Abstract
Early studies proposed that DNA methylation could have a role in regulating gene expression during development [Riggs, A.D. (1975) Cytogenet. Cell Genet. 14, 9–25]. However, some studies of DNA methylation in known tissue-specific genes during development do not support a major role for DNA methylation. In the results presented here, tissue-specific differentially methylated regions (TDMs) were first identified, and then expression of genes associated with these regions correlated with methylation status. Restriction landmark genomic scanning (RLGS) was used in conjunction with virtual RLGS to identify 150 TDMs [Matsuyama, T., Kimura, M.T., Koike, K., Abe, T., Nakao, T., Asami, T., Ebisuzaki, T., Held, W.A., Yoshida, S. & Nagase, H. (2003) Nucleic Acids Res. 31, 4490–4496]. Analysis of 14 TDMs by methylation-specific PCR and by bisulfite genomic sequencing confirms that the regions identified by RLGS are differentially methylated in a tissue-specific manner. The results indicate that 5% or more of the CpG islands are TDMs, disputing the general notion that all CpG islands are unmethylated. Some of the TDMs are within 5′ promoter CpG islands of genes, which exhibit a tissue-specific expression pattern that is consistent with methylation status and a role in tissue differentiation.
Keywords: DNA methylation, epigenetic, mouse, tissue differentiation
There is a history of inverse correlation between DNA methylation of CpG island promoter regions and gene expression. Almost all CpG islands on the inactive X chromosome are methylated, and monoallelic methylation of imprinted genes is associated with monoallelic gene expression (1–3). Moreover, programmed changes in DNA methylation are essential features of development, with disruption frequently resulting in aberrant development (4). Although early studies proposed that DNA methylation could have a role in regulating development (5, 6), more recent studies of DNA methylation in known tissue-specific genes during development did not support a major role for DNA methylation. Studies by Warnecke and Clark (7) found that the tissue-specific expression of the skeletal α-actin gene in the adult mouse does not correlate with the methylation state of the promoter. Walsh and Bestor (8) investigated the 5′ methylation status of seven tissue-specific genes and found no correlation with tissue-specific expression. In the study presented here, rather than examining the methylation status of known tissue-specific genes, tissue-specific differentially methylated regions (TDMs) were first identified, and then genes located near the TDMs were analyzed for tissue-specific expression.
Restriction landmark genomic scanning (RLGS) is a method for the two-dimensional display of end-labeled DNA restriction fragments (9–11) and can be used to scan for genomic DNA methylation (11). Because the NotI recognition site contains two CpGs and the great majority of NotI sites are within CpG islands, RLGS (with NotI as the restriction landmark) displays CpG islands and adjacent regions. If a NotI site is methylated, it will not be digested and will not be end-labeled, resulting in the absence of the spot in the RLGS profile. Global analysis of genomic DNA methylation of different tissues by using RLGS indicates that there are a relatively large number of differences in profiles suggesting TDMs (12–14). Numerous differences in the RLGS profiles of ES cells and differentiated tissues, such as kidney and brain, have been identified (13), but the DNA sequence of these genomic regions was not established, and it is not known whether these methylation differences are associated with tissue-specific gene expression.
In the results presented here, RLGS was used in conjunction with virtual RLGS to identify 150 TDMs (9, 10). vi-rlgs is a new computational software application that utilizes the mouse genome sequence information to generate a virtual RLGS image (10). Our results indicate that many CpG islands are differentially methylated in a tissue-specific fashion, which could be confirmed by methylation-specific PCR (MSP) and bisulfite genomic sequencing. Some of these TDMs are within 5′ promoter CpG islands of genes that are expressed only if the region is unmethylated.
Methods
DNA and RNA Preparations. The DNA for RLGS was isolated from tissues of 12-week-old C57BL/6J male mice by using the protocols described in refs. 9 and 15. TRIzol was used to extract both DNA for MSP or bisulfite sequencing and RNA from the same samples (16). The RNA was quantified by a spectrophotometer and aliquots were checked for integrity by electrophoresis in denaturing agarose gels (17).
RLGS. RLGS was performed according to published protocols (9, 10, 15). vi-rlgs is new computational software that displays a virtual RLGS image of mouse genome sequence information (10). The correspondence between the virtual and real RLGS spots for some of the loci were confirmed by PCR amplification of spot DNA eluted from RLGS gels by using primers derived from the sequences predicted by vi-rlgs (10). The identity between real and virtual spot sequences was further confirmed by sequence analysis of the PCR product.
Bisulfite Genomic Sequencing of TDMs. The bisulfite treatment of DNA and sequencing after PCR was done as described in ref. 10. The bisulfite PCR primers were designed by using the web program methprimer (18). The primers were chosen to include the NotI landmark within the amplification product and also as many CpG dinucleotides in the product as possible. The melting temperatures of the primer pairs were constrained to be between 50°C and 60°C. The bisulfite PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA). The PCR were carried out in either a Biometra T1 or an MJ Research PTC-100 thermocycler. The eluted PCR bands were sequenced at the Roswell Park Cancer Institute core facility's Applied Biosystems model 3100 sequencer. To compare the direct bisulfite sequencing results with those obtained by cloning the PCR products (see Fig. 4 and Fig. 6, which is published as supporting information on the PNAS web site), the PCR products amplified from bisulfite-treated tissue DNA were cloned into pGEM-T easy vector (Promega) according to the manufacturer's protocol. Plasmid DNA was purified with the DNA Purification system (Promega). Individual plasmids were then sequenced by using the Applied Biosystems model 3100 sequencer (Applied Biosystems).
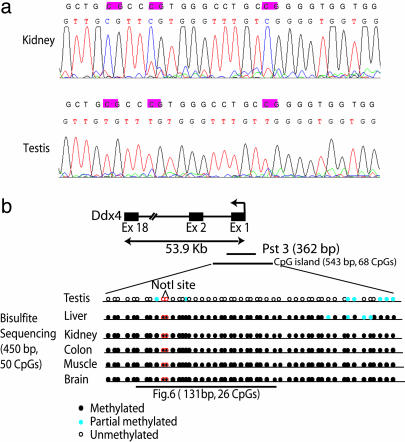
Fig. 4.
Bisulfite genomic sequencing of TDM Pst3. (a) Sequence traces obtained from the PCR products from bisulfite-treated DNA by using primers that would amplify a portion of the Pst3 locus. Because the PCR products were not cloned, the trace represents an approximation of the “average” methylation status at each CpG residue. The normal sequence and bisulfite sequence are shown. CpG sites are highlighted. (b) Diagrammatic representation of Pst3 within the 5′ promoter region of Ddx4 (DEAD-box protein 4) on chromosome 13. The CpG island includes the 5′ promoter, exon 1, and a portion of the first intron. The arrow indicates the direction of transcription. The methylation status of the CpGs (black circles, fully methylated; blue circles, partial methylation; white circles, unmethylated) within a 450-bp region of Pst3 is shown along with the location of the restriction landmark NotI site. Note the dense hypermethylation in liver, kidney, colon, muscle, and brain and the hypomethylation in testis. Fig. 6 shows the bisulfite sequence of a portion of the Pst3 region that was determined from cloned PCR products. The results are in general agreement, although the cloned sequences exhibit considerable heterogeneity in methylation. Fig. 10, which is published as supporting information on the PNAS web site, shows more detail as a larger version of this figure.
MSP. MSP was performed as described in ref. 19 by using the various bisulfite-treated tissue DNAs. The methylated and unmethylated primers were designed by using methprimer (18). The PCR (40 cycles) was performed by using the same conditions as for bisulfite genomic sequencing.
Quantitative, Real Time RT-PCR. Tissue-specific cDNAs were made by using the iScript cDNA kit from Bio-Rad according to the manufacturer's protocol. One microliter of each tissue cDNA was used per quantitative PCR. PCR primers were designed to bridge the exon–intron boundaries within the gene of interest to exclude possible contamination by genomic DNA (except for Hspa1l). The SYBR Green primers were designed by the web program primer3 (20) and purchased from Integrated DNA Technologies. A test RT-PCR was performed to check for a single PCR product before running the quantitative PCRs. The Assays on Demand primers sets were purchased from Applied Biosystems and used for the TaqMan assay. The quantitative PCRs were run on the Bio-Rad MyiQ Cycler for SYBR Green or iCycler for TaqMan according to the manufacturer's recommendations for each probe. The appropriate master mixes were used for each application. The resulting PCR cycle time (Ct) values were collected by using the software provided for the iCycler or MyiQ systems, and the data were then analyzed in Microsoft excel to determine ΔCt (ΔCt = test Ct - GAPDH standard Ct). The reverse transcription reactions were performed in triplicate with tissue RNA from three separate animals. The average value is presented with the standard deviation (Table 2). PCR products were analyzed by agarose gel electrophoresis after 40 cycles for correct product size. Melt curves and standard curves were performed for SYBR Green reactions. A list of all PCR primers used in these studies is provided in Table 3, which is published as supporting information on the PNAS web site.
Table 2. Tissue expression pattern of genes associated with TDMs: quantitative RT-PCR analysis.
| PCR, ΔCt ± SD
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | TDM location | Methyl. (tissue) | Gene | RT-PCR method | Brain | Kid | Liv | Col | Tes | Mus | GNF expression |
| Pst3 | 5′ promoter | U (testis) | Ddx4 | SYBR-G | 13.5 ± 0.6 | 14.6 ± 1.5 | 18.2 ± 1.7 | 14.6 ± 0.7 | -1.5 ± 0.3 | 17.4 ± 1.4 | Testis only/high expression |
| Pst6 | 3′ exon | U (testis) | Hsp-1-like | SYBR-G | 9.4 ± 1.4 | 7.6 ± 1.6 | 8.6 ± 1.2 | 5.9 ± 0.9 | -0.9 ± 0.1 | 10.7 ± 0.5 | Testis only/high expression |
| Pst6 | 3′ exon | U (testis) | Hsp1-like | TaqMan | 8.9 ± 1.3 | 10 ± 0.3 | 12.8 ± 1.1 | 10.7 ± 0.9 | -1.3 ± 1.1 | 6.7 ± 3.6 | Testis only/high expression |
| Pvu29 | 5′ promoter | U (testis) | RIKEN cDNA | SYBR-G | 7.2 ± 0.2 | 9.1 ± 0.4 | 8.8 ± 0.6 | 8.9 ± 0.5 | 2.7 ± 0.7 | 10.9 ± 0.3 | Broad pattern/high expression in testis |
| Pst61 | 5′ promoter | M (liver) | Gata2 | SYBR-G | 9.7 ± 0.4 | 7.5 ± 1.1 | 12.4 ± 0.9 | 11.1 ± 1.1 | 5.6 ± 0.4 | 10.2 ± 0.1 | Broad pattern/low expression in liver |
| Pst61 | 5′ promoter | M (liver) | Gata2 | TaqMan | 3.9 ± 1.5 | 1.6 ± 0.3 | 9.6 ± 1.8 | 6.4 ± 0.9 | 9.1 ± 0.6 | 7.6 ± 3.5 | Broad pattern/low expression in liver |
| Pvu2 | 3′ exon | U (testis) | Mct-like | SYBR-G | 8.4 ± 0.8 | 7.4 ± 0.4 | 8.1 ± 1.4 | 4.2 ± 0.9 | 7.5 ± 0.7 | 11.3 ± 1.6 | ND |
| Pvu66 | 3′ exon | M (liver) | Adra1b | SYBR-G | 8.1 ± 0.3 | 10.9 ± 1.1 | 6.9 ± 1.5 | 12.5 ± 2.5 | 11.8 ± 0.8 | 12.9 ± 5.7 | Broad pattern |
Genes associated with TDMs were identified by blat by using the University of California, Santa Cruz, Genome Bioinformatics database (http://genome.ucsc.edu). The methylation (methyl.; U, unmethylated; M, methylated) state (inferred by RLGS, MSP, and/or bisulfite sequencing) and location of the TDM relative to the gene are indicated. Although Pst6 is located in a 3′ exon, a CpG island 500 bp upstream of the promoter for Hspa11 had a methylation status similar to Pst6 (data not shown). Quantitative, real-time PCR was performed by using SYBR Green (SYBR-G) and TaqMan probes (see Methods). Expression levels were standardized to GAPDH by calculating ΔCt (ΔCt = Gene Ct – GAPDH Ct). The lower the ΔCt, the higher the mRNA level. ΔCt values with the highest mRNA level are shown in bold, and ΔCt values with the lowest mRNA level are shown in italics. A difference of one cycle is equivalent to an ≈2-fold difference in mRNA. Also indicated is the expression pattern determined from the GNF gene expression database (http://symatlas.gnf.org/SymAtlas). Kid, Kidney; Liv, liver; Col, colon; Tes, testis; Mus, muscle.
Results and Discussion
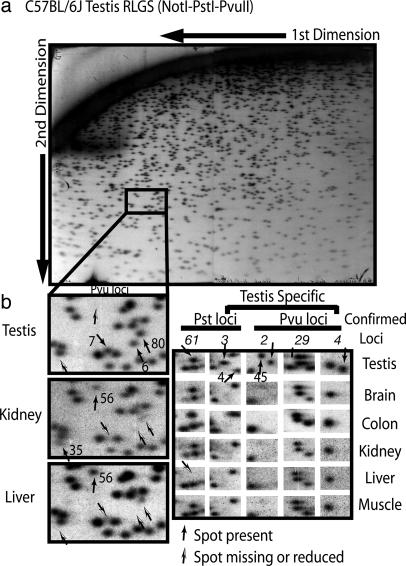
Identification of TDMs. RLGS profiles were performed for several different mouse tissues by using two restriction enzyme combinations (NotI–PstI–PvuII and NotI–PvuII–PstI, the same restriction enzymes but in a different order of digestion) that survey 3,200 potential sites of CpG methylation (NotI sites). Fig. 1 shows a portion of RLGS profiles from several tissues from C57BL/6J mice (12 weeks of age). In Fig. 1b Left, there are several spots present in testis that are absent in kidney and liver (Pvu6, Pvu7, and Pvu80), indicating that these regions are unmethylated in testis but methylated in kidney and liver. In contrast, Pvu56 is present in kidney and liver (unmethylated) but absent (methylated) in testis. Pvu35 was present only in kidney, but at less than full intensity, which indicates partial methylation. Additional TDMs are shown in Fig. 1b Right. RLGS analyses of tissues from different C57BL/6J mice indicate that the tissue-specific RLGS profiles are consistent (see Fig. 7, which is published as supporting information on the PNAS web site). vi-rlgs (10) identified 150 unique RLGS loci/spots that were present (unmethylated) in some tissues but absent (methylated) in others (Fig. 2; for a larger version of Fig. 2 see Fig. 8, which is published as supporting in formation on the PNAS web site). There was a large group (43 loci) that was unmethylated only in testis, but the majority (86 loci) were unmethylated in two to five of the tissues analyzed. blat analysis (University of California, Santa Cruz, Genome Bioinformatics database available at http://genome.ucsc.edu) of genomic sequences determined by vi-rlgs was used to identify genomic regions and genes associated with the TDMs. Approximately two-thirds of the loci (100) were located within CpG islands. Although 56% of the TDMs are located in apparent 5′ promoter regions, there are many that are located in exons, introns, or intergenic regions. The entire listing of TDMs identified by vi-rlgs can be found in Table 4, which is published as supporting information on the PNAS web site.
Fig. 1.
RLGS identification of TDMs. RLGS was performed with male C57BL/6J DNA (12 weeks of age) from testis, brain, colon, kidney, liver, and muscle by using two different RLGS restriction enzyme combinations (NotI–PstI–PvuII and NotI–PvuII–PstI). (a) The NotI-PstI–PvuII profile from testis is shown. (b) (Left) A section (enclosed section in a) from testis (Top), kidney (Middle), and liver (Bottom) is enlarged, and TDM loci are indicated. (Right)A portion of the RLGS profiles from six tissues showing additional TDM loci that were further confirmed (shown in italics) by MSP or bisulfite sequencing.
Fig. 2.
RLGS-inferred tissue methylation profiles for 150 TDMs identified by RLGS and virtual RLGS. Black squares indicate the RLGS spot was absent (methylated), purple squares indicate reduced intensity (partial methylation), and white squares indicate full diploid intensity (unmethylated). ND, the spot intensity could not be determined. Note that 64 of 150 loci were unmethylated in only one tissue and that 43 were unmethylated in testis only. Table 4 provides a complete listing of the TDMs and their locations in the genome.
Estimates of the Number of NotI Sites and CpG Islands That Are Differentially Methylated in the Mouse Genome. The mouse genome contains 6,057 NotI sites with 5,175 in nonrepetitive DNA (21). Because most of the NotI sites within repetitive DNA are methylated and not “visible,” RLGS can potentially resolve 5,175 NotI sites. The two restriction enzyme combinations used in this study display ≈3,200 RLGS spots. Although each NotI site produces two fragments, there is a <1% chance that both will be in the analyzed fragments (22). Also, there were no overlaps between NotI–PstI–PvuII and NotI–PvuII–PstI spot sequences identified by vi-rlgs. If one simplistically presumes that each RLGS spot corresponds to a single NotI site and that it was possible to identify 171 TDMs (150 identified by vi-rlgs) of 3,200 spots (171/3,200 = 5.3%), there would be 274 (5,175 × 0.053) NotI sites that are TDMs and 184 (274 × 0.67) that are in CpG islands that contain NotI sites [100 of 150 (67%) TDMs were in CpG islands]. There are estimated to be 15,500 CpG islands in the mouse genome (23), with 3,478 (22.4%) containing NotI sites (21). Therefore, one can estimate that 836 CpG islands contain TDMs (184/0.22), which suggests that ≈5.6% (836/15,500) of the CpG islands are TDMs. Because only six tissues were surveyed at only one developmental time point, it is quite likely that this calculation is an underestimate.
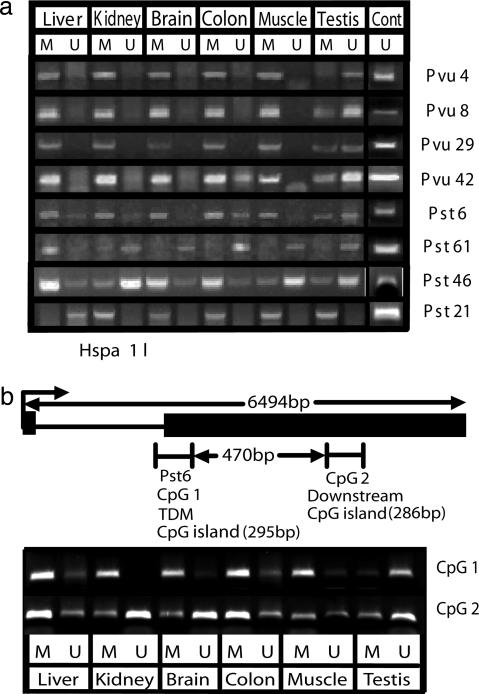
Confirmation of TDMs by MSP and Bisulfite Genomic Sequencing. Fourteen loci were analyzed more extensively by MSP (19) and/or bisulfite genomic sequencing (24) to confirm the RLGS results and determine the density of methylation in the TDM (Figs. 3 and 4, Table 1, and Figs. 6 and 9, which are published as supporting information on the PNAS web site, and data not shown). The MSP results (Fig. 3a) indicate loci that are unmethylated only in testis (Pvu4, Pvu8, and Pvu29), testis and colon (Pvu42 and Pst6), and methylated only in liver (Pst61). In contrast, Pst21 was methylated in all tissues except liver. Pst46 was unmethylated in kidney, muscle, and testis but was methylated in liver, brain, and colon. Note that both methylated and unmethylated sequences were detected in some tissues. This finding probably reflects heterogeneity of methylation and cell types within a tissue. These results are consistent with the methylation status inferred from the RLGS results (Fig. 2 and Table 1) except for the detection of some unmethylated Pst6 in colon by MSP that was not apparent by RLGS (see the legend for Fig. 3).
Fig. 3.
Tissue-specific DNA methylation. (a) MSP analysis of TDM loci. MSP primers that would amplify methylated (M) or unmethylated (U) genomic regions that contain or were close to the NotI restriction landmark site were designed by using methprimer (18). The presence of unmethylated and methylated products in some tissues probably reflects heterogeneity of cell types and methylation within a tissue. The detection of unmethylated Pst6 in colon by MSP that was not apparent by RLGS is probably due to the greater sensitivity of MSP. However, because MSP is not quantitative, it could represent a small fraction of the cells in colon. As a positive control for unmethylated primers, bacterial artificial chromosome genomic clones containing the genomic regions of interest (RP23-180J7, -285K3, -59D1, -430C5, -288H11, -467M14, -381B4, -278A5, and -446I5) were mixed, bisufite treated, and used as a template for MSP (Cont). (b) MSP analysis of the methylation status of Pst6 TDM and an adjacent CpG island. Genomic DNA was treated with sodium bisulfite and then amplified with primers specific for methylated and unmethylated sequences within the Pst6 TDM (CpG1) and a small CpG island just 3′ of Pst6 (CpG2). CpG1 is strongly methylated in all tissues except testis. In contrast, CpG2 is unmethylated in kidney and brain. See text for further explanation.
Table 1. TDM loci confirmed by MSP or bisulfite sequencing.
| RLGS
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci | Chr | Location, MB | CpG island | NotI site location | Gene | Function | Liv | Kid | Brain | Col | Mus | Tes |
| PvuII | ||||||||||||
| 2 | 11 | 114.98 | No | Exon | mRNA AK037416 | Mct transporter | 0 | 0 | 0 | 0 | 0 | 2 |
| 4 | 7 | 10.22 | Yes | 3′ exon | RefS D430030K24Rik | Ring Zn finger | 0 | 0 | 0 | 0 | 0 | 1 |
| 6 | 17 | 47.12 | Yes | 5′ promoter | RefS C330046L10Rik | Ubiquitin hydrolase | 0 | 0 | 0 | 0 | 0 | 1.5 |
| 8 | 4 | 58.76 | Yes | 5′ promoter | Est BE134335 | DNA J domain | 0 | 0 | 0 | 0 | 0 | 2 |
| 29 | 14 | 47.79 | Yes | 5′ promoter | RefS 4930548G07Rik | Chromatin binding | 0 | 0 | 0 | 0 | 0 | 1 |
| 42 | 1 | 16.79 | Yes | 5′ promoter | mRNA 6130401J04Rik | Ubiquitin-conj. E2 | 0 | 0.1 | 0.1 | 0.5 | 0 | 2 |
| 66 | 11 | 43.4 | Yes | 3′ exon | RefSeq Adra1b | G protein receptor | 0.1 | 2 | ND | 2 | 1 | 2 |
| PstI | ||||||||||||
| 3 | 13 | 111.46 | Yes | 5′ promoter | RefS Ddx4 | RNA helicase | 0 | 0 | 0 | 0 | 0 | 2 |
| 6 | 17 | 34.63 | Yes | 3′ exon | RefS HSP1-Like | Heat shock | 0 | 0 | 0 | 0 | 0 | 2 |
| 32 | 12 | 67.21 | Yes | 3′ exon | RefS Dact1 | Dsh signaling | 0 | 0.5 | 0 | 0 | 0 | 2 |
| 33 | 4 | ND | No | Exons | mRNA BC059845 | Unknown | 0 | 0.5 | 0 | 0 | 0 | 2 |
| 61 | 6 | 89.64 | Yes | 5′ promoter | mRNA GATA-2 | Transcription | 0 | 2 | 2 | 2 | 2 | 2 |
| 21 | 19 | 6.3 | No | Intron | Predicted gene | Unknown | 2 | 0.2 | 0 | 0 | 0 | 0 |
| 46 | 17 | 17.6 | No | Intron | Predicted gene | Unknown | 0 | 2 | 0 | 0 | 2 | 2 |
Shown is the chromosomal location (University of California, Santa Cruz, Bioinformatics Database, October 2003 assembly available at http://genome.ucsc.edu), whether the TDM was located within a designated CpG island, and the location of the TDM NotI site relative to a nearby gene. Gene and function information were also taken from links to the University of Califnornia, Santa Cruz, Genome Bioinformatics database. The RLGS spot intensity for each tissue is given, with 2 corresponding to a diploid single copy sequence, 1 indicating haploid spot intensity, and 0 indicating that the spot is absent. Values in bold indicate that the sequence is unmethylated (0.5–2 intensity). Values of <2 suggest that methylation is heterogeneous with the tissue. A complete listing of all TDM loci identified by VI-RLGS can be found in Table 4. MB, megabase pair; conj., conjugating enzyme; Mct, monocarboxylate transporter; Chr, chromosome; Liv, liver; Kid, kidney; Col, colon; Mus, muscle; Tes, testis.
Bisulfite genomic sequencing indicates the Pst3 CpG island sequence, found in the 5′ end of the Ddx4 gene, is methylated in kidney but not in testis, consistent with the RLGS results (Fig. 4a and Table 1). The more extensive bisulfite sequencing results shown diagrammatically in Fig. 4b (see also Fig. 6) indicate that almost the entire CpG island is densely methylated in all tissues except testis. Similar bisulfite sequencing results were found for Pvu2, located in exon 3 (National Center for Biotechnology Information accession no. AK037416) of a monocarboxylate transporter-like gene (Fig. 9), whereas for the Pvu66 CpG island in the 3′ end of the adrenergic receptor a 1b (Adra1b), methylation was demonstrated in the liver but not kidney and testis (data not shown). These results indicate that tissue-specific CpG methylation (TDMs) may be associated with both the 5′ and 3′ regions of genes as well as with internal exons.
MSP analysis of CpG island regions just upstream or downstream of TDMs indicates both similarities and differences in methylation status. For example, MSP analysis indicates a small CpG island (CpG2) 470 bp downstream of Pst6 is unmethylated in kidney and brain, whereas Pst6 (CpG1) is almost completely methylated in these tissues (Fig. 3b). However, both CpG1 and CpG2 are methylated in colon and muscle and unmethylated in testis. These results indicate that TDMs appear to be localized to small genomic regions, possibly a single CpG island. However, additional work is necessary to establish the precise boundaries of the TDMs.
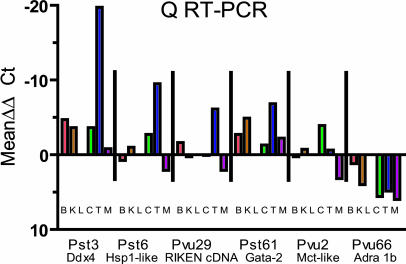
Tissue-Specific Expression of Genes Associated with TDMs. Promoter CpG island DNA methylation has frequently been associated with silencing of gene expression in cancer (25) and in normal cells (14). The dense methylation of the CpG island promoter region of DEAD-box protein 4 (Ddx4) (Fig. 4), associated with the Pst3 TDM, would suggest that Ddx4 is silenced in most tissues except testis. Quantitative, real time RT-PCR indicated a very high level of expression in testis (Fig. 5 and Table 2) and a low level expression in brain that was several hundred times less than testis. Furthermore, interrogation of the Genomics Institute of Novartis Research Foundation (GNF) gene expression database (available at http://symatlas.gnf.org/SymAtlas) (26) indicates that Ddx4 is expressed at very high levels in C57BL/6J testis and is either not expressed or expressed at much lower levels in other tissues. The targeted homozygous knockout of Ddx4 causes a deficiency in the proliferation and differentiation of mouse male germ cells resulting in sterility (27). Thus, there is a clear association between tissue-specific expression of Ddx4 and an unmethylated CpG island promoter region. Quantitative RT-PCR analysis and interrogation of the GNF database (26) of other genes associated with TDMs (Fig. 5 and Table 2) indicated that 5′ promoter methylation is frequently associated with silencing of gene expression. Although Pst6 was located in a 3′ exon, MSP indicated that a CpG island ≈500 bp upstream of the promoter for Hspa1l was also unmethylated in testis and methylated in the other tissues (data not shown). Quantitative RT-PCR (Fig. 5 and Table 2) and the GNF database (26) (data not shown) indicated essentially exclusive expression of Hspa1l in testis. The gene (4930548G07Rik) associated with the promoter region of Pvu29 is expressed at a relatively high level in testis (Fig. 5 and Table 2), although the GNF database (26) indicates expression in embryonic tissues as well. Pst61, which is located in a promoter CpG island associated with Gata2 and is methylated only in liver (Figs. 2 and 3) had the lowest level of expression in liver [GNF database (26)] (Fig. 5 and Table 2). Pvu2 and Pvu66 are located within nonpromoter region exons, and there is no apparent association between DNA methylation and tissue-specific silencing of genes associated with these TDMs. In fact, methylation of Pvu66 in liver is coupled with a higher level of expression of Adra1b (Fig. 5 and Table 2). These results suggest that tissue-specific DNA methylation in CpG island promoter regions is associated with tissue-specific gene repression. Fig. 5 shows the tissue-specific expression level of each of the genes relative to expression in liver [ΔΔCt = ΔCt - ΔCt(liver)], clearly indicating the testis-specific expression of genes associated with Pst3, Pst6, and Pvu29.
Fig. 5.
Graphical representation (SYBR Green RT-PCR) of tissue expression of genes associated with TDMs relative to expression in liver. Expression levels were standardized to GAPDH by calculating ΔCt (ΔCt = Gene Ct - GAPDH Ct). The mean ΔΔCt values were determined by subtracting the ΔCt for liver expression from the ΔCt of each of the other tissues. For most genes, liver expression was low or was the lowest of the tissues (e.g., Pst3 and Pst61).
TDMs. Based on RLGS analysis of six tissues, our results indicate that about 5% of the RLGS loci are TDMs. Previous RLGS analysis of mouse stem cells before and after differentiation, as well as somatic tissues (brain, kidney, placenta, and sperm) indicated that ≈16% of the RLGS loci exhibit differences (13). However these studies did not identify the DNA sequence of the RLGS loci or establish any association with gene expression. Strichman-Almashanu et al. (28) identified ≈40 CpG islands that are normally methylated in the human genome. It is presently not known whether any of these regions are associated with tissue-specific gene expression. Recently, Hattori et al. (29) identified 236 loci by virtual RLGS that are demethylated in Dnmt3a-/-,Dnmt3b-/- ES cells. Two of 40 loci that were further confirmed by methylation-sensitive real-time PCR correspond to TDM loci identified by virtual RLGS in our study (Pst13 and Pst42; see Table 4).
The results presented here indicate that the majority of the TDMs appear to be associated with 5′ promoter CpG islands and may have important roles in establishing or maintaining gene silencing during or after tissue differentiation. Other studies also support a role for DNA methylation in tissue-specific gene expression. Recent results indicate that the methylation of the maspin (SERPINB5) gene promoter contributes to the cell-type-specific expression pattern (30). Methylation of a critical CpG site within a signal transducer and activator of transcription-3-binding element of the glial fibrillary acidic protein promoter regulates expression in developing brain (31). Also, the promoter regions of a number of germ-line-specific genes, such as MAGE-type genes, are densely methylated in all tissues except testis, in which they are expressed (32). It is also possible that tissue-specific domains of active or inactive chromatin include several genes.
It is important that future studies establish the timing of DNA methylation at TDMs relative to alterations in gene expression during development and tissue differentiation. An unmethylated promoter region may be necessary but not sufficient for gene expression. Also, it is possible that silencing precedes DNA methylation. Although our results indicate that tissue-specific DNA methylation is associated with gene expression patterns, it is quite possible that there are other mechanistic consequences. In this context, it will be important to determine the significance of the nonpromoter TDMs. Almost half of the differentially methylated regions are not associated with 5′ promoter CpG islands and may have novel effects on genome structure and gene regulation. Preliminary analysis does not link the TDMs to any common repetitive sequence. Additional bioinformatics analysis of the sequence and genomic features of these regions should help to elucidate the critical roles of DNA methylation in development and disease.
Supplementary Material
Acknowledgments
We thank Michael Higgins and Dominic Smiraglia for critical comments on the manuscript. This research was supported by National Cancer Institute Grant CA102423 (to W.A.H.) and National Cancer Institute Core Center Grant CA16056 (to Roswell Park Cancer Institute).
Author contributions: S.F., J.F.S., M.T.K., H.N., and W.A.H. designed research; S.F., J.F.S., M.T.K., and A.D.M. performed research; T.M. and H.N. contributed new reagents/analytic tools; S.F., J.F.S., M.T.K., H.N., and W.A.H. analyzed data; and W.A.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TDM, tissue-specific differentially methylated region; RLGS, restriction landmark genomic scanning; MSP, methylation-specific PCR; Ct, cycle time; GNF, Genomics Institute of Novartis Research Foundation.
References
- 1.Bird, A. P. & Wolffe, A. P. (1999) Cell 99, 451-454. [DOI] [PubMed] [Google Scholar]
- 2.Norris, D. P., Brockdorff, N. & Rastan, S. (1991) Mamm. Genome 1, 78-83. [DOI] [PubMed] [Google Scholar]
- 3.Tilghman, S. M. (1999) Cell 96, 185-193. [DOI] [PubMed] [Google Scholar]
- 4.Li, E. (2002) Nat. Rev. Genet. 3, 662-673. [DOI] [PubMed] [Google Scholar]
- 5.Riggs, A. D. (1975) Cytogenet. Cell Genet. 14, 9-25. [DOI] [PubMed] [Google Scholar]
- 6.Holliday, R. & Pugh, J. E. (1975) Science 187, 226-232. [PubMed] [Google Scholar]
- 7.Warnecke, P. M. & Clark, S. J. (1999) Mol. Cell. Biol. 19, 164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh, C. P. & Bestor, T. H. (1999) Genes Dev. 13, 26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatada, I., Hayashizaki, Y., Hirotsune, S., Komatsubara, H. & Mukai, T. (1991) Proc. Natl. Acad. Sci. USA 88, 9523-9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama, T., Kimura, M. T., Koike, K., Abe, T., Nakao, T., Asami, T., Ebisuzaki, T., Held, W. A., Yoshida, S. & Nagase, H. (2003) Nucleic Acids Res. 31, 4490-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashizaki, Y., Hirotsune, S., Okazaki, Y., Hatada, I., Shibata, H., Kawai, J., Hirose, K., Watanabe, S., Fushiki, S., Wada, S., et al. (1993) Electrophoresis 14, 251-258. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe, S., Kawai, J., Hirotsune, S., Suzuki, H., Hirose, K., Taga, C., Ozawa, N., Fushiki, S. & Hayashizaki, Y. (1995) Electrophoresis 16, 218-226. [DOI] [PubMed] [Google Scholar]
- 13.Shiota, K., Kogok, Y., Ohgane, J., Imamura, T., Urano, A., Nishino, K., Tanaka, S. & Hattori, N. (2002) Genes Cells 7, 961-969. [DOI] [PubMed] [Google Scholar]
- 14.Shiota, K. (2004) Cytogenet. Genome Res. 105, 325-334. [DOI] [PubMed] [Google Scholar]
- 15.Hayashizaki, Y., Watanabe, S. (1997) (eds), in Restriction Landmark Genomic Scanning (RLGS) (Springer, Tokyo).
- 16.Chomczynski, P. (1993) BioTechniques 15, 532-537. [PubMed] [Google Scholar]
- 17.Sambrook, J. & Russel, D.W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 1, 3rd Ed.
- 18.Li, L. C. & Dahiya, R. (2002) Bioinformatics 18, 1427-1431. [DOI] [PubMed] [Google Scholar]
- 19.Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D. & Baylin, S. B. (1996) Proc. Natl. Sci. Acad. USA 93, 9821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawetz, S. & Misener, S., eds. Bioinformatics Methods and Protocols: Methods in Molecular Biology (Humana Press, Totowa, NJ), pp. 365-386.
- 21.Fazzari, M. J. & Greally, J. M. (2004) Nat. Rev. Genet. 5, 446-455. [DOI] [PubMed] [Google Scholar]
- 22.Costello, J. F., Fruhwald, M. C., Smiraglia, D. J., Rush, L. J., Robertson, G. P., Gao, X., Wright, F. A., Feramisco, J. D., Peltomaki, P., Lang, J. C. (2000) Nat. Genet. 24, 132-138. [DOI] [PubMed] [Google Scholar]
- 23.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 24.Frommer, M., McDonald, L., Millar, D., Collis, C., Watt, F., Grigg, G., Molloy, P. & Paul, C. (1992) Proc. Natl. Acad. Sci. USA 89, 1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herman, J. G. & Baylin, S. B. (2003) N. Engl. J. Med. 349, 2042-2054. [DOI] [PubMed] [Google Scholar]
- 26.Su, A. I., Cooke, M. P. Ching, K. A., Harkak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka, S. S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R. & Yokoyama, M. (2000) Genes Dev. 14, 841-853. [PMC free article] [PubMed] [Google Scholar]
- 28.Strichman-Almashanu, L. Z., Lee, R. S., Onyango, P. O., Perlman, E., Flam, F., Frieman, M. B. & Feinberg, A. P. (2002) Genome Res. 12, 543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hattori, N., Abe, T., Hattori, N, Suzuki, M., Matsuyama, T., Yoshida, S., Li, E. & Shiota, K. (2004) Genome Res. 14, 1733-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futscher, B. W., Oshiro, M. M., Wozniak, R. J., Holtan, N., Hanigan, C. L., Duan, H. & Domann, F. E. (2002) Nat. Genet. 31, 175-179. [DOI] [PubMed] [Google Scholar]
- 31.Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yangisawa, M., Fujita, N., Nakao, M. & Taga, T. (2001) Dev. Cell 1, 749-758. [DOI] [PubMed] [Google Scholar]
- 32.De Smet, C., Lurquin, C., Lethe, B., Martelange, V. & Boon, T. (1999) Mol. Cell. Biol. 19, 7327-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.