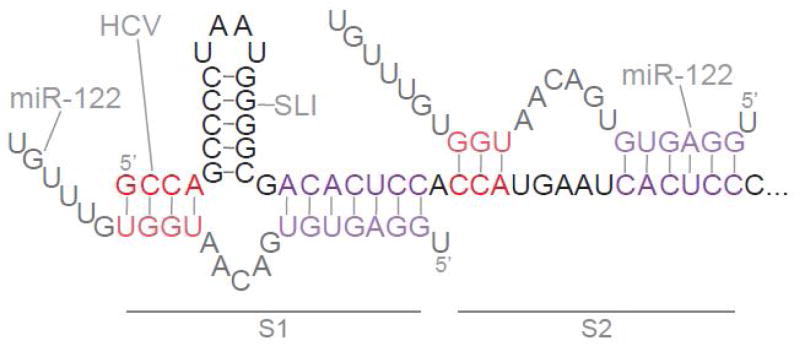Abstract
The single-stranded Hepatitis C Virus (HCV) genome adopts a set of elaborate RNA structures that are involved in every stage of the viral lifecycle. Recent advances in chemical probing, sequencing, and structural biology have facilitated analysis of RNA folding on a genome-wide scale, revealing novel structures and networks of interactions. These studies have underscored the active role played by RNA in every function of HCV and they open the door to new types of RNA-targeted therapeutics.
Introduction
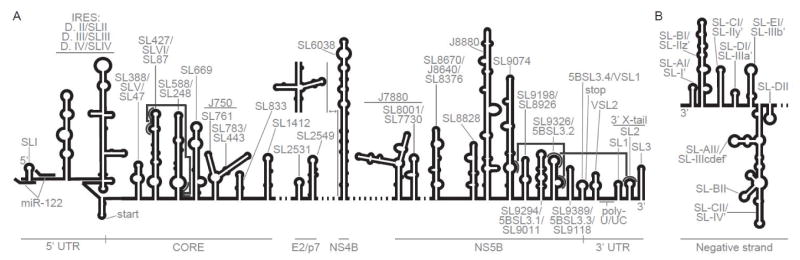
Hepatitis C Virus (HCV) is a positive-sense single-stranded RNA virus with a highly structured genome that is 9.6kb in length. This long RNA molecule includes 5′ and 3′ untranslated regions (UTRs), which flank an open reading frame that encodes a massive polyprotein that is cleaved into structural and replicative proteins. During replication, the viral RNA polymerase (NS5B) generates a negative-sense RNA that serves as the template for generating numerous copies of the positive-sense genome, which are then packaged into infectious particles. Like many other RNA molecules, the genome of HCV folds into complex structural elements that regulate and expand the functional repertoire of the virus (known structures are shown in Figure 1). Distinct RNA structural elements are scattered throughout all regions of the genomic RNA, and it has recently become possible to characterize them through a combination of structural biology and genetics. Previous analyses of HCV RNA structure have focused primarily on the 5′ and 3′ UTR of the positive-sense genome, along with regions at the 3′ end of the negative-sense strand (reviewed in [1]). In the past three years, new technological advances have enhanced our mechanistic and structural understanding of these elements, while revealing an abundance of new elements within the ORF that contribute to replication and infectivity. These studies have underscored the interplay between RNA and cellular factors in potentiating each phase of the viral lifecycle.
Figure 1.
Secondary, tertiary, and quaternary structural elements of the HCV genome. Labels at bottom indicate region of the genome where structures are located. A. Structures within the positive-sense strand of HCV genome. Long-range tertiary interactions are depicted in dark grey. Alternate stem-loop and clover-leaf structures of SL6038 depicted. B. Negative-sense strand 3′ end of HCV genome [49,50].
HCV infects three percent of the global human population, and chronic infection with this hepatotropic virus can lead to liver cirrhosis and hepatocellular carcinoma (HCC). The advent of direct-acting antiviral drugs that target encoded proteins has resulted in a marked improvement in patient outcomes [2]. However, drug development has been difficult due to the diversity of HCV genotypes found within patients (there are currently seven classified genotypes and 67 subtypes [3]) and the rapid development of drug resistance due to low fidelity of the RNA polymerase. Even within a single patient, the relative abundance of different HCV sequences can fluctuate drastically over time [4]. An improved understanding of the link between viral RNA sequences, their structures, and the roles these play in the virus will facilitate the development of improved therapeutics and more robust models for viral function.
Structural features of the 5′ UTR
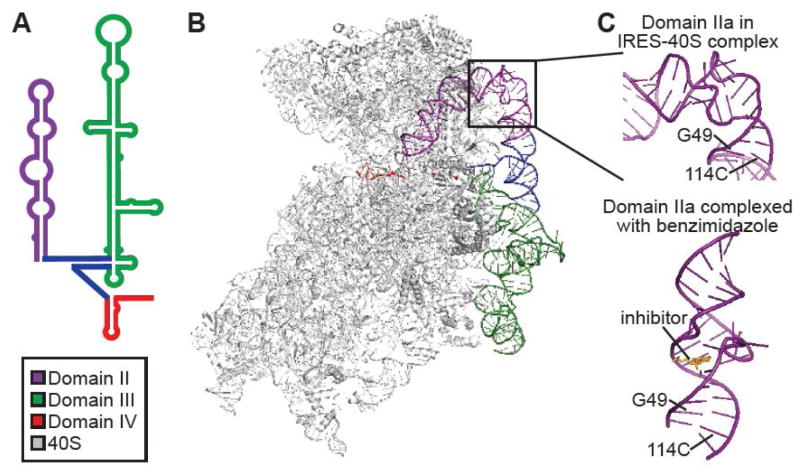
The architecture and molecular interaction networks of the HCV 5′ UTR are the best characterized of the genome. The 340 nucleotides of the 5′-UTR are grouped into four domains of relatively conserved sequence across all genotypes. Domain I comprises a short stem loop that is flanked by a miR-122 binding site (see below), and domains II–IV compose the internal ribosome entry site (IRES), which directs translation of the coding sequence (reviewed in [1]). Although the secondary structure of the IRES has been defined for two decades (Figure 2A), recent high resolution structural studies have revealed the three-dimensional tertiary structure of the IRES, both in isolation and when complexed with ribosomal subunits, thereby explaining how the IRES recruits and stabilizes the cellular translation machinery.
Figure 2.
Structural features of the HCV IRES. A. 2D structure of the HCV IRES. Domains of the IRES are color-coded according to the legend. B. IRES-40S interaction. Binary interaction between the IRES and human 40S ribosomal subunit. Adapted from [6], PDB 5A2Q. C. Benzimidazole straightens the bend in domain II. Structure of the bend region of domain II (domain IIa) complexed with 40S (left) and the corresponding region complexed with benzimidazole (right). Right adapted from [33], PDB 3TZR.
IRES
The HCV IRES exemplifies the ability of RNA molecules to balance structural rigidity with the capacity for dynamical rearrangement. For example, specific RNA structures in the IRES mediate interactions with other RNAs and proteins, but flexibility allows dynamic interplay among these interactions. Domains II and III of the IRES form large stem-loops that adopt extended structures, while domain IV forms a short, unstable stem that encompasses the start codon (Figure 2B). Domain III also contains a four-way junction that provides a stable platform for extending structural elements into solution, where they can be recognized by proteins. The flexibility of these domains are critical for function, as they enable the IRES to recruit the ribosomal 40S subunit and initiate translation of viral genes.
Advances in structural biology, particularly cryo-electron microscopy, have enabled us to visualize and interpret IRES interactions at high resolution. For example, the foundation for a strong IRES-40S interaction is the conserved base-pairing that forms between the 18S RNA of the 40S subunit and domain III of the IRES. Intriguingly, nucleotides involved in pairing to 18S RNA are required for IRES-mediated translation, but they are not required for canonical cellular translation [8], indicating that they have been co-opted by the virus. After the 40S subunit has been recruited, domain II reaches across the head of the 40S subunit, wedging open the mRNA binding tunnel and allow the HCV coding sequence to bind [7]. For this to occur, the weak stem-loop in domain IV must unfold, explaining why stabilizing mutations within this stem-loop are detrimental to HCV translation [9]. In addition to recruiting the 40S subunit, another function of domain III is to bind the cellular translation factor eIF3. Recent structural studies have demonstrated that this interaction displaces eIF3 from its canonical binding site near the exit tunnel of the 40S [10]. In this way, domain III removes eIF3 so that it does not interfere with IRES-40S binding. Finally, upon association of initiator tRNA, eIF2, and the 60S subunit, domain II is released from the 40S head, and translation of the HCV genome can begin [7].
miR-122
The interaction between HCV and the hepatocyte-specific microRNA miR-122 represents an important quaternary structure that forms during HCV infection [11]. There are two miR-122 binding sites at the terminus of the 5′-UTR, and recent functional and structural probing studies have explored the base-pairing requirements for these interactions (Figure 3) [12–14]. Both of the binding sites involve extensive base-pairing outside of the canonical “seed” sequence that is typically found in miRNA-mRNA interactions (Figure 3). The first binding site (S1) extends across the domain I stem loop and past the 5′ end of the genome, resulting in a 3′ overhang of six miRNA nucleotides. The second binding site for miR-122 (S2) is adjacent to S1, and the resulting duplexes are sufficiently close that they would be capable of coaxial stacking, particularly if pairing were maximized by flanking overhangs. The S2 interaction appears to involve weak duplexes that surround a large internal loop. Taken together, the S1 and S2 interactions may enable HCV RNA to sequester miR-122 from endogenous cellular mRNA targets [15]. Additionally, the 3′ overhang at the S1 site may protect the HCV genome from cellular exonucleases (reviewed in [16]). There is debate over whether miR-122 enhances translation, replication, or both, and structural probing has hinted at alterations in long-range interactions in and around the IRES upon miR-122 binding. However, a concrete mechanism for the dramatic effect of miR-122 on HCV infection remains unclear.
Figure 3.
Base-pairing interactions between the HCV 5′ UTR and miR-122. Two copies of miR-122 bind the 5′ end of the HCV genome via canonical seed-sequence interactions (purple) and additional interactions (red). The unpaired 3′ ends of miR-122 are depicted, as well as bulges formed in miR-122 and SLI/domain I of HCV. Adapted from [13].
The ORF contains important regulatory signals
It is becoming increasingly evident that RNA coding sequences adopt biologically significant structures (reviewed in [18]), but until recently, only a few HCV ORF substructures had been characterized. For example, it is well established that stem loops at the 5′-end of the ORF, within the Core-encoding region, are required for replication of the intact genome and that they promote translation [19,20]. At the 3′-end of the ORF, within the region encoding NS5B, there is a set of stem-loops that interact with RNA motifs in the 3′ UTR, resulting in network of RNA elements that are essential for HCV replication [21]. Interaction between NS5B and these same structures may explain why NS5B expression is required in cis [22].
Recent advances in methods for manipulating large RNA molecules, and for high-throughput structural probing of RNAs in-vitro and in-vivo have led to studies that reveal extensive, highly specific RNA structural features throughout the HCV ORF [23,24] (see Box). For example 90% of the Core-encoding sequence is involved in highly conserved base-pairings within well-defined RNA substructures, many of which are involved in elaborate secondary structures and long-range tertiary interactions [23]. These structures are not random, but rather they represent discrete folded units that contribute directly to numerous aspects of the viral lifecycle. Now that RNA structures can be rapidly identified via chemical probing, their roles are being tested through viral genetics and cell culture assays of viral function.
Box 1. A new era for genome-wide analysis of HCV structure.
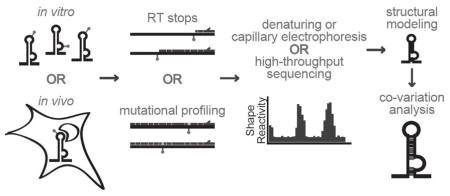
Historically, only the most highly conserved regions of the HCV genome have been structurally analyzed, and technical limitations only permitted examination of specific regions in the genome. However, a modern methodology known as SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) and related advances, particularly when coupled with high-throughput RNA sequencing, allows exploration of every base in the entire genome both in vitro andin vivo. This allows identification of structured regions throughout the genome and in the context of an intact genome. The powerful HCV genetic system can then be used to examine the phenotypic consequences of structures that are identified with SHAPE.
Basic workflow for SHAPE analysis: The SHAPE reagent modifies the 2′-OH groups on ribose sugars of unpaired nucleotides in an RNA structure. These modified nucleotides are revealed as stops or mutations by reverse transcriptase, enabling the calculation of relative SHAPE reactivity. Computational RNA folding programs then generate proposed structures based on thermodynamics and SHAPE reactivity constraints. Proposed structures can then be analyzed for co-variation of nucleotides to maintain base-pairing interactions. Finally, high-confidence structures can be analyzed for function.
Upstream regions of the HCV ORF are particularly rich in RNA structural elements, and functional analysis of stem-loops within in the Core region has shown that they form higher-order structures with specific mechanistic roles. For example, a long-range kissing-loop interaction between SL427 and 588 affects replication and is required for infectivity (Figure 1) [23]. Additionally, proper base-pairing of a stem loop within the sequence encoding the E1 protein (SL1412) is specifically required for production of infectious virus, while it has negligible impact on replication [23]. This structure may serve as an RNA packaging element that interacts with capsid proteins during particle assembly, as in the case of HIV RNA (reviewed in [25]).
Due to its location and high level of complexity, one of the most interesting downstream elements investigated to date is a large structural motif within the HCV NS4B-encoding sequence (SL6038). This region of RNA toggles between two distinct structural states: a long stem-loop and a cloverleaf conformation [23]. Genetic and functional analysis suggests that this region switches between different conformations during different phases of the HCV lifecycle, as locking the structure into the stem-loop conformation abolishes replication. Additional stem-loop structures have been identified in the NS5B-encoding sequence (J7880, J8880) and these impact viral replication [24]. The overall secondary structural organization of the HCV genome has implications for evasion of the human immune system, as 90% of helices are limited to seven base pairs or less, perhaps to avoid recognition by innate immune sensors that detect double-stranded RNA [24].
To date, most of the functionally-analyzed RNA structures within the ORF are conserved in all HCV genotypes. However, regions within the ORF sequence are divergent, and RNA structures formed by distinct genotypes may lead to differences in pathogenicity. Indeed, a parallel study of three different genotypes has identified regions of differing structure [24], and motifs found in the genotype 2a ORF are not maintained by co-varying mutations in other genotypes [23]. Therefore, RNA structures adopted by the ORF are likely to underlie shared and genotype-specific functions, such as interactions with viral and host factors, and it will be intriguing to uncover functional RNA elements that contribute to idiosynchratic features of viral subtypes.
RNA Elements within the 3′-UTR contribute to viral replication
Secondary structures within the 3′-UTR of the positive strand have been known for over a decade, and some of these are important for HCV replication (reviewed in [1]). The 3′-UTR has been classified into three regions: a variable region near the stop codon that forms two stem-loops, a flexible polyU/UC tract, and three highly conserved stem loops at the terminus known as the 3′ X-tail (Figure 1). These sections of RNA, like those at the 3′ end of the minus strand, are likely to direct the HCV RNA polymerase for replication. It is important to note that the NS5B polymerase initiates replication more efficiently from the negative-sense 3′-UTR than the positive-sense 3′-UTR [26], suggesting that the minus strand 3′-UTR contains structural features that promote formation of a particularly robust replication complex, thereby explaining the relative abundance of positive-sense genomes relative to negative-sense RNA in infected cells [27]. Characterization of structural features in the 3′-UTR, particularly on the negative strand, and understanding their interplay with the replication complex, remain rich areas for future investigation.
The potential of RNA-targeted antiviral therapeutics
Despite the recent success of HCV antiviral therapies, the development of resistance and the diversity of responses among viral genotypes necessitates an ongoing search for novel therapeutic strategies. An emerging alternative approach involves the inhibition of functional RNA structures. The most successful antibiotics ever developed are small molecules that target RNA structures within the bacterial ribosome [28], and there is increasing appreciation for the value of RNA as a target during treatment of diverse disease states [29]. Indeed, a recent unbiased screen for novel antibiotics has identified an RNA riboswitch as an effective drug target [30]. Drugs that bind HCV RNA elements already show therapeutic potential (discussed below), and a better understanding of HCV RNA structures will enhance the value of this approach.
Since the 5′-UTR is the best characterized section of the HCV genome, it is the first region that is being directly targeted for antiviral therapy. The exceptional conservation of the 5′ UTR suggests an evolutionary constraint on mutations, potentially reducing the likelihood for development of drug resistance. Furthermore, drugs targeting this relatively invariant region are likely to inhibit all HCV genotypes. Significant progress has already been made by using small molecules to target an RNA bulge substructure that is formed by the asymmetric loop within domain II of the IRES [31]. The flexible bend induced by this bulge is essential for IRES function, and several compounds have been shown to lock this region in a bent or extended conformation [32]. Crystallographic characterization of a benzimidazole-based inhibitor in complex with this region of domain II reveals a dramatic straightening of the bend, presumably inhibiting the ability of domain II to direct the 40S head-tilt [33] (Figure 2C). Structure-based design is being used to develop more drug-like small molecules with a similar mechanism of action [34].
Another approach for targeting the HCV genome involves the introduction of stable antisense oligonucleotides that base-pair with a target RNA, as in the case of Miravirsen, which is an antagomir that successfully targets miR-122 via complementarity to the seed sequence [35]. In a phase II clinical trial, treatment with Miravirsen reduced viral load in a dose-dependent manner and in certain cases cleared HCV [36]. The mode of action of Miravirsen is reminiscent of HCV RNA, which sequesters miR-122 and reduces its interactions with host mRNA targets [15]. Because loss of miR-122 leads to spontaneous development of HCC in mice [37,38] and is correlated with HCC progression in patients [39], the long-term effects of Miravirsen treatment should be carefully monitored. However, Miravirsen has demonstrated that modulating the interactions between genomic RNA and host factors can greatly affect viral load in patients. Therefore, exploring the molecular details of the HCV genome structure and its interplay with host factors will expand the repertoire of drug targets and uncover new mechanisms for the design of RNA inhibitors.
Implications for Flavivirus RNA structure
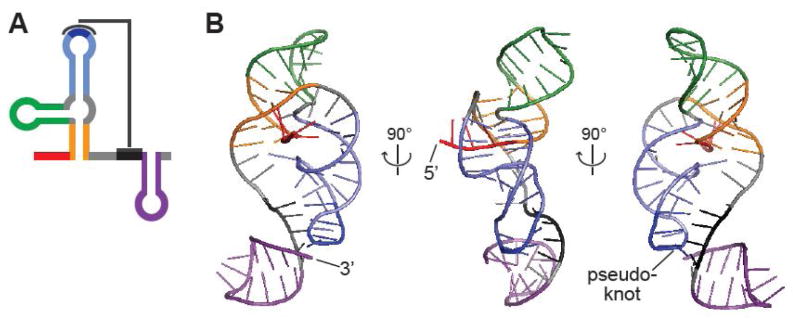
HCV is a member of the Flaviviridiae family, which also contains the Flavivirus genus that includes Dengue, West Nile, and Zika viruses. Unlike HCV, Flaviruses have capped 5′ ends and a shorter 5′ UTR that does not contain an IRES. A longer, highly structured 3′ UTR recruits translation factors, and genome circularization brings these factors into proximity to initiate translation of the viral polyprotein [40]. Despite these differences between HCV and flaviruses, structural elements identified in one type of virus can provide valuable mechanistic insights into potential mechanisms for the function of other viruses. For example, cyclization elements between the 5′ and 3′ UTR have been proposed for HCV [41]. A distinctive feature of Flavivirus structure is the sfRNA element in the 3′-UTR that blocks cellular exonucleases. Recent structural studies of Murray Valley Encephalitis and Zika Viruses have shown that the two stem-loops of sfRNA fold into intricate knot-like structures that are stabilized by formation of a pseudoknot and other long-range interactions (Figure 4) [42,43]. These knot-like structures inhibit RNA unfolding from the 5′ end while allowing RNA polymerase to unravel genomic RNA from the 3′ end. This example demonstrate how RNA structural elements impart an extra layer of function to viral genomes by modulating RNA folding. Interestingly, structures in the HCV 5′ UTR have recently been proposed to inhibit cellular exonuclease activity [44], but the details of this behavior are unknown. Thus, the common and unique RNA elements of Flaviviridiae provide insights into potential mechanisms for regulating function in diverse viruses.
Figure 4.
Structure of the Flavivirus sfRNA from Zika Virus. A. 2D structure of Zika sfRNA with stem-loops color-coded and pseudoknot depicted. B. Crystal structure of Zika sfRNA. Shown in orange and blue, the 3′ end of the first stem-loop wraps around the 5′ end of the RNA, inhibiting unwinding from the 5′ end. Adapted from [43], PDB 5TPY.
Perspective
Although the majority of available structural studies involve analysis of the HCV genome in-vitro, it is now possible explore the entire HCV genome within cells or in virus particles. It is anticipated that many HCV structural elements will be dynamic in vivo as the genome interacts transiently with RNA binding proteins and enzymes. Four of the ten encoded HCV proteins bind RNA, including the helicase NS3 and RNA polymerase NS5B, and these are expected to influence RNA structure. Modulation of genomic structure throughout infection is consistent with the differential requirements for structural motifs during the stages of translation, replication, and packaging. Targeting the inherent flexibility of RNA appears to be a promising approach for small molecule inhibitors, so exploration of HCV genome dynamics will be informative for future drug design. The concepts developed during studies of HCV structure and targeting can be applied to other members of the Flaviviridiae, underscoring the utility of HCV as a central model system.
An important and relatively unexplored area for future exploration of HCV and related viruses is the negative-sense genome, which is rich in structures that probably have functional roles. Finally, RNA modification by cellular enzymes will impact genome structure and function in ways that are only now being imagined. Recently, N6-methyladenosine (m6A) was found throughout the genomes of HCV and Flaviviruses, and HCV genome methylation has been shown to decrease viral infectivity [45,46]. Some sites of relatively high methylation overlap with specific RNA structural motifs in the HCV genome, suggesting that individual RNA elements are targeted by methyltransferases. Furthermore, it is likely that other RNA modifications will be identified within the HCV genome in cells. We are only beginning to understand and imagine how the structure of coding and non-coding viral RNA impacts function, but we can now navigate this new landscape with advanced new technologies.
Highlights.
Important RNA structures are found throughout the HCV genome, including the ORF
The HCV IRES adopts a dynamic structure that recruits the cellular translation machinery
RNA structural elements can be targeted for antiviral therapy
Acknowledgments
This work was supported by NIH RO1 AI089826 (to A.M.P.), NIH F32 AI126660 (to R.L.A.), and T32 GM07205 (to N.P.). A.M.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Sagan SM, Chahal J, Sarnow P. cis-Acting RNA elements in the hepatitis C virus RNA genome. Virus Res. 2015;206:90–98. doi: 10.1016/j.virusres.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter W, Connelly S, Struble K. Reinventing HCV Treatment: Past and Future Perspectives. J Clin Pharmacol. 2016 doi: 10.1002/jcph.830. [DOI] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatol Baltim Md. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghwani J, Rose R, Sheridan I, Lemey P, Suchard MA, Santantonio T, Farci P, Klenerman P, Pybus OG. Exceptional Heterogeneity in Viral Evolutionary Dynamics Characterises Chronic Hepatitis C Virus Infection. PLoS Pathog. 2016;12:e1005894. doi: 10.1371/journal.ppat.1005894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Pérard J, Leyrat C, Baudin F, Drouet E, Jamin M. Structure of the full-length HCV IRES in solution. Nat Commun. 2013;4:1612. doi: 10.1038/ncomms2611. This paper provided the solution structure of the full-length HCV IRES, demonstrating the flexibility of domain II and domain III. [DOI] [PubMed] [Google Scholar]
- 6.Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat Commun. 2015;6:7646. doi: 10.1038/ncomms8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, et al. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 2015;34:3042–3058. doi: 10.15252/embj.201592469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda D, Mauro VP. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc Natl Acad Sci U S A. 2014;111:15385–15389. doi: 10.1073/pnas.1413472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA N Y N. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 10•.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CUT, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. By cryo-EM, this paper demonstrated that domain III displaces eIF3 from its canonical binding site on the 40S subunit. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 12.Pang PS, Pham EA, Elazar M, Patel SG, Eckart MR, Glenn JS. Structural map of a microRNA-122: hepatitis C virus complex. J Virol. 2012;86:1250–1254. doi: 10.1128/JVI.06367-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Mortimer SA, Doudna JA. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013;41:4230–4240. doi: 10.1093/nar/gkt075. This paper provides a thorough biochemical analysis of the interaction between miR-122 and the HCV genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Luna JM, Scheel TKH, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. From a genome-wide RNA crosslinking and imuunoprecipitation experiment, this paper uncovers sequestration of miR-122 from cellular mRNA targets as a consequence of HCV infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson JA, Sagan SM. Hepatitis C virus and human miR-122: insights from the bench to the clinic. Curr Opin Virol. 2014;7:11–18. doi: 10.1016/j.coviro.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Díaz-Toledano R, Ariza-Mateos A, Birk A, Martínez-García B, Gómez J. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5′ region of hepatitis C virus RNA. Nucleic Acids Res. 2009;37:5498–5510. doi: 10.1093/nar/gkp553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 19.Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccalà G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci U S A. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You S, Rice CM. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J Virol. 2008;82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazakov T, Yang F, Ramanathan HN, Kohlway A, Diamond MS, Lindenbach BD. Hepatitis C virus RNA replication depends on specific cis- and trans-acting activities of viral nonstructural proteins. PLoS Pathog. 2015;11:e1004817. doi: 10.1371/journal.ppat.1004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol Cell. 2016;62:111–120. doi: 10.1016/j.molcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, Martin DP, Weeks KM. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci U S A. 2015;112:3692–3697. doi: 10.1073/pnas.1416266112. Through SHAPE analysis, the previous two studies uncover conserved structural elements within the HCV ORF. They also demonstrate phenotypic consequences of disrupting these structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reigadas S, Ventura M, Sarih-Cottin L, Castroviejo M, Litvak S, Astier-Gin T. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur J Biochem. 2001;268:5857–5867. doi: 10.1046/j.0014-2956.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 27.Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann T. Drugs targeting the ribosome. Curr Opin Struct Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Connelly CM, Moon MH, Schneekloth JS. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem Biol. 2016;23:1077–1090. doi: 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, et al. Selective small-molecule inhibition of an RNA structural element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. This paper identified a riboswitch as a drug target in an in vivo antibiotic screen, demonstrating that modification of RNA structure is a valid drug mechanism. [DOI] [PubMed] [Google Scholar]
- 31.Seth PP, Miyaji A, Jefferson EA, Sannes-Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ, et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 32.Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA, Castaldi MP, et al. Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J Med Chem. 2014;57:1694–1707. doi: 10.1021/jm401312n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc Natl Acad Sci U S A. 2012;109:5223–5228. doi: 10.1073/pnas.1118699109. Through x-ray crystallographic analysis, this paper demonstrates that a small molecule inhibitor of HCV IRES-mediated translation locks domain II in an extended conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charrette BP, Boerneke MA, Hermann T. Ligand Optimization by Improving Shape Complementarity at a Hepatitis C Virus RNA Target. ACS Chem Biol. 2016 doi: 10.1021/acschembio.6b00687. [DOI] [PubMed] [Google Scholar]
- 35.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 37.Hsu S-H, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai W-C, Hsu S-D, Hsu C-S, Lai T-C, Chen S-J, Shen R, Huang Y, Chen H-C, Lee C-H, Tsai T-F, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Blanco MA, Vasudevan SG, Bradrick SS, Nicchitta C. Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res. 2016;134:244–249. doi: 10.1016/j.antiviral.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Fricke M, Dünnes N, Zayas M, Bartenschlager R, Niepmann M, Marz M. Conserved RNA secondary structures and long-range interactions in hepatitis C viruses. RNA N Y N. 2015;21:1219–1232. doi: 10.1261/rna.049338.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi P-Y, Nix JC, Beckham JD, Kieft JS. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science. 2016;354:1148–1152. doi: 10.1126/science.aah3963. The previous two papers provide the x-ray crystal structure of two Flavivirus sfRNAs, demonstrating how they fold into a unique structure to inhibit unwinding from the 5′ end. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJT, Keene JD, Wilusz CJ, Bradrick SS, Wilusz J. XRN1 stalling in the 5′ UTR of Hepatitis C virus and Bovine Viral Diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog. 2015;11:e1004708. doi: 10.1371/journal.ppat.1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. This paper uncovers m6A modification of HCV and other Flaviviruses and the impact on viral replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA N Y N. 2002;8:824–841. doi: 10.1017/s1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu D, Ren S, Hu S, Wang WG, Subramanian A, Contreras D, Kanagavel V, Chung E, Ko J, Amirtham Jacob Appadorai RS, et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J Virol. 2013;87:5678–5696. doi: 10.1128/JVI.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster C, Isel C, Imbert I, Ehresmann C, Marquet R, Kieny MP. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J Virol. 2002;76:8058–8068. doi: 10.1128/JVI.76.16.8058-8068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith RM, Walton CM, Wu CH, Wu GY. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J Virol. 2002;76:9563–9574. doi: 10.1128/JVI.76.19.9563-9574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]