Abstract
Mitochondria are central to the efficient provision of energy for eukaryotic cells. The oxidative-phosphorylation system of mitochondria consists of a series of five major membrane complexes: NADH–ubiquinone oxidoreductase (commonly known as complex I), succinate–ubiquinone oxidoreductase (complex II), ubiquinol–cytochrome c oxidoreductase (cytochrome bc1 complex or complex III), cytochrome c–O2 oxidoreductase (complex IV), and F1F0–ATP synthase (complex V). Several lines of evidence have recently suggested that complexes I and III–V might interact to form supercomplexes. However, because of their fragility, the structures of these supercomplexes are still unknown. A stable supercomplex consisting of complex I and dimeric complex III was purified from plant mitochondria. Structural characterization by single-particle EM indicates a specific type of interaction between monomeric complex I and dimeric complex III in a 1:1 ratio. We present a model for how complexes I and III are spatially organized within the I+III2 supercomplex.
Keywords: Arabidopsis, oxidative phosphorylation, respiratory protein complexes, single-particle analysis, plant mitochondria
The mitochondrial respiratory chain consists of a series of sequentially acting electron carriers, most of which are integral membrane proteins with prosthetic groups capable of accepting and donating one or two electrons. The transfer of two electrons from NADH through the respiratory chain to molecular oxygen generates a proton-motive force across the inner mitochondrial membrane that drives the synthesis of ATP. Since the first isolation of respiratory-chain complexes >40 years ago (1), the concepts of how they are arranged within the membrane have changed. Evidence that mitochondrial electron-transfer complexes specifically interact to form supermolecular structures called supercomplexes came from recent work on the yeast Saccharomyces cerevisiae (2, 3), beef (3, 4), and plants (5–7). Several experimental findings indicate specific interactions between respiratory-chain complexes. Respiratory supercomplexes were shown to allow higher electron-transfer rates under in vitro and in organello conditions (3, 6). Inhibitor-titration experiments revealed evidence for III+IV supercomplexes in S. cerevisae (8) and a I+III2 supercomplex in beef mitochondria (9). Mutants in single genes encoding subunits of respiratory-chain complexes lead to combined enzyme-complex defects (10–12). Similar supermolecular structures were also described for the respiratory chains of bacteria (13–17). The roles that have been attributed to respiratory supercomplexes are substrate-channeling, catalytic enhancement, sequestration of reactive intermediates (3), stabilization of protein complexes (10), increasing the capacity of the inner mitochondrial membrane for protein insertion (2), and generating mitochondrial cristae morphology (18). Furthermore, the dynamic formation of supercomplexes is speculated to regulate alternative respiration in plants (5).
Solubilization of mitochondrial membranes with nonionic detergents and subsequent resolution of protein complexes by blue native (BN)/PAGE is the most popular experimental strategy for the analysis of the protein-complex composition of respiratory supercomplexes. Based on this procedure, beef mitochondria were reported to include I+III2 and I+III2+IV1–4 supercomplexes, which were proposed to be called “respirasomes,” and dimeric ATP synthase (3). Mitochondria of S. cerevisiae include III+IV1–2 supercomplexes. The ratio of these supercomplexes depends on growth conditions and the abundance of the supercomplexes on the cardiolipin content of the inner mitochondrial membrane (19, 20). Dimeric ATP synthase of S. cerevisiae was shown to include additional subunits that are absent in the monomers (2).
Higher-plant mitochondria are interesting systems for the study of supercomplex formation, in part, because of the indirect participation of mitochondria in photosynthesis (21). Mitochondria are able to decarboxylate large amounts of glycine during photorespiration. The respiratory chain of plant mitochondria is highly branched because of the presence of numerous so-called “alternative” oxidoreductases, which participate in electron transport without contributing to the proton gradient across the inner mitochondrial membrane (22). Respiratory-protein complexes are known to include side activities in plant mitochondria, such as the mitochondrial processing peptidase within complex III (23, 24) and carbonic anhydrases in complex I (25). BN/PAGE analysis allowed us to identify three types of respiratory supercomplexes in Arabidopsis thaliana. Complexes I and III form associations of I+III2 and I2+III4, and ATP synthase forms a dimeric supercomplex (5, 26). Other complex-IV-containing supercomplexes were identified in potato tuber and spinach mitochondria (6, 7). In plants, the I+III2 supercomplex proved to be especially stable. Upon solubilization of isolated mitochondria by digitonin, 50–90% of complex I forms part of this supercomplex in Arabidopsis, potato, bean, and barley (5).
Despite indirect biochemical and kinetic evidence for supercomplex formation in various organisms, direct evidence is lacking. Here, we describe the structure of a respiratory supercomplex. A stable I+III2 supercomplex, monomeric complex I, and dimeric complex III were purified from isolated Arabidopsis mitochondria by sucrose-density-gradient ultracentrifugation or affinity chromatography. Analysis by EM and single-particle analysis revealed the structures of all three complexes at a resolution of 18 Å. The structure of complex I from Arabidopsis exhibits two additional domains absent in the same respiratory-protein complex from fungi and animals, whereas dimeric complex III has a very similar structure in all three groups of organisms. Analysis of the I+III2 supercomplex revealed a lateral association of dimeric complex III to the tip of the membrane part of complex I. Functional implications are discussed below.
Materials and Methods
Purification of Mitochondrial I+III2 Supercomplex, Monomeric Complex I, and Dimeric Complex III from A. thaliana. Isolated mitochondria were solubilized by digitonin (5 mg of detergent per mg of mitochondrial protein), and protein complexes were subsequently resolved by sucrose-gradient ultracentrifugation [gradients of 0.3–1.5 M sucrose/15 mM Tris base, pH 7.0/20 mM KCl/0.2% digitonin (centrifugation at 150,000 × g for 20 h)]. Afterward, fractions were removed from the gradient from bottom to top. Protein complexes present in individual fractions were resolved by BN/PAGE and identified on the basis of their subunit compositions on second gel dimensions, which were carried out in the presence of SDS (5). Fractions close to the bottom of the gradients were found to include I+III2 supercomplex, whereas the fractions above included a mixture of complex I and I+III2 supercomplex. Dimeric complex III was purified by cytochrome-c-affinity chromatography as described by Weiss and Juchs (27) and Braun and Schmitz (28). Fractions eluted from the affinity column were analyzed by 1D SDS/PAGE. The peak of complex III was determined spectrophotometrically.
Electron Microscopy and Single-Particle Analysis. Samples of purified complexes were negatively stained by using the droplet method with 2% uranyl acetate on glow-discharged, carbon-coated copper grids. Electron microscopy was performed on a CM20FEG electron microscope (Philips, Eindhoven, The Netherlands) operated at 200 kV. Images were recorded with a 4000 SP 4K slow-scan camera (Gatan, Pleasanton, CA) at magnification ×66,850 with a pixel size (after binning the images) of 4.49 Å at the specimen level with grace software for semiautomated specimen selection and data acquisition (29). Single-particle analysis was performed with the groningen image processing (grip) software package on a PC cluster. Selected single-particle projections (128 × 128-pixel frame) were aligned by multireference and reference-free alignment procedures (29, 30). Particles were then subjected to multivariate statistical analysis followed by hierarchical ascendant classification (30). The resolution of the class averages was measured according to the procedure described in ref. 31.
Results and Discussion
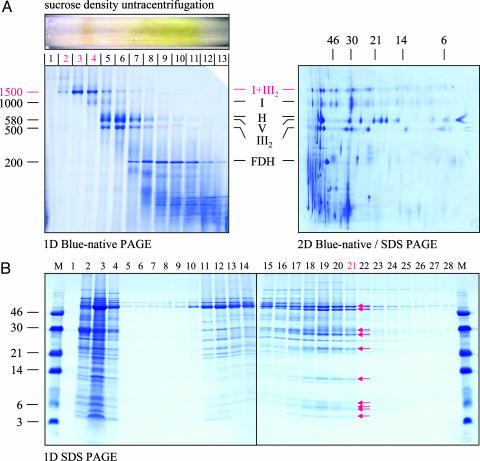
Purification of the I+III2 supercomplex with a mass of 1,500 kDa was accomplished from digitonin-treated mitochondrial fractions by sucrose-density ultracentrifugation (Fig. 1a). The isolated supercomplex proved to be exceptionally stable. To determine the structure of the supercomplex, a set of 5,000 single-particle EM projections was analyzed. The vast majority of projections of intact supercomplexes (fraction 2, Fig. 1a) turned out to represent one particular type of projection (Fig. 2a). A slightly smaller complex from fraction 3 (Fig. 1a) has a similar shape but appears to lack a large protein mass at its left side (Fig. 2b). Only a very small number of differently oriented particles was observed (Fig. 2c); these probably represent side views (see below).
Fig. 1.
Purification of mitochondrial I+III2 supercomplex, monomeric complex I, and dimeric complex III from A. thaliana. (A) Isolated mitochondria were solubilized by digitonin, and protein complexes were subsequently resolved by sucrose-gradient ultracentrifugation (Upper Left). BN/PAGE was carried out to analyze the protein-complex content of the fractions (Lower Left). Identities of the resolved complexes on the 1D gel were elucidated by a parallel 2D BN/SDS/PAGE of total mitochondrial protein (Right). I+III2, supercomplex formed of complex I and dimeric complex III; I, complex I; H, heat-stress protein 60 complex; V, ATP synthase complex; III2, dimeric complex III; FDH, formate dehydrogenase complex. The molecular masses (in kDa) of standard proteins are given to the left of the 1D gel and above the 2D gel. Fractions 2 and 3 of the sucrose gradient were used for EM analysis of the I+III2 supercomplex, and fraction 4 was used for EM analysis of monomeric complex I. (B) A mitochondrial-membrane fraction was solubilized by 3% Triton X-100, and complex III was subsequently purified by cytochrome-c-affinity chromatography (15). Proteins were eluted from the affinity column by a linear Tris/acetate gradient (20–200 mM) and analyzed by 1D SDS/PAGE (15). Fractions 1–4, flow-through; fractions 5–16, mitochondrial proteins not related to complex III; fractions 17–22, purified complex III (subunits are indicated by arrows). Fraction 21 was used for EM analysis of complex III. The molecular masses (in kDa) of standard proteins are given to the left.
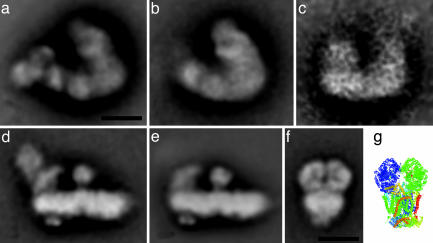
Fig. 2.
Projection maps at 18 Å of the Arabidopsis I+III2 supercomplex and its components obtained by single-particle averaging. (a) Averaged projection map of 1,073 top-view projections of I+III2 supercomplex particles, viewed from the matrix side of the membrane. (b) Average of the best 580 top views of a I+III2 supercomplex fragment. (c) Averaged side view of 22 projections of a I+III2 supercomplex fragment. (d) Average of 930 side-view projections of complex I. (e) Average of 2,100 side-view projections of complex I lacking NAD-oxidizing subunits. (f) Average of 1,014 projections of complex III in a side-view position. (g) View of an x-ray structural model of dimeric bovine complex III (24) in a position similar to that in the EM data, with the bulky matrix-exposed domains in the upper part. (Scale bar, 10 nm.)
For a precise structural assignment of the supercomplex map, we performed additional analyses of purified complexes I and III. The gross structure of mitochondrial complex I is known from single-particle EM of different organisms (32). It is L-shaped, with a hydrophobic arm residing in the membrane and a peripheral arm protruding into the matrix space. Analysis of >10,000 complex I projections indicated that the intact complex from Arabidopsis is also L-shaped (fraction 4 of Fig. 1a and Fig. 2d) and resembles complex I of the fungus Neurospora crassa in side-view position (33). However, it has two unique knob-like protein densities attached to either side of the hydrophobic arm not observed in any other complex I. There is biochemical evidence that complex I in Arabidopsis includes additional subunits: five structurally related proteins most likely representing γ-carbonic anhydrases (25) and an l-galactono-γ-lactone dehydrogenase (34), but it has yet to be established that these densities could represent one of these proteins. It is currently not possible to speculate on the localization of the extra subunits within complex I from plants. A substantial number of projections lack the upper part of the peripheral arm at the site where NAD-oxidizing subunits are located (Fig. 2e). On the other hand, the hydrophobic arm did not show variation. Interestingly, with its size of about 230 Å, the hydrophobic arm is the longest of all complex I structures analyzed up to now.
Investigation of single-particle projections of purified complex III showed only one type of projection (Fig. 2f). Assignment of the complex III projection was possible by a comparison with the x-ray structure from mammalian complex III (35), indicating that the EM projection is viewed from a side-view position, perpendicular to the twofold axis (Fig. 2g). The structures are similar but not identical. The bulky, matrix-exposed domains at the top appear to be somewhat larger in the EM projection, probably because the “core I and II” subunits constituting these domains are larger by ≈20 kDa in plants (36). In plants, these proteins represent the two subunits of the mitochondrial processing peptidase, which cleaves off the presequences of nuclear-encoded mitochondrial proteins upon their import into the organelle (23, 24). The middle part of the EM projection appears to be thicker because of the presence of a negative stain-excluding detergent boundary layer surrounding the single particles. The intermembrane exposed part of plant complex III is smaller than its mammalian counterpart, probably because of a smaller size of subunit 8 and some degradation in subunit 5.
The side-view maps of complex I and III indicate that the supercomplex maps represent top-view projections in the plane of the membrane. Comparison with known EM and x-ray structures of other large membrane proteins such as photosystem II indicated that the supercomplex is faced from the matrix side. Top views of complexes I and III suitable for direct comparison were not observed in our data sets. However, the comparison of the side-view EM data with the x-ray model of the complex III dimer suggests a rather similar overall shape (Fig. 2 f and g). Hence, we searched for an optimal fit of the latter structure within the supercomplex maps. Because negative-stain embedding of bulky objects, such as the investigated supercomplex, results in projections in which the stain mainly contrasts the protein parts closest to the support film, we considered fitting of the membrane-integrated parts of complex III. Fig. 3a indicates that the lower part of complex III fits snugly within the circumference of the upper “leg” of the supercomplex. This constrains the position of complex I to the lower leg, with the tip of the hydrophobic arm pointing to the right. The variability within the supercomplex, which is restricted to its left part, could then be explained by the presence or absence of the NAD-oxidizing unit of the peripheral arm of complex I, as depicted in Fig. 2 d and e.
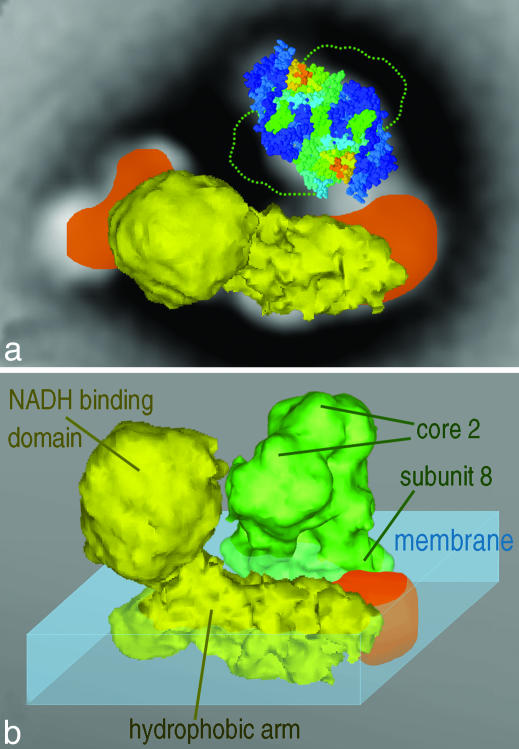
Fig. 3.
Model for the structure of the I+III2 supercomplex from Arabidopsis, incorporating the x-ray structure of complex III and the EM 3D density of complex I from beef heart. (a) Fit within the supercomplex top view of the complex I structure (yellow) and the membrane-embedded lower half of complex III (space-filling structure) seen from the matrix side. The projections of additional complex I densities present in Arabidopsis are in orange, and the outline of the hydrophilic domains of complex III within the matrix is indicated by a green dotted line. (b) Overview of the fitting of complex I and complex III (green), seen from an angle of 45° out of the membrane plane. Components of complexes I and III and the membrane outside the supercomplex (blue) are indicated. Note that the x-ray structure was truncated to 18 Å by using routines from the eman package (41) and displayed with vis5d software (www.ssec.wisc.edu/~billh/vis5d.html).
Because negative stain does not penetrate the hydrophobic-membrane parts, the 2D projection map of the I+III2 supercomplex indicates an interaction of complexes I and III within the plane of the membrane. We generated a 3D model of the supercomplex by fitting a 22-Å 3D volume of mammalian complex I (37) and an 18-Å truncated x-ray structure of complex III (35) within the supercomplex. The model shows that the tip of the hydrophilic arm, which is 40–50 Å longer in Arabidopsis than in mammalian complex I, is essential for the formation of the I+III2 supercomplex. It is not known which subunits of complex I constitute this tip; however, modeling indicates that subunit 9 of complex III is closest to the interface. It appears that the very bulky matrix-exposed domains of both complexes are in one another's vicinity but probably do not (strongly) interact (Fig. 3b). Despite a shortened membrane arm of complex I, the I+III2 supercomplex was also described for beef (3), indicating that its architecture might not be fully conserved.
Our results clearly suggest one specific orientation of the two respiratory-chain complexes within a supercomplex. EM studies on >100,000 projections of various complexes of photosystem I and its iron-stress-inducible antenna protein, IsiA, indicate that a nonspecific binding between photosystem I and IsiA subunits coincides with a strong loss in detail, well above the 18-Å resolution obtained in the supercomplex map (38). Thus, the mitochondrial I+III2 supercomplex is considered to be a structurally and functionally specific component of the respiratory chain.
It was previously shown for the A. thaliana chloroplast membrane that a supercomplex of photosystem II and the light-harvesting complex II were essential for proper functioning (39). Our results now offer insights into the functional role of the mitochondrial I+III2 supercomplex. The location of the ubiquinone-binding domain of complex I is not precisely known but is supposed to lie on the membrane arm directly beneath the starting point of the peripheral arm, which includes the NADH-binding domain (40). A direct role of the supercomplex in ubiquinone-channeling is unlikely, because the ubiquinone-binding domain of complex I is not in close proximity to the complex I–complex III interphase. However, even without direct ubiquinone channeling, electron-transfer rates on the basis of the I+III2 supercomplex are most likely higher than on the basis of freely diffusing single complexes. The main portion of the membrane arm of complex I is believed to play a role in proton translocation from the mitochondrial matrix to the intermembrane space. It is possible that complex III is essential for this function of complex I. Furthermore, association of these two complexes might be important for other reasons, such as stabilization of the single complexes, mitochondrial cristae morphology, or protein-packing within the inner mitochondrial membrane. Because of its exceptional stability, the Arabidopsis I+III2 supercomplex represents an ideal starting point for future investigations into supercomplex physiology.
Acknowledgments
We thank Dr. N. Grigorieff (Brandeis University, Waltham, MA) for providing the complex I structure. This work was supported by Deutsche Forschungsgemeinschaft Grant BR 1829 7/1 (to H.-P.B.) and an open-competition grant from the Netherlands Organization for Chemical Research, Council for Chemical Sciences (to E.J.B.).
Author contributions: E.J.B. and H.-P.B. designed research; N.V.D., H.E., and W.K. performed research; N.V.D., H.E., W.K., E.J.B., and H.-P.B. analyzed data; and N.V.D., E.J.B., and H.-P.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: BN, blue native.
References
- 1.Hatefi, Y. (1985) Annu. Rev. Biochem. 54, 1015-1069. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, I., Pfeiffer, K., Neupert, W., Stuart, R. A. & Schägger, H. (1998) EMBO J. 17, 7170-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schägger, H. & Pfeiffer, K. (2000) EMBO J. 19, 1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schägger, H. & Pfeiffer, K. (2001) J. Biol. Chem. 276, 37861-37867. [DOI] [PubMed] [Google Scholar]
- 5.Eubel, H., Jänsch, L. & Braun, H. P. (2003) Plant Physiol. 133, 274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eubel, H., Heinemeyer, J. & Braun, H. P. (2004) Plant Physiol. 134, 1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krause, F., Reifschneider, N. H., Vocke, D., Seelert, H., Rexroth, S. & Dencher, N. A. (2004) J. Biol. Chem. 279, 48369-48375. [DOI] [PubMed] [Google Scholar]
- 8.Boumans, H., Grivell, L. A. & Berden, J. A. (1998) J. Biol. Chem. 273, 4872-4877. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi, C., Genova, M. L., Parenti Castelli, G. & Lenaz, G. (2004) J. Biol. Chem. 279, 36562-36569. [DOI] [PubMed] [Google Scholar]
- 10.Acin-Perez, R., Bayona-Bafaluy, M. P., Fernandez-Silva, P., Moreno-Loshuertos, R., Perez-Martos, A., Bruno, C., Moraes, C. T. & Enriquez, J. A. (2004) Mol. Cell 13, 805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grad, L. I. & Lemire, B. D. (2004) Hum. Mol. Genet. 13, 303-314. [DOI] [PubMed] [Google Scholar]
- 12.Ugalde, C., Janssen, R. J., van den Heuvel, L. P., Smeitink, J. A. M. & Nijtmans, L. G. J. (2004) Hum. Mol. Genet. 13, 659-667. [DOI] [PubMed] [Google Scholar]
- 13.Berry, E. A. & Trumpower, B. L. (1985) J. Biol. Chem. 260, 2458-2467. [PubMed] [Google Scholar]
- 14.Sone, N., Sekimachi, M. & Kutoh, E. (1987) J. Biol. Chem. 262, 15386-15391. [PubMed] [Google Scholar]
- 15.Iwasaki, T., Matsuura, K. & Oshima, T. (1995) J. Biol. Chem. 270, 30881-30892. [DOI] [PubMed] [Google Scholar]
- 16.Niebisch, A. & Bott, M. (2003) J. Biol. Chem. 278, 4339-4346. [DOI] [PubMed] [Google Scholar]
- 17.Stroh, A., Anderka, O., Pfeiffer, K., Yagi, T., Finel, M., Ludwig, B. & Schägger, H. (2004) J. Biol. Chem. 279, 5000-5007. [DOI] [PubMed] [Google Scholar]
- 18.Paumard, P., Vaillier, J., Coulary, B., Schaeffer, J., Soubannier, V., Mueller, D. M., Brethes, D., di Rago, J. P. & Velours, J. (2002) EMBO J. 21, 221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, M., Mileykovskaya, E. & Dowhan, W. (2002) J. Biol. Chem. 277, 43553-43556. [DOI] [PubMed] [Google Scholar]
- 20.Pfeiffer, K., Gohil, V., Stuart, R. A., Hunte, C., Brandt, U., Greenberg, M. L. & Schägger, H. (2003) J. Biol. Chem. 278, 52873-52880. [DOI] [PubMed] [Google Scholar]
- 21.Douce, R., Bourguignon, J., Neuburger, M. & Rebeille, F. (2001) Trends Plant Sci. 6, 167-176. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie, S. & McIntosh, L. (1999) Plant Cell 11, 571-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun, H. P., Emmermann, M., Kruft, V. & Schmitz, U. K. (1992) EMBO J. 11, 3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun, H. P. & Schmitz, U. K. (1995) Trends Biochem. Sci. 20, 171-175. [DOI] [PubMed] [Google Scholar]
- 25.Parisi, G., Perales, M., Fornasari, M. S., Colaneri, A., Gonzalez-Schain, N., Gómez-Casati, D., Zimmermann, S., Brennicke, A., Araya, A., Ferry, J. G., et al. (2004) Plant Mol. Biol. 55, 193-207. [DOI] [PubMed] [Google Scholar]
- 26.Eubel, H., Heinemeyer, J., Sunderhaus, S. & Braun, H. P. (2004) Plant Physiol. Biochem. 134, 1450-1459. [Google Scholar]
- 27.Weiss, H. & Juchs, B. (1978) Eur. J. Biochem. 88, 17-28. [DOI] [PubMed] [Google Scholar]
- 28.Braun, H. P. & Schmitz, U. K. (1992) Eur. J. Biochem. 208, 761-767. [DOI] [PubMed] [Google Scholar]
- 29.Penczek, P., Radermacher, M. & Frank, J. (1992) Ultramicroscopy 40, 33-53. [PubMed] [Google Scholar]
- 30.van Heel, M., Gowen, B., Matadeen, R., Orlova, E. V., Finn, R., Pape, T., Cohen, D., Stark, H., Schmidt, R., Schatz, M. & Patwardhan, A. (2000) Q. Rev. Biophys. 33, 307-369. [DOI] [PubMed] [Google Scholar]
- 31.Van Heel, M. (1987) Ultramicroscopy 21, 95-100. [DOI] [PubMed] [Google Scholar]
- 32.Yagi, T. & Matsuno-Yagi, A. (2003) Biochemistry 42, 2266-2274. [DOI] [PubMed] [Google Scholar]
- 33.Guenebaut, V., Schlitt, A., Weiss, H., Leonard, K. & Friedrich, T. (1998) J. Mol. Biol. 276, 105-112. [DOI] [PubMed] [Google Scholar]
- 34.Millar, A. H., Mittova, V., Kiddle, G., Heazlewood, J. L., Bartoli, C. G., Theodoulou, F. L. & Foyer, C. H. (2003) Plant Physiol. 133, 443-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata, S., Lee, J. W., Okada, K., Lee, J. K., Iwata, M., Rasmussen, B., Link, T. A., Ramaswamy, S. & Jap, B. K. (1998) Science 281, 64-71. [DOI] [PubMed] [Google Scholar]
- 36.Berry, E. A., Guergova-Kuras, M., Huang, L. S. & Crofts, A. R. (2000) Annu. Rev. Biochem. 69, 1005-1075. [DOI] [PubMed] [Google Scholar]
- 37.Grigorieff, N. (1998) J. Mol. Biol. 277, 1033-1046. [DOI] [PubMed] [Google Scholar]
- 38.Yeremenko, N., Kouril, R., Ihalainen, J. A., D'Haene, S., van Oosterwijk, N., Andrizhiyevskaya, E. G., Keegstra, W., Dekker, H. L., Hagemann, M., Boekema, E. J., et al. (2004) Biochemistry 43, 10308-10313. [DOI] [PubMed] [Google Scholar]
- 39.Ruban, A. V., Wentworth, M., Yakushevska, A. E., Andersson, J., Lee, P. J., Keegstra, W., Dekker, J. P., Boekema, E. J., Jansson, S. & Horton, P. (2003) Nature 421, 648-652. [DOI] [PubMed] [Google Scholar]
- 40.Friedrich, T. & Böttcher, B. (2004) Biochim. Biophys. Acta 1608, 1-9. [DOI] [PubMed] [Google Scholar]
- 41.Ludtke, S. J., Baldwin, P. R. & Chiu, W. (1999) J. Struct. Biol. 128, 82-97. [DOI] [PubMed] [Google Scholar]