Abstract
Accumulating evidence indicates that various classes of non-coding RNAs (ncRNAs) including microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs) and long non-coding RNAs (lncRNAs) play important roles in normal state as well as the diseases of the CNS. Interestingly, ncRNAs have been shown to interact with messenger RNA, DNA and proteins, and these interactions could induce epigenetic modifications and control transcription and translation, thereby adding a new layer of genomic regulation. The ncRNA expression profiles are known to be altered after acute CNS injuries including stroke, traumatic brain injury and spinal cord injury that are major contributors of morbidity and mortality worldwide. Hence, a better understanding of the functional significance of ncRNAs following CNS injuries could help in developing potential therapeutic strategies to minimize the neuronal damage in those conditions. The potential of ncRNAs in blood and CSF as biomarkers for diagnosis and/or prognosis of acute CNS injuries has also gained importance in the recent years. This review highlighted the current progress in the understanding of the role of ncRNAs in initiation and progression of secondary neuronal damage and their application as biomarkers after acute CNS injuries.
Keywords: Non-coding RNA, microRNA, lncRNA, piRNA, ischemic stroke, hemorrhagic stroke, traumatic brain injury, spinal cord injury
Introduction
In the post-genome sequencing era, it has become increasingly evident that a large portion of the transcriptional output (~98%) do not code for proteins, but instead constitutes various classes of housekeeping non-coding RNAs (ncRNAs) such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and small nuclear RNAs (snRNAs), as well as regulatory ncRNAs including microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs) and long non-coding RNAs (lncRNAs). While the essential nature of rRNAs and tRNAs in protein translation is known for several decades, the physiologic functions of other ncRNAs are mostly unknown and hence they were previously considered as transcriptional noise. However, recent studies show that they are an outcome of pervasive transcription (Wade and Grainger, 2014; Birney et al., 2007) and control many functions including epigenetic modifications, transcriptional and translational regulation, and RNA and protein scaffolding, and thus adds a new layer of genomic regulation (Schmitz et al., 2016; Peschansky and Wahlestedt, 2014; Rinn and Chang, 2012; Qureshi and Mehler, 2012). Recent studies also showed that both acute and chronic injuries to CNS alter ncRNA expression and function (Dharap et al., 2009; Dharap et al., 2010; Dharap et al., 2011; Dharap et al., 2012; Nelson and Wang 2010; Thome et al., 2016; Wang et al., 2011b). This review briefly highlights the current knowledge of the role of ncRNAs in the pathophysiology of acute CNS injuries.
Acute CNS injuries including stroke rapidly changes the expression of protein-coding genes that regulate various processes such as excitotoxicity, oxidative stress, endoplasmic reticulum stress, inflammation, autophagy, apoptosis, neurogenesis and angiogenesis that synergistically promote neuronal death and/or plasticity after an injury (Lu et al., 2004; Schmidt-Kastner et al., 2002; Sharp et al., 2011). Secondary brain damage and neurologic dysfunction seen after other acute CNS injuries like traumatic brain injury (TBI), spinal cord injury (SCI) and subarachnoid hemorrhage (SAH) are also linked to many of these pathologic mechanisms. Hence, a better understanding of the roles of various classes of ncRNAs mediating the pathophysiological mechanisms associated with acute CNS injuries can help in designing much effective therapeutic agents for treating them.
MicroRNAs
The miRNAs are a highly conserved class of ncRNAs, ~22 nucleotide long and function by binding to the 3' untranslated regions (3'UTRs) of mRNAs by complementary base pairing. This interaction allows inhibition of protein synthesis by either mRNA degradation by complete base pairing or transient translational arrest by incomplete base pairing. The miRNA-dependent post-transcriptional gene silencing is complex as individual miRNAs can target many mRNAs and an individual mRNA may contain binding sites for multiple miRNAs. This unique feature allows a strong control of gene expression networks to regulate almost any biological process including cellular differentiation and maintenance, neurogenesis, apoptosis, inflammation and oxidative stress during development and diseases (Neo et al., 2014; Zhao et al., 2013a; Zeng et al., 2014a; Chen et al., 2014; Jadhav et al., 2014; Liu et al., 2015c; Varga et al., 2013). Interestingly, studies have shown that gene disruption of the miRNA processing machinery such as Dicer and DGCR8 results in lethality showing the essential nature of miRNAs for life (Bernstein et al., 2003; Wang et al., 2007).
MicroRNAs and post-stroke pathophysiology
We and others have shown that transient focal ischemia in experimental rodents leads to changes in global expression of cerebral and vascular miRNAs (Zhu et al., 2014; Lusardi et al., 2014, Gubern et al., 2013; Dharap et al., 2009; Jeyaseelan et al., 2008; Liu et al., 2010). Bioinformatic analysis showed that the miRNAs altered after stroke target the translation of proteins that modulate inflammation, excitotoxicity, oxidative stress, endoplasmic reticulum stress, autophagy and apoptosis after experimental stroke (Dharap et al., 2009; Liu et al., 2015a).
Apoptosis plays a major role in tissue loss in ischemic lesions. The existence of a therapeutic time window between the onset of stroke and loss of brain tissue surrounding the ischemic site due to apoptosis makes it a very attractive target for stroke therapy (Woodruff et al., 2011; Broughton et al., 2009). Many studies evaluated the role of miRNAs in post-stroke neuronal apoptosis (Fig. 1). For example, miR-210 was found to mediate neuroprotective actions of vagus nerve stimulation (VNS) in a rodent model of transient middle cerebral artery occlusion (MCAO) by targeting anti-oxidant and anti-apoptotic components (Jiang et al., 2015). In a hypoxia-ischemia (HI) rat model (ligation of the right common carotid artery followed by a 2.5h exposure to hypoxia), miR-139-5p was shown to target human growth and transformation dependent protein (HGTD-P) which is a promoter of neuronal apoptosis (Qu et al., 2014) (Fig. 1). Downregulation of miR-139-5p was also observed in rat cortical neurons subjected to oxygen-glucose deprivation (OGD) as well as in rat brain after HI (Qu et al., 2014). Treatment with agomiR-139-5p decreases cerebral HGTD-P expression and infarct volume in adult rats following HI (Qu et al., 2014). The widely known anti-apoptotic protein B-cell lymphoma-2 (Bcl2) was found to be a target of miR-181a following stroke in a rodent model of forebrain ischemia (Ouyang et al., 2012). Pre-treatment with antagomiR-181a led to a significant decrease in hippocampal neuronal death along with an increase in Bcl-2 levels following forebrain ischemia (Moon et al., 2013). Replenishing the levels of miR-9 which were downregulated in the brains of adult mice following transient MCAO was shown to prevent neuronal apoptosis by targeting proapoptotic Bcl-2-like 11 (Bcl2l11) leading to reduced behavioral deficits, smaller infarction and decreased edema (Wei et al., 2015) (Fig. 1). Another study found that there is a decrease in the levels of miR-99a in the plasma of stroke patients, and increasing its level decreases the secondary brain damage in a mouse MCAO model by preventing apoptosis (Tao et al., 2015). The neuroprotective potential of miR-99a was further validated by in vitro studies that showed that miR-99a alleviates hydrogen peroxide-induced oxidative stress and apoptosis. Further studies linked the neuroprotection afforded by miR-99a to targeting and decreasing the levels of cyclin D1 and cyclin-dependent kinase 6 (CDK6) expression in both in vitro and in vivo ischemia conditions that attenuates cell cycle progression leading to anti-apoptotic actions (Tao et al., 2015) (Fig. 1). The miR-479 has also been identified as an apoptosis-related miRNA that inhibits the expression of anti-apoptotic proteins Bcl-2 and Bcl-w and contribute to post-ischemic neuronal damage in mouse N2A cells after OGD. Inhibition of this particular miRNA increases the levels of Bcl-2 and Bcl-w leading to a decline in ischemic damage indicated by a decreased infarct volume and neurological deficit score (Yin et al., 2010a).
Fig. 1.
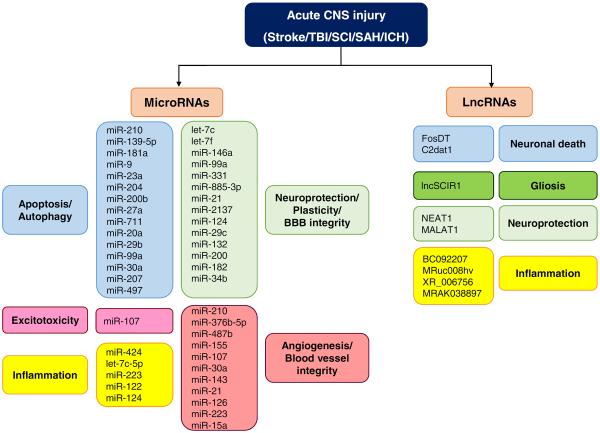
NcRNAs mediate common molecular mechanisms associated with the different acute CNS injuries. Different studies have shown that ncRNAs like miRNAs and lncRNAs contribute to the pathophysiology of acute CNS injuries by mediating common pathophysiological mechanisms like apoptosis, inflammation and glutamate excitotoxicity as well as neuroprotective mechanisms like neuroprotection, neuroplasticity and angiogenesis.
Several miRNAs were also shown to influence autophagy after stroke (Fig. 1). In particular, miR-30a was found to be significantly downregulated following 1h MCAO/24h reperfusion in mouse cerebral cortex and in mouse N2A neuroblastoma cells after 1h OGD/6–48h reoxygenation with a concomitant increase in levels of Beclin-1, an autophagy-signature protein. Treatment with antagomiR-30a decreased OGD-induced cell death in vitro and post-ischemic infarction and neurological deficits in vivo and thus an increase in Beclin-1 expression was linked with improvement in post-stroke outcome in both in vitro and in vivo conditions (Wang et al., 2014a). The miR-207 was shown to control autophagic cell death after ischemia by downregulating the expression of lysosomal-associated membrane protein 2 (LAMP2). Treatment with a miR-207 mimic decreased LAMP2 expression, lysosome count, infarct volume and improved the neurological function in rats after transient focal ischemia (Tao et al., 2015).
Glutamate excitotoxicity is another major promoter of ischemic brain damage (Kostandy et al., 2012). Focal ischemia in adult rats increased miR-107 expression and repression of its target glutamate transporter-1 (GLT-1) (Yang et al., 2014). This decrease in GLT-1 levels leads to a delay in the clearance of the released glutamate from the synaptic cleft leading to excitotoxic neuronal death. Further studies showed that treatment with miR-107 inhibitor attenuated the downregulation of GLT-1 protein expression in cultured neural cells subjected to hypoxia/reoxygenation (Yang et al., 2014) (Fig. 1).
Microglial activation is a widely observed response after stroke that promotes inflammation and thus aggravates ischemic brain damage (Taylor et al., 2013). A recent study showed that intracerebroventricular administration of miR-424 mimic prevented microglial activation leading to decreased infarct volume and brain edema after transient MCAO in adult mice (Zhao et al., 2013b) (Fig. 1). Decreased levels of let-7c-5p was observed in the plasma of stroke patients as well as in the plasma and brain of mice subjected to transient focal ischemia (Ni et al., 2015). When let-7c-5p was overexpressed, levels of caspase-3 were decreased leading to reduced microglial activation, decreased infarct volume and neurological deficits after focal ischemia (Ni et al., 2015) (Fig. 1). The lesser known role of caspase-3 in regulating microglial activation without triggering cell death through a protein kinase C dependent pathway might have played a role in this particular response (Burguillos et al., 2011). Replenishing miR-122 levels by intravenous administration of a miR-122 mimic was shown to reduce neurological deficits, infarct volume and ICAM-1 expression together with downregulation of direct and indirect target genes and maintenance of vascular integrity in a rat model of focal ischemia by acting on blood leukocytes rather than the brain tissue directly (Liu et al., 2016a) (Fig. 1). A combination therapy of VELCADE and tissue plasminogen activator was found to provide neuroprotection in aged rats after stroke by simultaneously upregulating miR-146a and inactivating toll-like receptor signaling pathway (Zhang et al., 2012a) (Fig. 1).
Our lab had identified miR-29c to directly target DNA methyltransferase 3a (DNMT3a) which was found to mediate neuronal cell death in PC12 cells subjected to OGD. Also, treatment with premiR-29c and DNMT3a siRNA decreased infarct volume post stroke indicating the importance of this miRNA-target relationship in regulating stroke-associated neuronal cell death (Pandi et al., 2013). Histone deacetylase inhibition by valproic acid is known to protect the post-stroke brain (Wang et al., 2011c). Two miRNAs, miR-331 and miR-885-3p were shown to be responsible for valproic acid-induced neuroprotection after ischemia by modulating their predicted targets associated with several networks including immune cell trafficking, neuronal cell death, synaptic depression and branching of neurites (Hunsberger et al., 2012). Although not directly related to stroke, Duan et al. (2014) showed that hyperglycemia leads to decreased expression of platelet miR-223 and miR-146a, and this in turn triggers platelet dysfunction and subsequent risk for onset of ischemic stroke. Many miRNAs were also shown to protect the brain and promote plasticity and regeneration by modulating neurogenesis and angiogenesis (Madathil et al., 2011; Selvamani et al. 2012; Shi et al., 2010) (Fig. 1). Inhibiting let-7f with an antagomiR was shown to protect the post-ischemic brain by increasing insulin-like growth factor-1 (IGF-1) levels (Selvamani et al. 2012). IGF-1 is a potent promoter of neurogenesis in normal and post-stroke brain. MiR-210 was found to increase post-stroke angiogenesis in a rat model of transient focal ischemia by activating the Notch signaling pathway involved in blood vessel formation (Lou et al., 2012). MiR-210 was upregulated following ischemic stroke and overexpression of this miRNA in vitro showed an increase in angiogenesis as well as the levels of Notch-1. The other target genes of miR-210 like ephrin A3 and hypoxia-inducing factor-1α might also be involved in blood vessel formation (Lou et al., 2012) (Fig. 1). A similar study indicated the role of miR-376b-5p in regulating the HIF-1α-mediated vascular endothelial growth factor (VEGF)/Notch1 signaling pathway following permanent MCAO in rats (Li et al., 2014a) (Fig. 1). MiR-487b that was found to be increased in the plasma of ischemic stroke patients is also a modulator of angiogenesis. Transfection of miR-487b in human umbilical vein endothelial cells increased their proliferation, migration, invasion and tube formation and expression of thrombospondin-1, which is an endogenous inhibitor of angiogenesis (Feng et al., 2015a) (Fig. 1). A similar link between a stroke-related miRNA and angiogenesis was shown in case of miR-155. Increased stabilization of blood vessels was observed to be regulated by miR-155 through its target protein Ras homolog enriched in brain in a distal MCAO mouse model (Caballero-Garrido et al., 2015) (Fig. 1). Another recent study showed that miR-107 induced in the ischemic boundary zone after permanent MCAO targets Dicer-1 leading to translational resulting in upregulation of endothelial cell-derived VEGF (VEGF165/VEGF164) that contributes to enhanced angiogenesis (Li et al., 2015). The neuroprotective action of peroxisome proliferator-activated receptor δ (PPARδ) in controlling ischemia-induced cerebrovascular damage was found to be an outcome of its inhibition of the pro-apoptotic miR-15a. Decrease in miR-15a expression in turn leads to increase in the levels of anti-apoptotic Bcl-2 that protects blood vessel integrity indicated by a reduction in caspase-3 activity, decreased Golgi fragmentation and reduced cell death (Yin et al., 2010b) (Fig. 1).
MicroRNAs and ischemic preconditioning
A short duration ischemia that doesn't kill neurons induces tolerance against a subsequent damaging ischemic insult. This phenomenon is known as preconditioning (PC)-induced ischemic tolerance is a consequence of multiple molecular mechanisms that include induction of protein chaperones, anti-oxidant enzymes, anti-inflammatory and anti-cell death pathways simultaneously to synergistically prevent ischemic cell death (Thompson et al., 2013; Stetler et al., 2014; Sisalli et al., 2015). In addition, ncRNAs are also thought to contribute to the PC-induced epigenetic reprogramming that convert ischemic intolerance to ischemic tolerance (Dharap et al., 2010; Lusardi et al., 2010). Many studies have shown that PC alters miRNA expression in rodent brain (Dharap and Vemuganti., 2010; Lee et al., 2010; Lusardi et al., 2010; Liu et al., 2012; Cao et al., 2012; Shi et al., 2013; Peng et al., 2013; Tripathi et al., 2014; Shin et al., 2014; Sun et al., 2015a; Vartanian et al., 2015; Feng et al., 2015b; Keasey et al., 2016). The window of maximal tolerance after ischemic PC is known to be 3 days. Our lab showed that many miRNAs alter in adult rat brain as early as 6h after PC and several of the changes sustain at least up to 3 days indicating their significance in promoting ischemic tolerance (Dharap and Vemuganti, 2010). Our studies showed upregulation of miRNAs like miR-374, miR-98, miR-340-5p, miR-21 and downregulation of miRNAs like miR-466c, miR-292-5p, miR-328, miR-873 following PC that modulate pathways like Notch signaling, ubiquitin-proteasomal system, gap junction proteins and nicotinamide adenine dinucleotide (NAD) metabolism that promote ischemic tolerance.
A previous study showed differential cerebral miRNA expression profiles between PC (15 min MCAO), ischemia (60 min MCAO) and ischemia followed by PC (15 min MCAO followed by a 60 min MCAO after 3 days) in adult mice (Lusardi et al., 2010). This study showed that PC decreases the levels of miR-132 concomitantly increasing the levels of its target methyl-CpG binding protein 2 (MeCP2) which is a transcriptional regulator. MeCP2 is thought to promote the PC-induced ischemic tolerance as MeCP2 knockout mice showed increased neuronal death compared to wild type controls when subjected to focal ischemia following PC (Lusardi et al., 2010). OGD-induced PC in hippocampal neurons showed. Increased expression of miRNAs miR-9, miR-21, miR-29b and miR-132 that were thought to be responsible for the ischemic tolerance (Keasey et al., 2016). This study is in contrast with the in vivo study by Lusardi et al., (2010). These authors thought that the upregulation of miR-132 mediates neuronal survival probably by targeting p250GAP, a Rho GTPase that decreases neuronal survival and by targeting MeCP2-mediated suppression of the expression of the neuroprotective BDNF (Keasey et al., 2016).
Hibernation in rodents is known to provide ischemic tolerance and during this phase expression levels of the miRNAs of miR-200 and miR-182 families were shown to be decreased leading to increased expression of the small ubiquitin-like modifier (SUMO) proteins which belongs to the ubiquitin-like protein modifier (ULM) family that promotes ischemic tolerance (Lee et al., 2012). Furthermore, inhibiting these miRNAs increased global protein SUMOylation that is thought to be critical for ischemic tolerance (Lee et al., 2012). Many pharmacological agents including 3-nitropropionic acid, lipopolysaccharide, estrogen, resveratrol and volatile anesthetics like isoflurane and sevoflurane are known to induce ischemic tolerance (Bracko et al., 2014; Vartanian et al., 2011; Raval et al., 2016; Koronowski et al., 2015; Lopez et al., 2016; Yan et al., 2016; Wang et al., 2011a). MiRNAs were thought to mediate the ischemic tolerance by some of these agents. Neuroprotection induced by morphine PC preceding an ischemic insult in primary cortical neurons was linked to the downregulation of miR-134 (Meng et al., 2016). Similarly, sevoflurane PC against a 6h hypoxic injury in PC12 cells led to altered expression of 14 miRNAs and of those downregulation of miR-101a and upregulation of miR-34b were linked to sevoflurane PC. Overexpression of miR-101a increased apoptosis. In contrast, overexpression of miR-34b increased cell viability and this was further confirmed when inhibition of miR-34b increased the number of the apoptotic cells (Sun et al., 2015b). Interestingly, a recent study from our lab showed that resveratrol PC does not alter miRNAs in mouse brain (Lopez et al., 2016).
Non-coding RNAs and hemorrhagic stroke
Hemorrhagic stroke occurs due to bleeding arising from the rupture of blood vessels that damages the surrounding brain tissue through activation of the secondary injury cascades including inflammation (Kumar et al., 2016, Sacco et al., 2013, Smith and Eskey, 2011). Hemorrhagic stroke can be due to intracerebral hemorrhage (ICH) or subarachnoid hemorrhage (SAH) (Sacco et al., 2013). The molecular mechanisms responsible for the onset and progression of either of the subtypes of hemorrhagic stroke are not completely understood. However, recent studies used antagomiRs and mimics to show the involvement of miRNAs in the pathophysiology of both types of hemorrhagic stroke (Yang et al., 2015, Kim et al., 2014, Muller et al., 2015) (Fig. 1). MiR-223 was found to target the nucleotide-binding domain, leucine-rich-containing family, pyrin domain-containing-3 (NLRP3) which contributes to microglial activation and neuronal injury following ICH in mice (Yang et al., 2015). The levels of miR-223 decreased after ICH and treatment with a miR-223 mimic reduced microglial activation, edema and inflammatory cytokine expression leading to decreased neuronal damage and neurological deficits (Yang et al., 2015). MiR-223 was also found to inhibit the inflammatory responses by reducing the levels of NLRP3, caspase-1 and IL-1β after ICH (Yang et al., 2015) (Fig. 1). Furthermore, reducing the levels of let-7c, which was observed to be highly upregulated after ICH in rats led to increased levels of its pro-survival targets like insulin like growth factor receptor 1 and p-Akt that in turn led to decreased edema, less neurological deficits and curtailed apoptosis (Kim et al., 2014). It was recently shown that miR-30a and miR-143 were significantly upregulated in cerebral arteries following SAH indicating that they might control vascular changes after SAH by targeting the connective tissue growth factor (Muller et al., 2015).
Non-coding RNAs and TBI
TBI in adult rats significantly alters the hippocampal miRNAome between 3h and 24h following injury and the bioinformatics analysis of TBI-responsive miRNAs predicted that they target mRNAs that modulate processes like cell differentiation and proliferation, signal transduction, transcriptional regulation and protein kinase activity (Redell et al., 2009). Altered hippocampal miRNA profiles following TBI in rodents between 1h to 7 days after injury was subsequently confirmed and bioinformatics analysis showed that the TBI-responsive miRNAs target mRNAs that control processes like response to stress, metabolism, protein binding, endocytic vesicle and synaptosomes that might be related to TBI pathophysiology (Liu et al., 2014b). Importantly, three of the miRNAs altered after TBI, miR-144, miR-153 and miR-340-5p target calcium/calmodulin-dependent serine protein kinase (CASK), nuclear factor erythroid 2-related factor 2 (NRF2) and α-synuclein (SNCA) which play significant roles in post-injury pathophysiology (Liu et al., 2014b). While CASK modulates synaptic function and dendritic spine formation (Hsueh 2006; Chao et al., 2008), NRF2 is a transcription factor that induces anti-oxidant gene expression and hence prevents neurodegeneration (Ma 2013, Calkins et al., 2009). SNCA is highly concentrated in presynaptic terminals and its abnormal accumulation is a defining pathological hallmark of neurodegeneration in Parkinson's disease (Cabin et al., 2002; Barker and Williams-Gray, 2016). Hence, these miRNAs altered after TBI might be functionally related to neurodegeneration seen after an acute injury. The miRNA profiles were also shown to be altered in the cerebral cortex in a rat model of fluid-percussion injury (Lei et al., 2009). Following TBI, significant alterations in the expression of several miRNAs were observed between 6h to 3 days, and one of those miRNAs called miR-21 was altered at all time points tested (Lei et al., 2009). The miR-21 was shown to have anti-apoptotic activity. Treatment with a miR-21 agomiR was shown to target the expression of apoptosis and angiogenesis related proteins leading to better neuronal survival after TBI in rodents (Ge et al., 2014). Increasing the levels of miR-21 by treatment with its agomiR was also shown to alleviate the BBB damage leading to better neurological functional recovery after TBI (Ge et al., 2015). Mechanistically, miR-21 was shown to arrest the loss of tight junction proteins like occludin and claudin-5 and increase the levels of Angiopoietin-1 and its receptor Tie-2 proteins which contribute to the BBB integrity (Ge et al., 2015) (Fig. 1). MiR-21 was also shown to be a molecular mediator that improves the cognition in mice subjected to running wheel exercise after TBI (Hu et al., 2015b; Miao et al., 2015). The protective potential of miR-21 might also due to its ability to block apoptosis by acting on the phosphatase and tensin homolog (PTEN)-Akt signaling pathway (Han et al., 2014).
Altered miRNA profiles were also shown following controlled cortical impact injury in adult mice (Meissner et al., 2015). The miR-2137 is one of the miRNAs that was found to be highly upregulated in the penumbral tissue surrounding the contusion at 6h following TBI, and was thought to be a marker of injury as it is not known to be expressed in normal brain (Meissner et al., 2015). Possible targets of miR-2137 include solute carrier family 12 member 5, pleckstrin and Sec7 domain containing protein that are involved in chloride homeostasis in neurons and development and maintenance of dendritic spines (Meissner et al., 2015; Funk et al., 2008; Choi et al., 2006). Sabirzhanov et al. (2016) showed that miR-711 upregulated after TBI in mouse cortex silences pro-survival Akt leading to neuronal death and administration of miR-711 hairpin inhibitor into injured cortex decreased the post-injury lesion volume, neuronal loss and behavioral deficits. TBI was also shown to downregulate miR-23a and miR-27a in mouse cortex following moderate CCI injury leading to an increase in their targets Noxa, Puma and Bax (Sabirzhanov et al., 2014). Treatment with antagomiRs for these miRNAs blocked the expression of these pro-apoptotic proteins leading to decreased neuronal death (Sabirzhanov et al., 2014). Jadhav et al. (2014) showed that TBI downregulates miR-200b in microglia leading to derepression of its target c-Jun N-terminal kinase that in turn mediates increased pro-inflammatory cytokine expression leading to neuronal death. MiRNA profiling of postmortem human cerebellar tissue from patients who had died from severe frontal cortex injuries also indicated significant upregulation of 13 miRNAs (Schober et al., 2015).
Non-coding RNAs and SCI
The lack of a definitive cure or treatment strategy for SCI is similar to that seen in TBI (Silva et al., 2014). This in turn has led to research efforts to better understand the molecular basis of secondary injury progression after SCI that has been attributed to cellular and molecular events such as oxidative stress, inflammation, apoptosis, glutamate excitotoxicity and endoplasmic reticulum stress (Jia et al., 2012; Donnelly and Popovich., 2008; Zhang et al., 2012b; Liu et al., 1999; Ohri et al., 2011). As ncRNAs can control all these mechanisms, studying their functional significance in regulating target proteins after SCI will provide better strategies to design therapies. Following SCI in adult rats, miRNAome was shown to alter in 3 different temporal patterns viz., upregulation, downregulation and lastly early upregulation at 4h followed by downregulation at 1 to 7 days (Liu et al., 2009). Bioinformatics analysis showed that miRNAs altered after SCI target genes involved in inflammation, oxidative stress and apoptosis and thus modulate the outcome after SCI (Liu et al., 2009). A subsequent study showed that miRNAs including miR210, miR300-3p, miR-325-5p, miR-487b and miR-16 altered after SCI might regulate the enzymes associated with cholesterol-metabolism like isopentenyl diphosphate isomerase-1 and farnesyl-diphosphate farnesyltransferase-1 (Chen et al., 2015b). This is understandable as cholesterol metabolism is known to be decreased after SCI (Gilbert et al., 2014; Myers et al., 2007) and SCI-responsive miRNAs might play a role in this. Using bioinformatics analysis, Liu et al. (2016b) showed that miRNAs altered after SCI target genes that promote plasticity and repair including those belonging to the neurotrophin signaling pathway. Expression of miR-124 was shown to be reduced in the perilesional region within 7 days following SCI (Zhao et al., 2015) and chitosan polyplex mediated delivery of miR-124 was shown to regulate the post-SCI microglial activation that aggravates the secondary injury (Louw et al., 2016) (Fig. 1). The miR-21 was shown to be significantly upregulated in corticospinal neurons following treatment with docosahexaenoic acid in parallel with enhancement of neural plasticity following SCI (Liu et al., 2015e). The increased expression of miR-21 might improve the functional recovery by targeting PTEN that has been linked to SCI pathophysiology (Liu et al., 2015e).
The role of miRNAs in neuronal apoptosis was reiterated in a recent study that showed that post-SCI apoptosis can be controlled by modulating miR-20a and miR-29b, and thus their targets myeloid cell leukemia 1 (anti-apoptotic) and BH3-only protein (pro-apoptotic) (Liu et al., 2015d) (Fig. 1). MiRNAs also modulate the post-SCI angiogenesis that helps plasticity. MiR-126 was shown to increase angiogenesis and reduce inflammation in a contusion SCI model in adult rodents by targeting Sprouty-related EVH1 domain-containing protein 1, phosphoinositol-3 kinase regulatory subunit 2, and also modulate inhibitors of angiogenic and cell survival signals in response to VEGF and vascular cell adhesion molecule 1 that promote vascular inflammation after injury (Hu et al., 2015a). Liu et al. (2015b) showed that miR-223 improves functional recovery in rats after SCI by promoting angiogenesis and blocking neuronal apoptosis by decreasing the expression of Bax and cleaved caspase-3 and increasing the levels of Bcl2 and GRIA1.
MicroRNAs as potential biomarkers for acute CNS injuries
There is an immediate need for identifying novel biomarkers in the body fluids that can inform the incidence, severity and response to treatment after an acute CNS injury. MiRNAs can fit this need as they are known to be released into both blood and CSF and remain stable for days. Hence, many studies evaluated their usefulness as biomarkers for identifying the onset of stroke, TBI and SCI (Reid et al., 2011; Tan et al., 2011; Wang et al., 2013).
MicroRNAs as stroke biomarkers
Stroke was shown to rapidly alter the miRNA expression profiles in blood in both rodents and humans (Sepramaniam et al., 2014; Selvamani et al., 2014; Liu et al., 2010; Tan et al., 2009; Jeyaseelan et al., 2008). In some studies, expression profiling of blood was conducted in parallel with brain (Liu et al., 2010; Jeyaseelan et al., 2008) and CSF (Sørensen et al., 2014) that showed overlapping patterns of miRNA levels between blood/brain and blood/CSF. The miRNAs that showed altered levels in blood after stroke include miR-107 (Yang et al., 2016b), miR-128b (Yang et al., 2016b), miR-153 (Yang et al., 2016b), miR-124 (Weng et al., 2011; Laterza et al., 2009), miR-210 (Zeng et al., 2011), miR-145 (Gan et al., 2012), miR-21 (Zhou and Zhang, 2014; Tsai et al., 2013), miR-221 (Tsai et al., 2013), miR-30a (Long et al., 2013), miR-126 (Chen et al., 2015a; Long et al., 2013), let-7b (Long et al., 2013), miR-223 (Wang et al., 2014d) and miR-24 (Zhou and Zhang., 2014). Importantly, Yang et al. (2014) showed that miR-107 was elevated in the plasma of both rats and humans after stroke. Zeng et al. (2013) highlighted the utility of using miRNAs together with protein biomarkers to identify ischemic stroke by demonstrating that a combination of an inflammatory cytokine, a hemostasis protein called fibrin degradation product and a repair-related miR-210 has a high sensitivity for predicting stroke recovery in humans. Furthermore, miR-122 was shown to be reduced in the blood after stroke in both rats and humans (Liu et al., 2010; Jickling et al., 2014). These studies illustrate the usefulness of blood miRNAs as stroke biomarkers.
Circulating miRNAs have also been studied as diagnostic biomarkers for diabetes-related stroke (Yang et al., 2016a; Duan et al., 2014). Decreased expression of miR-223 and miR-146a was observed in both the plasma and platelets in diabetic stroke patients compared to non-diabetic stroke patients and was correlated with higher blood glucose concentration and platelet activation rates (Duan et al., 2014). Increased miR-223 and decreased levels of miR-144 in platelets and plasma was thought to indicate a risk for ischemic stroke in type-2 diabetics (Yang et al., 2016a). Post-stroke depression is a common complication that is associated with increased morbidity and mortality (Whyte and Mulsant, 2002). A recent study that blood miRNAs can be used as biomarkers to predict the onset of post-stroke depression at an early stage (Zhang et al., 2016b). A correlation between increased serum miR-132 levels and post-stroke cognitive impairment was also shown recently in stroke patients (Huang et al., 2016).
The usefulness of circulating miRNAs as biomarkers to differentiate the stroke subtypes was also demonstrated in animals and humans. Liu et al. (2010) showed that the blood miRNA expression signatures between rats subjected to ischemic stroke and ICH are significantly different, but there is an overlap between brain and blood expression profile within a stroke subtype. Leung et al. (2014) showed that miR-124-3p levels were significantly higher while miR-16 levels were significantly lower in the plasma from hemorrhagic stroke patients compared to ischemic stroke patients indicating that blood miRNA profiles can predict the stroke subtypes in humans as well. A set of 30 inflammation-related miRNAs including miR-494, miR-1471 and miR-874 (upregulated) and miR-301a, miR-144 and miR-122 (downregulated) were observed to be significantly altered in plasma microvesicles after ICH in both male and female patients (Guo et al., 2013). Hematoma formation after ICH can be identified by miRNA profiling. Increased levels of circulating miRNAs like miR-522, miR-122, and miR-29c were observed in the plasma of ICH patients with hematoma compared to ICH patients without hematoma (Zheng et al., 2012). Blood miRNA profiling also indicated the possibility of identifying perihematoma edema (PHE) after ICH in both humans and rats (Zhu et al., 2015; Wang et al., 2016a). A correlation was observed between miR-126 levels in blood and the PHE volume in ICH patients (Zhu et al., 2015). The rat model of ICH indicated that increased levels of miR-29c and miR-122 in plasma correlates with PHE formation (Wang et al., 2016a). Rupture of intracranial aneurysms (IAs) is a major cause of hemorrhagic stroke, especially SAH (Krings et al., 2011). Changes in circulating miRNA profiles were shown to correlate with the presence of IAs (Jin et al., 2013; Li et al., 2014b). Jin et al. (2013) showed altered expression of 86 miRNAs (69 up- and 17 downregulated) in the serum of patients with aneurysms compared to control subjects. One of the miRNAs identified to be upregulated in this study (miR-25) was also shown by Li et al. (2014b). Another profiling study showed altered expression of 157 miRNAs (72 up- and 85 downregulated) in the intracranial aneurysmal tissue compared to normal superficial temporal arteries (Liu et al., 2014a).
MicroRNAs as TBI and SCI biomarkers
Usefulness of miRNAs as TBI biomarkers was evaluated in humans and animal models of acute CNS injuries. Plasma levels of miR-16 and miR-92a were shown to be elevated in humans following mild or severe TBI compared to controls (Redell et al., 2010). In mice, serum miRNA profiles were shown to be altered as early as 3h following a mild closed head injury compared to controls regardless of the severity of mild injury (Sharma et al., 2014). In adult rats, levels of let-7i were shown to be increased in both serum and CSF as early as 3h following blast TBI (Balakathiresan et al., 2012). Circulating miRNAs were also shown to act as biomarkers of pituitary dysfunction after TBI (Taheri et al., 2016). Serum levels of miR-126-3p and miR-3610 showed a correlation with hypopituitarism between day 1 to day 28 as well as at 5 years following TBI (Taheri et al., 2016). The miRNAs were shown to serve as biomarkers of injury severity and recovery after acute SCI in rodents (Hachisuka et al., 2014). Serum miRNA profiling in a mouse SCI model showed increased levels of miR-9*, miR-219 and miR-384-5p that correlated with the severity of SCI (Hachisuka et al., 2014).
Other non-coding RNAs altered following acute CNS injuries
Human and rodent genomes are replete with many types of ncRNA genes. Importantly, ~68% of the genes expressed in humans are lncRNAs making them the largest class of ncRNAs (Iyer et al., 2015). The functions of lncRNAs are not fully deciphered, but they were shown to play diverse roles in regulating cell proliferation, survival and migration, and maintaining the genomic stability by acting as guides, decoys, scaffolds and signaling molecules (Kung et al., 2013; Rinn and Chang., 2012; Mercer et al., 2009). The lncRNAs possess multiple domains that enable them to bind to DNA, RNA and protein to modify chromatin states, transcription and translation, to ultimately control the gene expression (Mercer and Mattick, 2013). By definition, lncRNAs are >200 nucleotides long, transcribed by RNA polymerases II or III, capped and polyadenylated similar to mRNAs, but lack the protein-coding potential due to truncated open reading frames (Kung et al., 2013; Mercer et al., 2009). They are sub-classified as long intergenic RNAs (lincRNAs) and transcribed ultraconserved regions (T-UCRs). LincRNAs transcribed from the intergenic regions of the genome regulate the expression of neighboring as well as distant genes (Guttman et al., 2011). Whereas, the functions of T-UCRs are not yet deciphered. While lincRNAs are least conserved, T-UCRs are most conserved between mammals and rodents (Bejerano et al., 2004).
Our lab showed that focal ischemia in adult rats extensively alters the cerebral lncRNA expression profiles during the acute reperfusion period (Dharap et al., 2012). Functional significance of the lncRNAs in post-stroke outcome is currently not known, however they might play a significant role in modifying the post-stroke epigenetic landscape. In support of this notion, a study from our lab showed that binding of many stroke-responsive lncRNAs to chromatin-modifying proteins (CMPs) Sin3A and coREST increases significantly after focal ischemia (Dharap et al., 2013). These 2 CMPs are co-repressors of the transcription factor REST which is known to promote ischemic neuronal death (Noh et al., 2012; Pandi et al., 2013; Hwang et al., 2014; Mehta et al., 2015). A recent study also showed that binding of an lncRNA named Fos Downstream Transcript (FosDT) upregulated after stroke is important for the post-ischemic regulation of REST target genes NF-κB2, GRIA2 (encodes the ionotropic glutamate receptor AMPA type subunit 2) and GRIN1 (encodes the ionotropic glutamate receptor NMDA type subunit 1) which play a significant role in the post-stroke brain damage (Mehta et al., 2015) (Fig. 1). Another recent study showed that increased expression of lncRNA CAMK2D-associated transcript 1 (C2dat1) promotes the expression of calcium/calmodulin-dependent kinase II (CaMKII) that mediates ischemic neuronal death (Xu et al., 2016) (Fig. 1). Further, silencing C2dat1 led to reduced expression of CaMKII and thus blocked the downstream NF-κB signaling that is known to kill neurons in the post-ischemic brain (Xu et al., 2016). Another lncRNA called Malat1 was shown to be involved in the protection of cerebral microvessels after an ischemic insult. Malat1 levels increased in cultured endothelial cells following OGD and in cerebral microvessels in mice subjected to transient MCAO (Zhang et al., 2016a). Furthermore, loss of Malat1 function induced by an LNA-GapmeR significantly increased OGD-induced endothelial damage (Yuan et al., 2015).
Role of lncRNAs was also studied after other acute CNS injuries including TBI, SCI and SAH. A recent study showed the expression of several lncRNAs was altered in mouse cerebral cortex at 24h after TBI (Zhong et al., 2016). This study also evaluated the effect of TBI on mRNA expression simultaneously. Bioinformatics analysis of the co-expression networks of mRNAs and lncRNAs altered after TBI showed that a majority of them are associated with inflammatory and immunological activity, metabolism, neuronal and vascular networks and cellular function (Zhong et al., 2016) (Fig. 1). Transplantation of human adipose-derived stem cells (hADSCs) and treatment with the conditioned media from the hADSCs improved the motor and cognitive functions, and reduced secondary lesion volume following TBI in young rats (Tajiri et al., 2014). These beneficial effects were attributed to lncRNAs NEAT1 and MALAT1 secreted from the hADSCs (Tajiri et al., 2014) (Fig. 1). A recent study also showed that an lncRNA called lncSCIR1 was downregulated following SCI in rats and its inhibition contributes to gliosis by selectively targeting the expression of adrenomedullin (Adm), bone morphogenetic protein 7 (BMP7), α-synuclein (α-syn) and wingless-type MMTV integration site family, member 3 (Wnt3) (Wang et al., 2015) (Fig. 1). While Adm activates pro-inflammatory cytokines, BMP7 prevents axonal growth, α-syn promotes post-SCI neuronal death and Wnt3 is essential for neural regeneration after SCI (Zeng et al., 2014b; Matsuura et al., 2008; Busch and Morgan, 2012; Yin et al., 2008). SAH was also shown to significantly alter the lncRNA expression profiles in adult rat brain (Zheng et al., 2015). Bioinformatics analysis of their co-expressed mRNAs indicated that pathways associated with inflammation, neuroactive ligand-receptor interaction, calcium signaling and antigen processing and presentation were probably controlled by these lncRNAs in promoting early brain injury following SAH (Zheng et al., 2015).
PiRNAs are a large class of ncRNAs which are 24 to 31 nucleotides in length and lack sequence conservation between species (O'Donell et al., 2007). Although their functional role and method of biogenesis are not clearly established, they are thought to control post-transcriptional silencing of retrotransposons to prevent mutations and thus maintains genome integrity (Halic and Moazed., 2009; Iwasaki et al., 2015). A study from our lab showed that stroke significantly alters piRNA expression profiles in rat brain (Dharap et al., 2011). Bioinformatics analysis showed that the stroke-responsive piRNAs target various classes of transposons and thus might be involved in maintaining genetic equilibrium and a set of transcription factors redundantly control the piRNA gene promoters after stroke (Dharap et al., 2011).
Conclusion
The molecular mechanisms responsible for the high morbidity and mortality associated with acute CNS injuries like ischemic stroke, TBI and SCI are not yet completely understood. Furthermore, no therapeutic agents are currently available to control the secondary brain damage and neurological dysfunction after acute CNS insults. Various studies discussed in this review indicate that ncRNAs play crucial roles in neurological dysfunction after acute CNS insults and may pave ways to devise newer molecular therapeutic options by modulating them. As ncRNAs target multiple pathologic mechanisms, they are attractive to control post-injury brain damage efficiently. Studies in both animal models and humans have also showed altered expression profiles of circulating ncRNAs following acute CNS injuries indicating their potential to be used as diagnostic and prognostic biomarkers.
Highlights.
-
(1)
Greater than 98% of the transcriptional output in mammals is noncoding RNAs.
-
(2)
Noncoding RNAs control transcription and translation.
-
(3)
Acute injuries to CNS that include stroke and traumatic brain injury significantly alters noncoding RNA expression and function.
-
(4)
Post-injury brain damage can be altered by modulating noncoding RNAs like microRNAs and long noncoding RNAs.
-
(5)
Noncoding RNAs in blood can be used as prognostic and diagnostic biomarkers for acute CNS injuries.
Acknowledgements
The authors are supported by grants from American Heart Association, National Institute of Health and Veterans Administration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest The authors declare that there are no conflicts of interest.
REFERENCES
- Balakathiresan N, Bhomia M, Chandran R, Chavko M, McCarron RM, Maheshwari RK. MicroRNA let-7i is a promising serum biomarker for blast-induced traumatic brain injury. J Neurotrauma. 2012;29:1379–1387. doi: 10.1089/neu.2011.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker RA, Williams-Gray CH. Review: The spectrum of clinical features seen with alpha synuclein pathology. Neuropathol Appl Neurobiol. 2016;42:6–19. doi: 10.1111/nan.12303. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O, Di Pietro V, Lazzarino G, Amorini AM, Tavazzi B, Artmann J, Wong EC, Buxton RB, Weller M, Luft AR, Wegener S. 3-Nitropropionic acid-induced ischemia tolerance in the rat brain is mediated by reduced metabolic activity and cerebral blood flow. J Cereb Blood Flow Metab. 2014;34:1522–1530. doi: 10.1038/jcbfm.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton BR, Reutens DC, Sobey CG. Apoptotic mechanisms after cerebral ischemia. Stroke. 2009;40:e331–339. doi: 10.1161/STROKEAHA.108.531632. [DOI] [PubMed] [Google Scholar]
- Burguillos MA, Deierborg T, Kavanagh E, Persson A, Hajji N, Garcia-Quintanilla A, Cano J, Brundin P, Englund E, Venero JL, Joseph B. Caspase signalling controls microglia activation and neurotoxicity. Nature. 2011;472:319–324. doi: 10.1038/nature09788. [DOI] [PubMed] [Google Scholar]
- Busch DJ, Morgan JR. Synuclein accumulation is associated with cell-specific neuronal death after spinal cord injury. J Comp Neurol. 2012;520:1751–1771. doi: 10.1002/cne.23011. [DOI] [PubMed] [Google Scholar]
- Caballero-Garrido E, Pena-Philippides JC, Lordkipanidze T, Bragin D, Yang Y, Erhardt EB, Roitbak T. In Vivo Inhibition of miR-155 Promotes Recovery after Experimental Mouse Stroke. J Neurosci. 2015;35:12446–12464. doi: 10.1523/JNEUROSCI.1641-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabin DE, Shimazu K, Murphy D, Cole NB, Gottschalk W, McIlwain KL, Orrison B, Chen A, Ellis CE, Paylor R, Lu B, Nussbaum RL. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha-synuclein. J Neurosci. 2002;22:8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins MJ, Johnson DA, Townsend JA, Vargas MR, Dowell JA, Williamson TP, Kraft AD, Lee JM, Li J, Johnson JA. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid Redox Signal. 2009;11:497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Feng C, Li L, Zuo Z. Contribution of microRNA-203 to the isoflurane preconditioning-induced neuroprotection. Brain Res Bull. 2012;88:525–528. doi: 10.1016/j.brainresbull.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HW, Hong CJ, Huang TN, Lin YL, Hsueh YP. SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J Cell Biol. 2008;182:141–155. doi: 10.1083/jcb.200712094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Du Y, Esposito E, Liu Y, Guo S, Wang X, Lo EH, Xing C, Ji X. Effects of Focal Cerebral Ischemia on Exosomal Versus Serum miR126. Transl Stroke Res. 2015a;6:478–484. doi: 10.1007/s12975-015-0429-3. [DOI] [PubMed] [Google Scholar]
- Chen G, Fang X, Yu M. Regulation of gene expression in rats with spinal cord injury based on microarray data. Mol Med Rep. 2015b;12:2465–2472. doi: 10.3892/mmr.2015.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xu J, Li L, Li H, Mao S, Zhang F, Zen K, Zhang CY, Zhang Q. MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis. 2014;5:e1132. doi: 10.1038/cddis.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee JR, Lee HW, Kim K, Chung HS, Kim H, Kim E. ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci. 2006;26:4811–4819. doi: 10.1523/JNEUROSCI.4182-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012;43:2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Pokrzywa C, Vemuganti R. Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST. ASN Neuro. 2013;5:283–289. doi: 10.1042/AN20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Vemuganti R. Ischemic pre-conditioning alters cerebral microRNAs that are upstream to neuroprotective signaling pathways. J Neurochem. 2010;113:1685–1691. doi: 10.1111/j.1471-4159.2010.06735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly DJ, Popovich PG. Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp Neurol. 2008;209:378–388. doi: 10.1016/j.expneurol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Zhan Q, Song B, Zeng S, Zhou J, Long Y, Lu J, Li Z, Yuan M, Chen X, Yang Q, Xia J. Detection of platelet microRNA expression in patients with diabetes mellitus with or without ischemic stroke. J Diabetes Complications. 2014;28:705–710. doi: 10.1016/j.jdiacomp.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Feng N, Wang Z, Zhang Z, He X, Wang C, Zhang L. miR-487b promotes human umbilical vein endothelial cell proliferation, migration, invasion and tube formation through regulating THBS1. Neurosci Lett. 2015a;591:1–7. doi: 10.1016/j.neulet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Feng Y, Li W, Wang JQ. microRNA-33A expression is reduced in cerebral cortex in a rat model of ischemic tolerance. Cell Mol Biol (Noisy-le-grand) 2015b;61:24–29. [PubMed] [Google Scholar]
- Funk K, Woitecki A, Franjic-Wurtz C, Gensch T, Mohrlen F, Frings S. Modulation of chloride homeostasis by inflammatory mediators in dorsal root ganglion neurons. Mol Pain. 2008;4:32. doi: 10.1186/1744-8069-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan CS, Wang CW, Tan KS. Circulatory microRNA-145 expression is increased in cerebral ischemia. Genet Mol Res. 2012;11:147–152. doi: 10.4238/2012.January.27.1. [DOI] [PubMed] [Google Scholar]
- Ge X, Han Z, Chen F, Wang H, Zhang B, Jiang R, Lei P, Zhang J. MiR-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain Res. 2015;1603:150–157. doi: 10.1016/j.brainres.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, Li SH, Jiang RC, Kang CS, Zhang JN. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. 2014;4:6718. doi: 10.1038/srep06718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert O, Croffoot JR, Taylor AJ, Nash M, Schomer K, Groah S. Serum lipid concentrations among persons with spinal cord injury - a systematic review and meta-analysis of the literature. Atherosclerosis. 2014;232:305–312. doi: 10.1016/j.atherosclerosis.2013.11.028. [DOI] [PubMed] [Google Scholar]
- Gubern C, Camos S, Ballesteros I, Rodriguez R, Romera VG, Canadas R, Lizasoain I, Moro MA, Serena J, Mallolas J, Castellanos M. miRNA expression is modulated over time after focal ischaemia: up-regulation of miR-347 promotes neuronal apoptosis. FEBS J. 2013;280:6233–6246. doi: 10.1111/febs.12546. [DOI] [PubMed] [Google Scholar]
- Guo D, Liu J, Wang W, Hao F, Sun X, Wu X, Bu P, Zhang Y, Liu Y, Liu F, Zhang Q, Jiang F. Alteration in abundance and compartmentalization of inflammation-related miRNAs in plasma after intracerebral hemorrhage. Stroke. 2013;44:1739–1742. doi: 10.1161/STROKEAHA.111.000835. [DOI] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachisuka S, Kamei N, Ujigo S, Miyaki S, Yasunaga Y, Ochi M. Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal Cord. 2014;52:596–600. doi: 10.1038/sc.2014.86. [DOI] [PubMed] [Google Scholar]
- Halic M, Moazed D. Transposon silencing by piRNAs. Cell. 2009;138:1058–1060. doi: 10.1016/j.cell.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Chen F, Ge X, Tan J, Lei P, Zhang J. miR-21 alleviated apoptosis of cortical neurons through promoting PTEN-Akt signaling pathway in vitro after experimental traumatic brain injury. Brain Res. 2014;1582:12–20. doi: 10.1016/j.brainres.2014.07.045. [DOI] [PubMed] [Google Scholar]
- Hsueh YP. The role of the MAGUK protein CASK in neural development and synaptic function. Curr Med Chem. 2006;13:1915–1927. doi: 10.2174/092986706777585040. [DOI] [PubMed] [Google Scholar]
- Hu J, Zeng L, Huang J, Wang G, Lu H. miR-126 promotes angiogenesis and attenuates inflammation after contusion spinal cord injury in rats. Brain Res. 2015a;1608:191–202. doi: 10.1016/j.brainres.2015.02.036. [DOI] [PubMed] [Google Scholar]
- Hu T, Zhou FJ, Chang YF, Li YS, Liu GC, Hong Y, Chen HL, Xiyang YB, Bao TH. miR-21 is Associated with the Cognitive Improvement Following Voluntary Running Wheel Exercise in TBI Mice. J Mol Neurosci. 2015b;57:114–122. doi: 10.1007/s12031-015-0584-8. [DOI] [PubMed] [Google Scholar]
- Huang S, Zhao J, Huang D, Zhuo L, Liao S, Jiang Z. Serum miR-132 is a risk marker of post-stroke cognitive impairment. Neurosci Lett. 2016;615:102–106. doi: 10.1016/j.neulet.2016.01.028. [DOI] [PubMed] [Google Scholar]
- Hunsberger JG, Fessler EB, Wang Z, Elkahloun AG, Chuang DM. Post-insult valproic acid-regulated microRNAs: potential targets for cerebral ischemia. Am J Transl Res. 2012;4:316–332. [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Kaneko N, Noh KM, Pontarelli F, Zukin RS. The gene silencing transcription factor REST represses miR-132 expression in hippocampal neurons destined to die. J Mol Biol. 2014;426:3454–3466. doi: 10.1016/j.jmb.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav SP, Kamath SP, Choolani M, Lu J, Dheen ST. microRNA-200b modulates microglia-mediated neuroinflammation via the cJun/MAPK pathway. J Neurochem. 2014;130:388–401. doi: 10.1111/jnc.12731. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- Jia Z, Zhu H, Li J, Wang X, Misra H, Li Y. Oxidative stress in spinal cord injury and antioxidant-based intervention. Spinal Cord. 2012;50:264–274. doi: 10.1038/sc.2011.111. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Li L, Tan X, Liu B, Zhang Y, Li C. miR-210 mediates vagus nerve stimulation-induced antioxidant stress and anti-apoptosis reactions following cerebral ischemia/reperfusion injury in rats. J Neurochem. 2015;134:173–181. doi: 10.1111/jnc.13097. [DOI] [PubMed] [Google Scholar]
- Jickling GC, Ander BP, Zhan X, Noblett D, Stamova B, Liu D. microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS One. 2014;9:e99283. doi: 10.1371/journal.pone.0099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Li C, Ge H, Jiang Y, Li Y. Circulating microRNA: a novel potential biomarker for early diagnosis of intracranial aneurysm rupture a case control study. J Transl Med. 2013;11:296. doi: 10.1186/1479-5876-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasey MP, Scott HL, Bantounas I, Uney JB, Kelly S. MiR-132 is Upregulated by Ischemic Preconditioning of Cultured Hippocampal Neurons and Protects them from Subsequent OGD Toxicity. J Mol Neurosci. 2016;59:404–410. doi: 10.1007/s12031-016-0740-9. [DOI] [PubMed] [Google Scholar]
- Kim JM, Lee ST, Chu K, Jung KH, Kim JH, Yu JS, Kim S, Kim SH, Park DK, Moon J, Ban J, Kim M, Lee SK, Roh JK. Inhibition of Let7c microRNA is neuroprotective in a rat intracerebral hemorrhage model. PLoS One. 2014;9:e97946. doi: 10.1371/journal.pone.0097946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronowski KB, Dave KR, Saul I, Camarena V, Thompson JW, Neumann JT, Young JI, Perez-Pinzon MA. Resveratrol Preconditioning Induces a Novel Extended Window of Ischemic Tolerance in the Mouse Brain. Stroke. 2015;46:2293–2298. doi: 10.1161/STROKEAHA.115.009876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostandy BB. The role of glutamate in neuronal ischemic injury: the role of spark in fire. Neurol Sci. 2012;33:223–237. doi: 10.1007/s10072-011-0828-5. [DOI] [PubMed] [Google Scholar]
- Krings T, Mandell DM, Kiehl TR, Geibprasert S, Tymianski M, Alvarez H, terBrugge KG, Hans FJ. Intracranial aneurysms: from vessel wall pathology to therapeutic approach. Nat Rev Neurol. 2011;7:547–559. doi: 10.1038/nrneurol.2011.136. [DOI] [PubMed] [Google Scholar]
- Kumar A, Aakriti, Gupta V. A review on animal models of stroke: An update. Brain Res Bull. 2016;122:35–44. doi: 10.1016/j.brainresbull.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, Glaab WE. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- Lee ST, Chu K, Jung KH, Yoon HJ, Jeon D, Kang KM, Park KH, Bae EK, Kim M, Lee SK, Roh JK. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Johnson KR, Hallenbeck JM. Global protein conjugation by ubiquitin-like-modifiers during ischemic stress is regulated by microRNAs and confers robust tolerance to ischemia. PLoS One. 2012;7:e47787. doi: 10.1371/journal.pone.0047787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- Leung LY, Chan CP, Leung YK, Jiang HL, Abrigo JM, Wang de F, Chung JS, Rainer TH, Graham CA. Comparison of miR-124-3p and miR-16 for early diagnosis of hemorrhagic and ischemic stroke. Clin Chim Acta. 2014;433:139–44. doi: 10.1016/j.cca.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Li LJ, Huang Q, Zhang N, Wang GB, Liu YH. miR-376b-5p regulates angiogenesis in cerebral ischemia. Mol Med Rep. 2014a;10:527–535. doi: 10.3892/mmr.2014.2172. [DOI] [PubMed] [Google Scholar]
- Li P, Zhang Q, Wu X, Yang X, Zhang Y, Li Y, Jiang F. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J Am Heart Assoc. 2014b;3:e000972. doi: 10.1161/JAHA.114.000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mao L, Gao Y, Baral S, Zhou Y, Hu B. MicroRNA-107 contributes to post-stroke angiogenesis by targeting Dicer-1. Sci Rep. 2015;5:13316. doi: 10.1038/srep13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Peng Z, Zhang N, Yu L, Han S, Li D, Li J. Identification of differentially expressed microRNAs and their PKC-isoform specific gene network prediction during hypoxic pre-conditioning and focal cerebral ischemia of mice. J Neurochem. 2012;120:830–841. doi: 10.1111/j.1471-4159.2011.07624.x. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhao L, Han S, Li J, Li D. Identification and Functional Analysis of MicroRNAs in Mice following Focal Cerebral Ischemia Injury. Int J Mol Sci. 2015a;16:24302–24318. doi: 10.3390/ijms161024302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Han L, Wu X, Yang X, Zhang Q, Jiang F. Genome-wide microRNA changes in human intracranial aneurysms. BMC Neurol. 2014a;14:188. doi: 10.1186/s12883-014-0188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Huang Y, Jia C, Li Y, Liang F, Fu Q. Administration of antagomir-223 inhibits apoptosis, promotes angiogenesis and functional recovery in rats with spinal cord injury. Cell Mol Neurobiol. 2015b;35:483–491. doi: 10.1007/s10571-014-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Xu GY, Pan E, McAdoo DJ. Neurotoxicity of glutamate at the concentration released upon spinal cord injury. Neuroscience. 1999;93:1383–1389. doi: 10.1016/s0306-4522(99)00278-x. [DOI] [PubMed] [Google Scholar]
- Liu da Z, Jickling GC, Ander BP, Hull H, Zhan X, Cox C, Shroff N, Dykstra-Aiello C, Stamova B, Sharp FR. Elevating microRNA-122 in blood improves outcomes after temporary middle cerebral artery occlusion in rats. J Cereb Blood Flow Metab. 2016a;36:1374–1383. doi: 10.1177/0271678X15610786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sun T, Liu Z, Chen X, Zhao L, Qu G, Li Q. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS One. 2014b;9:e103948. doi: 10.1371/journal.pone.0103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zhao H, Wang R, Wang P, Tao Z, Gao L, Yan F, Liu X, Yu S, Ji X, Luo Y. MicroRNA-424 protects against focal cerebral ischemia and reperfusion injury in mice by suppressing oxidative stress. Stroke. 2015c;46:513–519. doi: 10.1161/STROKEAHA.114.007482. [DOI] [PubMed] [Google Scholar]
- Liu XJ, Zheng XP, Zhang R, Guo YL, Wang JH. Combinatorial effects of miR-20a and miR-29b on neuronal apoptosis induced by spinal cord injury. Int J Clin Exp Pathol. 2015d;8:3811–3818. [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Han N, Li Q, Li Z. Bioinformatics Analysis of microRNA Time-Course Expression in Brown Rat (Rattus norvegicus): Spinal Cord Injury Self-Repair. Spine (Phila Pa 1976) 2016b;41:97–103. doi: 10.1097/BRS.0000000000001323. [DOI] [PubMed] [Google Scholar]
- Liu ZH, Yip PK, Adams L, Davies M, Lee JW, Michael GJ, Priestley JV, Michael-Titus AT. A Single Bolus of Docosahexaenoic Acid Promotes Neuroplastic Changes in the Innervation of Spinal Cord Interneurons and Motor Neurons and Improves Functional Recovery after Spinal Cord Injury. J Neurosci. 2015e;35:12733–12752. doi: 10.1523/JNEUROSCI.0605-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G, Wang F, Li H, Yin Z, Sandip C, Lou Y, Wang Y, Chen C, Wang DW. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013;13:178. doi: 10.1186/1471-2377-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MS, Dempsey RJ, Vemuganti R. Resveratrol preconditioning induces cerebral ischemic tolerance but has minimal effect on cerebral microRNA profiles. J Cereb Blood Flow Metab. 2016;36:1644–1650. doi: 10.1177/0271678X16656202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YL, Guo F, Liu F, Gao FL, Zhang PQ, Niu X, Guo SC, Yin JH, Wang Y, Deng ZF. miR-210 activates notch signaling pathway in angiogenesis induced by cerebral ischemia. Mol Cell Biochem. 2012;370:45–51. doi: 10.1007/s11010-012-1396-6. [DOI] [PubMed] [Google Scholar]
- Louw AM, Kolar MK, Novikova LN, Kingham PJ, Wiberg M, Kjems J, Novikov LN. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine. 2016;12:643–653. doi: 10.1016/j.nano.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Lu XC, Williams AJ, Yao C, Berti R, Hartings JA, Whipple R, Vahey MT, Polavarapu RG, Woller KL, Tortella FC, Dave JR. Microarray analysis of acute and delayed gene expression profile in rats after focal ischemic brain injury and reperfusion. J Neurosci Res. 2004;77:843–857. doi: 10.1002/jnr.20218. [DOI] [PubMed] [Google Scholar]
- Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, Simon RP, Saugstad JA. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi TA, Murphy SJ, Phillips JI, Chen Y, Davis CM, Young JM, Thompson SJ, Saugstad JA. MicroRNA responses to focal cerebral ischemia in male and female mouse brain. Front Mol Neurosci. 2014;7:11. doi: 10.3389/fnmol.2014.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madathil SK, Nelson PT, Saatman KE, Wilfred BR. MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays. 2011;33:21–26. doi: 10.1002/bies.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Taniguchi J, Hata K, Saeki N, Yamashita T. BMP inhibition enhances axonal growth and functional recovery after spinal cord injury. J Neurochem. 2008;105:1471–1479. doi: 10.1111/j.1471-4159.2008.05251.x. [DOI] [PubMed] [Google Scholar]
- Mehta SL, Kim T, Vemuganti R. Long Noncoding RNA FosDT Promotes Ischemic Brain Injury by Interacting with REST-Associated Chromatin-Modifying Proteins. J Neurosci. 2015;35:16443–16449. doi: 10.1523/JNEUROSCI.2943-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner L, Gallozzi M, Balbi M, Schwarzmaier S, Tiedt S, Terpolilli NA, Plesnila N. Temporal Profile of MicroRNA Expression in Contused Cortex after Traumatic Brain Injury in Mice. J Neurotrauma. 2016;33:713–720. doi: 10.1089/neu.2015.4077. [DOI] [PubMed] [Google Scholar]
- Meng F, Li Y, Chi W, Li J. Morphine Preconditioning Downregulates MicroRNA-134 Expression Against Oxygen-Glucose Deprivation Injuries in Cultured Neurons of Mice. J Neurosurg Anesthesiol. 2016;28:195–202. doi: 10.1097/ANA.0000000000000204. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Miao W, Bao TH, Han JH, Yin M, Yan Y, Wang WW, Zhu YH. Voluntary exercise prior to traumatic brain injury alters miRNA expression in the injured mouse cerebral cortex. Braz J Med Biol Res. 2015;48:433–439. doi: 10.1590/1414-431X20144012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JM, Xu L, Giffard RG. Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J Cereb Blood Flow Metab. 2013;33:1976–1982. doi: 10.1038/jcbfm.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AH, Povlsen GK, Bang-Berthelsen CH, Kruse LS, Nielsen J, Warfvinge K, Edvinsson L. Regulation of microRNAs miR-30a and miR-143 in cerebral vasculature after experimental subarachnoid hemorrhage in rats. BMC Genomics. 2015;16:119. doi: 10.1186/s12864-015-1341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J, Lee M, Kiratli J. Cardiovascular disease in spinal cord injury: an overview of prevalence, risk, evaluation, and management. Am J Phys Med Rehabil. 2007;86:142–152. doi: 10.1097/PHM.0b013e31802f0247. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer's disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neo WH, Yap K, Lee SH, Looi LS, Khandelia P, Neo SX, Makeyev EV, Su IH. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J Biol Chem. 2014;289:20788–20801. doi: 10.1074/jbc.M113.525493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Wang X, Chen S, Liu H, Wang Y, Xu X, Cheng J, Jia J, Zhen X. MicroRNA let-7c-5p protects against cerebral ischemia injury via mechanisms involving the inhibition of microglia activation. Brain Behav Immun. 2015;49:75–85. doi: 10.1016/j.bbi.2015.04.014. [DOI] [PubMed] [Google Scholar]
- Noh KM, Hwang JY, Follenzi A, Athanasiadou R, Miyawaki T, Greally JM, Bennett MV, Zukin RS. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A. 2012;109:E962–971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri SS, Maddie MA, Zhao Y, Qiu MS, Hetman M, Whittemore SR. Attenuating the endoplasmic reticulum stress response improves functional recovery after spinal cord injury. Glia. 2011;59:1489–1502. doi: 10.1002/glia.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandi G, Nakka VP, Dharap A, Roopra A, Vemuganti R. MicroRNA miR-29c down-regulation leading to de-repression of its target DNA methyltransferase 3a promotes ischemic brain damage. PLoS One. 2013;8:e58039. doi: 10.1371/journal.pone.0058039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Li J, Li Y, Yang X, Feng S, Han S, Li J. Downregulation of miR-181b in mouse brain following ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1. J Neurosci Res. 2013;91:1349–1362. doi: 10.1002/jnr.23255. [DOI] [PubMed] [Google Scholar]
- Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Wu J, Chen D, Zhao F, Liu J, Yang C, Wei D, Ferriero DM, Mu D. MiR-139-5p inhibits HGTD-P and regulates neuronal apoptosis induced by hypoxia-ischemia in neonatal rats. Neurobiol Dis. 2014;63:184–93. doi: 10.1016/j.nbd.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval AP, Bramlett H, Perez-Pinzon MA. Estrogen preconditioning protects the hippocampal CA1 against ischemia. Neuroscience. 2006;141:1721–1730. doi: 10.1016/j.neuroscience.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redell JB, Moore AN, Ward NH, 3rd, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microRNA levels. J Neurotrauma. 2010;27:2147–2156. doi: 10.1089/neu.2010.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80:193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanov B, Stoica BA, Zhao Z, Loane DJ, Wu J, Dorsey SG, Faden AI. miR-711 upregulation induces neuronal cell death after traumatic brain injury. Cell Death Differ. 2016;23:654–668. doi: 10.1038/cdd.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C, Dorsey SG, Faden AI. Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci. 2014;34:10055–10071. doi: 10.1523/JNEUROSCI.1260-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco RL, Kasner SE, Broderick JP, Caplan LR, Connors JJ, Culebras A, Elkind MS, George MG, Hamdan AD, Higashida RT, Hoh BL, Janis LS, Kase CS, Kleindorfer DO, Lee JM, Moseley ME, Peterson ED, Turan TN, Valderrama AL, Vinters HV. An updated definition of stroke for the 21st century: a statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2013;44:2064–2089. doi: 10.1161/STR.0b013e318296aeca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Kastner R, Zhang B, Belayev L, Khoutorova L, Amin R, Busto R, Ginsberg MD. DNA microarray analysis of cortical gene expression during early recirculation after focal brain ischemia in rat. Brain Res Mol Brain Res. 2002;108:81–93. doi: 10.1016/s0169-328x(02)00516-8. [DOI] [PubMed] [Google Scholar]
- Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober K, Ondruschka B, Dressler J, Abend M. Detection of hypoxia markers in the cerebellum after a traumatic frontal cortex injury: a human postmortem gene expression analysis. Int J Legal Med. 2015;129:701–707. doi: 10.1007/s00414-014-1129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamani A, Sathyan P, Miranda RC, Sohrabji F. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS One. 2012;7:e32662. doi: 10.1371/journal.pone.0032662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamani A, Williams MH, Miranda RC, Sohrabji F. Circulating miRNA profiles provide a biomarker for severity of stroke outcomes associated with age and sex in a rat model. Clin Sci (Lond) 2014;127:77–89. doi: 10.1042/CS20130565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepramaniam S, Tan JR, Tan KS, DeSilva DA, Tavintharan S, Woon FP, Wang CW, Yong FL, Karolina DS, Kaur P, Liu FJ, Lim KY, Armugam A, Jeyaseelan K. Circulating microRNAs as biomarkers of acute stroke. Int J Mol Sci. 2014;15:1418–1432. doi: 10.3390/ijms15011418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Chandran R, Barry ES, Bhomia M, Hutchison MA, Balakathiresan NS, Grunberg NE, Maheshwari RK. Identification of serum microRNA signatures for diagnosis of mild traumatic brain injury in a closed head injury model. PLoS One. 2014;9:e112019. doi: 10.1371/journal.pone.0112019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp FR, Jickling GC, Stamova B, Tian Y, Zhan X, Liu D, Kuczynski B, Cox CD, Ander BP. Molecular markers and mechanisms of stroke: RNA studies of blood in animals and humans. J Cereb Blood Flow Metab. 2011;31:1513–1531. doi: 10.1038/jcbfm.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Li P, Liang W, Chen J, Gao Y. Mechanisms of microRNA-mediated regulation of angiogenesis. Front Biosci (Elite Ed) 2010;2:1304–1319. doi: 10.2741/e191. [DOI] [PubMed] [Google Scholar]
- Shi H, Sun BL, Zhang J, Lu S, Zhang P, Wang H, Yu Q, Stetler RA, Vosler PS, Chen J, Gao Y. miR-15b suppression of Bcl-2 contributes to cerebral ischemic injury and is reversed by sevoflurane preconditioning. CNS Neurol Disord Drug Targets. 2013;12:381–391. doi: 10.2174/1871527311312030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Park YM, Kim DH, Moon GJ, Bang OY, Ohn T, Kim HH. Ischemic brain extract increases SDF-1 expression in astrocytes through the CXCR2/miR-223/miR-27b pathway. Biochim Biophys Acta. 2014;1839:826–836. doi: 10.1016/j.bbagrm.2014.06.019. [DOI] [PubMed] [Google Scholar]
- Silva NA, Sousa N, Reis RL, Salgado AJ. From basics to clinical: a comprehensive review on spinal cord injury. Prog Neurobiol. 2014;114:25–57. doi: 10.1016/j.pneurobio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Sisalli MJ, Annunziato L, Scorziello A. Novel Cellular Mechanisms for Neuroprotection in Ischemic Preconditioning: A View from Inside Organelles. Front Neurol. 2015;6:115. doi: 10.3389/fneur.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Eskey CJ. Hemorrhagic stroke. Radiol Clin North Am. 2011;49:27–45. doi: 10.1016/j.rcl.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Sorensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl Stroke Res. 2014;5:711–718. doi: 10.1007/s12975-014-0364-8. [DOI] [PubMed] [Google Scholar]
- Stetler RA, Leak RK, Gan Y, Li P, Zhang F, Hu X, Jing Z, Chen J, Zigmond MJ, Gao Y. Preconditioning provides neuroprotection in models of CNS disease: paradigms and clinical significance. Prog Neurobiol. 2014;114:58–83. doi: 10.1016/j.pneurobio.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Yamashita T, Shang J, Liu N, Deguchi K, Feng J, Abe K. Time-dependent profiles of microRNA expression induced by ischemic preconditioning in the gerbil hippocampus. Cell Transplant. 2015a;24:367–376. doi: 10.3727/096368915X686869. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li Y, Liu L, Wang Y, Xia Y, Zhang L, Ji X. Identification of miRNAs Involved in the Protective Effect of Sevoflurane Preconditioning Against Hypoxic Injury in PC12 Cells. Cell Mol Neurobiol. 2015b;35:1117–1125. doi: 10.1007/s10571-015-0205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri S, Tanriverdi F, Zararsiz G, Elbuken G, Ulutabanca H, Karaca Z, Selcuklu A, Unluhizarci K, Tanriverdi K, Kelestimur F. Circulating MicroRNAs as Potential Biomarkers for Traumatic Brain Injury-Induced Hypopituitarism. J Neurotrauma. 2016;33:1818–1825. doi: 10.1089/neu.2015.4281. [DOI] [PubMed] [Google Scholar]
- Tajiri N, Acosta SA, Shahaduzzaman M, Ishikawa H, Shinozuka K, Pabon M, Hernandez-Ontiveros D, Kim DW, Metcalf C, Staples M, Dailey T, Vasconcellos J, Franyuti G, Gould L, Patel N, Cooper D, Kaneko Y, Borlongan CV, Bickford PC. Intravenous transplants of human adipose-derived stem cell protect the brain from traumatic brain injury-induced neurodegeneration and motor and cognitive impairments: cell graft biodistribution and soluble factors in young and aged rats. J Neurosci. 2014;34:313–326. doi: 10.1523/JNEUROSCI.2425-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JR, Koo YX, Kaur P, Liu F, Armugam A, Wong PT, Jeyaseelan K. microRNAs in stroke pathogenesis. Curr Mol Med. 2011;11:76–92. doi: 10.2174/156652411794859232. [DOI] [PubMed] [Google Scholar]
- Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Liu W, Shang G, Zheng Y, Huang J, Lin R, Chen L. MiR-207/352 regulate lysosomal-associated membrane proteins and enzymes following ischemic stroke. Neuroscience. 2015;305:1–14. doi: 10.1016/j.neuroscience.2015.07.064. [DOI] [PubMed] [Google Scholar]
- Taylor RA, Sansing LH. Microglial responses after ischemic stroke and intracerebral hemorrhage. Clin Dev Immunol. 2013;2013:746068. doi: 10.1155/2013/746068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome AD, Harms AS, Volpicelli-Daley LA, Standaert DG. microRNA-155 Regulates Alpha-Synuclein-Induced Inflammatory Responses in Models of Parkinson Disease. J Neurosci. 2016;36:2383–2390. doi: 10.1523/JNEUROSCI.3900-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JW, Dave KR, Young JI, Perez-Pinzon MA. Ischemic preconditioning alters the epigenetic profile of the brain from ischemic intolerance to ischemic tolerance. Neurotherapeutics. 2013;10:789–797. doi: 10.1007/s13311-013-0202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi AK, Dwivedi A, Pal MK, Rastogi N, Gupta P, Ali S, Prabhu MB, Kushwaha HN, Ray RS, Singh SK, Duggal S, Narayan B, Mishra DP. Attenuated neuroprotective effect of riboflavin under UV-B irradiation via miR-203/c-Jun signaling pathway in vivo and in vitro. J Biomed Sci. 2014;21:39. doi: 10.1186/1423-0127-21-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PC, Liao YC, Wang YS, Lin HF, Lin RT, Juo SH. Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease. J Vasc Res. 2013;50:346–354. doi: 10.1159/000351767. [DOI] [PubMed] [Google Scholar]
- Varga ZV, Kupai K, Szucs G, Gaspar R, Paloczi J, Farago N, Zvara A, Puskas LG, Razga Z, Tiszlavicz L, Bencsik P, Gorbe A, Csonka C, Ferdinandy P, Csont T. MicroRNA-25-dependent up-regulation of NADPH oxidase 4 (NOX4) mediates hypercholesterolemia-induced oxidative/nitrative stress and subsequent dysfunction in the heart. J Mol Cell Cardiol. 2013;62:111–121. doi: 10.1016/j.yjmcc.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Vartanian K, Stenzel-Poore M. Toll-like receptor tolerance as a mechanism for neuroprotection. Transl Stroke Res. 2010;1:252–260. doi: 10.1007/s12975-010-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian KB, Stevens SL, Marsh BJ, Williams-Karnesky R, Lessov NS, Stenzel-Poore MP. LPS preconditioning redirects TLR signaling following stroke: TRIF-IRF3 plays a seminal role in mediating tolerance to ischemic injury. J Neuroinflammation. 2011;8:140. doi: 10.1186/1742-2094-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Grainger DC. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol. 2014;12:647–653. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- Wang H, Lu S, Yu Q, Liang W, Gao H, Li P, Gan Y, Chen J, Gao Y. Sevoflurane preconditioning confers neuroprotection via anti-inflammatory effects. Front Biosci (Elite Ed) 2011a;3:604–615. doi: 10.2741/e273. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu B, Cao F, Sun S, Zhang Y, Zhu Q. Down regulation of lncSCIR1 after spinal cord contusion injury in rat. Brain Res. 2015;1624:314–320. doi: 10.1016/j.brainres.2015.07.052. [DOI] [PubMed] [Google Scholar]
- Wang MD, Wang Y, Xia YP, Dai JW, Gao L, Wang SQ, Wang HJ, Mao L, Li M, Yu SM, Tu Y, He QW, Zhang GP, Wang L, Xu GZ, Xu HB, Zhu LQ, Hu B. High Serum MiR-130a Levels Are Associated with Severe Perihematomal Edema and Predict Adverse Outcome in Acute ICH. Mol Neurobiol. 2016a;53:1310–1321. doi: 10.1007/s12035-015-9099-0. [DOI] [PubMed] [Google Scholar]
- Wang P, Liang J, Li Y, Li J, Yang X, Zhang X, Han S, Li S, Li J. Down-regulation of miRNA-30a alleviates cerebral ischemic injury through enhancing beclin 1-mediated autophagy. Neurochem Res. 2014a;39:1279–1291. doi: 10.1007/s11064-014-1310-6. [DOI] [PubMed] [Google Scholar]
- Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011b;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Yang GY. MicroRNAs in Cerebral Ischemia. Stroke Res Treat. 2013;2013:276540. doi: 10.1155/2013/276540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Huang J, Chen X, Gu X, Wang Y, Zeng L, Yang GY. Increase of circulating miR-223 and insulin-like growth factor-1 is associated with the pathogenesis of acute ischemic stroke in patients. BMC Neurol. 2014b;14:77. doi: 10.1186/1471-2377-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Leng Y, Tsai LK, Leeds P, Chuang DM. Valproic acid attenuates blood-brain barrier disruption in a rat model of transient focal cerebral ischemia: the roles of HDAC and MMP-9 inhibition. J Cereb Blood Flow Metab. 2011c;31:52–57. doi: 10.1038/jcbfm.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Xiao L, Xue R, Zhang D, Zhou J, Ren H, Guo S, Xu J. MicroRNA-9 Mediates the Cell Apoptosis by Targeting Bcl2l11 in Ischemic Stroke. Mol Neurobiol. 2015;11:11. doi: 10.1007/s12035-015-9605-4. [DOI] [PubMed] [Google Scholar]
- Weng H, Shen C, Hirokawa G, Ji X, Takahashi R, Shimada K, Kishimoto C, Iwai N. Plasma miR-124 as a biomarker for cerebral infarction. Biomed Res. 2011;32:135–141. doi: 10.2220/biomedres.32.135. [DOI] [PubMed] [Google Scholar]
- Whyte EM, Mulsant BH. Post stroke depression: epidemiology, pathophysiology, and biological treatment. Biol Psychiatry. 2002;52:253–264. doi: 10.1016/s0006-3223(02)01424-5. [DOI] [PubMed] [Google Scholar]
- Woodruff TM, Thundyil J, Tang SC, Sobey CG, Taylor SM, Arumugam TV. Pathophysiology, treatment, and animal and cellular models of human ischemic stroke. Mol Neurodegener. 2011;6:11. doi: 10.1186/1750-1326-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Deng F, Xing Z, Wu Z, Cen B, Xu S, Zhao Z, Nepomuceno R, Bhuiyan MI, Sun D, Wang QJ, Ji A. Long non-coding RNA C2dat1 regulates CaMKIIdelta expression to promote neuronal survival through the NF-kappaB signaling pathway following cerebral ischemia. Cell Death Dis. 2016;7:e2173. doi: 10.1038/cddis.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Chen Z, Chen J, Chen H. Isoflurane preconditioning protects rat brain from ischemia reperfusion injury via up-regulating the HIF-1alpha expression through Akt/mTOR/s6K activation. Cell Mol Biol (Noisy-le-grand) 2016;62:38–44. [PubMed] [Google Scholar]
- Yang S, Zhao J, Chen Y, Lei M. Biomarkers Associated with Ischemic Stroke in Diabetes Mellitus Patients. Cardiovasc Toxicol. 2016a;16:213–222. doi: 10.1007/s12012-015-9329-8. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhong L, Xian R, Yuan B. MicroRNA-223 regulates inflammation and brain injury via feedback to NLRP3 inflammasome after intracerebral hemorrhage. Mol Immunol. 2015;65:267–276. doi: 10.1016/j.molimm.2014.12.018. [DOI] [PubMed] [Google Scholar]
- Yang ZB, Li TB, Zhang Z, Ren KD, Zheng ZF, Peng J, Luo XJ. The Diagnostic Value of Circulating Brain-specific MicroRNAs for Ischemic Stroke. Intern Med. 2016b;55:1279–1286. doi: 10.2169/internalmedicine.55.5925. [DOI] [PubMed] [Google Scholar]
- Yang ZB, Zhang Z, Li TB, Lou Z, Li SY, Yang H, Yang J, Luo XJ, Peng J. Up-regulation of brain-enriched miR-107 promotes excitatory neurotoxicity through down-regulation of glutamate transporter-1 expression following ischaemic stroke. Clin Sci (Lond) 2014;127:679–689. doi: 10.1042/CS20140084. [DOI] [PubMed] [Google Scholar]
- Yin ZS, Zu B, Chang J, Zhang H. Repair effect of Wnt3a protein on the contused adult rat spinal cord. Neurol Res. 2008;30:480–486. doi: 10.1179/174313208X284133. [DOI] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010a;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010b;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Zhang J, Chen YE, Yin K-J. Abstract 72: Long Non-coding RNAs Mediate Cerebrovascular Endothelial Pathologies in Ischemic Stroke. Stroke. 2015;46:A72–A72. [Google Scholar]
- Zeng L, He X, Wang Y, Tang Y, Zheng C, Cai H, Liu J, Wang Y, Fu Y, Yang GY. MicroRNA-210 overexpression induces angiogenesis and neurogenesis in the normal adult mouse brain. Gene Ther. 2014a;21:37–43. doi: 10.1038/gt.2013.55. [DOI] [PubMed] [Google Scholar]
- Zeng L, Liu J, Wang Y, Wang L, Weng S, Chen S, Yang GY. Cocktail blood biomarkers: prediction of clinical outcomes in patients with acute ischemic stroke. Eur Neurol. 2013;69:68–75. doi: 10.1159/000342896. [DOI] [PubMed] [Google Scholar]
- Zeng L, Liu J, Wang Y, Wang L, Weng S, Tang Y, Zheng C, Cheng Q, Chen S, Yang GY. MicroRNA-210 as a novel blood biomarker in acute cerebral ischemia. Front Biosci (Elite Ed) 2011;3:1265–1272. doi: 10.2741/e330. [DOI] [PubMed] [Google Scholar]
- Zeng X, Lin MY, Wang D, Zhang Y, Hong Y. Involvement of adrenomedullin in spinal glial activation following chronic administration of morphine in rats. Eur J Pain. 2014b;18:1323–1332. doi: 10.1002/j.1532-2149.2014.493.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE, Yin KJ. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol. 2016a;277:162–70. doi: 10.1016/j.expneurol.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chopp M, Liu X, Teng H, Tang T, Kassis H, Zhang ZG. Combination therapy with VELCADE and tissue plasminogen activator is neuroprotective in aged rats after stroke and targets microRNA-146a and the toll-like receptor signaling pathway. Arterioscler Thromb Vasc Biol. 2012a;32:1856–1864. doi: 10.1161/ATVBAHA.112.252619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yin Y, Xu SJ, Wu YP, Chen WS. Inflammation & apoptosis in spinal cord injury. Indian J Med Res. 2012b;135:287–296. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng L, Chen Y, Yang GY, Liu J, Zeng L. Clinical predictor and circulating microRNA profile expression in patients with early onset post-stroke depression. J Affect Disord. 2016b;193:51–8. doi: 10.1016/j.jad.2015.12.061. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Ye P, Li S, Shi Y. MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci Rep. 2013a;3:1329. doi: 10.1038/srep01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Wang J, Gao L, Wang R, Liu X, Gao Z, Tao Z, Xu C, Song J, Ji X, Luo Y. MiRNA-424 protects against permanent focal cerebral ischemia injury in mice involving suppressing microglia activation. Stroke. 2013b;44:1706–1713. doi: 10.1161/STROKEAHA.111.000504. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang H, Zhang D, Yu CY, Zhao XH, Liu FF, Bian GL, Ju G, Wang J. Loss of microRNA-124 expression in neurons in the peri-lesion area in mice with spinal cord injury. Neural Regen Res. 2015;10:1147–1152. doi: 10.4103/1673-5374.156983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liu H, Wang R, Xu S, Liu Y, Wang K, Hou X, Shen C, Wu J, Chen X, Wu P, Zhang G, Ji Z, Wang H, Xiao Y, Han J, Shi H, Zhao S. Expression signatures of long non-coding RNAs in early brain injury following experimental subarachnoid hemorrhage. Mol Med Rep. 2015;12:967–973. doi: 10.3892/mmr.2015.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng HW, Wang YL, Lin JX, Li N, Zhao XQ, Liu GF, Liu LP, Jiao Y, Gu WK, Wang DZ, Wang YJ. Circulating MicroRNAs as potential risk biomarkers for hematoma enlargement after intracerebral hemorrhage. CNS Neurosci Ther. 2012;18:1003–1011. doi: 10.1111/cns.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Jiang L, Cheng C, Huang Z, Zhang H, Liu H, He J, Cao F, Peng J, Jiang Y, Sun X. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 2016;1646:589–600. doi: 10.1016/j.brainres.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang J. Identification of miRNA-21 and miRNA-24 in plasma as potential early stage markers of acute cerebral infarction. Mol Med Rep. 2014;10:971–976. doi: 10.3892/mmr.2014.2245. [DOI] [PubMed] [Google Scholar]
- Zhu F, Liu JL, Li JP, Xiao F, Zhang ZX, Zhang L. MicroRNA-124 (miR-124) regulates Ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. J Mol Neurosci. 2014;52:148–155. doi: 10.1007/s12031-013-0155-9. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang JL, He ZY, Jin F, Tang L. Association of Altered Serum MicroRNAs with Perihematomal Edema after Acute Intracerebral Hemorrhage. PLoS One. 2015;10:e0133783. doi: 10.1371/journal.pone.0133783. [DOI] [PMC free article] [PubMed] [Google Scholar]