Abstract
RNA granules are ensembles of specific RNA and protein molecules that mediate localized translation in eukaryotic cells. The mechanisms for formation and selectivity of RNA granules are unknown. Here we present a model for assembly of one type of RNA granule based on experimentally measured binding interactions among three core multivalent molecular components necessary for such assembly: specific RNA molecules that contain a cis-acting sequence called the A2 response element (A2RE), hnRNP A2 proteins that bind specifically (with high affinity) to A2RE sequences or nonspecifically (with lower affinity) to other RNA sequences, and heptavalent protein cytoskeleton-associated protein 5 (CKAP5, an alternative name for TOG protein) that binds both hnRNP A2 molecules and RNA. Non-A2RE RNA molecules (RNA without the A2RE sequence) that may be recruited to the granules through nonspecific interactions are also considered in the model. Modeling multivalent molecular interactions in granules is challenging because of combinatorial complexity in the number of potential molecular complexes among these core components and dynamic changes in granule composition and structure in response to changes in local intracellular environment. We use a hybrid modeling approach (deterministic-stochastic-statistical) that is appropriate when the overall compositions of multimolecular ensembles are of greater importance than the specific interactions among individual molecular components. Modeling studies titrating the concentrations of various granule components and varying effective site pair affinities and RNA valency demonstrate that interactions between multivalent components (TOG and RNA) are modulated by a bivalent adaptor molecule (hnRNP A2). Formation and disruption of granules, as well as RNA selectivity in granule composition are regulated by distinct concentration regimes of A2. Our results suggest that granule assembly is tightly controlled by multivalent molecular interactions among RNA molecules, adaptor proteins, and scaffold proteins.
Introduction
A major challenge in understanding and modeling cellular biological systems is posed by the fact that interactions among multivalent reactants can lead to formation of large, dynamic molecular ensembles, which are known in some cases to be associated with phase transitions. Examples include Nephrin-Nck-NWasp complex formation during maintenance of kidney filtration barrier (1, 2), P granule assembly during embryonic development (3), localized protein expression in RNA granules in neuronal cells (4), postsynaptic density formation in the postsynaptic compartment (5), assembly and dynamics of focal adhesions during cell migration (6), structural integrity of cytoskeleton (7), and transduction of extracellular input by transmembrane receptor signaling platforms (8, 9). Mayer et al. (10) introduced the term “pleomorphic molecular ensembles” to distinguish multimolecular complexes with dynamic and variable composition from “molecular machines”, which are assemblies of strongly interacting molecules with uniquely defined stoichiometry and shape (11). Here we will refer to such structures as “molecular ensembles”.
Traditional biochemical approaches are insufficient to provide a comprehensive understanding of molecular ensembles that arise through interactions between large numbers of molecules to form extended networks. Theoretical and computational modeling approaches provide complementary tools to test hypotheses and guide interpretation of experimental observations of biochemical interactions. Analytical and numerical modeling of multimeric complexes has a rich history, originating with the classical theories of Flory and Stockmayer (12, 13, 14, 15). These theories make use of binding probabilities to provide insight into ensemble organization at specific time points. However, such analytical treatments are restricted to simple systems consisting of one or two types of binding sites and have fundamental limitations in handling intermolecular reactions among multiple different molecules, which can undergo phase transitions. Many biological systems are characterized by multivalency and high diversity of binding sites, which introduce further combinatorial complexity into the system.
Rule-based modeling, which was introduced to overcome some of the combinatorial complexity of networks of interacting molecules (16, 17, 18), relies on either building a completely defined network (BioNetGen software (16)) that could become infinite, or agent-based versions (e.g. NFSim software (19) and Kappa software (20)), that keep track of the interaction topology at each time step and account for each individual binding and unbinding event, incurring high computational cost for intermolecular interactions of high frequency. Recently we extended the Flory-Stockmayer formalism to develop a hybrid approach (21) employing deterministic pre-computing of the probability of specific bond formation, stochastic assembly of complexes at time points of interest, and statistical analysis of ensemble organization. Our method and developed software allow investigation of formation and overall composition of pleomorphic dynamic molecular ensembles, without monitoring trajectories of individual molecular complexes within such ensembles.
Here, we apply our methodology (21) to the study of RNA granules, which are large (up to 0.5 μm in diameter) molecular ensembles containing multiple different molecular components including RNAs, RNA binding proteins, and multivalent scaffold proteins (22, 23, 24). RNA granules were first identified as fundamental intermediates in RNA trafficking (25), which is a mechanism for targeting RNAs encoding specific proteins to particular subcellular locations. Individual RNA granules are variable in size, composition, and structure, which distinguishes them from ordered stoichiometric macromolecular assemblies, such as ribosomes, proteasomes, or nuclear pore complexes (10). Our model handles differential binding properties of several interacting components encountered in RNA granules: each RNA molecule can potentially bind to hundreds of different multivalent RNA binding proteins (Fig. 1). Furthermore, the molecular composition and network of molecular interactions in each granule may be variable and flexible, and can undergo rapid remodeling. To our knowledge, the algorithm described here represents the first attempt to model granule composition and selectivity. “Composition” refers to the number of different protein and RNA molecules that are coassembled into each granule. “Selectivity” refers to the mechanism whereby some types of RNA molecules are segregated into different RNA granules.
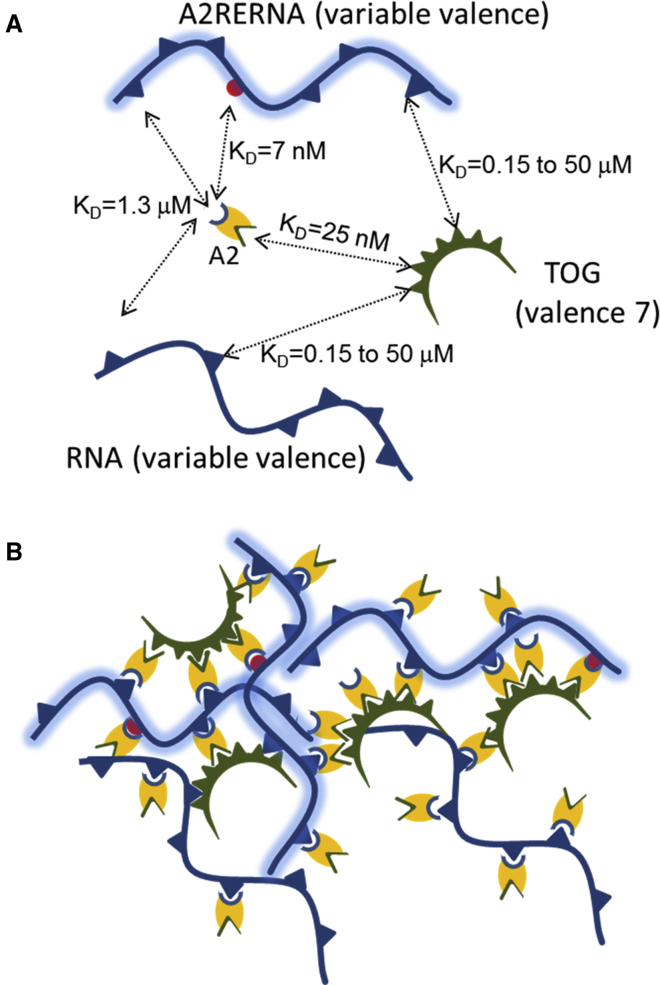
Figure 1.
Multivalent interactions in RNA granules. (A) Schematics of interactions among variable valency RNA (curved line), variable valency A2RE RNA (shaded curved line), heptavalent scaffold protein TOG (semicircles), and bivalent protein A2 (oval shapes with distinct sites for binding to TOG and A2RE RNA). Interactions among sites are shown with arrows with respective dissociation constants. A single spherical site in each A2RE RNA is the high specificity A2RE site for A2 protein. (B) An example of an aggregate resulting from interactions among these four types of molecules. To see this figure in color, go online.
We model a well-characterized type of RNA granule, the A2 granule (22, 25, 26, 27, 28, 29, 30), which contains multiple different RNA molecules, including A2RE RNAs with characteristic nucleotide sequences termed “A2 response elements” (A2RE) that are recognized by a cognate RNA binding protein called “hnRNP A2”. In vitro experiments indicate that high concentrations of purified RNA binding proteins (such as hnRNP A2) and RNA molecules can undergo phase transitions from solution to liquid droplet under certain conditions (31, 32). In this study, we explore the parameters that modulate this process and serve as determinants of granule composition and selectivity. We find that key parameters are concentration of multivalent RNA binding protein and effective valency of RNA, i.e., the number of sites in each RNA molecule that are not saturated by other proteins and are thus available for binding to either a scaffold molecule (such as the heptavalent TOG, introduced in the next section) or RNA binding proteins (such as the hnRNP A2) (29).
We explore the impact of a single high affinity A2RE site in A2RE RNA representing only a fraction (1–10%) of the total RNA valency. Our modeling studies demonstrate that high affinity binding between hnRNP A2 and scaffold protein, and between hnRNP A2 and A2RE RNA, allows hnRNP A2 to act as an adaptor and regulator in granule assembly that can promote or inhibit interactions between the other two multivalent components depending on its concentration. Low affinity binding between scaffold molecule and RNA molecules accurately recapitulates the composition (33) of granules in live cells and inhibits aggregation of RNA into very large complexes.
Materials and Methods
Model components
A2RE RNAs contain characteristic 11-nucleotide cis-acting sequences termed “A2 response elements” (A2RE). The A2RE sequence was first identified (34) by deletion analysis of RNA encoding myelin basic protein (MBP). Deletion of the A2RE sequence from MBP RNA prevented assembly of MBP RNA into granules, and insertion of the A2RE sequence into non-A2RE RNAs facilitated assembly of those RNAs into granules. A2RE-containing RNA molecules encoding functionally related neuronal proteins required for learning and memory (αCamKII, neurogranin, ARC) are colocalized in the same granules in dendrites (24, 35).
The A2RE sequence is recognized by a cognate RNA binding protein called “hnRNP A2” (36). Specific binding of hnRNP A2 to A2RE sequences and nonspecific binding of hnRNPA2 to nonspecific sequences has been characterized by affinity pull-down (36), surface plasmon resonance (SPR), and fluorescence correlation spectroscopy (FCS) (29, 35).
In addition to binding A2RE RNA, hnRNP A2 also binds to a multivalent scaffold protein called “cytoskeleton-associated protein 5” (CKAP5; also called TOG protein, standing for the protein encoded by the tumor overexpressed gene), as shown by yeast two-hybrid analysis (28). Binding of hnRNP A2 to different domains of TOG has been experimentally characterized by SPR and FCS (J.H.C., V. Tatavarty, and G. Korza, unpublished data). Granule assembly is prevented in TOG knock-out neurons and rescued by full-length TOG but not by a mixture of individual TOG domains (30), indicating that granules are formed through multivalent interactions and heptavalent TOG molecules are required for granule formation (Fig. 1).
Anti-sense knockdown of hnRNP A2 (26, 35) or injection of function-blocking isoform-specific antibodies (29, 35) both result in reduced numbers of A2RE RNA granules in processes of oligodendrocytes and diffuse RNA distribution in the perikaryon. Originally this was interpreted to mean that hnRNP A2 was necessary for transport of A2RE RNA granules from the perikaryon to the processes. However, in light of the molecular interactions between hnRNP A2 and TOG, an alternative explanation is that hnRNP A2 is necessary for A2RE RNA granule formation. Here, we use our model to investigate potential roles of hnRNP A2 in granule formation via its bivalent interaction with TOG and RNA.
RNA granules contain multiple RNA molecules and RNA binding proteins that can potentially affect granule assembly through competing interactions with A2RE RNA, hnRNP A2, or TOG protein. Nonspecific RNA binding proteins in granules may occupy non-A2RE sites on individual RNA molecules, thereby decreasing the effective valency of each RNA molecule. The hnRNP A2 molecules can potentially interact with multiple other binding partners, including telomeres (31), splicing factors (32), and nuclear transport factors (37), which may compete with RNA or TOG protein, thereby decreasing the effective hnRNP A2 concentration available for binding to TOG protein and RNA. TOG protein in the cell can also interact with multiple other binding partners, including microtubules (38), microfilaments (39), and kinesin (40), which may reduce the effective concentration of TOG scaffold available for binding to hnRNP A2 or RNA. The effects of such competing interactions on granule assembly are represented in the model by reducing the number of nonspecific binding sites available for binding on each RNA molecule, reducing the effective concentration of hnRNP A2 or reducing the effective concentration of TOG protein.
Model parameters
In our model, we used pairwise molecular affinities between individual granule components (A2RE and non-A2RE RNA, TOG, and hnRNP A2 molecules) measured in vitro, and adjusted these parameters to recapitulate in vivo experimental observations. For simplicity, we refer to hnRNP A2 as simply “A2” from now on.
Table 1 shows pairwise affinities between interacting sites, measured in vitro by SPR analysis or FCS analysis. Data on specific binding of A2 to A2RE sequences and nonspecific RNA sequences has been reported in Gao et al. (35). The values in Table 1 for A2 binding to A2RE and non-A2RE sequences represent averages for multiple different A2RE sequences. Data on binding of TOG domains to A2 and RNA molecules is yet unpublished (J.H.C., V. Tatavarty, and G. Korza, unpublished data). The values in Table 1 for TOG domain binding to A2 or RNA represent averages for the different TOG domains. Importantly, the affinities of TOG domains for A2 are greater than their affinities for RNA, and the affinity of A2 for A2RE sites is greater than its affinity for non-A2RE sites in RNA.
Table 1.
Dissociation Constant KD between Sites and Corresponding References
| A2 | A2RE RNA Site | Nonspecific RNA Site | |
|---|---|---|---|
| A2 | (7 ± 3) nM (35)a | 1300 nM (35)b | |
| TOG | (25 ± 9) nMc,d | 150 nMb,c | 150 nMb,c |
SD for affinity differences among A2RE sequences in different RNAs (MBP, αCamKII, NG, ARC, PKMζ).
SD not available because only a single RNA sequence was analyzed (ARC A2RE anti-sense).
J.H.C., V. Tatavarty, and G. Korza, unpublished data; values obtained using SPR, and consistent with FCS measurements. Specific and nonspecific binding constants were resolved by fitting SPR data to a heterogeneous two-ligand model.
SD for affinity differences among individual TOG domains (D1–D7).
Model simulation
Here we model the system of RNA granule assembly (Fig. 1) with competing multivalent RNA molecules containing a large number of identical (or equivalent) sites using the approach developed in Falkenberg et al. (21). We consider a system with four types of molecules: 1) A2RE RNA with one A2RE high affinity site for A2 protein and up to 100 low affinity nonspecific sites for either A2 or TOG proteins; 2) non-A2RE RNA with up to 100 low affinity nonspecific sites for A2 or TOG; 3) A2 protein with one site that binds RNA (specifically or nonspecifically) and a second site that binds TOG; and 4) TOG protein with seven equivalent TOG domains, each of which can bind either RNA (nonspecifically) or A2 protein.
Our algorithm is based on the assumption that all interactions among sites are independent of each other (i.e., the principle of equal reactivity). Experimental evidence is not sufficient to suggest any plausible hypothesis on cooperativity or allostery at this point. The algorithm begins by deconstructing each multivalent molecule into a set of individual monovalent binding sites. In Fig. 1, each A2 molecule (oval) is deconstructed into two sites (one specific to TOG and one specific to A2RE sequence), each with the same total concentration as A2. TOG molecule is deconstructed into seven identical sites, which is the same as defining a single site with total concentration sevenfold the concentration of TOG. Nonspecific RNA is deconstructed into a single nonspecific site with concentration being equal to concentration of RNA times its valency. Specific RNA is deconstructed into a set of two sites—one specific A2RE sequence with concentration equal to A2RE RNA concentration, and a nonspecific site with concentration being equal to the concentration of RNA times its valency. All possible interactions for each individual site type are identified, and the reactions between individual binding sites are evaluated deterministically as a function of time, using the binding and unbinding rate constants and concentrations of individual sites. This deterministic step is accomplished by solving the system of ordinary differential equations (ODEs) described in the Supporting Material (nomenclature and parameters in Tables S1 and S2), until steady state is reached (it is assumed that the granules are present in the cell body long enough for the system to reach steady state, before they are transported to the periphery). The system of ODEs consists of equations for each type of binding site (six) and each type of site-site interaction (seven). The initial conditions for the system corresponds to the state when all molecules are unbound, so there are no bonds. Solving the system of ODEs provides the fraction of bound sites of each type. The bound and free states are then stochastically assigned among all corresponding sites in the system, and the molecules corresponding to each bond is identified. The aggregates are reconstructed by identifying all molecules that are directly and indirectly connected. The composition of each reconstructed aggregate in the simulation is then recorded. This stochastic procedure is repeated multiple times to generate distribution statistics for each molecular ensemble. Simulations are repeated for each different set of parameters to determine how each parameter set affects granule assembly.
Results
Dual functions of multivalent molecules: increasing concentration may result in either larger or smaller granules
The affinities and valencies for molecular interactions among A2RE RNA, A2, and TOG have been measured experimentally in vitro. However, the effective affinities and valencies of these molecules in live cells may be quite different. Each RNA molecule theoretically contains hundreds of potential binding sites corresponding to overlapping target sequences at each nucleotide position in the RNA molecule. In the cell, many of these potential binding sites may be occupied by RNA binding proteins or sterically occluded by RNA secondary structure, which may reduce the apparent affinity and valency for RNA binding to A2 or TOG in vivo. A2 is bivalent, with one RNA binding site that can bind to either the A2RE sequence with high affinity or to non-A2RE sequences with lower affinity, and one TOG binding site. In the following simulations, we have generally used experimentally determined affinities and valencies as starting conditions (Table 1) and adjusted the valencies to take into account potential competing interactions in live cells.
We used our algorithm (21) to perform multiple simulation runs for the model as described in the Materials and Methods, using fixed concentrations of RNA and TOG (corresponding to concentrations measured or estimated in live cells) and varying concentrations of A2, for specific values of affinity between RNA and TOG (Table 1). The mean number of RNA molecules per granule is defined as the total number of all RNA molecules in granules divided by the total number of granules , where is the number of complexes containing RNA molecules. A granule is defined as an ensemble containing at least two RNA molecules. The value of cannot exceed 200, which is the total number of RNA molecules in our simulated system (corresponding to 10 nM).
Our results (Fig. 2 A) demonstrate that for concentrations of A2 below a certain threshold, all RNA molecules are assembled into a single large granule. Note that 0.05 nM corresponds to a simulation with a single A2 molecule, i.e., A2 is virtually absent. Increasing A2 concentration leads to a decrease in the number of RNA molecules per granule. As both RNA and TOG become saturated with A2, RNA and TOG can no longer interact directly with each other. Further increasing A2 concentration makes it less and less likely that the same A2 molecule simultaneously binds to RNA and TOG. This suggests that low valency molecules such as A2 can function as modulators of granule size, by inhibiting direct interaction between high valency components such as TOG and RNA.
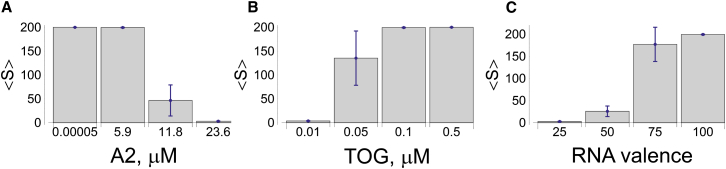
Figure 2.
Effect of protein concentrations and RNA valency on the mean number of RNAs in a granule . The volume is defined so that 1 μM = 2 × 104 molecules. The amount of RNA is 0.01 μM (200 molecules, non-A2RE RNA only), thus = 200 corresponds to all RNA sequestered. (A) For RNA with valency = 100, A2 concentrations <5.9 μM results in 100% of the RNA molecules assembled into a single granule. However, increasing A2 concentration disrupts granule formation. (B) All RNA is assembled into a single granule for TOG concentrations >0.1 μM. (C) Decreasing RNA valency results in smaller granules. Results correspond to 100 simulations, with 0.01 μM RNA, 5.9 μM A2, and 0.1 μM TOG, unless otherwise noted. To see this figure in color, go online.
Varying TOG concentration has an opposite effect on granule size relative to A2. As the concentration of TOG increases above a certain threshold (100 nM) (corresponding to the total concentration of TOG in cytoplasm), all RNA molecules are sequestered into a single large granule (Fig. 2 B). Thus, assembly of small granules is possible only for TOG concentrations below this threshold. For TOG concentration equal to 10 nM, the average granule size is significantly reduced = 3.7 ± 0.5 RNA molecules), consistent with the experimental findings that TOG is essential for granule assembly (30).
We investigated the effect of RNA valency on granule size. For RNA valency ≥100, all RNA molecules are assembled into a single granule (Fig. 2 C). Decreasing the effective valency of RNA (due to occupancy by other RNA binding proteins or occlusion by RNA secondary structure, for example) reduces the mean size of the granules. RNA with large valency leads to assembly of all RNAs into a single granule for all physiological concentrations of A2 and TOG. If the effective valency of RNA is reduced to 25, the average granule size decreases to 26 ± 12 RNA molecules per granule, which corresponds approximately to the number of RNA molecules per granule measured in cells (33).
In summary, for the affinities in Table 1, TOG promotes formation of granules (number of RNA molecules per granule increases with increasing TOG concentration), while A2 disrupts formation of granules (number of RNA molecules per granule decreases with increasing A2 concentration).
Affinity between TOG and multivalent RNA modulates the assembly of RNA into granules in absence of A2
Fig. 2 A suggests that interactions among RNA and TOG alone, may drive assembly of all RNA molecules into a single large granule, given the large granule size when A2 concentration is negligible. In cells, RNA molecules are distributed in multiple separate granules, each containing a finite number of RNA molecules (22, 23, 25, 26, 29, 33, 35). Here we investigate interaction between RNA and TOG molecules, to identify parameter regimes that modulate transition from systems with large numbers of granules containing few RNA molecules per granule, to systems with few granules containing large numbers of RNA molecules.
The Flory-Stockmayer theory (12, 13, 41) can be used to investigate the likelihood of formation of persistent large complexes containing a significant fraction of all molecules in the system (gelation).
We aim to identify system parameter regimes that simulate the type of granule formation observed in live cells. While TOG is heptavalent, the effective valency of RNA is not known. It can potentially interact with multiple different RNA binding proteins and assume complicated secondary structures, either of which could reduce its effective valency for binding to TOG protein. The effective in vivo affinity between TOG and RNA is also unknown because TOG can also bind to microtubules and microfilaments in cells. Accordingly, for this analysis, the concentrations for TOG and RNA were adjusted from at least onefold below to onefold above their estimated in vivo concentrations of 0.1 and 0.01 μM, respectively. The site pair affinity between TOG and RNA was adjusted over three orders of magnitude, and the RNA valency was adjusted from 5 to 100. Fig. 3 demonstrates that tight binding between TOG and RNA leads to formation of large molecular ensembles containing many RNAs. Decreasing affinity shifts the system from large ensembles to smaller complexes with fewer RNA molecules per complex even if the effective RNA valency is larger. Specifically, site pair affinity between TOG and RNA, using the experimentally measured value of KD = 150 nM, would result in formation of large ensembles for effective RNA valencies as low as five, in the absence of A2 and for rather low concentrations of TOG and RNA. Decreasing affinities between RNA and TOG allows formation of smaller ensembles with fewer RNA molecules per complex when the effective valency of RNA is increased (Fig. 3 B and C). In the cell, the effective affinity between TOG and RNA may be reduced because both TOG and RNA have alternate potential molecular partners that may compete for binding.
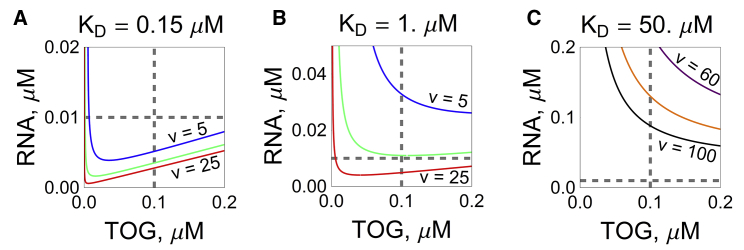
Figure 3.
The effect of site pair affinity (dissociation constant KD) between RNA and TOG sites and RNA valency v on gelation in the absence of A2. Each line represents the Flory-Stockmayer gelation curve for RNA valencies 5 (top curve), 10 (middle curve), 25 (bottom curve) in (A) and (B); and 60 (top curve), 80 (middle curve), and 100 (bottom curve) in (C). Concentration pairs located above each curve would result in gelation-sequestering of almost all RNAs into few large complexes. (A) For KD = 0.15 μM, the measured concentrations of 0.1 μM of TOG and 0.01 μM of RNA (shown with dashed lines) would result in gelation for RNA effective valency as low as 5 (top curve). (B) For KD = 1 μM, an effective RNA valency ≤10 prevents gelation for measured concentrations. (C) Further increasing of KD up to 50 μM enables formation of small complexes for valencies as high as 100 (bottom curve). To see this figure in color, go online.
These simulation results lead us to further explore the system under the assumption that the effective interactions between TOG and RNA in the cell may be of relatively low affinity (high KD). Otherwise, in the absence of A2, it seems that a significant fraction of RNA molecules could be sequestered in a single large complex. We hypothesize that this is unlikely because it could potentially interfere with dispersion of RNA granules to distal dendritic sites via transport through narrow dendritic processes.
A2 may facilitate or disrupt formation of RNA granules
Next, we analyzed the role of the adaptor molecule A2, RNA valency, and affinity between RNA and TOG on granule size . To perform this study for A2RE granules, we ran simulations with an equal mix of non-A2RE and A2RE RNA (0.01 μM of each) of effective valency of 10 (Fig. 4 A, where A2RE RNA has one A2RE sequence and nine nonspecific sites) or 100 (Fig. 4 B, where A2RE RNA has one A2RE sequence and 99 nonspecific sites). In both simulations, we assume a concentration of 0.1 μM for TOG and varying concentrations of A2. We varied the dissociation constant (KD) between TOG and RNA from 0.15 to 50 μM.
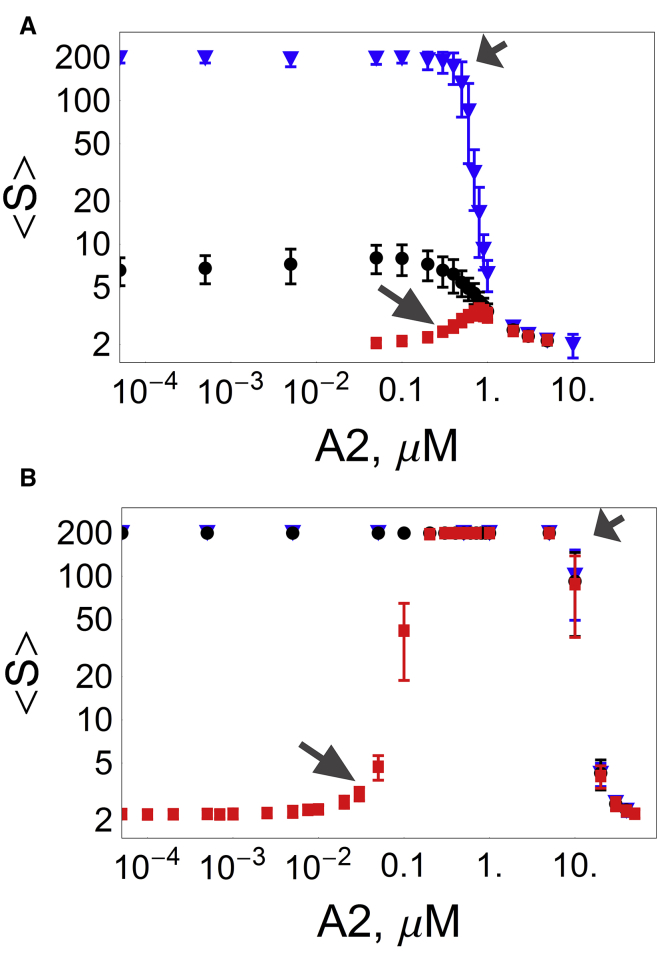
Figure 4.
The mean number of RNA molecules per granule as a function of A2 concentration. (A) Mix of 0.005 μM of non-A2RE and A2RE RNA (100 molecules each) of effective valency of 10, and 0.1 μM TOG (2000 molecules). For dissociation constant between TOG sites and RNA sites KD = 0.15 μM (solid triangles), increasing A2 concentrations at >1 μM leads to formation of small granules. For lower KD (1 μM, solid circles; 50 μM, solid squares) small granules are always more prevalent than large aggregates, but there is an A2 concentration that maximizes for (0.1 and 0.8 μM, respectively). (B) Simulations for RNA valency = 100. For KD = 0.15 μM (solid triangles) and KD = 1 μM (solid circles), A2 concentration has to be above a certain threshold to enable formation of small granules only. For KD = 50 μM (solid squares), the average granule size increases with A2 concentration on the order of 0.1 μM, reaching sequestration of all RNA into a single aggregate for A2 concentrations on the order of 1 μM. Further increase of A2 concentration on the order of 10 μM leads to disruption of the large aggregate into smaller granules. Variations are shown for averages of 100 simulations. (Large arrows) Beginning of the A2 range facilitating formation of granules (for KD = 50 μM); (short arrows) beginning of the A2 range disrupting formation of granules (for KD between 0.15μM and 50 μM). To see this figure in color, go online.
For RNA of valency 10 and A2 concentrations smaller than 1 μM, the average granule size is larger for higher effective affinities (lower KD) between TOG and RNA (Fig. 4 A).
We then analyzed the effect of A2 on . Consistent with the results in the previous section, high affinity between TOG and RNA (KD = 0.15 μM, solid triangles in Fig. 4) results in large aggregates when A2 concentration is negligible. For RNA of either low valency 10 (Fig. 4 A) or high valency 100 (Fig. 4 B), increasing A2 concentration disrupts formation of granules (highlighted by short arrow).
For low affinity between TOG and RNA (KD = 50 μM, solid squares in Fig. 4), A2 has dual functions: it may act to facilitate formation of granules (the transition from small to large granules as A2 increases is shown by large arrows) or to disrupt formation of granules (the transition from large to small granules as A2 increases is shown by small arrows). This effect is the same for both low valency of RNA (Fig.4 A) and high valency of RNA (Fig. 4 B), but the width of the sensitivity of to A2 is different: 0.1–1 μM for low valency and 0.1–10 μM for high valency RNA.
While the A2 concentration regime for facilitating formation of granules seems to be dependent on both RNA valency and the affinity between TOG and RNA, the concentration regime for disrupting formation of granules seems to be dependent on RNA valency alone. First, we look at facilitation of granule formation for fixed valency 10 (Fig. 4 A) and analyze the effect of affinity, as follows: for KD = 1 μM (dots), the maximum average granule size = 8 ± 2 is reached when A2 concentrations are between 0.05 and 0.1 μM, while for KD = 50 μM (squares) the maximum average granule size = 3.4 ± 0.4 is reached when A2 concentration is 0.8 μM. The effect of valency becomes explicit by looking at KD = 50 μM (squares) for RNA valency 10 (Fig. 4 A) and 100 (Fig. 4 B): higher valency makes the system more sensitive to A2, with noticeable disruption of granule formation at lower concentrations.
The threshold concentration of A2 for disruption of granule formation is on the order of 1 μM for all affinities when RNA valency is 10 (Fig. 4 A) and on the order of 10 μM for all affinities when RNA valency is 100 (Fig. 4 B).
To analyze the impact of RNA valency on the average granule size , we fixed the affinity between TOG and RNA to KD = 50 μM and performed simulations for RNA valencies 10, 20, 50, and 100 (Fig. S1 in Supporting Material). Higher valencies are more sensitive to A2 for facilitation of granule formation (noted at lower A2 concentrations) and least sensitive for disruption of granule formation (larger concentrations of A2 are required).
In summary, A2 may disrupt formation of granules when in excess, saturating the RNA and TOG molecules and preventing their interaction. However, when affinities between RNA and TOG are insufficient for granule formation, A2 may function as an adaptor, enhancing the chances of assembling TOG and RNA molecules into the same complex. The simulation results showing that A2 is required for granule formation are also consistent with experimental observations that reducing A2 expression or interfering with A2 function disrupts granule formation in cells (26, 29, 35).
A2 as a regulator of granule selectivity
In the previous section, we examined the effect of A2 concentration on the average number of RNA molecules per granule, and discovered that in some systems, two distinct A2 concentrations can result in the same mean number of RNA molecules per granule . Here we examine the effect of A2 concentration on granule selectivity (whether specific A2RE RNA molecules are more effectively assembled into granules compared to non-A2RE RNA molecules).
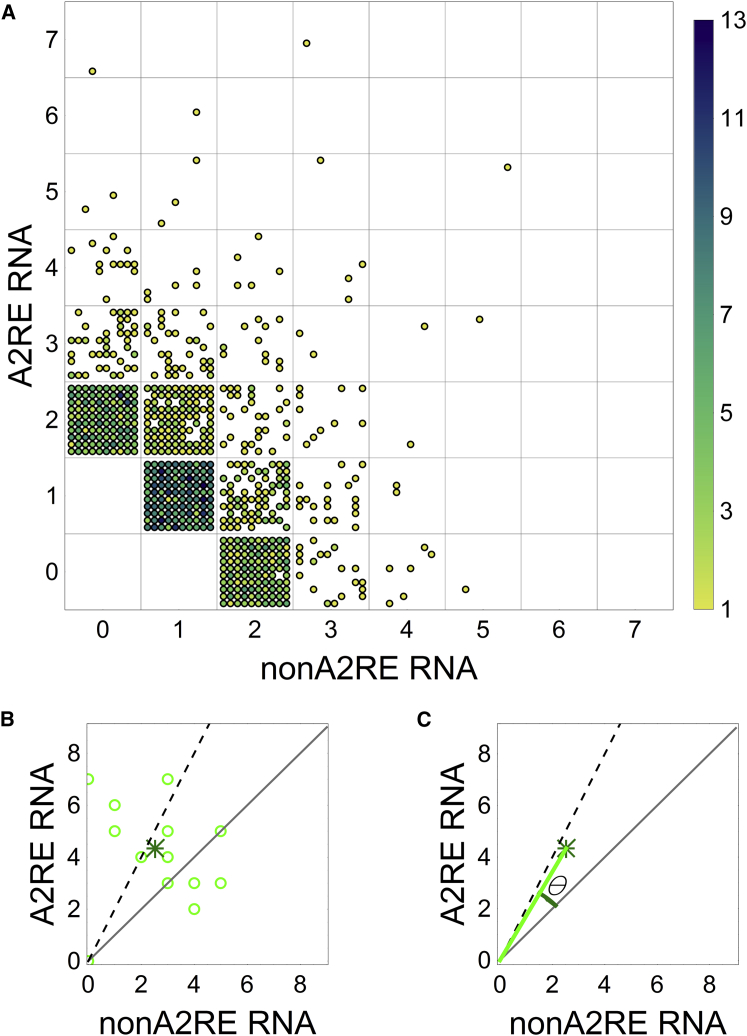
For each A2 concentration, 100 simulations were performed with 0.005 μM of A2RE RNA (100 molecules), 0.005 μM non-A2RE RNA (100 molecules), 0.1 μM TOG (2000 molecules), RNA valency = 100, and KD = 50 μM between the sites of RNA and TOG. We use simulations with A2 concentration = 0.01 μM to illustrate how selectivity was computed (Fig. 5). Each circle in Fig. 5 A represents a molecular ensemble containing specific numbers of A2RE RNA and non-A2RE RNA molecules as indicated by their position in the gridded mesh. The different locations within a square in the mesh correspond to different simulations. The number of occurrences of each composition in the same simulation is represented by the color intensity of the circle. In this set of simulations, the maximum occurrence per simulation for granules of any specific composition was 13.
Figure 5.
Identification of composition and definition of selectivity of RNA granules. (A) All RNA ensembles formed during 100 simulations. The position of each circle indicates the granule composition (as labeled on the axes) and simulation number (each mesh unit has 10 × 10 locations, totaling 100 simulations). The color intensity of each circle indicates the number of occurrences in the same simulation. (B) (Open circle) Average composition of visible granules (at least six RNAs) in each simulation. (Asterisk) Average composition of visible granules in all simulations. (C) The angle θ between the line that connects the origin to the asterisk and the diagonal (y = x) can be used as a measure for selectivity, tanθ. (Diagonal) Equivalent numbers of specific and nonspecific RNAs (selectivity 0); (dashed line) ratio of 1 nonspecific to 2 specific RNAs (selectivity 0.33). All simulations were performed for RNA of valency 100 and KD = 50 μM between TOG sites and RNA sites, concentrations 0.005 μM of non-A2RE and A2RE RNA each, 0.1 μM TOG. To see this figure in color, go online.
To analyze selectivity in granules that are formed during different simulation runs, we define the average granule composition for visible granules in each simulation as an average of specific and nonspecific RNAs among visible granules (Fig. 5 B, circles). Here we assume that the majority of granules that are visible by fluorescent microscopy contain at least six RNA molecules per granule. Such criterion is dependent on the experimental setup and resolution (fluorescence intensity or single molecule imaging), and different criteria will be discussed at the end of this section. Granules that have larger numbers of A2RE RNA molecules than non-A2RE RNA molecules appear above the diagonal y = x, and are considered selective. For reference, the dashed line indicates the location of granules with twice as many A2RE RNA molecules compared to non-A2RE RNA molecules. We define the “average composition” (asterisk) as an average over multiple simulations. Next, we determine the angle θ between the line from the origin to the average composition (asterisk) and the diagonal y = x (Fig. 5 C). We define the selectivity score as tan θ, where θ is defined by the equation . The selectivity score varies from −1 to 1, with maximum selectivity = positive 1 for granules containing only A2RE RNA molecules, selectivity = negative 1 for granules containing only non-A2RE RNA molecules, and selectivity = 0 for granules with equal numbers of non-A2RE RNA and A2RE RNA molecules. If the average granule composition coincides with y = 2 x (meaning that the granules have on average twice as many A2RE RNA molecules as non-A2RE RNA molecules), then the selectivity score = 0.333.
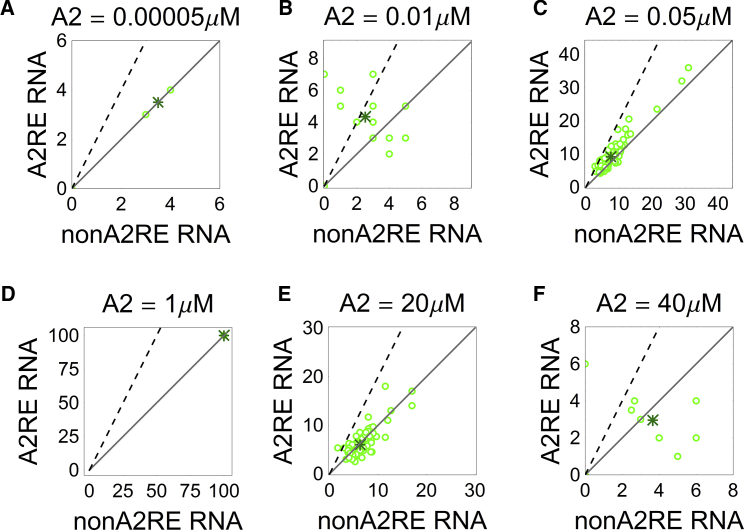
Averages of individual runs and average compositions (100 simulations) for several concentrations of A2 are shown in Fig. 6. Selectivity scores are reported in Fig. 7 A.
Figure 6.
RNA granule composition as a function of A2 concentration. (Open circle) Average composition of visible granules in each simulation. (Asterisk) Average composition for 100 simulations. (A) Negligible A2 concentrations seldom result in visible granules. (B) Low concentrations promote formation of selective granules. (C) Selectivity is reduced as more A2 molecules are added and larger granules occur. (D) Concentrations on the order of 1 μM result in sequestration of all RNA molecules in a large molecular complex, resulting in no selectivity. (E and F). As more A2 molecules become available, the RNA sites become saturated and are excluded from granules. Due to the A2RE binding site, A2RE RNAs are excluded more often than non-A2RE RNA, resulting in a negative selectivity score. All simulations were performed for the system with equal concentrations of specific and nonspecific RNAs (100 molecules each, RNA valency 100 and KD = 50 μM between the sites of RNA and TOG; concentrations 0.005 μM of non-A2RE and A2RE RNA each, 0.1 μM TOG). To see this figure in color, go online.
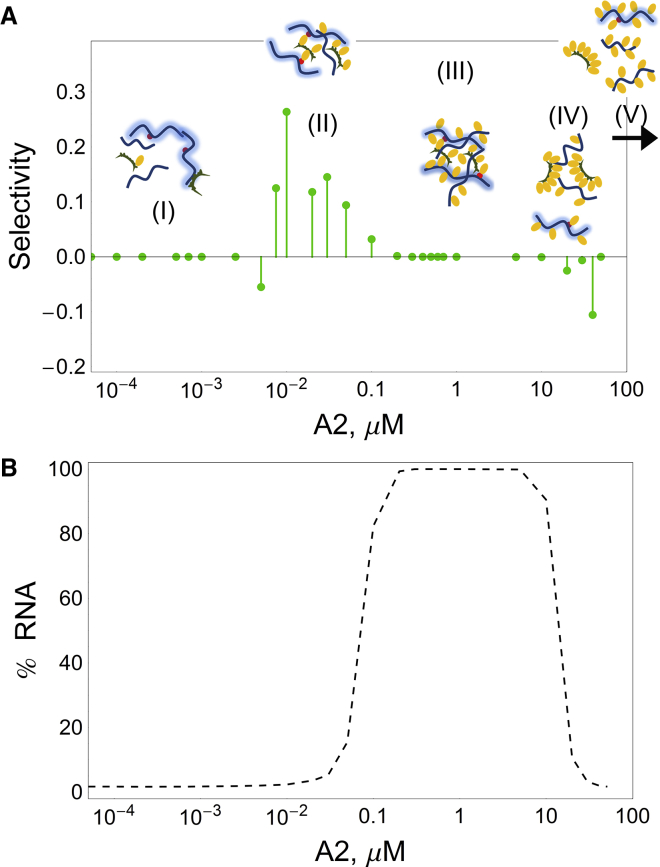
Figure 7.
Impact of A2 concentration on selectivity and assembly. (A) Five different regimes of A2 effect of selectivity, with illustrative cartoons. Regime (V) includes A2 concentrations beyond the plot (represented by the arrow). (B) Fraction of total RNA that is sequestered by the largest granule. Granule components as in Fig. 1: RNA (curved line), A2RE RNA (shaded curved line), A2 (oval), and TOG (semicircle). Average of 100 simulations, RNA valency 100 and KD = 50 μM between the sites of RNA and TOG; concentrations 0.005 μM of non-A2RE and A2RE RNA each, 0.1 μM TOG. To see this figure in color, go online.
Based on multiple numeric simulations, our model identifies five qualitatively different regimes of granule composition and selectivity as a function of A2 concentration (Figs. 6 and 7), as follows:
-
1)
For very low concentrations of A2 and TOG, RNAs rarely interact and visible granules rarely form (Fig. 6 A and Fig. 7 A, region I).
-
2)
As concentration of A2 increases, interaction between A2RE RNA and TOG increases and visible selective granules are formed (Fig. 7 A, region II). Note in Fig. 6 B that, for most simulations, the average visible granule is small and selective.
-
3)
When there are sufficient A2 molecules to promote significant aggregation of both A2RE RNA and nonspecific RNA into the largest granule, selectivity is lost (Fig. 6 D and Fig. 7 A, region III). Fig. 7 B shows that loss of selectivity is due to assembly of all RNA molecules into a single large complex.
-
4)
When there are too many A2 molecules, the likelihood that the same A2 molecule binds both RNA and TOG starts to decrease, and the granule size decreases (Fig. 6, E and F, and Fig. 7 A, region IV). Selective A2RE RNA molecules may be excluded from granules and negative selectivity may occur. However, the negative selectivity is not robust for lower valencies (Figs. S2 and S3).
-
5)
Finally, when A2 concentration is too high, all RNA and TOG molecules become saturated with A2, and the likelihood that the same A2 molecule is bound to both RNA and TOG becomes negligible (Fig. 7 A, region V). Consequently, high concentrations of A2 abrogate formation of visible granules.
Selectivity in granule assembly has been analyzed experimentally by measuring colocalization of different RNAs in individual granules in neurons using image correlation analysis (35). Pairwise combinations of A2RE RNAs exhibit cross-correlation coefficients >0.6, indicating a high level of coassembly of A2RE RNAs into the same granules, whereas pairwise combinations of non-A2RE and A2RE RNAs exhibit reduced cross-correlation coefficients ∼0.2, indicating a lower level of coassembly of A2RE RNA and non-A2RE RNAs into the same granules. These results reveal moderate selectivity in granule formation in vivo, consistent with the moderate selectivity in granule formation observed in our model. The analysis reported in Fig. 7 A was repeated using different resolution for visible granules (minimum number of RNA molecules 2, 3, and 4, in comparison to 6), with consistent results: selective granules are expected for A2 concentration regimes when A2 functions as a facilitator of granule formation, while mild negative selectivity may occur when A2 functions as a disruptor of granule formation (Fig. S4).
Discussion
One of the best-characterized types of RNA granules is the A2 granule, where multivalent A2RE RNA molecules bind to the cognate bivalent RNA binding protein A2 and are linked by the heptavalent scaffold protein TOG (4, 22, 26, 28, 29, 30, 33, 34). High resolution in situ hybridization studies of Drosophila embryos indicate that most RNAs in the Drosophila exome have characteristic localization patterns (42), suggesting that there are many different types of RNA granules, each containing a different combination of RNA molecules, RNA binding proteins, and scaffold proteins. Thus, the computational modeling approach described here may be applicable to many different types of RNA granules if the RNA specificity, cognate RNA binding proteins, and cognate scaffold proteins are known.
We used modeling and simulations to predict and analyze granule composition for large ranges of multiple parameters. Our model identified parameter ranges that lead to formation of excessively large or excessively small granules. It has been observed that decreased A2 expression resulted in a reduced number of granules in the cell processes while RNA remained diffuse in the perikaryon (26, 27, 29, 35). This has been interpreted to mean that A2 is required for transport of RNA granules in dendrites (26). However, our results suggest two alternative hypotheses: 1) if TOG and RNA have high affinity (apparent KD in the cell on the order of hundreds of nanoMolar), the reduced concentration of A2 leads to the formation of a gel-like large aggregate that may sequester most RNA molecules, and the granule size may prevent transport through narrow processes; or 2) if TOG and RNA have low effective affinity (KD on the order of 50 μM), A2 may act to facilitate granule formation at low concentrations (see Figs. 4 and S1 for range as a function of RNA valency). For the latter hypothesis, our results also identify two distinct A2 concentration regimes that may determine granule selectivity (shown as II and IV in Fig. 7). In the first regime (lower A2 concentration, II), A2 facilitates formation of selective granules. In the second regime (higher A2 concentration, IV), the granules may have moderately negative selectivity. The model observations correspond to hypotheses yet to be tested experimentally.
Because A2 is involved in several cell processes, experimentally induced changes in its concentration may impact other aspects of the live cell physiology, representing a challenge for model validation. Mathematical and numerical models always require assumptions and simplifications in representing the real biological system. Our analysis is based on populations of granules that would be visible under fluorescent microscopy (here, we assumed that at least six RNA molecules would be necessary). Nevertheless, smaller granules may also be functional. We showed that regardless of the experimental setup for resolving individual granules, positive selectivity is expected in the A2 concentration regime where it acts as a facilitator of granule formation, and mild negative selectivity may occur in the A2 concentration regime where it acts as a disruptor of granule formation (Fig. S4).
Our model considers three key molecular components of granules, each of which is necessary for A2 granule formation. In the cell, these molecules may also interact with other molecular components. Here we account for such effects by considering variable valencies and affinities. Another limitation of our approach is applying a principle of equal reactivity, where individual binding sites of the same type have the same affinity for interactions. In reality, formation of some bonds may affect affinity for further interactions. However, relevant experimental data is currently unavailable and therefore corresponding assumptions are yet to be formulated.
Our model can be easily extended to treat larger systems and new experimental scenarios as data becomes available. Constant concentration of molecules was assumed, representing a steady state. Due to the limited number of RNA molecules in the cell, the effect of stochasticity may be significant as revealed by the large number of different simulations. When the output of interest is the average granule size, similar results and deviations are obtained for 10 or 100 simulations. However, a large number of repetitions becomes relevant when performing more detailed analysis of the simulation results. For example, Fig. 5 A illustrates a situation where granules with at least six RNAs are not formed in every simulation, while granules with at least two RNA molecules are formed in every simulation. From this observation, it follows that the resolution criteria used when computing selectivity affects the number of simulations required for computing the statistical result when interested in selectivity, as illustrated in Fig. 5 A.
Finally, the results presented here reveal, to our knowledge, a novel and potentially generalizable concept that low valency molecules (in this case, bivalent hnRNP A2) can function as adapters between multivalent molecules (in this case, A2RE RNA and TOG) to tune the size and specificity of multimolecular aggregates (in this case, RNA granules).
Author Contributions
C.V.F., J.H.C., and M.L.B. designed research; C.V.F. performed research; C.V.F. contributed numeric and analytic tools; C.V.F., J.H.C., and M.L.B. analyzed data; and C.V.F., J.H.C., and M.L.B. wrote the article.
Acknowledgments
We thank Dhruv Vig and Rene Rovozzo for many helpful discussions regarding granule composition and performing experimental measurements.
M.L.B. was supported by National Institutes of Health (NIH) grants No. NIH NIGMS P41-GM103313 and R01-GM095485. J.H.C. was supported by NIH grants No. NS15190 and NS099730.
Editor: Reka Albert.
Footnotes
Materials and Methods, four figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30159-5.
Contributor Information
Cibele Vieira Falkenberg, Email: falkenberg@auburn.edu.
Michael L. Blinov, Email: blinov@uchc.edu.
Supporting Material
References
- 1.Li P., Banjade S., Rosen M.K. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones N., Blasutig I.M., Pawson T. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature. 2006;440:818–823. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- 3.Brangwynne C.P., Eckmann C.R., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 4.Carson J.H., Gao Y., Barbarese E. Multiplexed RNA trafficking in oligodendrocytes and neurons. Biochim. Biophys. Acta. 2008;1779:453–458. doi: 10.1016/j.bbagrm.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng W., Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- 6.Schiller H.B., Fässler R. Mechanosensitivity and compositional dynamics of cell-matrix adhesions. EMBO Rep. 2013;14:509–519. doi: 10.1038/embor.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditlev J.A., Mayer B.J., Loew L.M. There is more than one way to model an elephant. Experiment-driven modeling of the actin cytoskeleton. Biophys. J. 2013;104:520–532. doi: 10.1016/j.bpj.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray D., Levin M.D., Morton-Firth C.J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein B., Perelson A.S. Equilibrium theory for the clustering of bivalent cell surface receptors by trivalent ligands. Application to histamine release from basophils. Biophys. J. 1984;45:1109–1123. doi: 10.1016/S0006-3495(84)84259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer B.J., Blinov M.L., Loew L.M. Molecular machines or pleiomorphic ensembles: signaling complexes revisited. J. Biol. 2009;8:81. doi: 10.1186/jbiol185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suderman R., Deeds E.J. Machines vs. ensembles: effective MAPK signaling through heterogeneous sets of protein complexes. PLOS Comput. Biol. 2013;9:e1003278. doi: 10.1371/journal.pcbi.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flory P.J. Cornell University Press; Ithaca, NY: 1953. Principles of Polymer Chemistry. [Google Scholar]
- 13.Stockmayer W. Theory of molecular size distribution and gel formation in branched-chain polymers. J. Chem. Phys. 1943;11:45–55. [Google Scholar]
- 14.Cohen R., Benedek G. Equilibrium and kinetic-theory of polymerization and the sol-gel transition. J. Phys. Chem. 1982;86:3696–3714. [Google Scholar]
- 15.Falk M., Thomas R. Molecular-size distribution in random polyfunctional condensation with or without ring formation—computer simulation. Can. J. Chem. 1974;52:3285–3295. [Google Scholar]
- 16.Blinov M.L., Faeder J.R., Hlavacek W.S. BioNetGen: software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–3291. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 17.Faeder J.R., Blinov M.L., Hlavacek W.S. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol. Biol. 2009;500:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 18.Hlavacek W.S., Faeder J.R., Fontana W. Rules for modeling signal-transduction systems. Sci. STKE. 2006;2006:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- 19.Sneddon M.W., Faeder J.R., Emonet T. Efficient modeling, simulation and coarse-graining of biological complexity with NFsim. Nat. Methods. 2011;8:177–183. doi: 10.1038/nmeth.1546. [DOI] [PubMed] [Google Scholar]
- 20.Feret J., Danos V., Fontana W. Internal coarse-graining of molecular systems. Proc. Natl. Acad. Sci. USA. 2009;106:6453–6458. doi: 10.1073/pnas.0809908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkenberg C.V., Blinov M.L., Loew L.M. Pleomorphic ensembles: formation of large clusters composed of weakly interacting multivalent molecules. Biophys. J. 2013;105:2451–2460. doi: 10.1016/j.bpj.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbarese E., Koppel D.E., Carson J.H. Protein translation components are colocalized in granules in oligodendrocytes. J. Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- 23.Carson J.H., Worboys K., Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskeleton. 1997;38:318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Carson J.H., Cui H., Barbarese E. RNA trafficking in oligodendrocytes. Results Probl. Cell Differ. 2001;34:69–81. doi: 10.1007/978-3-540-40025-7_5. [DOI] [PubMed] [Google Scholar]
- 25.Ainger K., Avossa D., Carson J.H. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro T.P., Magee R.J., Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- 27.Shan J., Munro T.P., Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosturko L.D., Maggipinto M.J., Barbarese E. The microtubule-associated protein tumor overexpressed gene binds to the RNA trafficking protein heterogeneous nuclear ribonucleoprotein A2. Mol. Biol. Cell. 2005;16:1938–1947. doi: 10.1091/mbc.E04-08-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han S.P., Friend L.R., Smith R. Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms. Traffic. 2010;11:886–898. doi: 10.1111/j.1600-0854.2010.01072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbarese E., Ifrim M.F., Carson J.H. Conditional knockout of tumor overexpressed gene in mouse neurons affects RNA granule assembly, granule translation, LTP and short term habituation. PLoS One. 2013;8:e69989. doi: 10.1371/journal.pone.0069989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran-Jones K., Wayman L., Smith R. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 2005;33:486–496. doi: 10.1093/nar/gki203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shilo A., Ben Hur V., Karni R. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA. 2014;20:505–515. doi: 10.1261/rna.042259.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatavarty V., Ifrim M.F., Carson J.H. Single-molecule imaging of translational output from individual RNA granules in neurons. Mol. Biol. Cell. 2012;23:918–929. doi: 10.1091/mbc.E11-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ainger K., Avossa D., Carson J.H. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y., Tatavarty V., Carson J.H. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol. Biol. Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoek K.S., Kidd G.J., Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 37.Carson J.H., Blondin N., Korza G. Rules of engagement promote polarity in RNA trafficking. BMC Neurosci. 2006;7(Suppl. 1):S3. doi: 10.1186/1471-2202-7-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimaldi A.D., Zanic M., Kaverina I. Encoding the microtubule structure: allosteric interactions between the microtubule +TIP complex master regulators and TOG-domain proteins. Cell Cycle. 2015;14:1375–1378. doi: 10.1080/15384101.2015.1026521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsvetkov A.S., Samsonov A., Popov S.V. Microtubule-binding proteins CLASP1 and CLASP2 interact with actin filaments. Cell Motil. Cytoskeleton. 2007;64:519–530. doi: 10.1002/cm.20201. [DOI] [PubMed] [Google Scholar]
- 40.Holmfeldt P., Zhang X., Gullberg M. CaMKIIγ-mediated inactivation of the Kin I kinesin MCAK is essential for bipolar spindle formation. EMBO J. 2005;24:1256–1266. doi: 10.1038/sj.emboj.7600601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flory P. Molecular size distribution in three dimensional polymers. I. Gelation. J. Am. Chem. Soc. 1941;63:3083–3090. [Google Scholar]
- 42.Lécuyer E., Yoshida H., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.