Abstract
The paramyxovirus family comprises major human and animal pathogens such as measles virus (MeV), mumps virus (MuV), the parainfluenzaviruses, Newcastle disease virus (NDV), and the highly pathogenic zoonotic hendra (HeV) and nipah (NiV) viruses. Paramyxovirus particles are pleomorphic, with a lipid envelope, nonsegmented RNA genomes of negative polarity, and densely packed glycoproteins on the virion surface. A number of crystal structures of different paramyxovirus proteins and protein fragments were solved, but the available information concerning overall virion organization remains limited. However, recent studies have reported cryo-electron tomography-based reconstructions of Sendai virus (SeV), MeV, NDV, and human parainfluenza virus type 3 (HPIV3) particles and a surface assessment of NiV-derived virus-like particles (VLPs), which have yielded innovative hypotheses concerning paramyxovirus particle assembly, budding, and organization. Following a summary of the current insight into paramyxovirus virion morphology, this review will focus on discussing the implications of these particle reconstructions on the present models of paramyxovirus assembly and infection.
Introduction
Together with the rhabdo-, filo-, borna- and pneumoviruses, the paramyxoviruses form the order mononegavirales that features enveloped virions with single-stranded, non-segmented RNA genomes of negative polarity. Common to all members of the paramyxovirus family are two membrane glycoprotein complexes, the attachment (H, HN, or G) and the fusion (F) proteins, that are responsible for receptor binding and cell entry through fusion of the viral envelope with target cell membranes, respectively [1] (Figure 1). The RNA genome is encapsidated by the viral nucleocapsid (N) protein, resulting in the formation of a helical ribonucleoprotein (RNP) complex that serves as the template for the viral RNA-dependent RNA-polymerase complex composed of the viral phospho- (P) and large (L) proteins. The matrix (M) protein organizes particle assembly through interaction with both N proteins in the RNP complex and the membrane-embedded glycoprotein complexes. Some members of the family such as pathogens of the rubulavirus genus contain a small hydrophobic (SH) transmembrane protein in addition to these six structural proteins. Only J paramyxovirus encode a fourth integral membrane protein, transmembrane (TM), that stimulates cell-to-cell fusion but not viral entry [2].
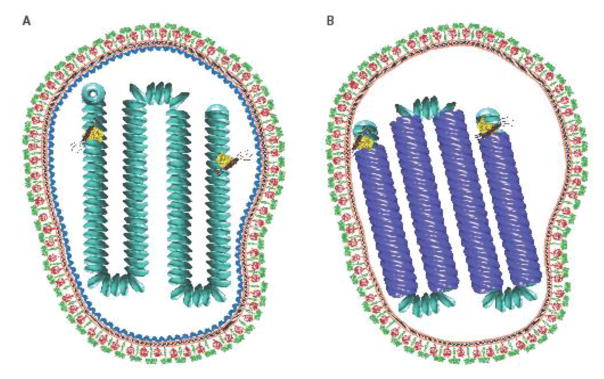
Figure 1.
A) Model of an MeV virion in which the matrix protein coats the nucleocapsid protein. The viral envelope is shown in orange. The nucleocapsid proteins are shown in cyan. The fusion protein trimers are shown in red. The attachment glycoprotein tetramers are shown as green. Matrix protein depicted is from Newcastle disease virus (PDB 4g1g). Viral glycoproteins are based on PIV5 (PDB: 4gip for the F protein; PDB ID: 4jf7 and 3tsi for the PIV5 HN ectodomain and stalk, respectively. The vesicular stomatitis virus L protein structure (yellow) was used to represent the unknown paramyxovirus L conformation (PDB ID: 5a22). The phosphoprotein (brown) was modeled using the oligomerization domain of measles P (PDB ID: 3zdo). B) A model of an MeV virion in which a matrix protein array is located at the inner leaflet of the viral membrane. The matrix protein is shown in blue. The matrix-coated nucleocapsids were created using Chimera [92] using electron density maps EMD-1973 and EMD-1974 [43]. PDB structures were created in PyMOL [93].
While electron microscopy has established a basic framework for the paramyxovirus virion organization [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23] and a number of crystal structures of paramyxovirus proteins and protein fragments have been solved (for instance [6,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42], the reconstruction of the 3D ultrastructures of paramyxovirus particles is impaired by particle size and the pleomorphic nature of the virions, which prevents single particle reconstruction approaches. To overcome the problem, recent studies have applied cryo-electron tomography (cryo-ET) to the analysis of paramyxovirus particles. By providing insight into the 3D structures of SeV, MeV, and NDV particles, and the NiV glycoprotein organization displayed on VLPs [6,25,43,44], this work has returned two highly unexpected new proposals concerning paramyxovirus particle assembly and organization: i) coating of the MeV RNPs by M protein tubes, which may spotlight an alternative particle assembly mechanism; and ii) an organized assembly of NiV F protein trimers into a hexameric ring-like assembly that may contribute to F conformation stability and concerted triggering for efficient viral entry.
Virion morphology and substructures
Overall particle morphology appears to vary considerably depending on the paramyxovirus genus investigated. For instance, tomograms of SeV, a member of the respirovirus genus that also includes the human parainfluenzaviruses type 1 and 3, showed predominantly spherical particles [44], while reconstructions of NDV virions, a member of the avulavirus genus, revealed shapes ranging from spherical to elongated ellipsoidal [6], and particles of the morbillivirus type species MeV, schematically shown in figure 1A, showed a multitude of different configurations [43]. Independent of the predominant particle shape and consistent with previous negative-stain EM-based visualization of paramyxoviruses, each of these studies spotlighted large variations in particle size, ranging from approximately 110–540 nm in diameter for SeV, 100–250 nm diameter for largely spherical NDV particles, and 50–510 nm in the case of MeV. In particular elongated to filamentous particles frequently exceed 500 nm in length, resembling in appearance the largely filamentous organization of respiratory syncytial virus particles of the related pneumovirus family [45].
All paramyxovirus RNP genomes show a characteristic herringbone-like structure when examined by negative-stain EM [46]. Reconstructions of the nucleocapsids revealed a left-handed helical arrangement with a pitch varying from 4.6 to 7 nm, depending on the paramyxovirus family member analyzed [7,8,28,43,47,48]. Cross-sections through the nucleocapsids showed an inner diameter of 4–5 nm and an overall tube diameter of approximately 20 nm [6,43]. Virion reconstructions and functional studies have demonstrated that multiple genome copies can be packaged by a single particle [44,49], likely reflecting a poorly ordered assembly process. Like all other negative-polarity RNA viruses, isolated paramyxovirus genomes are not infectious and only the viral RNBA-dependent RNA-polymerase (RdRp) complex is capable of transcribing and replicating the RNP genomes [46]. In addition to the nucleocapsid, infectious particles must therefore package and deliver RdRp complexes to target cells to initiate a new infectious cycle.
Paramyxovirus entry is mediated under neutral pH conditions by a concerted action of the viral attachment and F glycoproteins. Cryo-electron tomograms of unstained paramyxovirus particles have revealed a dense array of glycoprotein spikes on the virion surface, but no higher order organization was apparent and the identification of individual glycoprotein oligomers was prevented by the tight packaging of the complexes [6,43,44,50].
Matrix protein assemblies
A crystal structure of the NDV M protein was recently solved and revealed a dimeric organization with 4-fold symmetry [6]. In addition to interacting with the N protein and the glycoprotein tails, the M protein contains positive charge patches on the surface that allow interaction with lipid membranes. Through multimerization of M dimers into a grid-like protein array with a 6° angle between the individual dimers, the M protein can introduce membrane curvature that is thought to promote virus budding [51,52]. Transient expression of M was shown to be sufficient in several cases to induce paramyxovirus VLPs formation [53,54,55,56,57,58,59,60,61,62]. However, this observation does not apply to members of the rubulavirus genus such as mumps virus, which require co-expression of M protein with the viral NP and glycoproteins to induce efficient particle production. In the case of the rubulaviruses, the M-N interaction not only recruits RNP to the sites of particle assembly, but is also thought to trigger particle release [21,63]. The ability to only release viral particles that contain RNP would be an advantage; allowing the virus to limit the release of noninfectious, empty virions. In addition to the M protein, also the glycoproteins, especially the F protein, have been implicated in modulating assembly and budding of at least some paramyxoviruses such as MuV [63], SeV [56], PIV5 [21] and NiV [59,64]. For NiV, autonomous formation of virus-like particles (VLP) by the F protein, and to a much lesser extent the G protein, was observed in addition to the more conventional M protein-mediated budding [59,64]. Although rare among the paramyxoviruses, autonomous F-induced VLP formation was also described for SiV and MeV [54,55,56]. However, efficient virion assembly requires in all of these cases the presence of the M protein and the biological function of F-mediated VLPs is still unclear.
Interaction of the M protein with the RNP complex
In NDV and SeV particle reconstructions, a distinct M layer of approximately 5 nm was observed below the envelope membrane, but only in a minority of virions examined and mostly covering only parts of the luminal membrane surface [6,44]. Interestingly, in membrane areas with detectable M protein layer, the NDV glycoproteins appeared to follow the pattern of the M array, positing the cytoplasmic tails of the HN and F protein complexes in the gaps between the M protein dimers. The low abundance of M protein arrays in the tomograms was hypothesized to represent disassembly of the arrays after budding is complete to facilitate subsequent membrane fusion and particle uncoating [6], but could alternatively also originate from specimen preparation, storage, and/or cryo-preservation. While the presence of functional M protein and intact glycoprotein cytoplasmic tails can down-modulate cell-to-cell fusion activity of some paramyxoviruses [65,66,67], it is unclear whether F refolding is indeed suppressed through F tail contact with intact M protein arrays as was speculated [6]. However, it is difficult to envision the subsequent introduction of extreme negative membrane curvature required for lipid merger and opening of a fusion pore [68] in the presence of an intact M protein lattice, necessitating the partial or complete breakdown of the arrays at some point prior to infection. The molecular driving force for disassembly of the ordered M layer is currently unknown. Receptor binding by the attachment protein was suggested as a possible impetus [6], but evidence is lacking that receptor binding translates to conformational rearrangements of the cytosolic tails and even if these occur it is not apparent how they could be sufficient to disturb highly ordered M protein arrays.
Unlike the matrix protein patches found below the envelope membrane in some of the NDV and SeV particle reconstructions, no significant protein density was detected in density profiles obtained from MeV tomograms. Rather, tubular structures with a diameter of approximately 30 nm were noted in some virions in addition to the 20 nm herringbone-like nucleocapsids (figure 1B) [43]. While the 20 nm tubes adhered to both anti-N and anti-M immunosorbent EM grids, the 30 nm tubes were precipitated only onto the latter grids, suggesting that the larger diameter structures consist of RNPs wrapped into M sheaths. The 30 nm M tubules formed a left-handed helix like the MeV RNP itself (figure 2), albeit with a pitch of 7.2 nm versus 6.4 nm calculated for the nucleocapsid. Within virions, the tubes packed into tight bundles that stood in lateral contact with the viral envelope. Interestingly, tubes of either diameter precipitated poorly onto anti-P grids, ruling out that assembled nucleocapsids are decorated by default with a high content of P molecules interacting with individual N protomers. By contrast, cylindrical M arrangements or structures equivalent to the 30 nm MeV tubes were not found in NDV particles [6]; unfortunately, SeV reconstructions lacked sufficient resolution of the RNPs to test for the presence of different types of tubular structures [44]. M tubes were likewise absent from HPIV3 particles [5]. However, the interpretation of the HPIV3 data is challenging, since none of the reconstructed HPIV3 particles that featured RNP and/or glycoprotein spikes contained any M density, while a particle proposed to contain M arrays lacked genome, glycoproteins, and was substantially smaller in size.
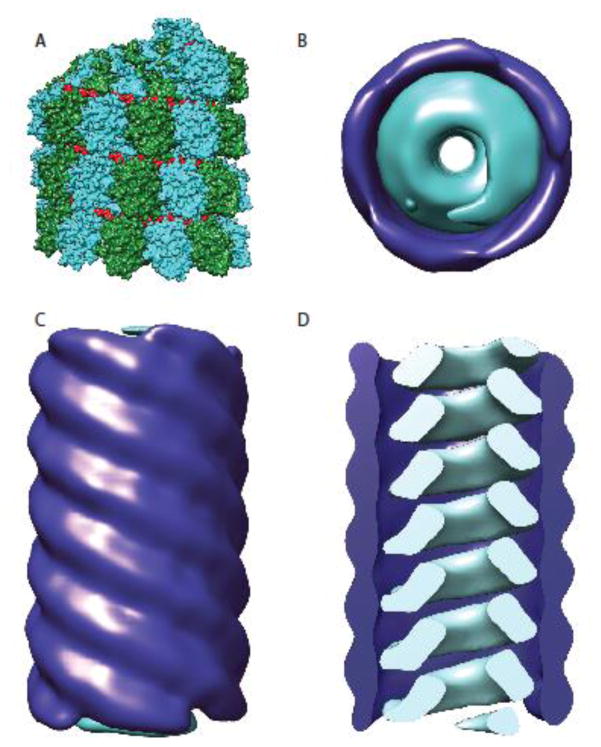
Figure 2.
Organization of the paramyxovirus nucleocapsid. A) Nucleocapsid proteins for all paramyxoviruses form helical assemblies of N proteins encapsidating the viral RNA (shown in this model are MeV RNPs, N proteins are depicted in cyan and forest green, the RNA is colored in red) (PDB ID: 4UFT). B–D) While RNPs for 20 nm diameter tubules, MeV RNPs were also found M protein wrapped in larger diameter 30 nm tubules [43]. Matrix proteins are shown in dark blue and nucleocapsids in cyan. Top view (B) and side view (C) of the 30 nm tubules depicting the distinct cylindrical M complex surrounding the MeV nucleocapsid. D) A clipped model of the 30 nm tubule structure. The matrix coated nucleocapsids were created using Chimera [92] based on electron density maps EMD-1973 and EMD-1974 [43].
The differences in results in particular between reconstructed NDV and MeV particles bring up the question of whether MeV nucleocapsids wrapped into M tubes represent a sample preparation artifact, dead-end complexes of a particle assembly process gone catastrophically wrong, or a physiologically relevant stage of the replication cycle of morbilliviruses, and perhaps of even a broader subset of paramyxovirus genera. Without further sightings of these M structures in additional MeV particle reconstructions that ideally follow particles through different stages of assembly and budding, this issue cannot be addressed definitively. However, we can examine the general plausibility of a physiological role of these M tubules by considering three basic questions: i) does a driving force exist for the formation of M tubules under native conditions; ii) is a physiological role of M tubules in the MeV life cycle conceivable; and iii) does the available phenotypic information support the existence of MeV M tubules?
The paramyxovirus N protein is composed of an N-terminal Ncore domain that is responsible for N homo-oligomerization and RNA encapsidation and a C-terminal Ntail that extends outwards from the RNP assembly. While Ntail is structurally intrinsically disordered, areas of high sequence conservation were identified that engage in multiple protein-protein interactions [69]. A molecular recognition element (MoRE) mediates transient binding of the P-L polymerase complex [70], and the terminal residues of the Ntail were demonstrated to mediate specific contacts with the MeV M protein [71], promoting genome packaging into nascent virions. Consequently, a high density of M binding sites is displayed on the surface of assembled MeV RNPs, which should be sufficient to trigger M multimerization around the nucleocapsid in infected cells. Disorder of the central Ntail section was furthermore proposed to provide the necessary flexibility to negotiate the different symmetries of the 20 nm inner RNP tubes and the 30 nm outer M cylinders [43]. However, a recent study has demonstrated that removal of the disordered tail region and relocation of the MoRE domain into Ncore does not abrogate replication of recombinant MeV, provided the M-binding terminal residues are added to the Ntail stump [72]. Although these MeV recombinants were not tested for the presence of the 30 nm tubes, they demonstrate that structural flexibility provided by the central Ntail section does not constitute a requirement for productive MeV particle assembly.
A number of candidate physiological effects of M tubules around the RNPs is conceivable. Certainly, RNP wrapping would prevent RdRp access and/or migration along the genome, blocking both genome transcription and replication. The tight arrangements of the 30 nm tubules in tomograms furthermore suggests that the wrapping may increase genome density, and it may ensure that genomes are packaged into nascent virions by recruiting the pre-wrapped nucleocapsids to budding sites. While all of these effects are poised to enhance the assembly of infectious particles, RNP wrapping by M tubules appears incompatible with a central step of paramyxovirus budding models, the introduction of membrane curvature through M protein arrays below the lipid membrane. Although tomograms of MeV particles showed association of the 30 nm tubules with the luminal surface of the viral envelope, large planar arrays rather than a cylindrical arrangement should be required to introduce sufficient membrane curvature for effective formation of progeny virions. On the other hand, MeV budding is inefficient [73], particle shape is more diverse than reported for members of some other paramyxovirus genera [6,43,44], and progeny virions remain largely cell-associated rather than being released into culture supernatants [74]. Also, it was suggested that MeV M could play different roles in RNP wrapping and the introduction of membrane curvature [43], although this would require additional interfaces between M proteins engaged in tubular and planar arrangements. In addition, tight spatial and temporal regulation of RNP wrapping would be essential, since the bulk of paramyxovirus proteins is generated through secondary transcription of progeny genomes [46] and premature shut-down of the transcriptase through genome wrapping would be catastrophic for the infection cycle. Equally important, the M tubules must disassemble to allow polymerase access to the genome after infection, and the molecular basis for both regulation of M tube formation and the subsequent induction of M depolymerization is unknown. A direct role of receptor binding, via conformational changes of the cytosolic glycoprotein tails, appears even less likely in the case of M tubules than for planar M arrays as was suggested for SeV uncoating.
Three lines of phenotypic evidence are consistent with a physiological role of M tubules in the MeV life cycle. Expression of the MeV M protein reduces genome transcription and replication [71,75] but no such effect was noted for SeV M [76]; intracellular transport of nucleocapsids of recombinant MeV harboring M proteins with reduced half life was inefficient in the absence of M accumulation at intracellular membranes [57]; and MeV N proteins lacking the disordered central region of the tail domain altered viral mRNA expression in the context of virus infection but not in polycistronic minigenome reporter assays [72]. Since M protein is only present in the infection but not the minigenome experiments, it is conceivable that removing the disordered tail section affects the frequency with which the tubular M structures form and thus the timing of shut-down of genome access by the RdRp.
Glycoprotein organization
The physiological oligomer of the paramyxovirus attachment protein is the tetramer, consisting of a dimer of homotypic dimers, whereas the F protein, a type I viral fusion protein, assembles into homotrimers [46]. The attachment proteins feature a globular head domain with the beta propeller fold characteristic for sialidases that attach to the transmembrane domain and cytosolic tails through a helical stalk domain [68]. A large body of evidence supports that the protein interface region responsible for specific hetero-oligomerization with homotypic F protein trimers resides in this stalk [50,77,78,79,80,81]. Upon receptor binding by the globular head domain, exposure of [79,82] and/or a conformational change in the membrane-proximal attachment protein stalk domain [77,83,84] is considered to trigger major conformational changes in the metastable prefusion F trimer, resulting in propelling of a membrane attack group or fusion peptide towards the opposing membrane, hairpin formation, and the assembly of a thermodynamically highly stable fusion core or six-helix bundle structure, which induces extreme local negative membrane curvature and brings the F trimer transmembrane domains and fusion peptides, and thus viral envelope and target membranes, into close proximity [68].
Although attachment and F glycoprotein oligomers could not be definitely separated in reconstructions of unstained paramyxovirus particles, tomograms of HPIV3 particles [5] and recombinant MeV particles displaying stalk-elongated attachment proteins allowed the identification of the attachment protein globular head domains in radial density distribution plots [50]. The MeV attachment proteins in particular were engineered to extend the stalk length by approximately 4 nm, which is equivalent to nearly 50% of the length of the unmodified MeV H stalk, while maintaining bioactivity. These reconstructions confirmed a spatial arrangement of functional fusion complexes in which the attachment protein head domain is positioned membrane distal and above the prefusion F trimers [78].
However, the tomograms failed to decipher the H to F oligomer stoichiometry in physiological fusion complexes. Multiple studies investigating different type I viral membrane fusion proteins have concluded that a concerted action of several fusion protein complexes is required to induce sufficient negative curvature in the viral envelope and cellular membrane to trigger local lipid disarray in the outer leaflets of the approaching bilayers, allowing merger of the disordered monolayers at the fusion tip and ultimately opening of a fusion pore [85,86,87,88,89,90,91]. If individual paramyxovirus glycoprotein homo-oligomers interact with each other for membrane fusion, each attachment protein tetramer would be sterically able to contact two F protein trimers in parallel. A recent study proposed an alternative F protein organization based on crystal structures of recombinant soluble NiV F protein trimers and tomograms of NiV glycoprotein-coated VLPs that both suggest a hexamer of F trimers ring-like assembly [25] (figure 3). In this arrangement, each F trimer contacts with its head domain two neighboring trimers, leaving only a single priming site available for interaction with the attachment protein. This F organization is provocative, since it may be able to reinforce the metastable prefusion conformation of the individual F trimers engaged in the ring structure prior to receptor binding, while activation of a single associated attachment protein tetramer may be sufficient to trigger the spatially and temporally highly coordinated refolding of all six F trimers locked into the ring structure. Since some F trimers appeared to be part of more than one hexameric structure, one could even envision a highly effective concerted wave-like refolding of numerous F complexes present on a viral particle after receptor binding by a small number of attachment protein tetramers. However, since hexameric F rings have so far been proposed only for NiV fusion proteins and NiV VLPs lacked the corresponding NiV G attachment proteins, it is unclear at present whether this F arrangement is germane only to the henipavirus genus and whether the presence of the attachment protein tetramers would alter the spatial organization of the F trimers.
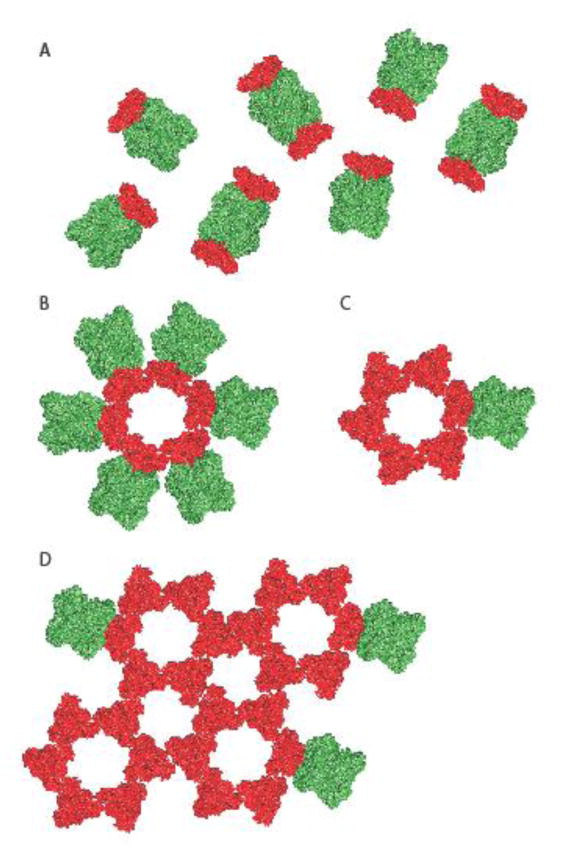
Figure 3.
Models of alternative spatial organizations of the paramyxovirus glycoproteins on the virion surface. A) The overall organization of the paramyxovirus glycoproteins was thought to be random with an undetermined relative stoichiometry of individual fusion protein trimers (red) and attachment protein tetramers (green). B, C, D) In a recent study [25], an arrangement of NiV F protein trimers into hexamers of trimers and higher order complexes was proposed. Schematics of hexameric F trimer arrangements in contact with one (A) or multiple (B) attachment protein tetramers and higher order F assemblies consisting of interacting hexamers of trimers (C). Different hypothetical contacts of the F assemblies with attachment protein tetramers are shown, but the stoichiometry and positioning of the attachment protein oligomers relative to the F protein complexes has not yet been defined.
Conclusions
Although only a limited number of paramyxovirus virion reconstructions is available at present, the insight gained from these studies has substantially impacted the current view of particle organization, assembly, and budding. A number of unexpected and potentially paradigm-changing hypotheses concerning the possible roles of the matrix protein in particle formation, genome packaging, and transcription control and of higher order glycoprotein organization in mediating efficient viral entry has emerged from these reconstructions. In addition to spotlighting previously unappreciated features of the paramyxovirus life cycle, these studies have also reinforced that central parts of the paramyxovirus replication, assembly, and uncoating machinery are currently mechanistically not understood. Since a detailed structural and mechanistic understanding of these central steps of the viral life cycle will inform the targeted development of much needed improved prophylactic and therapeutic anti-paramyxoviruses strategies, obtaining additional high-resolution particle structures and substructures is a high priority to test the physiological relevance of individual observations and assess their applicability to clinically-relevant pathogens of different genera within the family.
Highlights.
Recent cryo-electron tomography reconstructions of Sendai virus, Newcastle disease virus, measles virus, and Nipah virus-derived virus like particles have yielded novel insight into paramyxovirus particle assembly and organization
The Newcastle disease virus matrix protein formed distinct layers below the viral envelope and the viral envelope glycoproteins followed the pattern of this matrix array, while tubular matrix protein assemblies were observed in measles virus particles that wrap the viral genome into matrix sheaths
The physiological role of tubular matrix sheaths is unclear but could involve mediating transcription and replication shut-off at the time of particle assembly
Nipah virus-based virus like particles have revealed a previously unappreciated hexameric ring-like organization of the viral fusion protein that if present on native virions may set the stage for efficient viral entry through concerted refolding of multiple F complexes
Acknowledgments
We are grateful to members of the Plemper lab for helpful discussions when preparing this review and to A.L. Hammond for critical reading of the manuscript. This work was supported, in part, by public health service grants AI071002 and HD079327 from the NIH/NIAID and NIH/NICHD (to R.K.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Yanagi Y, Takeda M, Ohno S, Seki F. Measles virus receptors and tropism. Jpn J Infect Dis. 2006;59:1–5. [PubMed] [Google Scholar]
- 2.Li Z, Hung C, Paterson RG, Michel F, Fuentes S, et al. Type II integral membrane protein, TM of J paramyxovirus promotes cell-to-cell fusion. Proc Natl Acad Sci U S A. 2015;112:12504–12509. doi: 10.1073/pnas.1509476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce LA, Yu M, Waddington LJ, Barr JA, Scoble JA, et al. Structural characterization by transmission electron microscopy and immunoreactivity of recombinant Hendra virus nucleocapsid protein expressed and purified from Escherichia coli. Protein Expr Purif. 2015;116:19–29. doi: 10.1016/j.pep.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Gutsche I, Desfosses A, Effantin G, Ling WL, Haupt M, et al. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348:704–707. doi: 10.1126/science.aaa5137. High-resolution reconstruction of the measles virus genome assembly that provides important structural insight into the viral nucleoprotein-RNA interaction. [DOI] [PubMed] [Google Scholar]
- 5.Gui L, Jurgens EM, Ebner JL, Porotto M, Moscona A, et al. Electron tomography imaging of surface glycoproteins on human parainfluenza virus 3: association of receptor binding and fusion proteins before receptor engagement. MBio. 2015;6:e02393–02314. doi: 10.1128/mBio.02393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, et al. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci U S A. 2012;109:13996–14000. doi: 10.1073/pnas.1210275109. First cryo-electron tomography reconstruction of Newcastle disease virus particles combined with X-ray crystallography of analysis of the viral matrix protein reveals pseudotetrameric matrix arrays that generate the membrane curvature required for virus budding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoehn G, Mavrakis M, Albertini A, Wade R, Hoenger A, et al. The 12 A structure of trypsin-treated measles virus N-RNA. J Mol Biol. 2004;339:301–312. doi: 10.1016/j.jmb.2004.03.073. [DOI] [PubMed] [Google Scholar]
- 8.Bhella D, Ralph A, Yeo RP. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J Mol Biol. 2004;340:319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 10.Chua KB, Wang LF, Lam SK, Crameri G, Yu M, et al. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology. 2001;283:215–229. doi: 10.1006/viro.2000.0882. [DOI] [PubMed] [Google Scholar]
- 11.Chua KB, Wang LF, Lam SK, Eaton BT. Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Arch Virol. 2002;147:1323–1348. doi: 10.1007/s00705-002-0815-5. [DOI] [PubMed] [Google Scholar]
- 12.Curry A, Appleton H, Dowsett B. Application of transmission electron microscopy to the clinical study of viral and bacterial infections: present and future. Micron. 2006;37:91–106. doi: 10.1016/j.micron.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldsmith CS, Ksiazek TG, Rollin PE, Comer JA, Nicholson WL, et al. Cell culture and electron microscopy for identifying viruses in diseases of unknown cause. Emerg Infect Dis. 2013;19:886–891. doi: 10.3201/eid1906.130173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsmith CS, Miller SE. Modern uses of electron microscopy for detection of viruses. Clin Microbiol Rev. 2009;22:552–563. doi: 10.1128/CMR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall WW, Martin SJ. Purification and characterization of measles virus. J Gen Virol. 1973;19:175–188. doi: 10.1099/0022-1317-19-2-175. [DOI] [PubMed] [Google Scholar]
- 16.Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- 17.Kvellestad A, Dannevig BH, Falk K. Isolation and partial characterization of a novel paramyxovirus from the gills of diseased seawater-reared Atlantic salmon (Salmo salar L) J Gen Virol. 2003;84:2179–2189. doi: 10.1099/vir.0.18962-0. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Yu M, Zhang H, Magoffin DE, Jack PJ, et al. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology. 2006;346:219–228. doi: 10.1016/j.virol.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 19.Mast J, Demeestere L. Electron tomography of negatively stained complex viruses: application in their diagnosis. Diagn Pathol. 2009;4:5. doi: 10.1186/1746-1596-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philbey AW, Kirkland PD, Ross AD, Field HE, Srivastava M, et al. Infection with Menangle virus in flying foxes (Pteropus spp.) in Australia. Aust Vet J. 2008;86:449–454. doi: 10.1111/j.1751-0813.2008.00361.x. [DOI] [PubMed] [Google Scholar]
- 21.Schmitt AP, Leser GP, Waning DL, Lamb RA. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol. 2002;76:3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terrier O, Rolland JP, Rosa-Calatrava M, Lina B, Thomas D, et al. Parainfluenza virus type 5 (PIV-5) morphology revealed by cryo-electron microscopy. Virus Res. 2009;142:200–203. doi: 10.1016/j.virusres.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig K, Schade B, Bottcher C, Korte T, Ohlwein N, et al. Electron cryomicroscopy reveals different F1+F2 protein States in intact parainfluenza virions. J Virol. 2008;82:3775–3781. doi: 10.1128/JVI.02154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong JJ, Paterson RG, Lamb RA, Jardetzky TS. Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form. Proc Natl Acad Sci U S A. 2016;113:1056–1061. doi: 10.1073/pnas.1523303113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Xu K, Chan YP, Bradel-Tretheway B, Akyol-Ataman Z, Zhu Y, et al. Crystal Structure of the Pre-fusion Nipah Virus Fusion Glycoprotein Reveals a Novel Hexamer-of-Trimers Assembly. PLoS Pathog. 2015;11:e1005322. doi: 10.1371/journal.ppat.1005322. First report that Nipah fusion protein trimers may exist in higher order oligomer assemblies on the surface of viral particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guryanov SG, Liljeroos L, Kasaragod P, Kajander T, Butcher SJ. Crystal Structure of the Measles Virus Nucleoprotein Core in Complex with an N-Terminal Region of Phosphoprotein. J Virol. 2015;90:2849–2857. doi: 10.1128/JVI.02865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yabukarski F, Lawrence P, Tarbouriech N, Bourhis JM, Delaforge E, et al. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol. 2014;21:754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 28•.Cox R, Pickar A, Qiu S, Tsao J, Rodenburg C, et al. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci U S A. 2014;111:15208–15213. doi: 10.1073/pnas.1413268111. Study presents evidence that the viral phosphoprotein may mediate local de-encapsidation of the mumps virus RNP genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, et al. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog. 2013;9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan P, Paterson RG, Leser GP, Lamb RA, Jardetzky TS. Structure of the ulster strain newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulence. PLoS Pathog. 2012;8:e1002855. doi: 10.1371/journal.ppat.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012;109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, et al. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, et al. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 2011:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson K, Wen X, Leser GP, Paterson RG, Lamb RA, et al. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402:372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 36.Bowden TA, Crispin M, Harvey DJ, Aricescu AR, Grimes JM, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. First high-resolution structure of a paramyxovirus fusion protein in a prefusion conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 39.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 42.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 43••.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci U S A. 2011;108:18085–18090. doi: 10.1073/pnas.1105770108. First reconstruction of measles virus particles, revealing unexpected tubular matrix protein assemblies around the viral RNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Loney C, Mottet-Osman G, Roux L, Bhella D. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of sendai virus. J Virol. 2009;83:8191–8197. doi: 10.1128/JVI.00693-09. First cryo-electron tomography-based reconstruction of Sendai virus particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 47.Bhella D, Ralph A, Murphy LB, Yeo RP. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J Gen Virol. 2002;83:1831–1839. doi: 10.1099/0022-1317-83-8-1831. [DOI] [PubMed] [Google Scholar]
- 48.Egelman EH, Wu SS, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rager M, Vongpunsawad S, Duprex WP, Cattaneo R. Polyploid measles virus with hexameric genome length. Embo J. 2002;21:2364–2372. doi: 10.1093/emboj/21.10.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, et al. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol. 2013;87:11693–11703. doi: 10.1128/JVI.01945-13. First demonstration that induced tetramerization of morbillivirus attachment protein stalk domains restores intracellular transport competence and fusion triggering activity in the absence of the head domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Najjar F, Schmitt AP, Dutch RE. Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production. Viruses. 2014;6:3019–3054. doi: 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol. 2010;42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riedl P, Moll M, Klenk HD, Maisner A. Measles virus matrix protein is not cotransported with the viral glycoproteins but requires virus infection for efficient surface targeting. Virus Res. 2002;83:1–12. doi: 10.1016/s0168-1702(01)00379-3. [DOI] [PubMed] [Google Scholar]
- 54.Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol. 2007;88:1243–1250. doi: 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 55.Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, et al. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology. 2004;325:1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 56.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. Role of matrix and fusion proteins in budding of Sendai virus. J Virol. 2001;75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Runkler N, Pohl C, Schneider-Schaulies S, Klenk HD, Maisner A. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol. 2007;9:1203–1214. doi: 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 58.Ciancanelli MJ, Basler CF. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol. 2006;80:12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patch JR, Crameri G, Wang LF, Eaton BT, Broder CC. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol J. 2007;4:1. doi: 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun W, McCrory TS, Khaw WY, Petzing S, Myers T, et al. Matrix proteins of Nipah and Hendra viruses interact with beta subunits of AP-3 complexes. J Virol. 2014;88:13099–13110. doi: 10.1128/JVI.02103-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol. 2006;80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coronel EC, Murti KG, Takimoto T, Portner A. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol. 1999;73:7035–7038. doi: 10.1128/jvi.73.8.7035-7038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M, Schmitt PT, Li Z, McCrory TS, He B, et al. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol. 2009;83:7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnston GP, Contreras EM, Dabundo J, Henderson BA, Matz KM, et al. Cytoplasmic Motifs in the Nipah Virus Fusion Protein Modulate Virus Particle Assembly and Egress. J Virol. 2017:91. doi: 10.1128/JVI.02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, et al. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 1998;17:3899–3908. doi: 10.1093/emboj/17.14.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cathomen T, Naim HY, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72:1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sawatsky B, Bente DA, Czub M, von Messling V. Morbillivirus and henipavirus attachment protein cytoplasmic domains differently affect protein expression, fusion support and particle assembly. J Gen Virol. 2016;97:1066–1076. doi: 10.1099/jgv.0.000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plemper RK. Cell Entry of Enveloped Viruses. Curr Opin Virol. 2011;1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sourimant J, Plemper RK. Organization, Function, and Therapeutic Targeting of the Morbillivirus RNA-Dependent RNA Polymerase Complex. Viruses. 2016:8. doi: 10.3390/v8090251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Longhi S. Nucleocapsid structure and function. Curr Top Microbiol Immunol. 2009;329:103–128. doi: 10.1007/978-3-540-70523-9_6. [DOI] [PubMed] [Google Scholar]
- 71•.Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, et al. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol. 2009;83:10374–10383. doi: 10.1128/JVI.01056-09. First mapping of a measles virus matrix protein binding domain near the carboxy-terminus of the nucleocapsid protein tail domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Cox RM, Krumm SA, Thakkar VD, Sohn M, Plemper RK. The structurally disordered paramyxovirus nucleocapsid protein tail domain is a regulator of the mRNA transcription gradient. Sci Adv. 2017;3:e1602350. doi: 10.1126/sciadv.1602350. First demonstration that productive interaction of the viral polymerase complex with the RNP genome does not require a previously postulated fly-casting mechanism mediated by the intrinsically unstructured paramyxovirus nucleoprotein tail domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Griffin DE. Measles Virus. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2007. pp. 1551–1585. [Google Scholar]
- 74.McChesney MB, Miller CJ, Rota PA, Zhu YD, Antipa L, et al. Experimental measles. I. Pathogenesis in the normal and the immunized host. Virology. 1997;233:74–84. doi: 10.1006/viro.1997.8576. [DOI] [PubMed] [Google Scholar]
- 75•.Suryanarayana K, Baczko K, ter Meulen V, Wagner RR. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J Virol. 1994;68:1532–1543. doi: 10.1128/jvi.68.3.1532-1543.1994. Early demonstration that the measles virus matrix protein suppresses transcription of the viral genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mottet-Osman G, Iseni F, Pelet T, Wiznerowicz M, Garcin D, et al. Suppression of the Sendai virus M protein through a novel short interfering RNA approach inhibits viral particle production but does not affect viral RNA synthesis. J Virol. 2007;81:2861–2868. doi: 10.1128/JVI.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Brindley MA, Chaudhury S, Plemper RK. Measles Virus Glycoprotein Complexes Preassemble Intracellularly and Relax during Transport to the Cell Surface in Preparation for Fusion. J Virol. 2015;89:1230–1241. doi: 10.1128/JVI.02754-14. Comprehensive assessment of the maturation cascade of the measles virus glycoprotein oligomers, demonstrating tight intracellular pre-assembly of the fusion and attachment proteins prior to surface expression and incorporation into viral particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, et al. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83:10480–10493. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bose S, Song AS, Jardetzky TS, Lamb RA. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol. 2014;88:3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80•.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, et al. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A. 2012;109:E2625–2634. doi: 10.1073/pnas.1213813109. First report demonstrating efficient triggering of a paramyxovirus fusion protein complex by an attachment protein mutant lacking the receptor binding head domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Q, Bradel-Tretheway B, Monreal AI, Saludes JP, Lu X, et al. Nipah virus attachment glycoprotein stalk C-terminal region links receptor binding to fusion triggering. J Virol. 2015;89:1838–1850. doi: 10.1128/JVI.02277-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, et al. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 2013;9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brindley MA, Takeda M, Plattet P, Plemper RK. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci U S A. 2012;109:E3018–3027. doi: 10.1073/pnas.1210925109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84•.Brindley MA, Plemper RK. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J Virol. 2010;84:12174–12184. doi: 10.1128/JVI.01222-10. First biochemical conformation that receptor binding-induced structural changes in the measles virus attachment protein lead to fusion triggering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci U S A. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Freed EO, Delwart EL, Buchschacher GL, Jr, Panganiban AT. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci U S A. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown JC, Newcomb WW, Lawrenz-Smith S. pH-dependent accumulation of the vesicular stomatitis virus glycoprotein at the ends of intact virions. Virology. 1988;167:625–629. [PubMed] [Google Scholar]
- 89.Lee KK. Architecture of a nascent viral fusion pore. Embo J. 2010 doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gui L, Ebner JL, Mileant A, Williams JA, Lee KK. Visualization and Sequencing of Membrane Remodeling Leading to Influenza Virus Fusion. J Virol. 2016;90:6948–6962. doi: 10.1128/JVI.00240-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Calder LJ, Rosenthal PB. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat Struct Mol Biol. 2016;23:853–858. doi: 10.1038/nsmb.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 93.The PyMOL Molecular Graphics System, Version 1.8. Schrödinger, LLC; 2010. [Google Scholar]