Abstract
DNA double-strand breaks (DSBs) are dangerous lesions that if not properly repaired can lead to genomic change or cell death. Organisms have developed several pathways and have many factors devoted to repairing DSBs, which broadly occur by homologous recombination that relies on an identical or homologous sequence to template repair, or nonhomologous end-joining. Much of our understanding of these repair mechanisms has come from the study of induced DNA cleavage by site-specific endonucleases. In addition to their biological role, these cellular pathways can be co-opted for gene editing to study gene function or for gene therapy or other applications. While the first gene editing experiments were done more than 20 years ago, the recent discovery of RNA-guided endonucleases has simplified approaches developed over the years to make gene editing an approach that is available to the entire biomedical research community. Here, we review DSB repair mechanisms and site-specific cleavage systems that have provided insight into these mechanisms and led to the current gene editing revolution.
Keywords: Double-strand break repair, homologous recombination, homology-directed repair, nonhomologous end-joining, gene editing, CRISPR-Cas9, HO endonuclease, I-SceI endonuclease
The most deleterious form of DNA damage is a double-strand break (DSB), whose repair is essential to maintain genome integrity. DSBs are repaired primarily by two classes of mechanisms, homologous recombination (HR), sometimes called homology-directed repair, and nonhomologous end-joining (NHEJ). We owe much of our understanding of these repair mechanisms to the study of induced DNA cleavage by site-specific endonucleases. More recently, the development of a number of different site-specific cleavage systems, in particular CRISPR-Cas9, has made it possible to efficiently modify the genome of virtually any organism. This “democratization” of gene editing is revolutionizing both the basic and biomedical sciences. The use of site-specific endonucleases for DSB repair studies and gene editing is the focus of this review.
Homologous recombination: DSB repair and gene targeting
Gene targeting
Until recently, HR between chromosomal DNA and linear DNA fragments introduced into cells - gene targeting - had been the major approach for modifying genomes (Fig. 1A). Budding yeast is remarkable in that DNA introduced into cells will almost always integrate by HR [1, 2], leading it to become a favored organism for reverse genetic studies. In mammalian cells and other organisms, transfected DNA typically integrates at a nonhomologous genomic location, such that extensive screening or selection tricks have been necessary to isolate gene-targeted clones [3–5]. However, when gene targeting is successful in mouse embryonic stem cells, these targeted cells can be used to generate precisely modified animals [6].
Figure 1.
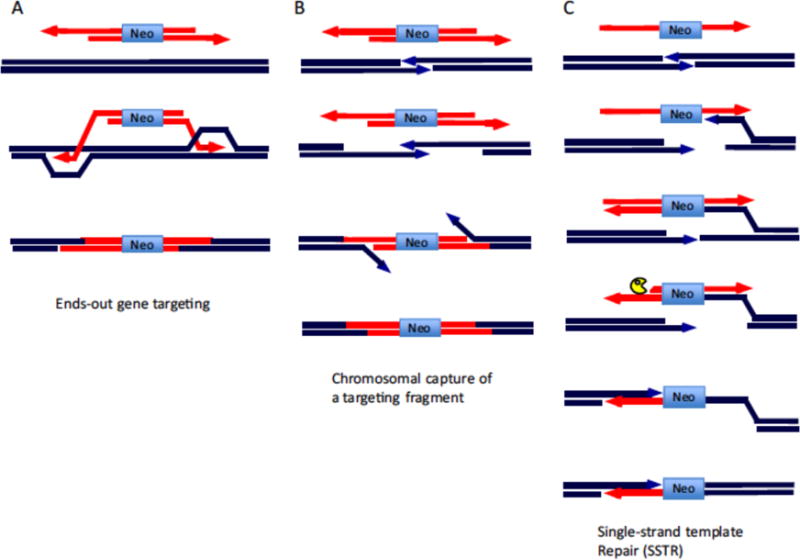
Gene targeting. A. Ends-out gene targeting. A DNA fragment carrying a selectable marker (neo) recombines by homologous recombination with an intact chromosome. B. A DSB introduced into the target site on a chromosome can incorporate a linear DNA fragment by annealing of 5′ to 3′ resected ends of both the DSB and the fragment. Incorporation of the fragment often proceeds by gene conversion as illustrated in Fig. 2A. C. Single-strand template repair of a chromosomal DSB.
The key discovery which guided the study of eukaryotic DSB repair by HR was carried out by Orr-Weaver et al. using budding yeast [7]. They observed that a plasmid with two distinct regions of homology to the yeast genome could integrate at either locus, but making a DSB within one of the two regions on the plasmid targeted integration at its homologous site in the genome. Subsequently, it became evident in yeast that a site-specific DSB created in a chromosome by a rare-cutting endonuclease would trigger repair HR with another genomic locus [8–10]. Further, in mammalian cells a DSB in the genome was found to trigger HR with introduced plasmid DNA and as well to induce NHEJ [11], a paradigm for current gene editing approaches.
HR as a DSB repair mechanism
DSB repair by HR in mitotic cells is necessary for the repair of spontaneously arising-lesions, for example those arising during DNA replication [12], given the slow growth or lethality of cells/mice in which HR genes are mutated [13]. Repair of a DSB repair by HR requires that the ends of the broken chromosome be able to invade into, and use an intact copy of the same (or similar) sequences as a template. In mitotic cells the repair template most typically is the sister chromatid, but the homologous chromosome or an ectopically located repeated sequence can also be used.
HR can occur by several pathways [14]. In a simple gene conversion event, a small amount of DNA is copied from the template to produce a short patch of new DNA at the repaired site (Fig. 2A,B). HR repair by gene conversion is generally regarded as the most precise type DSB of repair, although it is not completely error-free. In budding yeast mutations arise during gene conversion 1000 times more often than during replication of the same sequences [15]. Most noncrossover gene conversion events arise from a synthesis-dependent strand annealing mechanism (Fig. 2A), whereas an alternative pathway results in the formation of a pair of Holliday junctions that can be resolved with or without crossing-over [2](Fig. 2B) where the short patch of new DNA is flanked by sequences from the two recombining molecules. While such crossovers are genetically silent if they involve the identical sequence on the sister chromatid (sister chromatid exchange), crossovers involving a homologous chromosome (homolog) as template can lead to loss of heterozygosity (LOH), which can reveal deleterious recessive alleles [16]. Further, crossing-over involving an ectopically-located repeated sequence (non-allelic HR, or NAHR) can lead to genome rearrangements, including deletions, duplications, and translocations. Then, it is not surprising that in mitotic cells (the focus of this review) crossing-over is typically suppressed. Sequence divergence between repeats also suppresses HR (e.g., [17, 18]).
Figure 2.
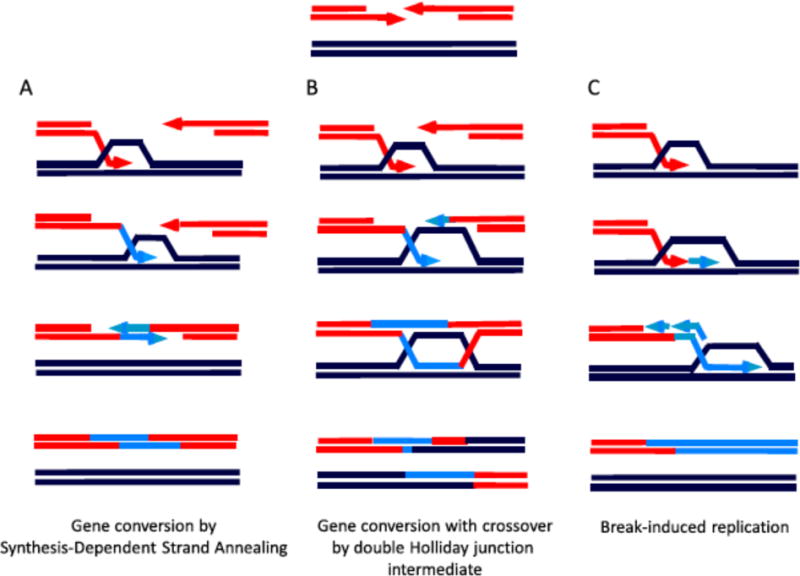
Homologous recombination of chromosomal DSBs. A. Noncrossover gene conversion by synthesis-dependent strand annealing (SDSA). B. Crossover-associated gene conversion through a double Holliday junction (dHJ) intermediate. The dissolution of the dHJ intermediate can also lead to a noncrossover outcome. C. Break-induced replication occurs when only one end of a DSB shares homology with a genomic template.
BIR
An alternative mechanism is break-induced replication (BIR) (Fig. 2C), which is a recombination-dependent DNA replication process in which one end of the DSB locates a template that can then be copied all the way to the telomere (typical in yeast) or, presumably, until the BIR replication bubble encounters a converging replication fork. In mammalian cells BIR is typically completed by NHEJ [19, 20]. Although BIR involves the participation of all three major DNA polymerases, this mode of replication is distinctly different from normal chromosomal replication. New DNA synthesis begins as primer extension from the 3′ end of the invading single strand, but only much later, when a long ssDNA has been displaced by a moving D-loop does the second strand get copied [21, 22] (Fig. 1C). BIR is prevented by deletion of the nonessential DNA polymerase δ subunit, Pol32 [23] or the 5′ to 3′ helicase Pif1, although an initial phase of DNA synthesis can occur [21]. The lack of tight coordination between leading and lagging strand synthesis may account for the high level of mutagenesis associated with BIR, especially mutations that would be expected to occur in newly copied single-stranded regions (e.g., cytosine deamination) [24]. In addition, the moving replication bubble appears to be highly unstable, allowing remarkably frequent template jumps, either between homologous chromosomes [25] or between nonallelic sites [26].
Meiotic recombination
Unlike DSB repair in most mitotic cells, DSB repair in meiotic cells involves the repair of programmed DSBs, typically using the homolog as a repair template, and with crossing over as a critical outcome [27, 28]. More than 100 DSBs are purposely introduced into the genome during meiosis by the SPO11-TOPOVIBL transesterase [29]. HR between homologous chromosomes is initially necessary for homolog pairing; crossing over is critical later for homolog segregation at the first meiotic division. Meiotic DSBs typically occur in promoter regions in yeast [30], whereas in mammals they are directed toward specific sequences determined by the PRDM9 protein [31] which recognizes DNA through a variable number of different zinc finger DNA binding domains [32]. In yeast, meiotic DSBs can be directed to new locations by fusing Spo11 to sequence-specific DNA binding domains, including zinc fingers [33, 34]. In mice, meiotic DSBs have been targeted to new locations by swapping the mouse PRDM9 zinc finger domain with that from the human [35].
Nonhomologous end-joining: canonical, alternative, and microhomology-mediated
The predominance of nonhomologous integration of transfected DNA in mammalian cells suggested the presence of relatively efficient “illegitimate” recombination pathways. NHEJ was first described as a DSB repair mechanism using assays for the recircularization of linearized plasmids, in which DNA ends are brought together without any homology or with a few nucleotides of homology, termed microhomology [36]. Similar joining events are also observed in yeast [37]. NHEJ was further recognized for its role in antigen receptor rearrangement in the immune system when gene segments incurring RAG recombinase-induced DSBs are brought together [38]. Later, NHEJ involvement in class switch recombination was also uncovered [39].
The canonical NHEJ factors KU70/80, DNA ligase IV/XRCC4/XLF, DNA-PKcs were initially identified using mammalian cell lines and mice that were both defective in antigen receptor rearrangement and sensitive to X-rays [40–43], and new factors continue to be discovered [44]. Most of these components were subsequently identified in budding and fission yeasts as well [45, 46]. The conserved Mre11 complex also plays a critical role in many NHEJ outcomes because of its DNA end tethering activity [47, 48]. Although it is not absolutely required for antigen receptor rearrangement, most likely because the RAG recombinase is able to hold ends together for repair [49], tethering may assist in that process as well [50].
In mammalian cells, NHEJ is important for the repair of spontaneously-arising and DNA damaging-agent induced DSBs throughout the cell cycle, whereas HR is more restricted because it is disfavored in G1 [51]. By contrast in budding yeast, HR genes such as Rad51 (Radiation) contribute most of the X-ray resistance, while NHEJ plays only a minor role unless an HR gene is already mutated. However, in HR-deficient yeast, canonical NHEJ factors become critical for cell survival even for a single DSB [47, 52].
NHEJ does not always require canonical NHEJ factors, in which case it is called alternative NHEJ (alt-NHEJ) or microhomology-mediated end joining (MMEJ), although the physiologic relevance of alt-NHEJ/MMEJ is not clear [53]. While these pathways are typically observed in the absence of canonical NHEJ factors, e.g., in plasmid recircularization in the absence of Ku [54–56], they can also be observed in the presence of canonical factors, e.g., chromosomal translocations in mouse cells [57]. Breakpoint junctions often appear to have arisen by annealing of complementary strands of DSB ends at microhomologies [58, 59]. It is difficult to determine whether any particular junction has arisen by canonical NHEJ or MMEJ, since the DNA end structure (e.g., long overhangs) and precise DNA sequences near ends (e.g., microhomology of several nucleotides) can impact whether microhomology is used, even by the canonical NHEJ pathway [57, 60, 61].
A number of factors have been proposed to be involved in alt-NHEJ, and MMEJ in particular, in mammalian cells, in particular including DNA ligase III and DNA polymerase theta ([62, 63] and references therein), both of which are lacking in budding or fission yeast. In budding yeast, only a fraction of MMEJ events are DNA ligase IV-dependent, so DNA ligase I almost certainly can serve as back-up. DNA ligase I has also been shown to play a minor role in mammalian alt-NHEJ [62].
Meganucleases used to study DSB repair
HO
Two endonucleases derived from budding yeast have been particularly important for DSB repair studies: HO and I-SceI. The normal role of HO endonuclease is to create a DSB that catalyzes yeast mating-type (MAT) gene switching, from MATa to MATα or vice versa [8] as often as every cell division (reviewed in [64]). Once a MATa cell conjugates with MATα, the expression of co-repressors expressed by these two loci turns off the switching process by repressing expression of the HO endonuclease. MATa differs from MATα by the presence of different ~ 700 bp Ya or Yα sequences; conversion from one to the other involves the induction of a site-specific DSB by the HO endonuclease at a 22-bp degenerate cleavage site that creates a DSB just adjacent to either Ya or Yα, leading to the removal of the Y sequence at MAT and its replacement by gene conversion from one of two templates, HMLα or HMRa, located near the two ends of the same chromosome containing MAT. HML and HMR are maintained in a transcriptionally silent state by the Sir2 histone deacetylase complex that creates a highly positioned array of nucleosomes that prevent HO endonuclease from cleaving either donor locus.
I-Scel
The I-SceI endonuclease is encoded within a self-splicing intron found in the mitochondrial rDNA locus (termed ω+). In perhaps the first example of gene drive I-SceI creates a DSB within an intronless rDNA gene (ω−), encountered after conjugation, leading to the insertion of the intron encoding the endonuclease ([9, 10]). Thus, the ω+ intron encodes I-SceI that cleaves the ω− gene at the precise position at which the intron is to be inserted; the DSB induces gene conversion to copy the intron from the ω+ gene into the ω− gene such that it now becomes ω+ as well. This system is so efficient that when conjugation is initiated between equal populations of ω+ and ω− cells, 99% of the population will become ω− within a few generations. I-SceI recognizes a well-defined 18 bp site [65]; since there are no perfect matches to this site in mammalian cells, its constitutive expression is not toxic [66].
The use of I-SceI depended upon converting the coding sequence from its mitochondrial origin (where UGA encodes tryptophan) to a nuclear/cytoplasmic-compatible sequence (where UGA would be a stop codon) [67]. The adoption of I-SceI for DSB repair studies - especially in early mammalian cell work - was facilitated by the commercial availability of the enzyme. Although crude HO assays were developed [68], the enzyme was never purified or commercially exploited. Like HO, I-SceI can catalyze efficient gene conversion in the nucleus of yeast [69].
VDE
Another less used yeast endonuclease is VDE (also termed PI-SceI), which arises from an intein present in the nuclear genome in the first known example of protein splicing [70]. The VMA1 gene encodes VDE, which will create a DSB during meiosis within an inteinless version of the gene containing a VDE recognition site on the homologous chromosome. Repair of the DSB by HR leads to insertion of the intein with high efficiency [71]. I-SceI and VDE are both considered “homing” endonucleases because they cleave a genomic site that does not encode them to induce copying of their coding sequence into the site.
The catalytic motifs of HO, I-SceI, and VDE are related, such that each gives rise to a 4-base 3′ overhang [72, 73]. The sequences of the recognition sites are quite distinct, however, although each is asymmetric and long, giving rise to the term “meganuclease”. Meganucleases are not related to the restriction enzymes that are involved in bacterial immunity.
Meganuclease I-AniI and I-PpoI and restriction enzyme AsiSI
Three other nucleases have been used in DSB repair studies: two meganucleases involved in intron homing, I-AniI (from Apergillis nidulans) [74] and I-PpoI (from Physarum polycephalum) [75], and restriction enzyme AsiSI (from Arthrobacter) [76]. I-PpoI cleaves 28S rDNA as well as a number of other sites in the human genome due to its degenerate site and has been used for chromatin immunoprecipitation studies to assess protein dynamics at a number of DSBs [75, 77]. Given its 8-bp recognition site, AsiSI has an even larger number of sites but it is blocked by CpG methylation and is inducibly expressed [76]; the large number of sites provides the opportunity for genome-wide analysis, for example, leading to the identification of chromatin domains that affect DSB repair [78].
Mechanistic studies of DSB repair
In budding yeast, the creation in yeast of a galactose-inducible version of HO [79] made it possible to initiate HO cleavage synchronously and led to the first physical monitoring of a DSB repair event [80]. HO cleavage is nearly complete within 20–30 min but repair takes >1 hr. During this interval it is possible to establish a sequence of slow events that culminate in HR repair, which relies on the Rad51 recombination protein and its auxiliary protein partners [15, 81–83]. The key steps in gene conversion (Fig. 1A,B) include the regulation of 5′ to 3′ resection of DSB ends to create 3′-ended single-stranded DNA (ssDNA) tails that are bound by Rad51, the search for homology to locate a donor sequence for strand invasion, the initiation of new DNA synthesis, and the capture of the second DSB end by the newly-copied DNA.
Even with the same galactose-inducible promoter, I-SceI is less efficient than HO in creating synchronously-induced DSBs in yeast [69], so that most of the kinetic analysis of different aspects of DSB repair have utilized HO. Another advantage of HO is its intrinsic instability; it harbors an amino acid sequence that promotes its own degradation so that there is essentially no activity 30 min after transferring cells from galactose to glucose medium. I-SceI apparently persists long after induction is turned off. Nonetheless, I-SceI has had utility in yeast and has been indispensible for establishing gene editing [11] and elucidating HR mechanisms and factors in mammalian cells [16], including roles of the breast cancer suppressors BRCA1 and BRCA2 [84, 85].
Studies with HO and I-SceI have led to a number of insights, especially in delineating several HR pathways once strand invasion has initiated. In synthesis-dependent strand annealing, the newly synthesized strand can be displaced to “return” to the other DSB end, commonly leading to noncrossover gene conversion in which all the newly copied DNA is in the recipient [86]. Less frequent formation of a Holliday junction intermediate can lead to resolution by a number of resolvases giving rise to a crossover outcome [87]. Alternatively, strand invasion can lead to capture of the second DSB end to form a double Holliday junction intermediate. The two Holliday junctions can migrate towards each other to be “dissolved” by a topoisomerase, specifically the BLM-TOP3-RMI1–RMI2 complex in mammals [87, 88] and the Sgs1/Top3 complex in budding yeast [89]. By contrast, the double Holliday junction can be resolved to a crossover. In meiosis, where crossovers are essential to meiotic progression, specific factors stabilize double Holliday junctions [90].
The use of site-specific DSBs in budding yeast has revealed that NHEJ comprises a complex set of functions that have different requirements and different outcomes, depending on the assay. Because HO is rapidly degraded when its expression is turned off, it is possible to show that the efficient re-ligation of the 4-base overhangs that are generated is dependent on canonical NHEJ factors. When HO is continually expressed, only cells that create a mutated cleavage site are able to survive. There are at least three distinct canonical NHEJ pathways to repair this specific type of overhang [47]: perfect re-ligation of overhanging ends; mis-alignment and filling in of ends to create small, templated indels; and misalignment with deletion. In addition yeast, like mammals, can used alt-NHEJ/MMEJ [91, 92]. As in mammalian cells, the requirements for MMEJ have been explored by using HO-induced substrates that have short (<12 bp) repeats flanking the DSB. Joining of these sequences is distinct from the Rad52-dependent single-strand annealing process [93].
It is important to note that in cycling mammalian cells and yeast the relative use of HR and NHEJ is significantly different, despite similarities in many of the components and pathways: in yeast HR wins out over NHEJ in repairing a chromosomal DSB by about 10:1 [94], whereas in mouse cells, NHEJ is about twice as efficient as HR in repairing an I-SceI-generated break [11, 95].
HR vs NHEJ pathway choice and the importance of resection
A major determinant of whether HR will occur is cell cycle phase, as many HR factors are not expressed efficiently or active in G1/G0 cells and sister chromatids are lacking. Because canonical NHEJ is inefficient on substrates with long ssDNA tails [45, 96], the initial 5′ to 3′ resection step is a critical regulatory step. Several methods to assess the extent of 5′ to 3′ resection have shown that HO-induced DSBs are resected at a rate of about 4 kb/h [97]. If there is no donor sequence to allow HR repair, resection will continue at a constant rate for at least 24 h. For much of this time, the 3′-ended ssDNA is resistant to cleavage or degradation [98, 99]. This protection is dependent on the abundance of the single-strand DNA binding protein, RPA. In mammalian cells, resection appears to be restrained by a feedback mechanism involving RPA recruitment of a helicase (HELB) to ssDNA [100].
Several distinct resection “machines” come into play to process an HO-induced DSB. The initial resection is carried out by the Mre11 complex (Mre11-Rad50-Xrs2 or MRX), in conjunction with Sae2 [101]. Except when yeast cells are blocked in G2/M, deletion of these proteins reduces but does not stop resection; in G2/M deletion of Mre11 or Rad50 (and likely Xrs2) completely stops resection [102]; whereas removing Sae2 has less impact. In mammals, Xrs2 is replaced by NBS1 in the MRN complex and the Sae2 homolog, CtIP plays a decisive role in resection in cycling cells [103]. In budding yeast, extensive resection, beyond a few hundred bp, depends on two competing resection processes [97, 104, 105]. Exo1 likely removes single nucleotides as it progresses; its accessibility to a DSB end is impaired by yeast’s Ku70–Ku80 complex. The second pathway requires the Sgs1 complex (Sgs1-Rmi1-Top3) and the endonuclease function of the Dna2 protein, which also acts as a flap nuclease during DNA replication. Deletion of either Exo1 or Sgs1 slows resection to some extent, but a double sgs1Δ exo1Δ mutant is severely impaired in resection. Resection is also blocked when yeast cells are arrested in the G1 phase of the cell cycle, where the key cell division kinase, Cdk1, is inactive [106, 107]. This block can be overcome by creating a phosphomimetic mutation in the Sae2 protein (a target of Cdk1) or by removing Ku, allowing Exo1 to act, even though CDK is still “off” [108]. When resection of an HO DSB is blocked, nonhomologous end-joining rates go up [106].
Mammals use the same resection machines as yeast except that Xrs2 is replaced by NBS1 (MRN complex) and the Sae2 homolog is CtIP [103, 109, 110]. Inactivating CtIP more profoundly impairs resection than deleting Sae2 in yeast. It is not yet clear how the rate or extent of resection in mammals compares with that in yeast, but specific assays of this rate in vivo are being developed [111]. Several factors are at odds with each other with respect to the control of resection and pathway choice in mammalian cells [112, 113]. Most notably, BRCA1 promotes resection, while 53BP1 (and interacting proteins) inhibits resection [114, 115]. In cells lacking both proteins, resection occurs and HR is restored. Recently, BRCA1 has been found to be involved in a second control point in the cell cycle regulation of pathway choice, in this case involving PALB2, another breast cancer suppressor, which acts as a bridge between BRCA1 and BRCA2 [13]. The BRCA1-PALB2 interaction in G1 is suppressed by ubiquitylation involving the PALB2-interacting protein KEAP1 [116]. Thus, HR can be activated in a tumor cell line in G1 if resection is activated by a CtIP phosphomimetic combined with 53BP1 loss, and the BRCA1–PALB2 interaction is restored by blocking ubiquitylation to promote BRCA2 and hence RAD51 recruitment. HELB regulates both EXO1 and BLM-DNA2-dependent resection [100].
Gene editing
The paradigm for gene editing - modification of the genome through repair of a chromosomal DSB - was developed using the I-SceI meganuclease. The I-SceI recognition site was integrated into the mouse genome and DNA fragments homologous to sequences flanking the DSB were introduced together with an I-SceI expression vector [11]. HR of the chromosome with the plasmid fragments was elevated several orders of magnitude, indicating that the introduction of a chromosomal DSB is a viable way to increase gene targeting in organisms refractory to spontaneous gene targeting [117]. Subsequent studies confirmed gene editing by HR in embryonic stem cells and other cell types [118–122]. Alternatively, DNA ends generated by I-SceI were rejoined by NHEJ leading to small indels at the DSB site [11]. Precise joining events between two DSBs could also be detected, in this case leading to a several kb deletion. Two I-Scel-generated DSBs were also found to give rise to translocations [123, 124]
In yeast, a chromosomal DSB was also found to induce gene editing by NHEJ, both in HR-deficient cells [37] and wild-type cells [47]. Not surprisingly, a DSB in the chromosome in yeast was also found to induce gene editing by HR [125, 126]. In HR events, an I-SceI-generated DSB could be efficiently repaired by small double-stranded oligonucleotides sharing fewer than 50 bp homology on either side of a DSB or even a pair of DSBs separated by many kb of DNA. Repair could occur by gene conversion with the introduced fragment as the donor (Fig. 2A) but could also result from the annealing of complementary single strands created by 5′ to 3′ resection of the ends of the DSB and of the fragment (Fig. 1B). More surprising, an I-SceI-induced DSB could be repaired by a single-stranded oligonucleotide that was complementary to the resected end of one side of the DSB [125]. Such single-strand template repair requires several additional steps (Fig. 1C): first, DNA synthesis from the 3′end of one side of the DSB would create a dsDNA intermediate, and second, resection would then create an end complementary to the second, resected DSB end. Of particular note, suppressing strand invasion by RAD51 mutation, increased gene editing, in keeping with a mechanism that involved annealing of single strands. The approach was modified further by tethering the oligonucleotide directly to I-SceI, leading to an increase in HR which was also observed in mammalian cells [127].
RNA-templated DNA repair
An even more provocative finding was that single-stranded RNA could be used as a genetargeting template, albeit inefficiently compared to DNA [128]. These findings, made first with yeast, have been verified in mammalian cells [129]. More recently, endogenous RNA transcripts have been found to participate in HR repair of an HO-induced DSB, suggesting that endogenous RNAs may participate as templates in genome modification, possibly even in non-dividing cells [130]. One model is that in actively transcribing DNA nascent RNA generated just prior to DSB formation can be used as a template for reverse transcriptase-dependent extension of broken DNA ends.
An emerging range of endonucleases
Meganucleases
Experiments with I-SceI endonuclease made it clear that gene editing could be readily achieved by efficient introduction of a DSB at a target locus [117]; however, engineering meganucleases like I-SceI to recognize other DNA sequences is challenging, given the complex interactions with DNA. For example, I-SceI makes 56 contacts with DNA, 27 of which are with base pairs in the recognition site [131]. Despite these challenges, progress has been made in redesigning meganuclease like I-SceI [132–134]. Although unlikely to be developed as generalized cleavage reagents for research purposes, meganucleases are small and single chain (I-SceI is only 235 amino acids) which facilitates their delivery to cells. Thus, for particular applications, for example in gene therapy or biotechnology, engineering a meganuclease could still provide advantages [135].
ZFNs
Because its cleavage domain is distinct from its DNA binding domain, the restriction enzyme FokI provided an entry point for creating endonucleases with alternative cleavage specificities [136, 137]. To create a chimeric nuclease, the FokI cleavage domain was first successfully fused to a homeobox DNA binding domain [138]. However, the modularity of zinc finger DNA binding domains clearly presented a more flexible route to engineer endonucleases with desired recognition specificities, leading to the creation of the first zinc finger nuclease (ZFN) [139]. In its simplest form each zinc finger interacts with three base pairs [140–142], such that three or more “fingers” recognizing distinct triplets could be assembled and fused to two FokI dimeric cleavage domains to cleave a unique site in a complex genome [143]. Although initially created to expand the repertoire of restriction endonucleases, the use of ZFNs for gene editing was readily apparent, given the success of the I-SceI. Gene editing with a ZFN was first reported at an endogenous locus in Drosophila at the scoreable yellow gene [144]. Gene editing of an endogenous gene using a homologous plasmid for HR was first accomplished at the diseaserelevant IL-2Rγ gene in human cells [145]. What was particularly remarkable from the Urnov et al study was the bi-allelic targeting could be achieved and at relatively high frequency.
TALENs
While ZFNs were successfully used in a number of studies [143], the assembly of zinc finger modules that recognize DNA with high specificity is difficult because each zinc finger is not completely independent of its neighbors and zinc fingers to each of the 64 triplets are not readily available. The discovery of the simple DNA recognition code of TAL effector proteins from plant pathogens essentially solved this problem: two amino acids within each module recognize a single base pair and each module essentially binds independently [146–149]. As in ZFNs, fusions of TAL effector domains are made to two FokI cleavage domains to generate nucleases (TALENs) [150]. The discovery of the simple DNA binding code of TAL effectors revolutionized gene editing by making it broadly usable within the biomedical research community [151]. However, TALEN expression vectors may take several days or a week to construct and require the assembly of a number of repeats, which can be unstable when propagated in bacteria.
Democratization of genome editing: CRISPR-Cas9
In principle, Watson-Crick base pairing represents the simplest form of DNA sequence recognition that could be applied for generating cleavage reagents for gene editing [117]. Approaches developed in the 1990s to make use of base pairing included oligonucleotides with an incorporated chemical cleaving moiety coated with the bacterial strand exchange protein RecA [152]. However, little progress was made in adapting these approaches to in vivo settings.
The CRISPR-Cas9 nuclease provides the simple Watson-Crick base pairing code for DNA recognition, making it an ideal programmable nuclease, but with the twist of relying on RNA:DNA rather than DNA:DNA recognition [153, 154]. This simple design from nature, which is critical for bacterial adaptive immunity [155], has truly revolutionized gene editing. The approach relies on the Cas9 nuclease from Streptococcus pyogenes that recognizes a short NGG motif in the target DNA, together with a Cas9-bound guide RNA that forms 20 base pairs with the DNA sequences adjacent to the NGG motif. DNA cleavage occurs 3 bases upstream of the NGG motif on the guide RNA-bound DNA strand and typically 3 bases upstream on the non-bound strands to generate blunt-ended DSBs, although there can be some flexibility in cleavage on the non-bound strand.
Because of the simplicity of modifying a single RNA sequence, CRISPR-Cas9 was readily adapted to gene editing [156, 157] and has become the approach of choice for researchers for genome modification for many organisms and applications [158]. The principles established with other nucleases hold for Cas9, including the most fundamental feature that Cas-induced DSBs can be repaired by either HR or NHEJ. Importantly as well, Cas-induced DSBs can lead to efficient biallelic modification by either HR or NHEJ. NHEJ typically leads to a variety of indels, which is suitable for many applications requiring the gene knockouts. Although HR is typically less efficient, it is often more desirable because it can lead to a precise genome modification, including the knockin of fluorescent markers or loxP sites to make conditional alleles. The donor DNA for HR can be introduced from plasmids or as a single-stranded oligonucleotide [159]; the latter approach has recently been improved by taking advantage of the asymmetric release of one DNA end from the strand not bound to the RNA [160] and by using modified (phosphorothioate) single-stranded oligonucleotide which are more stable in cells [161]. Cas9 and the gRNA can be expressed in cells from expression vectors or the Cas9 mRNA and guide RNA can be directly transfected. Alternatively, or the Cas9-guide RNA complex can be directly delivered to cells [162].
The programmable 20 nucleotides in the guide RNA are long enough that Cas9 can be used to specify cleavage of complex genomes, the only requirement being the presence of an NGG motif. However, several modifications to the approach have already been undertaken. With Cas9, genomic sites that are not identical to the guide RNA sequence may be recognized at some frequency, leading to off-target cleavage, which is a concern for any of the nuclease platforms, given the efficiency at which indel formation can occur. To address this, Cas9 from S. pyogenes has recently been modified to reduce off-target cleavage while maintaining specific cleavage [163–165]. The modifications leading to enhanced, high fidelity cleavage are based on structural determinations of the Cas9 nuclease with guide RNA with the DNA target and the premise that reducing non-specific Cas9 contacts with the DNA backbone in the strand complementary to the guide RNA [163] or with the non-complementary strand in a basic groove at the interface of the nuclease domains [164] would force a greater reliance on correct RNA:DNA base pairing.
Cas9 binding to the NGG motif is critical to its initial interaction with DNA, however, the requirement for this motif, termed a PAM, restricts cleavage to sequences that contain a close by GG dinucleotide. This is not a burden in most gene editing experiments, however, it can be problematic when targeting AT-rich genomes or in HDR experiments, given that gene conversion tracts are often very short [166]. Investigators have approached this by developing S. pyogenes Cas9 mutants that have altered recognition motifs (PAMs) [167], for example, NGA [168]. This latter study also identified a Cas9 variant with enhanced specificity to the NGG motif. In addition to S. pyogenes Cas9, Staphylococcus aureus and other RNA-guided nucleases are being investigated with somewhat different properties, including recognition of different PAM motifs and generation of different overhangs [168–171].
As with ZFNs and TALENs, fusions with a catalytically inactive Cas9 have proved useful for non-gene editing purposes, including modulating endogenous gene transcription with activation or repressor domains or fluorescently labeling of endogenous loci [158]. A major advance possible with a guide RNA-based DNA recognition system but not protein-based systems is the ability to multiplex and for genome wide screens.
Nicking and Paired Nicking
Nicking
Gene editing by HR is often a more desirable approach than NHEJ because it leads to precise genome modification. However, even when a homologous template is provided, DSBs are efficiently repaired by NHEJ. One approach to specifically induce HR is to create a nick in the genome rather than a DSB since a nick is not a substrate for NHEJ [156, 157, 172–175]. Meganucleases, ZFNs, and Cas9 have been successfully converted into nickases by mutating catalytic residues. I-SceI is a monomer with two overlapping catalytic sites; mutation of either of two conserved, pseudosymmetric lysine residues adjacent to the catalytic aspartate residues modifies activity to result in nicking of opposite strands ([176]; see also, [177]). Another monomeric meganuclease, I-AniI, has also been converted into a nickase [74]. ZFNs have two FokI cleavage domains that dimerize; mutation of the catalytic site of one of the domains converts it into a nickase [173, 174]. Although TALEN-based nickases have been reported [178], the two FokI cleavage domains may have more flexibility than in ZFNs, such that it may be difficult to prevent DSB formation. Finally, Cas9 contains two distinct nuclease domains, such that mutation of the catalytic site in either domain converts it to a nickase [153, 154].
Because nicks can be readily sealed, they are typically not as robust as DSBs for inducing HR [74, 173, 174, 179–181]. Nickases have been used in a number of studies to determine mechanisms of nick-induced HR. Genetic differences between the results induced by a DSB and a nick suggest that nicks are recombinogenic without being converted to DSBs by replication [180, 181]. The canonical HR factors RAD51 and BRCA2 are required for nick-induced HR with an intact plasmid or chromosome sequence [179, 181], but unlike DSB-induced HR, NHEJ factors do not suppress nick-induced HR [181]. Interestingly, an alternative HR pathway has been described in nick repair in which canonical HR factors actually suppress HR [179]. This pathway appears to involve strand assimilation (annealing) rather than strand invasion and can operate with single-stranded oligonucleotide donor DNA.
Paired nicking
Cas9 off-target cleavage is a concern in many gene-editing applications. In addition to using enhanced Cas9s, another route is to reducing off-target effects is to use nickases to create two nicks on opposite strands to generate a DSB [182, 183]. In the case of a Cas9 nickase, specificity is increased because of the requirement for two guide RNAs to bind in close proximity. Interestingly, the distance between nicks can be hundreds of base pairs [181–183], indicating efficient unwinding or degradation of DNA between the nicks. The canonical NHEJ pathway appears to be involved when nicks are offset to generate 5′ overhangs [60, 181].
Prospects
Decades of research in DSB repair have provided the basis for current gene editing approaches. Gene editing was made feasible for the biomedical research community by the identification of the simple DNA recognition code of TAL effectors and the development of TALENs. However, within a couple of years of their development, TALENs were mostly supplanted by CRISPR-Cas9 for gene editing and other applications that rely on readily programmable DNA target site recognition. The simple RNA:DNA recognition code of Cas9 and other CRISPR nucleases makes it is difficult to imagine that as yet undiscovered systems will substantially advance gene editing approaches beyond what is currently achievable from CRISPR systems. Nonetheless, it remains possible that other discoveries in the future will impact genome modification approaches to further alter the course of biomedical research.
Acknowledgments
We thank the members of the Jasin lab at MSK and the Haber lab at Brandeis for their contributions to DSB repair research throughout the years, as well as our many colleagues in the field. Funding: R01 GM054668 to M.J.; NIH/NCI Cancer Center Support Grant P30 CA008748 to MSK. NIH grants R37 GM20056, R01 GM76020 and P01 GM105473 to J.E.H
Abbreviations
- DSB
double-strand break repair
- HR
homologous recombination
- HDR
homology-directed repair
- BIR
break-induced replication
- NHEJ
nonhomologous end-joining
- Alt-NHEJ
alternative-NHEJ
- MMEJ
microhomology-mediated end-joining
- ssDNA
single-stranded DNA
References
- 1.Hinnen A, Hicks JB, Fink GR. Transformation of yeast. Proc Natl Acad Sci U S A. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 3.Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 4.Jasin M, Berg P. Homologous integration in mammalian cells without target gene selection. Genes Dev. 1988;2:1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- 5.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal ß-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 6.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 7.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strathern JN, Klar AJ, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 9.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 10.Zinn AR, Butow RA. Kinetics and intermediates of yeast mitochondrial DNA recombination. Cold Spring Harb Symp Quant Biol. 1984;49:115–121. doi: 10.1101/sqb.1984.049.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 13.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci U S A. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson A, Hendrix M, Jinks-Robertson S, Crouse GF. Regulation of mitotic homeologous recombination in yeast. Functions of mismatch repair and nucleotide excision repair genes. Genetics. 2000;154:133–146. doi: 10.1093/genetics/154.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott B, Jasin M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol Cell Biol. 2001;21:2671–2682. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013 doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 24.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 26.Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28:2394–2406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 2010;24:1201–1207. doi: 10.1101/gad.1944710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Peterson SE, Jasin M, Keeney S. Mechanisms of germ line genome instability. Semin Cell Dev Biol. 2016 doi: 10.1016/j.semcdb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Robert T, Vrielynck N, Mezard C, de Massy B, Grelon M. A new light on the meiotic DSB catalytic complex. Semin Cell Dev Biol. 2016 doi: 10.1016/j.semcdb.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 30.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, Hochwagen A, Keeney S. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pecina A, Smith KN, Mezard C, Murakami H, Ohta K, Nicolas A. Targeted stimulation of meiotic recombination. Cell. 2002;111:173–184. doi: 10.1016/s0092-8674(02)01002-4. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas A. Modulating and targeting meiotic double-strand breaks in Saccharomyces cerevisiae. Methods Mol Biol. 2009;557:27–33. doi: 10.1007/978-1-59745-527-5_3. [DOI] [PubMed] [Google Scholar]
- 35.Davies B, Hatton E, Altemose N, Hussin JG, Pratto F, Zhang G, Hinch AG, Moralli D, Biggs D, Diaz R, Preece C, Li R, Bitoun E, Brick K, Green CM, Camerini-Otero RD, Myers SR, Donnelly P. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530:171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of doublestrand breaks in Saccharomyces cerevisiae are repaired by similar ftdD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth DB, Nakajima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 40.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 41.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Fulop GM, Phillips RA. The scid mutation in mice causes a general defect in DNA repair. Nature. 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 43.Waters CA, Strande NT, Wyatt DW, Pryor JM, Ramsden DA. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst) 2014;17:39–51. doi: 10.1016/j.dnarep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, Blundell TL, Jackson SP. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ. Evolutionary and functional conservation of the DNA nonhomologous end-joining protein, XLF/Cernunnos. J Biol Chem. 2006;281:37517–37526. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- 47.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 50.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siede W, Friedl AA, Dianova I, Eckardt SF, Friedberg EC. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics. 1996;142:91–102. doi: 10.1093/genetics/142.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 55.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 56.Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 60.Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colleaux L, D’Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colleaux L, d’Auriol L, Betermier M, Cottarel G, Jacquier A, Galibert F, Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986;44:521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 68.Kostriken R, Heffron F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- 69.Plessis A, Perrin A, Haber JE, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gimble FS, Thorner J. Purification and characterization of VDE, a site-specific endonuclease from the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:21844–21853. [PubMed] [Google Scholar]
- 71.Nogami S, Fukuda T, Nagai Y, Yabe S, Sugiura M, Mizutani R, Satow Y, Anraku Y, Ohya Y. Homing at an extragenic locus mediated by VDE (PI-SceI) in Saccharomyces cerevisiae. Yeast. 2002;19:773–782. doi: 10.1002/yea.872. [DOI] [PubMed] [Google Scholar]
- 72.Haber JE, Wolfe KH. Evolution and function of HO and VDE endoncucleases in fungi. In: Belfort B, Derbyshire V, Stodddard B, Wood D, editors. Homing Endonucleases and Inteins. Springer Verlag; 2005. pp. 161–175. [Google Scholar]
- 73.Dujon B. Homing Endonucleases and the Yeast Mitochondrial w Locus — A Historical Perspective. In: Belfort B, Derbyshire V, Stodddard B, Wood D, editors. Homing Endonucleases and Inteins. Springer Verlag; 2005. pp. 11–31. [Google Scholar]
- 74.McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ, Jr, Stoddard BL. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci U S A. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monnat RJ, Jr, Hackmann AF, Cantrell MA. Generation of highly site-specific DNA doublestrand breaks in human cells by the homing endonucleases I-PpoI and I-CreI. Biochem Biophys Res Commun. 1999;255:88–93. doi: 10.1006/bbrc.1999.0152. [DOI] [PubMed] [Google Scholar]
- 76.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berkovich E, Monnat RJ, Jr, Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc. 2008;3:915–922. doi: 10.1038/nprot.2008.54. [DOI] [PubMed] [Google Scholar]
- 78.Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, Legube G. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jensen R, Sprague GF, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983;80:3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Connolly B, White CI, Haber JE. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 83.Wolner B, van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 84.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 85.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 86.Ira G, Satory D, Haber JE. Conservative inheritance of newly synthesized DNA in doublestrand break-induced gene conversion. Mol Cell Biol. 2006;26:9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 89.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, Hoog C, Cohen PE, Hunter N. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet. 2013;45:269–278. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Villarreal DD, Lee K, Deem A, Shim EY, Malkova A, Lee SE. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012;8:e1003026. doi: 10.1371/journal.pgen.1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valencia-Burton M, Oki M, Johnson J, Seier TA, Kamakaka R, Haber JE. Different mating-type-regulated genes affect the DNA repair defects of Saccharomyces RAD51, RAD52 and RAD55 mutants. Genetics. 2006;174:41–55. doi: 10.1534/genetics.106.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 97.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends in biochemical sciences. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 98.Vaze M, Pellicioli A, Lee S, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber J. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires srs2 helicase. Mol Cell. 2002;10:373. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 99.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tkac J, Xu G, Adhikary H, Young JT, Gallo D, Escribano-Diaz C, Krietsch J, Orthwein A, Munro M, Sol W, Al-Hakim A, Lin ZY, Jonkers J, Borst P, Brown GW, Gingras AC, Rottenberg S, Masson JY, Durocher D. HELB Is a Feedback Inhibitor of DNA End Resection. Mol Cell. 2016;61:405–418. doi: 10.1016/j.molcel.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 101.Mimitou EP, Symington LS. Sae2, Exol and Sgsl collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- 103.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & development. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. Embo J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. The EMBO journal. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daley JM, Chiba T, Xue X, Niu H, Sung P. Multifaceted role of the Topo IIIalpha-RMI1–RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection. Nucleic Acids Res. 2014;42:11083–11091. doi: 10.1093/nar/gku803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou Y, Paull TT. Direct measurement of single-stranded DNA intermediates in mammalian cells by quantitative polymerase chain reaction. Anal Biochem. 2015;479:4850. doi: 10.1016/j.ab.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 112.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, Xia B, Peter M, Durocher D. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 118.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci U S A. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA doublestrand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 125.Storici F, Snipe JR, Chan GK, Gordenin DA, Resnick MA. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol Cell Biol. 2006 doi: 10.1128/MCB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific doublestrand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci U S A. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ruff P, Koh KD, Keskin H, Pai RB, Storici F. Aptamer-guided gene targeting in yeast and human cells. Nucleic Acids Res. 2014;42:e61. doi: 10.1093/nar/gku101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shen Y, Nandi P, Taylor MB, Stuckey S, Bhadsavle HP, Weiss B, Storici F. RNA-driven genetic changes in bacteria and in human cells. Mutat Res. 2011;717:91–98. doi: 10.1016/j.mrfmmm.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 130.Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J Mol Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 132.Takeuchi R, Choi M, Stoddard BL. Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc Natl Acad Sci U S A. 2014;111:4061–4066. doi: 10.1073/pnas.1321030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J Am Chem Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 134.Arnould S, Delenda C, Grizot S, Desseaux C, Paques F, Silva GH, Smith J. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel. 2011;24:27–31. doi: 10.1093/protein/gzq083. [DOI] [PubMed] [Google Scholar]
- 135.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc Natl Acad Sci U S A. 1992;89:4275–4279. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 138.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci U S A. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 142.Nardelli J, Gibson TJ, Vesque C, Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991;349:175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- 143.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 144.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 146.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 147.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 148.Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Doyle EL, Stoddard BL, Voytas DF, Bogdanove AJ. TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 2013;23:390–398. doi: 10.1016/j.tcb.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Baliga R, Singleton JW, Dervan PB. RecA.oligonucleotide filaments bind in the minor groove of double-stranded DNA. Proc Natl Acad Sci U S A. 1995;92:10393–10397. doi: 10.1073/pnas.92.22.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 156.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jiang W, Marraffini LA. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu Rev Microbiol. 2015;69:209–228. doi: 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- 159.Chen F, Pruett-Miller SM, Davis GD. Gene editing using ssODNs with engineered endonucleases. Methods Mol Biol. 2015;1239:251–265. doi: 10.1007/978-1-4939-1862-1_14. [DOI] [PubMed] [Google Scholar]
- 160.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 161.Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, Perrouault L, Tesson L, Edouard J, Thinard R, Cherifi Y, Menoret S, Fontaniere S, de Croze N, Fraichard A, Sohm F, Anegon I, Concordet JP, Giovannangeli C. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 162.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Urnov F. Genome editing: The domestication of Cas9. Nature. 2016;529:468–469. doi: 10.1038/529468a. [DOI] [PubMed] [Google Scholar]
- 166.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Anders C, Bargsten K, Jinek M. Structural Plasticity of PAM Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9. Mol Cell. 2016;61:895–902. doi: 10.1016/j.molcel.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee MJ, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Metzger MJ, McConnell-Smith A, Stoddard BL, Miller AD. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res. 2011;39:926–935. doi: 10.1093/nar/gkq826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, Cathomen T, Scharenberg AM, Joung JK. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, Holmes MC. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22:1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Munoz MC, Yanez DA, Stark JM. An RNF168 fragment defective for focal accumulation at DNA damage is proficient for inhibition of homologous recombination in BRCA1 deficient cells. Nucleic Acids Res. 2014;42:7720–7733. doi: 10.1093/nar/gku421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Niu Y, Tenney K, Li H, Gimble FS. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J Mol Biol. 2008;382:188–202. doi: 10.1016/j.jmb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chan SH, Stoddard BL, Xu SY. Natural and engineered nicking endonucleases-from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res. 2011;39:1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu Y, Gao T, Wang X, Hu Y, Hu X, Hu Z, Pang J, Li Z, Xue J, Feng M, Wu L, Liang D. TALE nickase mediates high efficient targeted transgene integration at the human multicopy ribosomal DNA locus. Biochem Biophys Res Commun. 2014;446:261–266. doi: 10.1016/j.bbrc.2014.02.099. [DOI] [PubMed] [Google Scholar]
- 179.Davis L, Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc Natl Acad Sci U S A. 2014;111:E924–932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Katz SS, Gimble FS, Storici F. To nick or not to nick: comparison of I-SceI single- and double-strand break-induced recombination in yeast and human cells. PLoS One. 2014;9:e88840. doi: 10.1371/journal.pone.0088840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Vriend LE, Prakash R, Chen CC, Vanoli F, Cavallo F, Zhang Y, Jasin M, Krawczyk PM. Distinct genetic control of homologous recombination repair of Cas9-induced doublestrand breaks, nicks and paired nicks. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]


