), including interactions of basepairs, base stacking, and base-backbone; lists of nonredundant RNA-containing 3D structures; and the RNA 3D Motif Atlas (http://rna.bgsu.edu/rna3dhub/motifs). A recent focus is annotating RNA 3D structures in the mmCIF format and forming nonredundant lists by chain rather than by entire 3D structure files. This change adds 5S rRNA and tRNAs to the nonredundant lists. These lists along with the structural annotations are made available through the NDB site (http://ndbserver.rutgers.edu/).
A program for searching these motifs called JAR3D (pronounced “Jared”) has also been developed (11) (http://rna.bgsu.edu/jar3d/). JAR3D aims to find possible 3D geometries for hairpin and internal loops by matching loop sequences to motif groups from the RNA 3D Motif Atlas, when available. Otherwise, probabilistic scoring and other distance criteria are used for novel sequences. The scoring reflects the ability of the sequences to form the same pattern of interactions observed in 3D structures of the motif. The JAR3D web-server accepts one or more sequences of a single or multiple loops as input, and the output contains the 10 best-matching motif groups.
Ongoing efforts also focus on classification of RNA/protein interactions, including pseudo-pairs and RNA base/aromatic amino acid stacking, and possible extensions to simulating the dynamics of RNAs or docking ligands into solved RNAs.
As larger RNAs become targets of interest for experimentation and modelers, it is clear that such motif annotation programs will be valuable resources for the community. Annotations of pairwise interactions produced by different programs tend to be most consistent for high-quality structures. However, annotations from different resources that are not always in consonance with one another may nonetheless help advance the field.
Kink turns emerge as fundamental motifs in sculpting RNA, with exciting potential application to RNA design
Because it is conceivable that an RNA tertiary structure could be defined in terms of its constituent sequence/structure motifs, there have been many efforts to enumerate RNA’s structural repertoire, as discussed above and also below, in the context of graph representations (i.e., simplified graph representations of RNA secondary structures that provide good candidate predictions of global RNA scaffold and present RNA design targets that require automatic generation of atomic models). One outstanding example of such a sequence/motif signature has been the kink-turn (k-turn) motif, which has been thoroughly studied and applied by David Lilley's lab. K-turns are a widely distributed motif in large RNA/protein assemblies like the ribosome and in other RNAs like riboswitches, snoRNAs, and more (12, 13). They direct and sculpt the trajectory of helical segments within the RNA by forming unusually large angles of ∼50° between the two axes of the helical arms that are stabilized by two cross-strand A-minor interactions. Thus, the sequence signature of a standard k-turn involves a duplex RNA with a short bulge followed by G-A and A-G basepairs. The associated A-minor motifs involve nearby residues.
To understand these sequence requirements and the versatility of k-turns, Huang et al. (13) recently probed both the foldability and the nature of the final folded structure of the resulting k-turn as a function of sequence for variant residues in neighboring conformations by FRET experiments. Interestingly, they find that k-turns do not always form because of a delicate water/ion network that depends on the sequence. Subclasses of k-turns can also be identified.
These complexities in the folding states of RNAs, and their pathways, highlight general issues that complicate our ability to predict and understand tertiary structure formation in RNA. As discussed above in the subsection “Is RNA Folding Truly Hierarchical?”, the order of structure formation is also unclear, because critical ion, water, protein, and ligand interactions affect the foldability and function of RNAs. Further research that will carefully examine the folding trajectories of RNAs as a function of sequence and environment will be needed to dissect these difficult issues.
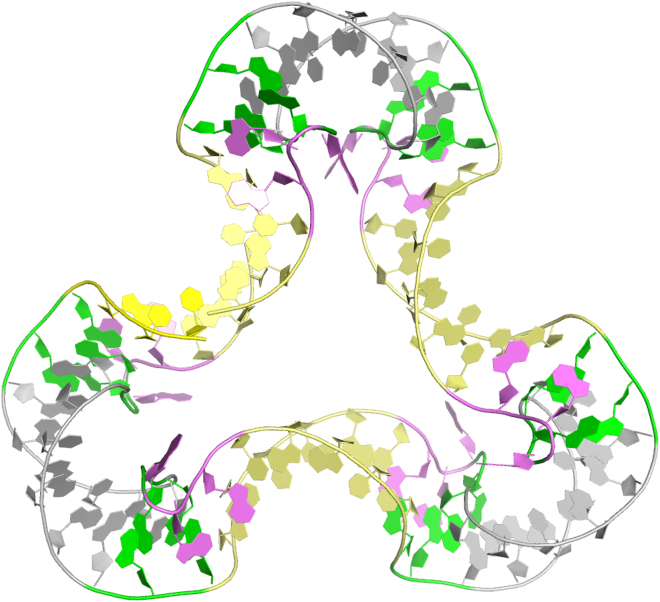
The complexity and versatility of RNA motifs also presents a natural opportunity for design of nano-objects (14) and therapeutic agents. This was also illustrated by Lilley, who showed recent work that exploits the k-turn motif to assemble nanostructures with 2–8 k-turns assemblies, some of which have been successfully crystallized (see Fig. 1) (15). These designs present inherent symmetries and binding pockets that could be imagined to bind ligands. Their design success suggests that k-turns could be used naturally as building blocks for RNA nanotechnology with potentially exciting applications.
Figure 1.
The structure of a quasi-cyclic duplex RNA molecule with six k-turn motifs (15). To see this figure in color, go online.
As our understanding of RNA’s structural repertoire increases, a combination of k-turns and other motifs will undoubtedly lead to exciting and novel structures and functions of RNA assemblies.
RNA structure quality
Quality control of the deposited files for resulting RNA structures remains an issue for RNAs (16, 17) and will likely increase as more large RNA structures are solved by cryo-electron microscopy (18). In many RNAs, there are inconsistencies and errors in specific regions such as the RNA backbone and the sugar pucker geometries. Such errors can complicate mechanistic interpretations of biological reactions and functions, as well as lead to problems in modeling based on those structures. Fortunately, the community is becoming more aware of these possible errors, and practitioners are using manual and automatic refinement procedures to correct them. For example, experimental validation utilities, such as real-space refinement statistics available for x-ray structures through the PDB or the program PHENIX (19), help address some of these issues.
Furthermore, because all these tools rely on structural resources, it is also important to have available updated databases like Rfam (rfam.xfam.org) and nonredundant RNA structure/motif datasets.
Chemical reactivity data for structure prediction and structural inference
Improved treatments are needed for utility of chemical probing data, to address reactivity of the states and protein binding
Because the diverse roles of RNA molecules are determined by their functional structures, many experimental and computational strategies have been developed to improve our ability to determine RNA structures. Chemical probing and cross-linking methods provide important foundations for structural interpretations because they report the structural state of each nucleotide as a function of cellular parameters like the ion concentration and temperature. Examples include hydroxyl radical footprinting, selective 2′-OH acylation by primer extension (SHAPE) chemical probing for RNA backbone flexibility (20, 21), and dimethylsulfate probing for hydrogen bonding (22). Yet computational frameworks are needed to fine-tune the interpretation of biochemical data (8). Moreover, how such data are best utilized in combination with structure prediction algorithms for 2D structure detection (8) remains an open problem. While advances in experimental, computational, and comparative analysis strategies have succeeded for many small RNAs, handling large RNAs remains a challenge. In particular, it is unclear how to fully incorporate the features of RNA structure that govern chemical reactivity and how to address the multiple conformational states.
Recent work from the David Mathews lab (8) suggests that computational efforts employing SHAPE measurements can be extended by considering that the biochemical probes measure an average of all conformations at equilibrium. Therefore, a new method based on stochastic sampling, which models Boltzmann ensembles of populations in a way that matches the chemical reactivity data, improves predictions overall for RNAs that sample multiple conformations. Importantly, such additional considerations can be incorporated with similar computational complexity as the original SHAPE-data-utilizing algorithms. Further challenges remain regarding the interpretation of SHAPE data when the RNA is bound to proteins, because the proteins can either protect the RNA or amplify the reactivity of the nearby residues.
Work from the lab of Kevin Weeks (21) illustrated how SHAPE data can be integrated with massively parallel sequencing using the approach of mutational profiling such that the very high throughput data are as accurate as those obtained with older quantitative but more laborious methods. The ability of SHAPE to measure the effects of the cellular environment was also highlighted. For free RNA, using SHAPE in conformational predictions appears to consistently lead to more accurate structural models. Remaining frontier challenges include modeling RNAs that interact extensively with proteins, and those that sample states that are more poorly defined than the highly structured conformations revealed from crystallography.
Incorporating chemical mapping contacts improves predictions of RNA tertiary structure using fragment-assembly knowledge-based techniques
The fragment assembly approach, based on empirical potentials derived from available experimental structures, has proven successful for predicting tertiary structures of proteins from sequence using the Rosetta program (23). Significantly, in recent studies even side chains can be predicted in atomic resolution. As evident from the superb performance in the RNA-Puzzles initiative (24), Rosetta’s adaptation for RNA by Rhiju Das and coworkers has benefited from the inclusion of chemical mapping information tailored for RNA.
Specifically, Tian and Das (25) has shown that tertiary structure predictions are far more successful and can approach atomic resolution when constraints are incorporated from classic phylogenetic analyses that can assess compensatory mutations (24). Namely, specific tertiary motifs (e.g., kissing loops) that sculpt the 3D folds can be better predicted, and features that are not often apparent from conventional chemical mapping data can emerge, particularly using newly established techniques for multidimensional chemical mapping (25). Such methods interrogate the environment of each nucleotide of the RNA in response to modifications for all other residues. This information, when properly incorporated into prediction models, can sense compensatory mechanisms when the local chemical environment is perturbed. Such extended chemical maps can also more readily treat multiple conformations and assess the effects of proteins or other ligands on the RNA conformations.
Because the computational complexity of these nucleotide/nucleotide contacts increases quadratically with the sequence size, the methods are not generally applicable to molecules larger than 200 nucleotides. New strategies are needed to address some of these limitations (25).
More generally, incorporation of chemical constraints more widely into other predictive efforts, as well as extensions to assess RNA motions and functions, will likely be productive.
Incorporating thermodynamics in addition to structural information challenges in functional inference of RNAs
Biochemical reactivity data can also be utilized to infer RNA functions. However, the extent of multiple conformational states and the approximations inherent in minimum free energy functions present challenges.
This was illustrated by the work of Alain Laederach, whose lab focuses on using SHAPE data to make quantitative functional predictions on the translation efficiency of specific messenger RNAs (26). One continuing challenge for broad applicability of RNA structure prediction algorithms is the functional interpretation of those results. Although it is often suggested that structure prediction will naturally inform function, such relationships are not always obvious or quantitative (27). This is particularly true in messenger RNAs, which are not expected to adopt single conformation but usually adopt an ensemble of conformations (26).
A quantitative model of 5′ UTR-mediated translation efficiency based on analysis of the Kozak sequence strength alone was shown to be far less accurate than a model that incorporates SHAPE-informed thermodynamic folding free energies. These results suggest that both thermodynamics and structural features are important for functional prediction. Furthermore, data comparing in vivo versus in vitro SHAPE experiments suggest that the thermodynamic parameters driving RNA structure formation are not fundamentally different in vitro and in the cell. These findings suggest new ways to make functional predictions using computational techniques (28).
Complementary modeling of RNAs at both atomic and coarse-grained levels
Atomistic RNA simulations have advanced from dynamics to chemical reactivity but remain elusive in general
Impressive advances in computational platforms and simulation algorithms have made it possible to simulate RNAs at atomic resolution. Such simulations can provide insights into ribosome motions (29) and chemical reactivity such as ribozyme catalysis, as recently described in Darrin York's lab for the twister ribozyme (30). However, issues in force field parameterization for RNA, divalent ion modeling, and others still plague practitioners (31, 32) and are frequently discussed in RNA meetings of modelers. These difficulties make successful all-atom RNA simulations viable for a select few experts who are working on well-tested systems. Thus, reliable protocols and programs for RNA simulations remain an important future goal in the field.
Simplified graph representations of RNA secondary structures that provide good candidate predictions of global RNA scaffold and present RNA design targets require automatic generation of atomic models
Complementary to these atomic-level simulations and modeling studies, various coarse-grained representations of RNA (e.g., (33, 34, 35, 36, 37)) have been shown to be effective in many applications, including configurational sampling, structure prediction, and RNA design. Simplified or coarse-grained representations of macromolecules are successful at capturing essential features of biomolecules while making computations accessible for a variety of applications due to a drastic reduction in the number of degrees of freedom. Historically, united-atom presentations for proteins were used to simulate their dynamics (e.g., McCammon et al. (38)), and recent coarse-grained models of chromatin have provided insights into chromatin architecture (39).
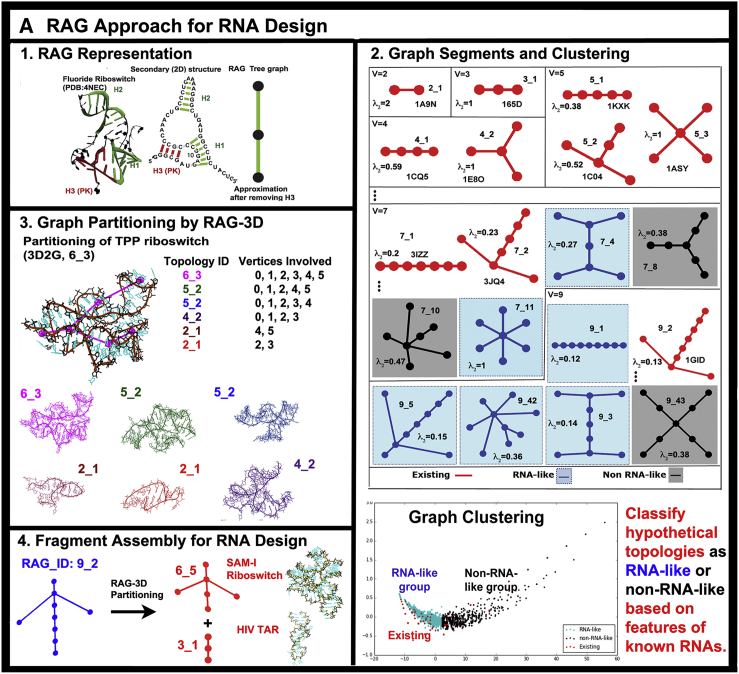
Graph models of RNAs have been developed as early as the 1970s by Tinoco, Nussinov, Waterman, Shapiro, and others, as reviewed in 2013 (40, 41). Different graph theoretical approaches have been developed and applied to RNA by Kim et al. (see Fig. 2 A). The advantage of graphical representations of RNA secondary structures is that all possible motifs can be described explicitly by graph enumeration methods (42).
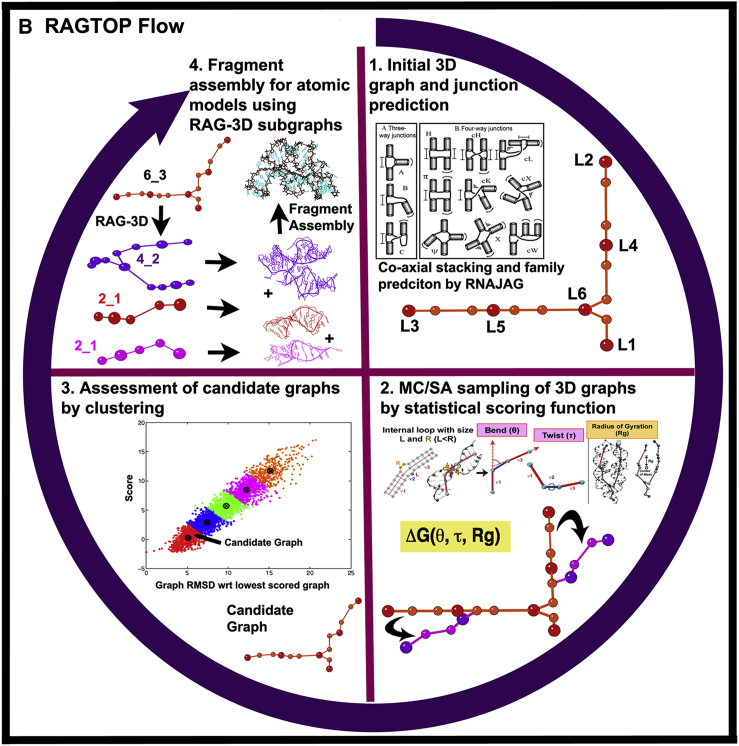
Figure 2.
RAG elements for RNA motif classification, prediction, partitioning, and design. (A) The RAG approach for RNA representation (42) and design (43, 45) is illustrated as: 1) tree graph of a riboswitch; 2) RAG tree graph catalog segments, organized by the second eigenvalue λ2 of the connectivity matrix (Laplacian) associated with the graph and classified by clustering into three groups: existing (red), RNA-like (blue), and non-RNA-like (black) motifs (see http://www.biomath.nyu.edu/rag/home for more information); 3) graph partitioning for a riboswitch by RAG-3D (44), which suggests modular RNA building blocks, for which PDB structures are available; and 4) fragment assembly of such subgraph fragments using the modular subunits. (B) RAGTOP for 3D structure prediction by a hierarchical MC sampling of tree graphs (46, 48, 49): 1) Initial junction topology prediction; 2) MC sampling of 3D graphs scored by a statistical scoring function with components for bend, twist, and radius-of-gyration; 3) clustering of generated graphs to identify candidate graph; and 4) determination of atomic models from the candidate graph using the fragment assembly based on RAG-3D subgraphs. To see this figure in color, go online.
The Schlick group has recently clustered all these enumerated motifs and, using information from the solved RNAs, predicted which among the hypothetical RNAs are more RNA-like in overall features (43). This recent assessment has shown that such structures provide good candidates for RNA design, better than those classified as non-RNA-like.
Furthermore, practical design strategies were developed using automatic graph partitioning methods and fragment-assembly methods. Specifically, a recently released program and web-server called “RAG-3D” (44) extends the RNA-As-Graphs (RAG) catalog to 3D graphs (http://www.biomath.nyu.edu/RAG3D/) and links solved PDB structures to these 3D graphs (Fig. 2 A). In response to a query RNA structure or PDB file, the RAG-3D program searches for similar blocks in the database and partitions any solved RNA, represented as graphs, into building blocks, or modular units. Because each subgraph has a corresponding known sequence (44), the pieces can be combined to build the candidate atomic model. Previous work on this concept proved its utility (45). Since 2014, half of the candidate models were solved experimentally, with significant correlations in the designed sequences (43). Related partitioning of 2D graphs have also been developed using graph theoretical methods, for tree graphs by the gap cut method, which leaves junctions intact (46), and for dual graphs, which can represent pseudoknots (L. Petingi and T. Schlick, Ninth Annual International Conference on Combinatorial Optimization and Applications, COCOA ’15, Dec. 18–20 2016, Houston, TX).
Recent work focuses on applying these partitioning and fragment assembly elements for RNA design to structure prediction from secondary structure using the ‘RAGTOP’ program (‘RNA-As-Graphs-Topology Prediction’ (47, 48, 49); see Fig. 2 B). In this hierarchical graph sampling approach, the coarse-grained representation of graphs is exploited for efficient sampling of the associated conformational space. Specifically, 2D graphs are made into 3D directed graphs that incorporate sequence lengths of the loops and junctions; a bioinformatics/data-mining tool called JunctionExplorer is then applied to this 3D tree graph to predict initial junction arrangements (50). This initial graph is then subjected to Monte Carlo sampling guided by a statistical scoring function, defined from bend and torsion angle orientations about loops, and a radius of gyration term. Finally, the candidate graph or graphs (deduced from ensemble clustering) are built into atomic models using a fragment assembly approach. This last component is now being automated.
Current results show promise in predicting RNA tertiary structures by the combined approach, including using a tailored kink-turn potential (51), extending previous results (48, 49). The junction assembly component helps bring essential tertiary elements in space into the right orientation. The challenge to automate generation of atomic models from graphs is being accomplished by using the partitioning of RAG-3D and fragment assembly, as used for design, in combination with structure refinement by geometry optimization methods like PHENIX (19) and all-atom force fields. The concepts are simple, but implementing the details successfully, especially in light of the many fragments available for building RNAs, is a technical challenge.
The above work underscores the importance of using reduced representations to solve problems in RNA structure and design. Chemical constraints, as implemented in the atomic fragment-assembly approach by Tian and Das (25), may also improve the predictions. In general, multiple approaches also contribute to our understanding of RNA architecture and assembly. As always, failures in computational approaches reveal knowledge gaps in our basic understanding of RNAs and raise important challenges for future work. There is also room for a combination of methods and approaches, because there is duplication of efforts in popular areas like motif annotation and RNA structure prediction. It will also be interesting to relate these approaches to one another and combine them or their parts in advantageous ways.
Large RNA challenges
Modeling large RNAs by combined experimental and computational paradigms define promising ways to solve large noncoding RNA structures but raise several database and program issues
Long noncoding RNA (lncRNA) structures are recognized as important agents in fundamental biological processes of gene expression and are involved in splicing, development, epigenetics, cancer, and much more. Yet only relatively few such large RNAs, which range from 200 to more than 20,000 nucleotides, are characterized crystallographically. As a well studied model for noncoding RNA architecture, the group II intron provides a useful guide for developing approaches to infer structure and function of such large RNAs (52).
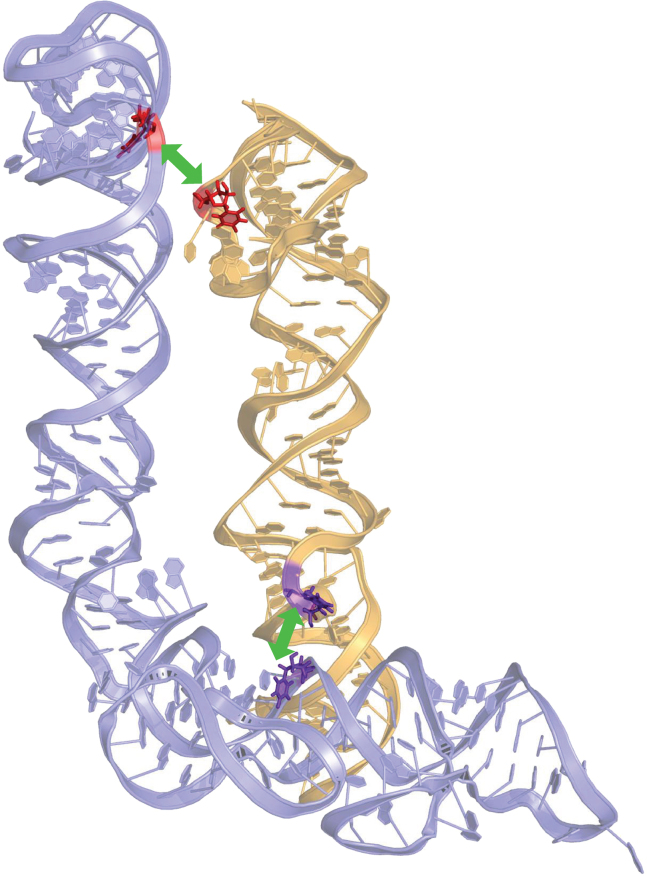
To determine these structures and infer their functions, an elaborate paradigm of experimental and computational strategies have been developed, as described by Pyle's lab (52, 53) for the HOTAIR, lincRNA-p21, and RepA noncoding RNAs involved in gene silencing (see Fig. 3), and separately by Sanbonmatsu (54) for the steroid receptor lncRNA, SRA-1. These strategies involve an iterative process of chemical probing to infer basepairing, phylogenetic analysis across genomes in combination with candidate 2D structures, model building with the aid of sequence alignment methods, covariance analysis to suggest helix positions, photo-crosslinking, and possible tertiary structure modeling and simulations.
Figure 3.
3D model of two subdomains in lncRNA RepA. The two spatially proximal subdomains in lncRNA RepA (shown in purple and yellow) were modeled with RNAComposer using the experimentally identified crosslinks as distance constraints (54). The nucleotides participating in the crosslinks are shown in purple and red. The potential tertiary interactions between the two subdomains are represented with green arrows. To see this figure in color, go online.
In the above protocols, the covariations in RNA alignments help deduce evolutionarily conserved RNA secondary structures that in turn help improve alignments and suggest function. However, quantifying the statistical significance of basepair covariations in RNA alignments is important for deducing evolutionary relationships. A quantitative tool developed for this purpose by Rivas et al. (55) called “RNA Structural Covariation Above Phylogenetic Expectation” aims to provide such information. However, program parameters used to infer evolutionary relationship require caution, as too stringent criteria may obfuscate possible relationships. Thus, numerous ongoing challenges for determining the structures and functions of lncRNAs using a combination of experimental data and computational tools remain. Because general computational programs for secondary and tertiary structure determination are not reliable for such large systems, it might be interesting to devise modular implementations, of smaller components of the long RNA molecules, that are well integrated with the available experimental data.
Conclusion
There remain many opportunities and challenges in understanding the basic biological and chemical processes of RNA (hierarchical folding, multiple conformational states, and functional inference), and in the associated computational approaches (2D and 3D structure prediction, clustering, atomic simulations, incorporation of biochemical data, reduced representations, and phylogenetic analysis). Other emerging issues occur in structure annotation (motif enumeration, nonredundant datasets, and various issues in RNA software) and design applications (combining structural motifs for therapeutic and technological applications, predicting RNA-like candidate motifs, and performing their design in silico). Many software aspects such as differences of handling various RNA motifs by available programs, quality checks on RNA structures, and the availability of updated RNA databases need to be addressed by the growing RNA community. As scientists from varied disciplines become familiar with these issues and drawn into the inherent challenges, new collaborative approaches and transformational technologies will undoubtedly emerge.
In particular, we have highlighted issues in structural assemblies that emerge as we grow to appreciate the flexibility and functional versatility of RNAs. The order of folding of RNA elements may be more complex than originally believed, and the organization/classification of RNA secondary structure substates is an emerging challenge. Annotating RNA motifs continues to be a work in progress, both in biological and practical terms. Namely, various motif descriptions and database approaches exist, and database updating, as well as structural accuracy and quality, are important issues to resolve. Biochemical mapping is enjoying enhanced usage in important areas of RNA analysis, such as structure prediction and functional inference. However, incorporating chemical reactivity data requires a better appreciation of the complexity of the cellular environment, especially the flexibility of RNA, echoing the above theme regarding our growing appreciation of the multiple conformational states of RNA. As modeling studies increase for RNA and RNA complexes, both detailed atomic-level simulations and coarse-grained approaches emerge as valuable and complementary. The former class can incorporate specific features such as ion and ligand binding and can help suggest and interpret dynamics mechanisms and chemical reactivity of RNAs. The latter approach can accelerate conformational sampling, help identify general structural patterns, and suggest modular motifs for intuitive RNA design. All these issues are relevant to the challenges present in predicting structures of large RNAs. Undoubtedly, the experimental and computational communities will continue to work together to analyze, interpret, predict, and design RNA molecules and their complexes and to pursue important biomedical and engineering applications.
Acknowledgments
We thank all meeting participants for their generous time in contributing to the exciting presentations and discussions related to the work discussed here.
We are grateful for partial support of the workshop by the National Institute of General Medical Sciences (NIGMS) award No. R13GM112216 to T.S. and A.M.P., and for the Telluride Science Research Center (TSRC) staff.
Editor: Brian Salzberg.
Contributor Information
Tamar Schlick, Email: schlick@nyu.edu.
Anna Marie Pyle, Email: anna.pyle@yale.edu.
References
- 1.Pyle A.M., Schlick T. Challenges in RNA structural modeling and design. J. Mol. Biol. 2016;428:733–735. doi: 10.1016/j.jmb.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brion P., Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 3.Tinoco I., Jr., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 4.Behrouzi R., Roh J.H., Woodson S.A. Cooperative tertiary interaction network guides RNA folding. Cell. 2012;149:348–357. doi: 10.1016/j.cell.2012.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H.T., Kilburn D., Woodson S.A. Molecular crowding overcomes the destabilizing effects of mutations in a bacterial ribozyme. Nucleic Acids Res. 2015;43:1170–1176. doi: 10.1093/nar/gku1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abeysirigunawardena S.C., Woodson S.A. Differential effects of ribosomal proteins and Mg2+ ions on a conformational switch during 30S ribosome 5′-domain assembly. RNA. 2015;21:1859–1865. doi: 10.1261/rna.051292.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgs P.G. RNA secondary structure: physical and computational aspects. Q. Rev. Biophys. 2000;33:199–253. doi: 10.1017/s0033583500003620. [DOI] [PubMed] [Google Scholar]
- 8.Sloma M.F., Mathews D.H. Improving RNA secondary structure prediction with structure mapping data. Methods Enzymol. 2015;553:91–114. doi: 10.1016/bs.mie.2014.10.053. [DOI] [PubMed] [Google Scholar]
- 9.Rogers E., Heitsch C. New insights from cluster analysis methods for RNA secondary structure prediction. Wiley Interdiscip. Rev. RNA. 2016;7:278–294. doi: 10.1002/wrna.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu X.-J., Bussemaker H.J., Olson W.K. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015;43:e142. doi: 10.1093/nar/gkv716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roll J., Zirbel C.L., Leontis N. JAR3D Webserver: scoring and aligning RNA loop sequences to known 3D motifs. Nucleic Acids Res. 2016;44(W1):W320–W327. doi: 10.1093/nar/gkw453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Daldrop P., Lilley D.M. The k-junction motif in RNA structure. Nucleic Acids Res. 2014;42:5322–5331. doi: 10.1093/nar/gku144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L., Wang J., Lilley D.M. A critical base pair in k-turns determines the conformational class adopted, and correlates with biological function. Nucleic Acids Res. 2016;44:5390–5398. doi: 10.1093/nar/gkw201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger L., Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Huang L., Lilley D.M.J. A quasi-cyclic RNA nano-scale molecular object constructed using kink turns. Nanoscale. 2016;8:15189–15195. doi: 10.1039/c6nr05186c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukasiak P., Antczak M., Blazewicz J. RNAssess--a web server for quality assessment of RNA 3D structures. Nucleic Acids Res. 2015;43(W1):W502–W506. doi: 10.1093/nar/gkv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain S., Richardson D.C., Richardson J.S. Computational methods for {RNA} structure validation and improvement, Chapter 7. In: Woodson S.A., Allain F.H.T., editors. Vol. 558. Academic Press; Cambridge, MA: 2015. pp. 181–212. (Methods in Enzymology, Structures of Large RNA Molecules and Their Complexes). [DOI] [PubMed] [Google Scholar]
- 18.Nogales E., Scheres S.H.W. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Mol. Cell. 2015;58:677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams P.D., Afonine P.V., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deigan K.E., Li T.W., Weeks K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegfried N.A., Busan S., Weeks K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stern S., Moazed D., Noller H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 23.Ovchinnikov S., Kim D.E., Baker D. Improved de novo structure prediction in CASP11 by incorporating co-evolution information into Rosetta. Proteins. 2016;84:67–75. doi: 10.1002/prot.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao Z., Adamiak R.W., Westhof E. RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA. 2015;21:1066–1084. doi: 10.1261/rna.049502.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian S., Das R. RNA structure through multidimensional chemical mapping. Q. Rev. Biophys. 2016;49:e7. doi: 10.1017/S0033583516000020. [DOI] [PubMed] [Google Scholar]
- 26.Ritz J., Martin J.S., Laederach A. Evolutionary evidence for alternative structure in RNA sequence co-variation. PLoS Comput. Biol. 2013;9:e1003152. doi: 10.1371/journal.pcbi.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somarowthu S. Progress and current challenges in modeling large RNAs. J. Mol. Biol. 2016;428:736–747. doi: 10.1016/j.jmb.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutchko K.M., Laederach A. Transcending the prediction paradigm: novel applications of SHAPE to RNA function and evolution. Wiley Interdiscip. Rev. RNA. 2016;8:e1374. doi: 10.1002/wrna.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitford P.C., Blanchard S.C., Sanbonmatsu K.Y. Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLoS Comput. Biol. 2013;9:e1003003. doi: 10.1371/journal.pcbi.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaines C.S., York D.M. Ribozyme catalysis with a twist: active states of twister ribozyme in solution predicted from molecular simulation. J. Am. Chem. Soc. 2016;138:3058–3065. doi: 10.1021/jacs.5b12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Šponer J., Banáš P., Otyepka M. Molecular dynamics simulations of nucleic acids. From tetranucleotides to the ribosome. J. Phys. Chem. Lett. 2014;5:1771–1782. doi: 10.1021/jz500557y. [DOI] [PubMed] [Google Scholar]
- 32.Kührová P., Best R.B., Banáš P. Computer folding of RNA tetraloops: identification of key force field deficiencies. J. Chem. Theory Comput. 2016;12:4534–4548. doi: 10.1021/acs.jctc.6b00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le S.Y., Nussinov R., Maizel J.V. Tree graphs of RNA secondary structures and their comparisons. Comput. Biomed. Res. 1989;22:461–473. doi: 10.1016/0010-4809(89)90039-6. [DOI] [PubMed] [Google Scholar]
- 34.Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 35.Jonikas M.A., Radmer R.J., Altman R.B. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X., Chen S.-J. Physics-based RNA structure prediction. Biophys. Rev. 2015;1:2–13. doi: 10.1007/s41048-015-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boniecki M.J., Lach G., Bujnicki J.M. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016;44:e63. doi: 10.1093/nar/gkv1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCammon J.A., Gelin B.R., Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 39.Ozer G., Luque A., Schlick T. The chromatin fiber: multiscale problems and approaches. Curr. Opin. Struct. Biol. 2015;31:124–139. doi: 10.1016/j.sbi.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim N., Fuhr N., Schlick T. Graph applications to RNA structure and function, Chapter 3. In: Russell R., editor. Biophysics of RNA Folding, Biophysics for the Life Sciences. Springer; New York, NY: 2013. pp. 23–51. [Google Scholar]
- 41.Kim N., Petingi L., Schlick T. Network theory tools for RNA modeling. WSEAS Trans. Math. 2013;9:941–955. [PMC free article] [PubMed] [Google Scholar]
- 42.Gan H.H., Pasquali S., Schlick T. Exploring the repertoire of RNA secondary motifs using graph theory; implications for RNA design. Nucleic Acids Res. 2003;31:2926–2943. doi: 10.1093/nar/gkg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba N., Elmetwaly S., Schlick T. Predicting large RNA-like topologies by a knowledge-based clustering approach. J. Mol. Biol. 2016;428(5 Pt. A):811–821. doi: 10.1016/j.jmb.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahran M., Bayrak C.S., Schlick T. RAG-3D: a search tool for RNA 3D substructures. Nucleic Acids Res. 2015;43:9474–9488. doi: 10.1093/nar/gkv823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim N., Shiffeldrim N., Schlick T. Candidates for novel RNA topologies. J. Mol. Biol. 2004;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 46.Kim N., Zheng Z., Schlick T. RNA graph partitioning for the discovery of RNA modularity: a novel application of graph partition algorithm to biology. PLoS One. 2014;9:e106074. doi: 10.1371/journal.pone.0106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laing C., Jung S., Schlick T. Predicting helical topologies in RNA junctions as tree graphs. PLoS One. 2013;8:e71947. doi: 10.1371/journal.pone.0071947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim N., Laing C., Schlick T. Graph-based sampling for approximating global helical topologies of RNA. Proc. Natl. Acad. Sci. USA. 2014;111:4079–4084. doi: 10.1073/pnas.1318893111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim N., Zahran M., Schlick T. Computational prediction of riboswitch tertiary structures including pseudoknots by RAGTOP: a hierarchical graph sampling approach. Methods Enzymol. 2015;553:115–135. doi: 10.1016/bs.mie.2014.10.054. [DOI] [PubMed] [Google Scholar]
- 50.Laing C., Wen D., Schlick T. Predicting coaxial helical stacking in RNA junctions. Nucleic Acids Res. 2012;40:487–498. doi: 10.1093/nar/gkr629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bayrak C.S., Kim N., Schlick T. Using sequence signatures and kink-turn motifs in knowledge-based statistical potentials for RNA structure prediction. Nuc. Acids Res. 2017 doi: 10.1093/nar/gkx045. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pyle A.M. Looking at LncRNAs with the ribozyme toolkit. Mol. Cell. 2014;56:13–17. doi: 10.1016/j.molcel.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 53.Liu F., Somarowthu S., Pyle A.M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017 doi: 10.1038/nchembio.2272. Jan 9. http://dx.doi.org/10.1038/nchembio.2272. PMID:28068310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanbonmatsu K. Towards structural classification of long non-coding RNAs. Biochim. Biophys. Acta. 2016;1859:41–45. doi: 10.1016/j.bbagrm.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivas E., Clements J., Eddy S.R. Lack of evidence for conserved secondary structure in long noncoding RNAs. Nat. Methods. 2017;14:45–48. doi: 10.1038/nmeth.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]