Abstract
We previously characterized nutrient-specific transcriptional changes in Escherichia coli upon limitation of nitrogen (N) or sulfur (S). These global homeostatic responses presumably minimize the slowing of growth under a particular condition. Here, we characterize responses to slow growth per se that are not nutrient-specific. The latter help to coordinate the slowing of growth, and in the case of down-regulated genes, to conserve scarce N or S for other purposes. Three effects were particularly striking. First, although many genes under control of the stationary phase sigma factor RpoS were induced and were apparently required under S-limiting conditions, one or more was inhibitory under N-limiting conditions, or RpoS itself was inhibitory. RpoS was, however, universally required during nutrient downshifts. Second, limitation for N and S greatly decreased expression of genes required for synthesis of flagella and chemotaxis, and the motility of E. coli was decreased. Finally, unlike the response of all other met genes, transcription of metE was decreased under S- and N-limiting conditions. The metE product, a methionine synthase, is one of the most abundant proteins in E. coli grown aerobically in minimal medium. Responses of metE to S and N limitation pointed to an interesting physiological rationale for the regulatory subcircuit controlled by the methionine activator MetR.
Keywords: Escherichia coli genomics, flagella, methionine regulation, nutrient metabolism, RpoS
We have previously explored global responses of Escherichia coli K12 to limitation for the nutrients nitrogen (N) or sulfur (S) on glass-slide DNA microarrays (1). To determine responses of a wild-type strain, we compared its transcriptional profiles under nutrient-limiting and -excess conditions and under conditions of rapid transition between the two, magnifying transcriptional responses (2). Homeostatic responses to N or S limitation entail increased assimilation of preferred N or S sources, respectively, and scavenging of alternative N or S sources from the medium (1, 3). Common responses to N and S limitation apparently occur as a consequence of slow growth. Here, we characterize the latter responses, both increases and decreases in transcription, and explore several of them biologically.
Materials and Methods
Bacterial Strains. An rpoS::Tn10 allele (4) and a Tn10 insertion at an innocuous locus (zih-102::Tn10) were transferred to NCM3722 by P1-mediated transduction to generate NCM3890 and control strain NCM3964, respectively. The same lesions were also transferred to MG1655 (CGSC 6300) [also known as NCM3105 (5)] to generate NCM3962 and NCM3963, respectively. NCM3876 [glnL(Up)] has been described (1), and NCM4022 was isolated as a spontaneous motile derivative of NCM3722 on an arginine swim plate.
Nutrient Shifts and Other Growth Experiments. For carbon (C) and N downshift experiments growth of rpoS and control strains was monitored at 37°C in N-C- minimal medium containing various C or N sources. For C downshift experiments, ammonium chloride (10 mM) was the N source, and the C sources were glucose (0.04%) plus glycerol (0.4%), or glucose (0.04%) plus lactose (0.2%). For N downshift experiments, glycerol (0.4%) was the C source, and the N sources were ammonium (1 mM) plus arginine (2.5 mM). For S downshift experiments, cells were grown in N–C–S– minimal medium with glycerol (0.4%) and ammonium (10 mM) as the C and N sources, respectively. The S sources were sulfate (0.02 mM) plus glutathione (0.25 mM). The glutathione stock (25 mM) was stored at -20°C, and each aliquot was thawed and used once. For C upshift experiments, growth was started on glycerol (0.4%) and shifted by addition of glucose (to 0.2%); for N upshift, growth was started on arginine (2.5 mM) and shifted by addition of ammonium chloride (to 10 mM); for S upshift, growth was started on glutathione (0.25 mM) and shifted by addition of sulfate (to 0.25 mM). Cultures used for inoculation were grown on the appropriate preshift medium and were inoculated at a ratio of 1:50 into large baffled tubes containing 5 ml of medium and incubated with rapid shaking. Experiments were performed at least three times each.
Lanthionine and l-djenkolic acid were purchased from TCI America (Portland, OR). The doubling time of strain NCM3722 on djenkolate appeared to be similar to that on glutathione. However, when NCM3722 was inoculated on l-djenkolate, it initially grew rapidly on what we presume to be a contaminant and then shifted to the slower growth rate. We were unable to obtain l-djenkolic acid from another commercial source.
Motility Tests. Motility was assessed by using swim agar plates (0.3% Difco agar) or by direct observation under the light microscope (Zeiss Axiophot). Details are provided in Supporting Methods, which is published as supporting information on the PNAS web site.
Immunoblots. After adaptation to a particular medium, cells were grown to mid-exponential phase (OD600 of 0.4–0.45) at 37°C in a tube roller. Whole-cell preparations were made by suspending cells (1 ml) grown with ammonium or arginine as the N source in 0.25 ml of SDS loading buffer [60 mM Tris·HCl, pH 6.8/2% SDS/0.7 mM 2-mercaptoethanol/0.1 mM phenylmethylsulfonyl fluoride/10% glycerol/0.001% Bromophenol blue] supplemented with n-octyl β-d-glucopyranoside (Sigma), and disrupted by incubation at 37°C for 30 min. Samples (10 or 15 μl) were run on acrylamide gels (12%) and blotted to nitrocellulose membranes (6). Polyclonal rabbit antibodies raised to the flagellin protein [prepared in the laboratory of the late R. MacNab (Yale University, New Haven, CT) and obtained from L. McCarter (University of Iowa, Iowa City)] were diluted 1:10,000 before use. Detection was performed with goat anti-rabbit IgG (1:20,000) conjugated to horseradish peroxidase.
Microarray Procedures and Data Analysis. Details of strain growth, microarray procedures, statistical analysis, and clustering methods were as described (1).
For more information, see Figs. 6, 7, 8, 9, 10, 11, which are published as supporting information on the PNAS web site.
Results
RpoS-Mediated Increases in Transcription (mRNA Levels). In a study of global responses of E. coli K12 strain NCM3722 to nutrient limitation, we found that expression of some genes was increased under both N- and S-limiting conditions, presumably in response to slow growth. Of the 23 genes whose transcription was assessed numerically to be increased upon both N and S downshift (and/or decreased upon the opposite shifts), approximately one-fourth (6 genes) were RpoS-controlled genes [based on the list of 64 genes compiled by Loewen et al. (7) plus another 7 genes from recent literature (8–10)]. Transcription of an additional 15 RpoS-controlled genes was increased under conditions of N downshift, whereas transcription of only an additional 4 genes was increased under conditions of S downshift (see figure 3C of ref. 1). Table 3, which is published as supporting information on the PNAS web site, shows the list of RpoS-regulated genes that responded. The doubling time of NCM3722 on arginine, the poor N source we used, is ≈230 min, whereas its doubling time on the poor S source glutathione is much shorter, at ≈120 min (1). We suspect that additional RpoS-mediated responses on arginine are being made largely to slow growth, whereas those on glutathione may involve responses to oxidative stress (see Discussion). Unfortunately, it is difficult to find N sources that allow a range of doubling times (11) and the same appears to be true for the S sources. The doubling time of NCM3722 on djenkolic acid, another poor S source, is similar to that on glutathione (data not shown; see Materials and Methods). Doubling times on the alternative N source ethanolamine and the alternative S sources taurine, Hepes, and lanthionine are short (at most 10–15 min longer than on the preferred N and S sources, ammonium and sulfate; strain NCM3964, Table 1).
Table 1. Doubling times (in minutes) of an rpoS strain in different backgrounds on poor N and S sources.
| Strain | Genotype | 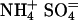 |
Arginine 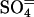
|
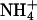 glutathione glutathione |
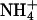 taurine taurine |
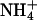 Hepes Hepes |
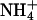 lanthionine lanthionine |
|---|---|---|---|---|---|---|---|
| NCM3964 | zih-102* | 60 | 230 | 120 | 70 | 70 | 55 |
| NCM3890 | rpoS* | 60 | 180 | 180 | 80 | 80 | 70 |
| NCM3963 | zih-102† | 75 | 630 | 90 | |||
| NCM3962 | rpoS† | 70 | 260 | 200 |
Cells were grown in N-C- or N-C-S- minimal medium with glycerol (0.4%) as the C source. The N sources (10 mM total N) and S sources (0.25 mM total S) were as indicated. Doubling times with glucose (0.2%) [or lactose (0.2%)] were: 50 min for NCM3964 and NCM3890 and 65 and 60 min, respectively, for NCM3963 and NCM3962.
The strain carries a Tn10 insertion in the background of NCM3722
The strain carries a Tn10 insertion in the background of MG1655 (CGSC 6300) [also known as NCM3105 (5)]
Growth of an rpoS Disruption Strain(s) Under Nutrient-Limiting Conditions. Observing RpoS-mediated transcriptional responses to N and S limitation led us to explore the consequences of disrupting rpoS. Surprisingly, the doubling time of strain NCM3890 (rpoS) on arginine was shorter than that of NCM3722 and an isogenic control strain, NCM3964 (zih-102::Tn10) (Table 1). This result was also true in the MG1655 background. By contrast, the doubling times of the rpoS strains on glutathione were longer than those of the appropriate control strains and the same was true for strain NCM3890, the only rpoS strain tested, on the alternative S sources taurine, Hepes, and lanthionine.
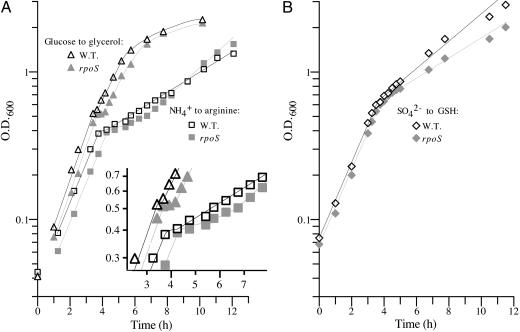
When the rpoS strains were subjected to downshift from ammonium to arginine, they had longer diauxic lags than control strains, despite the fact that they grew more rapidly after adaptation to arginine (Fig. 1A and data not shown). Strain NCM3890, the only rpoS strain tested, behaved similarly to its parental strain upon upshift from arginine to ammonium (data not shown). Because the rpoS disruption strains grew slower than control strains after adaptation to glutathione, it was hard to assess whether they also had longer diauxic lags upon downshift from sulfate to glutathione (Fig. 1B and data not shown). Strain NCM3890 (rpoS) behaved similarly to its parental strain upon upshift from glutathione to sulfate. Finally, the rpoS disruption strains had longer diauxic lags upon shift from glucose to glycerol (Fig. 1 A and data not shown), although the difference was subtle for strain NCM3962 (rpoS), possibly because it and its parental and control strains grow slowly on glucose (Table 1) (5). NCM3890 (rpoS) behaved similarly to its parental strain on upshift from glycerol to glucose. The rpoS disruption strains did not have noticeably longer diauxic lags than control strains upon shift from glucose to lactose. The doubling times of both parental and rpoS strains on lactose were as good as those on glucose (legend to Table 1).
Fig. 1.
Effect of an rpoS disruption on shifts from optimal to alternative nutrient sources. The strains were: NCM3964 (zih-102::Tn10), the control strain derived from NCM3722 (white symbols); and NCM3890 (rpoS::Tn10), which is congenic with it (gray symbols). In A, cultures were shifted from NH4+ (1 mM) to arginine (2.5 mM) (squares) or from glucose (0.02%) to glycerol (0.4%) (triangles). In B, cultures were shifted from sulfate (20 μM) to glutathione. Doubling times before and after shift were, respectively: ammonium to arginine, NCM3964, 65 and 245 min; and NCM3890, 65 and 180 min; glucose to glycerol, NCM3964, 50 and 80 min; and NCM3890, 55 and 85 min; sulfate to glutathione, NCM3964, 65 and 175 min; and NCM3890, 65 and 240 min.
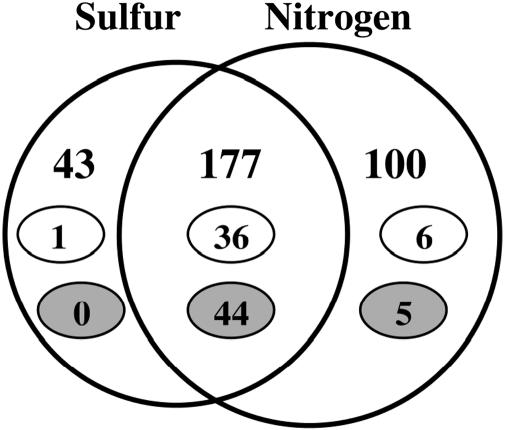
Decreased Transcription (mRNA Levels) of Genes Involved in Translation and Motility Under Nutrient-Limiting Conditions. Of 177 genes whose transcription was assessed numerically to be decreased in strain NCM3722 on both N and S limitation, one-fourth (44 genes) were genes coding for ribosomal proteins, and a comparable number (36 genes) were genes whose products are involved in motility and chemotaxis (overlapping portion of Fig. 2; small gray and white ovals, respectively). These represent large percentages (≈80% and ≈65%, respectively) of the total genes in these two categories (≈55 each; see refs. 12–15). Decreases in flagellar gene transcription would not have been detected in MG1655 because this strain and strain MC4100 express them poorly under all conditions (5, 16, 17). Transcription of only a few ribosomal or flagellar/chemotaxis genes was assessed numerically to be decreased only under N- or S-limiting conditions (Fig. 2).
Fig. 2.
Venn diagram summarizing numerical analysis of effects of N and S availability on transcription (mRNA levels) in strain NCM3722. Numbers in both circles indicate genes with lower expression under nutrient-limiting than nutrient-excess conditions or genes with higher expression on nutrient upshift. Numbers in small white ovals indicate genes whose products are involved in motility and chemotaxis and numbers in small gray ovals indicate genes encoding ribosomal proteins (15). The large circles and the region of overlap between them are not drawn to scale. Of the 100 genes listed as specific to N, 24 are S-related (table 1 of ref. 1). Of the 43 genes specific to S, 4 genes are members of the NtrC/Nac regulon.
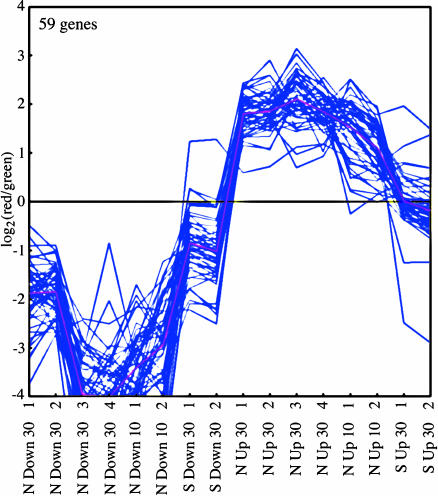
The single K-means cluster of 59 genes showing the largest average decrease in transcription under conditions of nutrient downshift and the largest average increase upon upshift (Fig. 3, cluster 1) contained 41 ribosomal and translation genes. Some additional genes in this category were present in two additional K-means clusters in which responses were of the same nature but lesser magnitude (cluster 2, 60 genes showing intermediate average response; cluster 3, 186 genes of small average response; Fig. 6). Flagellar genes (total of 36 genes) were split among the three K-means clusters, with the largest number (24 genes) being in cluster 3. Transcriptional responses of genes in these three K-means clusters were larger under conditions of nutrient downshift than nutrient upshift.
Fig. 3.
K-means cluster containing genes that are down-regulated on nutrient downshift and/or up-regulated on nutrient upshift. Experiments (from 16 genome prints) are indicated on the x axis and are described more fully in ref. 1. Numbers indicate the times after shift at which samples were taken. Log ratios of expression after shift to before shift are indicated on the y axis. Purple lines show data for individual genes and the pink line shows the average. Two additional K-means clusters with similar expression patterns are shown in Fig. 6.
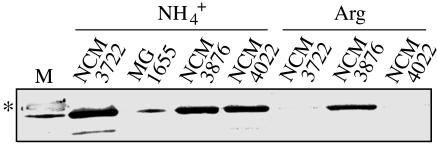
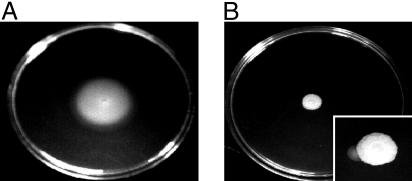
Decreased Transcription of Flagellar Genes Is Reflected in Decreased Motility. Whereas decreased expression of the translation apparatus under conditions of slow growth is one of the most exhaustively studied responses in the biology of enteric bacteria (18), decreased expression of genes required for motility under these conditions is little explored. Hence, we wanted to assess whether decreased transcription of flagellar genes under nutrient-limiting conditions was, in fact, accompanied by a decrease in flagella and motility. Western analysis indicated that strain NCM3722 had considerably lower levels of flagellin, the flagellar filament protein, when it was grown on arginine as the N source rather than ammonium (Fig. 4). Levels of flagellin on arginine were >10-fold lower than on ammonium (assessed by comparing dilutions of samples from ammonium-grown cells to samples from arginine-grown cells; data not shown). Whereas NCM3722 was motile on ammonium swim plates (13, 19), it was barely motile on arginine swim plates (Fig. 5 A vs. B). Motile derivatives of NCM3722 were observed on arginine (Fig. 5B Inset). One strain, NCM4022, which did not have increased flagellin levels on arginine (Fig. 4), was also more motile than NCM3722 on ammonium swim plates (data not shown). This strain may have a lesion that increases tumbling frequency and thus decreases the time of idling at barriers in soft agar (19) (see below). Strain NCM3876 [glnL(Up)], which grows rapidly on arginine, had higher levels of flagellin than NCM3722 when grown with this N source (Fig. 4). It was more motile than NCM3722 on arginine swim plates but appeared to be less motile on ammonium swim plates (data not shown). With glutathione as the S source, NCM3722 was somewhat motile on swim plates but apparently not as motile as on sulfate (Fig. 8). We did not pursue changes in motility under S-limiting conditions because they were not as well defined as under N-limiting conditions.
Fig. 4.
Western analysis of the relative amounts of flagellin (fliC product) in cells grown on ammonium or arginine as the N source. Cultures were grown and whole-cell samples were prepared, subjected to SDS/PAGE (12% gel), and analyzed as described in Materials and Methods. The nitrocellulose membrane was hybridized with rabbit polyclonal antiserum directed against E. coli flagellin (diluted 104-fold). For cells grown on ammonium (lanes 2–5), 10 μlof extract (≈3.5 μg of protein) was used, whereas for cells grown on arginine (lanes 6–8), 15 μl of extract (≈5 μg of protein) was used. Lanes 2 and 6, NCM3722; lane 3, MG1655 (CGSC 6300); lanes 4 and 7, NCM3876 [glnL(Up)]; and lanes 5 and 8, NCM4022. NCM4022 was isolated as a motile derivative of NCM3722 on an arginine swim plate (Fig. 5B Inset). Molecular mass standards (Benchmark Prestained, GIBCO/BRL) are in lane 1, and the standard at 50 kDa is marked with an asterisk. The band below it is flagellin. Strains NCM4026 and NCM4027, which carry fliC::Tn5 and fliA::Tn5, respectively, in the NCM3722 background, do not have this band. A Coomassie blue-stained gel showing that equal amounts of protein were loaded is shown in Fig. 7.
Fig. 5.
Swim plates showing behavior of NCM3722 on N-C- minimal medium with glycerol (0.4%) as the C source, and ammonium (10 mM; A) or arginine (2.5 mM; B) as the N source. Plates were inoculated with 5 μl of culture adapted to the corresponding medium and in mid-exponential phase (≈106 cells), and were incubated at 37°C for 2 (nutrient excess) to 4 days (nutrient limitation). (Inset) The plate was incubated for 5 days. Note the sector formed by a motile mutant.
When examined microscopically in growth medium, cells of strain NCM3722 or NCM4022 grown to mid-exponential phase on ammonium as the N source appeared to be swimming. Cells of the latter seemed to spin more frequently. Most cells of strain NCM3722 grown to mid-exponential phase on arginine did not appear to be swimming actively, and the same was true for NCM4022. The results were not changed if cells were separated from medium and suspended in a glycerol-containing buffer before being observed (see Materials and Methods). Cells of strain NCM3876 grown on ammonium were less motile than those of strains NCM3722 and NCM4022. However, motility of NCM3876 was not notably decreased when it was grown on arginine.
Decreased Transcription of Genes Involved in Metabolic Coordination Under N- and S-Limiting Conditions. Apart from large blocks of genes whose products are required for translation and motility, there is decreased transcription of a number of other genes under both N- and S-limiting conditions. Products of some are involved in macromolecular synthesis or protein folding and secretion. The genes for several of these products [e.g., prlA (b3300), rpoA (b3295), and priB (b4201), which are in K-means cluster 1 (Fig. 3)] are cotranscribed with genes whose products function in translation. Some down-regulated genes code for enzymes involved in small-molecule metabolism. Prominent among these are genes of glycerol catabolism, with glycerol being the carbon source we used. The glpQ and F genes (b2239 and b3927, respectively) are in K-means cluster 1, and genes in operons with them, glpT (b2240) and glpK (b3926), respectively, are in K-means cluster 2. Other genes whose mRNA levels are similarly regulated and that lie in K-means clusters 1 and 2 code for products involved in energy metabolism (e.g., components of the tricarboxylic acid cycle, the F1F0 ATPase, and the aerobic respiratory chain). Many additional genes in these operons are in K-means cluster 3. Other genes whose expression is prominently down-regulated under nutrient-limiting conditions are biosynthetic. They include a gene whose product is involved in gluconeogenesis [pckA (b3403) in cluster 2]; amino acid biosynthetic genes [e.g., ilvB and C (b3671 and b3774, respectively) in cluster 2]; genes whose products are involved in nucleotide biosynthesis and metabolism [e.g., ndk (b2518) in cluster 1 and purF (b2312), which codes for the first enzyme of purine biosynthesis, in cluster 2]; a gene whose product is required for synthesis of both amino acids and nucleotides [carA (b0032) in cluster 2]; and genes of fatty acid biosynthesis [fabG-acpP (b1093-b1094) and accBC (b3255-b3256) in cluster 2]. Many more genes of each sort are found in K-means cluster 3, which also contains nine genes coding for amino acyl t-RNA synthetases and ≈20% ORFs (38 of 186 genes total). Broadly similar observations were made by Chang et al. (20) for cells shifted from glucose to lactose or exposed to a pulse of H2O2.
Decreased Transcription of the metE Gene Under N- and S-Limiting Conditions. Unlike other methionine biosynthetic genes (namely, metA, B, C, F, K, and L), whose expression was increased under S-limiting conditions, expression of metE was profoundly decreased under these conditions (Table 2). As was true of the other met genes but more so, expression of metE was also decreased under N-limiting conditions. In fact, metE was a member of K-means cluster 1 (Fig. 3), whereas the other met genes were found in a K-means cluster containing genes specifically regulated by S availability (1). The met genes in the latter cluster, called the S cluster, are all repressed by the MetJ repressor in the presence of its ligand AdoMet (SAM). Other than metE, the only met biosynthetic gene not in the S cluster was metH, which is not regulated by MetJ, but only by the activator MetR. The metJ and metR regulatory genes were also missing from the S cluster. All three genes showed relatively small responses to S or N availability compared with the met genes discussed above.
Table 2. Ratios of mRNA levels for methionine biosynthetic genes under different conditions of S or N availability.
| S or N source
|
||||||
|---|---|---|---|---|---|---|
| Glutathione | S downshift | S upshift | Arginine | N downshift | N upshift | |
| Average met* | 2.35 | 2.47 | 0.54 | 0.91 | 0.58 | 2.12 |
| metE | 0.27 | 0.23 | 1.06 | 0.59 | 0.17 | 3.54 |
Values were determined relative to those on sulfate or ammonium as the S or N source, respectively, or relative to those before shift. Growth conditions were as described (1). For S shifts, samples were taken 30 min after shift. For N shifts, samples were taken 10 and 30 min after shift, and values were averaged. Effects were larger 10 min after shift, particularly for upshifts.
Average values for metA, B, C, F, K, and L
Discussion
Genomic studies yielded several major conclusions about changes in transcription that were common to limitation of either N or S. First, transcription of a number of RpoS-regulated genes and genes thought to be responsive to specific stresses was increased. Second, apart from well known decreases in transcription of ribosomal genes and others whose products function in translation, there were decreases in transcription of flagellar genes and genes whose products are required for motility and chemotaxis. Third, there was decreased transcription of many genes whose products are involved in catabolism and anabolism of small molecules. Finally, unlike the case for other met genes, transcription of the metE gene was markedly decreased under both S-and N-limiting conditions. These conclusions, documented numerically and by analysis of K-means clusters in Results, could also be drawn qualitatively by visual inspection of genome images (1). Genome images are representations of primary microarray data in genome order (21). We discuss each conclusion briefly below.
RpoS. The RpoS sigma factor is required for survival in stationary phase and is involved in stress responses under other conditions (7, 22, 23). Our most significant finding was that RpoS contributes to the ability to adapt to nutrient downshifts, not only for N and S but also for C (Fig. 1 and Table 1). Strains with a disruption of rpoS had increased diauxic lags in each case. That this fact was not known previously is surprising, given that RpoS-mediated changes in gene expression during the glucose-lactose diauxie have been studied extensively and the analogies between this period of adaptation to a poorer nutrient source and the stationary phase have been noted (20, 24). Which RpoS-regulated products are needed and how many are involved remain to be determined. Disruption of rpoS has no deleterious effects during nutrient upshifts.
After adaptation to N-limiting conditions, increased expression of RpoS-regulated genes, or RpoS itself, was inhibitory. The rpoS disruption strains grew more rapidly than their parents on arginine as the N source, as they did in ammonium-limited continuous culture (25). By contrast, they grew more slowly on the poor S source glutathione and several others. S limitation results in oxidative stress (reviewed in ref. 1), and RpoS may be required because products of several genes under its control (e.g., katE, katB, and dps) ameliorate oxidative stress (7, 26).**As reported by others (24), an rpoS disruption strain grew as well as its parent on glycerol or lactose as the C source. Hence, one cannot generalize about whether or not RpoS or RpoS-regulated gene products are required under a particular nutrient-limiting condition or whether they might be inhibitory. Results for C differed between continuous (25) and batch cultures, perhaps because growth rates in continuous cultures were slower. Moreover, as others have noted (27), the fact that expression of a gene or set of genes increases under a particular condition does not mean that the gene products are necessarily useful under that condition. They may be needed under comparable circumstances more reflective of natural environments or under conditions of transition to or from the one chosen for study.
Motility. As is true for the translation apparatus, >50 gene products are required to build flagella and make tactic responses (12, 13). Cells grown on glycerol with a limiting N or S source are poorly motile on swim plates. Levels of the flagellar filament protein are decreased by at least 10-fold in cells grown with arginine rather than ammonium as the N source. Although it seems counterintuitive that cells would be less motile under nutrient-limiting conditions, homeostatic responses to N limitation are largely scavenging responses and scavenging responses are also marked for S-limited cells (1). Hence, one can speculate that E. coli has evolved to concentrate low levels of many alternative N or S sources under nutrient-limiting conditions rather than to seek a particular one. To our knowledge, apart from peptides that might contain cysteine or methionine, no S-containing compound serves as a chemoattractant for E. coli. It will be of interest to define the molecular mechanisms mediating down-regulation of flagellar transcription under nutrient-limiting conditions and to determine whether decreased transcription reflects homogeneous changes in the population, i.e., whether all cells have fewer or possibly shorter flagella, or whether an increased proportion of them have no flagella.
Coordination of Metabolism. Transcription of a variety of genes involved in catabolism, e.g., of glycerol, the carbon source we used, and in biosynthesis of several classes of small molecules is decreased under both N- and S-limiting conditions. Such decreases, appearing to reflect coordinated changes in metabolism during slow growth, were difficult to document in adapted batch cultures but were amplified upon nutrient shifts. Details of the regulatory circuitry mediating coordination are not well defined. As reviewed in depth by others, the signal molecules guanosine tetra- and pentaphosphate almost certainly play an important role (18, 20, 28) and are known to be required for RpoS-mediated responses. However, it seems likely that additional regulatory mechanisms will be involved in propagating depletions of primary metabolite pools (glutamine, acetyl serine, and sulfide) to central regulatory signals (29).
The MetR Regulatory Subcircuit Controlling metE Expression. Pioneering work of Stauffer and colleagues (30–33) showed that transcription of the metE gene depends on the MetR activator (the nomenclature is confusing) complexed with its ligand homocysteine. Activation can be overridden by the MetJ repressor complexed with its ligand SAM, which can be considered the end product of the methionine biosynthetic pathway. With only one exception, all other methionine biosynthetic genes are controlled solely or largely by MetJ (34). (The exception is metH, which is activated by MetR in the absence of homocysteine, but is not repressed by MetJ.) Although one physiological rationale for MetR control of metE is known (see below), we would like to propose another.
The MetE protein is the non-corrinoid-dependent methionine synthase, which has a low turnover number and constitutes 5% of cell protein in sulfate-containing minimal medium under aerobic conditions (29). (The other methionine synthase is the MetH protein, whose activity depends on the presence of exogenous vitamin B12.) As shown in Results, transcription of metE is markedly decreased under S-limiting conditions, whereas transcription of MetJ-regulated genes is increased, presumably in response to a lowering of the free-pool concentration of SAM (1). By itself, relief of MetJ repression would also result in increased expression of metE, the opposite of what is seen. The increase is apparently prevented by the dependence of metE transcription on MetR complexed with homocysteine. It is reasonable to assume that the pool concentration of homocysteine, an S-containing amino acid that is a substrate for MetE and the coactivator for MetR, is also lowered under S-limiting conditions, and that lower pools of homocysteine account for decreased metE transcription. Although MetE contains a smaller percentage of S-containing amino acids than the average E. coli protein (1), it is a large protein, and any increase in its already very high rate of synthesis would represent an enormous investment of limiting S. In addition, there are a number of indications that S-limited E. coli is oxidatively stressed (see above, ref. 1, and references therein), and it has just been shown that MetE is quickly and reversibly inactivated under conditions of oxidative stress (35, 36). Hence, use of the MetR-homocysteine subcircuit in S-limited cells apparently prevents increased transcription of metE under conditions where it would be disadvantageous from at least two perspectives. Under N-limiting conditions, expression of metE is decreased more than that of other met genes (Table 2). Although pool concentrations of both homocysteine and SAM are probably higher under N-limiting than N-excess conditions, MetJ-mediated repression of metE presumably prevents MetR-mediated activation. Thus, dual control of metE by both MetR and MetJ allows its transcription to be reduced under N- and S-limiting conditions. In addition to these proposed rationales, the MetR subcircuit is known to mediate decreased expression of metE when vitamin B12 is available and the more efficient MetH enzyme can function (30, 32, 37).
Acknowledgments
We thank Kevin D. Loh and Eirene Markenscoff Papadimitriou for microscopic observations of cell motility and Western analysis of flagellin levels; Linda McCarter for advice on these assays and providing antiserum; and Laszlo Csonka, Rowena Matthews, and George Stauffer for critical review of the manuscript. This work was supported by National Institutes of Health Grant GM38361 and a grant from the Torrey Mesa Research Institute, Syngenta Research and Technology, La Jolla, CA (to S.K.).
Abbreviation: SAM, AdoMet.
Footnotes
Park, S., You, X. & Imlay, J. A., 104th ASM General Meeting, May 23–27, 2004, New Orleans, LA.
References
- 1.Gyaneshwar, P., Paliy, O., McAuliffe, J., Popham, D. L., Jordan, M. I. & Kustu, S. (2005) J. Bacteriol. 187, 1074-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingraham, J. L., Maaløe, O. & Neidhardt, F. C. (1983) Growth of the Bacterial Cell (Sinauer Associates, Sunderland, MA).
- 3.Zimmer, D. P., Soupene, E., Lee, H. L., Wendisch, V. F., Khodursky, A. B., Peter, B. J., Bender, R. A. & Kustu, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengge-Aronis, R. & Fischer, D. (1992) Mol. Microbiol. 6, 1877-1886. [DOI] [PubMed] [Google Scholar]
- 5.Soupene, E., van Heeswijk, W. C., Plumbridge, J., Stewart, V., Bertenthal, D., Lee, H., Prasad, G., Paliy, O., Charernnoppakul, P. & Kustu, S. (2003) J. Bacteriol. 185, 5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher, S., Winston, S. E., Fuller, S. A. & Hurrell, J. G. R. (1997) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), Vol. 2, pp. 10.18.11-10.18.21. [Google Scholar]
- 7.Loewen, P. C., Hu, B., Strutinsky, J. & Sparling, R. (1998) Can. J. Microbiol. 44, 707-717. [DOI] [PubMed] [Google Scholar]
- 8.Bouvier, J., Gordia, S., Kampmann, G., Lange, R., Hengge-Aronis, R. & Gutierrez, C. (1998) Mol. Microbiol. 28, 971-980. [DOI] [PubMed] [Google Scholar]
- 9.Lindqvist, A., Membrillo-Hernandez, J., Poole, R. K. & Cook, G. M. (2000) Antonie Leeuwenhoek 78, 23-31. [DOI] [PubMed] [Google Scholar]
- 10.Schellhorn, H. E., Audia, J. P., Wei, L. I. & Chang, L. (1998) J. Bacteriol. 180, 6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda, T. P., Shauger, A. E. & Kustu, S. (1996) J. Mol. Biol. 259, 589-607. [DOI] [PubMed] [Google Scholar]
- 12.Chilcott, G. S. & Hughes, K. T. (2000) Microbiol. Mol. Biol. Rev. 64, 694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacNab, R. M. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 123-145. [Google Scholar]
- 14.Noller, H. F. & Nomura, M. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 167-186. [Google Scholar]
- 15.Riley, M. & Labedan, B. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 2, pp. 2118-2202. [Google Scholar]
- 16.Wendisch, V. F., Zimmer, D. P., Khodursky, A., Peter, B., Cozzarelli, N. & Kustu, S. (2001) Anal. Biochem. 290, 205-213. [DOI] [PubMed] [Google Scholar]
- 17.Shi, W., Zhou, Y., Wild, J., Adler, J. & Gross, C. A. (1992) J. Bacteriol. 174, 6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cashel, M., Gentry, D. R., Hernandez, V. J. & Vinella, D. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 1458-1496. [Google Scholar]
- 19.Wolfe, A. J. & Berg, H. C. (1989) Proc. Natl. Acad. Sci. USA 86, 6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, D. E., Smalley, D. J. & Conway, T. (2002) Mol. Microbiol. 45, 289-306. [DOI] [PubMed] [Google Scholar]
- 21.Zimmer, D. P., Paliy, O., Thomas, B., Gyaneshwar, P. & Kustu, S. (2004) Genetics 167, 2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 1497-1512. [Google Scholar]
- 23.Hengge-Aronis, R. (2002) J. Mol. Microbiol. Biotechnol. 4, 341-346. [PubMed] [Google Scholar]
- 24.Fischer, D., Teich, A., Neubauer, P. & Hengge-Aronis, R. (1998) J. Bacteriol. 180, 6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notley-McRobb, L., King, T. & Ferenci, T. (2002) J. Bacteriol. 184, 806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao, G., Ceci, P., Ilari, A., Giangiacomo, L., Laue, T. M., Chiancone, E. & Chasteen, N. D. (2002) J. Biol. Chem. 277, 27689-27696. [DOI] [PubMed] [Google Scholar]
- 27.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Véronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., André, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 28.Gentry, D. R., Hernandez, V. J., Nguyen, L. H., Jensen, D. B. & Cashel, M. (1993) J. Bacteriol. 175, 7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, M. W. & Neidhardt, F. C. (1983) J. Bacteriol. 154, 344-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urbanowski, M. L. & Stauffer, G. V. (1987) J. Bacteriol. 169, 5841-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maxon, M. E., Redfield, B., Cai, X. Y., Shoeman, R., Fujita, K., Fisher, W., Stauffer, G., Weissbach, H. & Brot, N. (1989) Proc. Natl. Acad. Sci. USA 86, 85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urbanowski, M. L. & Stauffer, G. V. (1989) J. Bacteriol. 171, 3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byerly, K. A., Urbanowski, M. L. & Stauffer, G. V. (1990) J. Bacteriol. 172, 2839-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greene, R. C. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 542-560. [Google Scholar]
- 35.Hondorp, E. R. & Matthews, R. G. (2004) PLoS Biol. 2, e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leichert, L. I. & Jakob, U. (2004) PLoS Biol. 2, e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, W. F., Urbanowski, M. L. & Stauffer, G. V. (1992) J. Bacteriol. 174, 4833-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]